ABSTRACT
The degree to which surface-motile bacteria explore their surroundings is influenced by aspects of their local environment. Accordingly, the regulation of surface motility is controlled by numerous chemical, physical, and biological stimuli. The discernment of such regulation due to these multiple cues is a formidable challenge. Additionally, inherent ambiguity and variability from the assays used to assess surface motility can be obstacles to clear the delineation of regulated surface motility behavior. Numerous studies have reported single environmental determinants of microbial motility and lifestyle behavior, but the translation of these data to understand surface motility and bacterial colonization of human host or environmental surfaces is unclear. Here, we describe the current state of the field and our understanding of exogenous factors that influence bacterial surface motility.
KEYWORDS: flagella, gliding, rhamnolipid, sliding, surfactant, swarming, twitching, type IV pili
TEXT
Arguably, one major advantage for many bacteria is their motility; self-motile bacteria possess the ability to seek out favorable growth environments through exploration and movement toward nutrients or away from toxins, or by assembling complex communities which enable survival (1). Of particular interest is bacterial surface motility, which allows some species to quickly colonize surfaces. Four surface motility modes, originally termed by Henrichsen (2), appear regularly in the literature: swarming, twitching, gliding, and sliding. Nearly all known surface-motile bacteria are soil bacteria, eukaryotic pathogens, or both (Table 1). Interaction with surfaces is important to most bacterial species; thus, it is logical that many organisms have evolved motility strategies as part of their surface colonization. Motility, in turn, shapes the competition among species, potential virulence, biogeography, and biofilm development of a surface-associated population. Understanding the mechanisms that dictate how bacteria respond to environmental factors is of the utmost consequence, as they provide the springboard to recognizing the role of motility in survival and virulence and can provide the tools to modify these responses.
TABLE 1.
Surface motile bacteria grouped by motility mode
| Motility mode | Bacteria (references) |
|---|---|
| Swarming | Alicyclobacillus acidoterrestris (150), Azospirillum sp. (3), Bacillus subtilis (3, 12, 16), Clostridium sp. (6), Delftia acidovorans (152), Escherichia coli (17, 18), Klebsiella pneumoniae (104), Paenibacillus sp. (8, 68, 146), Proteus mirabilis (5, 12, 14, 103), Pseudomonas aeruginosa (15, 20, 66, 67, 82, 95, 107, 148), Pseudomonas veronii (152), Ralstonia taiwanensis (152), Rhizobium sp. (55, 156), Salmonella Typhimurium (11, 18, 72, 114), Serratia marcescens (12, 93), Serratia liquefaciens (13, 70), Serratia rubidaea (13), Sinorhizobium meliloti (131), Vibrio cholerae (3, 104), Vibrio parahaemolyticus (3, 9, 111, 157), Vibrio alginolyticus (158), Yersinia sp. (3) |
| Twitching | Acinetobacter baumannii (80, 81, 120, 121), Bacteroides sp. (27), Caulobacter sp. (26), Clostridium perfringens (98, 99), Geobacter sp. (26), Kingella kingae (126, 153), Myxococcus xanthus (35, 123, 154), Neisseria gonorrhoeae (30), Pseudomonas aeruginosa (26, 28, 97, 107, 118, 155), Ralstonia solanacearum (79), Vibrio sp. (26, 27), Xylella fastidiosa (127, 128) |
| Gliding | Flavobacterium sp. (5, 31), Mycoplasma sp. (5, 31, 34), Myxococcus sp. (5, 31, 36, 151) |
| Sliding | Bacillus subtilis (42), Escherichia coli (39), Legionella pneumophila (130), Mycobacterium smegmatis (40), Pseudomonas aeruginosa (23, 44), Salmonella Typhimurium (45), Sinorhizobium meliloti (39, 131), Staphylococcus aureus (41), Streptomyces sp. (100), Vibrio sp. (39) |
In addition to regulation of any required appendages and motors, surface motilities are greatly affected by environmental factors, such as humidity, nutrients, and self-produced compounds (Table 2). Most surface motility is also influenced by the actions and motion of their surrounding neighbors. While mechanisms that cease motility and initiate biofilm formation have received much attention, how environmental cues shape surface motility itself is poorly understood. Even more mysterious is how multiple external signals can be integrated by individual cells to produce a coordinated population response. Here, we review known environmental influences to bacterial surface motility.
TABLE 2.
Exogenous factors known to promote or impede bacterial surface motility
| Bacterium and motility mode | Physical factor (reference[s]) | Chemical factor (reference[s]) |
|---|---|---|
| Alicyclobacillus acidoterrestris: swarming | Low pH (4.5) inhibits swarming (and increases biofilm formation) (150) | |
| Bacillus subtilis | ||
| Swarming | Rich medium (16) | |
| Sliding | Potassium (42) | |
| Delftia acidovorans: swarming | Inhibited by zinc and cadmium (146, 152) | |
| Myxococcus xanthus | ||
| Twitching | Agar concn (35) | Calcium availability (123) |
| Gliding | Agar concn (35) | |
| Neisseria gonorrhoeae: twitching | Oxygen (159) | |
| Pseudomonas aeruginosa | ||
| Swarming | Agar concn (15), surface wetness (17), other appendages (148) | Carbon source (20, 95), nitrogen source (66, 101, 102), phosphate availability (105), iron availability (115, 119), mucin (82) |
| Twitching | Carbon source (97), phosphate (97, 107), calcium availability (28), osmolarity (155) | |
| Sliding | Rhamnolipid (23, 44) | Oxygen availability (23) |
| Proteus mirabilis: swarming | Glutamine, arginine, histidine, malate, and ornithine (103) | |
| Ralstonia solanacearum: swarming | Sugars and amino acids (160) | |
| Salmonella Typhimurium | ||
| Swarming | Hyperflagellation (surface sensing) (18) | Glucose (18), phosphorus (104), iron availability (114) |
| Sliding | Low magnesium (45) | |
| Streptomyces venezuelae: sliding | Trimethylamine (and low glucose) (100) | |
DEFINING SURFACE MOTILITY MODES
Swarming.
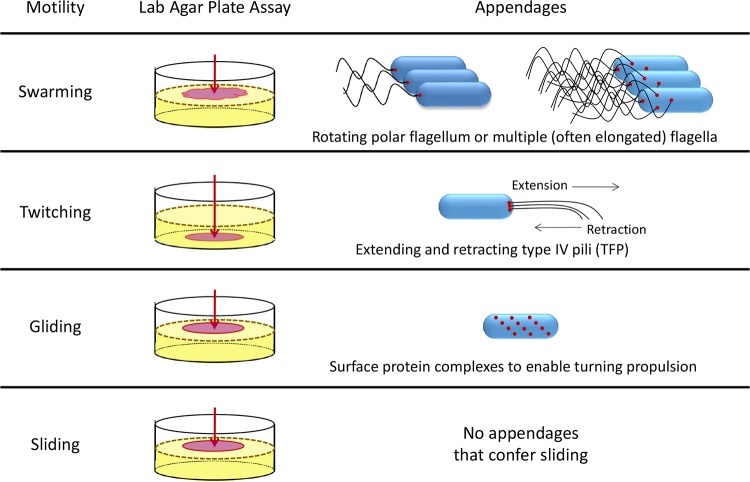
In his 1972 review of bacterial surface translocation, Henrichsen defines swarming motility as a “surface translocation produced through the action of flagella,” partly dependent on cell-cell interactions (2). We now appreciate the importance of surface wetness to allow swarming of flagellated bacteria through a thin liquid layer on a semisolid surface (Fig. 1). Both Gram-negative and Gram-positive species are known to swarm; however, swarming is most conserved in three bacterial families, Gammaproteobacteria, Alphaproteobacteria, and Firmicutes (3, 4). These families include species of Azospirillum, Bacillus, Clostridium, Paenibacillus, Proteus, Pseudomonas, Rhizobium, Rhodospirillum, Salmonella, Sinorhizobium, Vibrio, and Yersinia, among others; species from these genera are found in a variety of environments, including soil and eukaryotic hosts (2, 3, 5–8). Many swarming species exhibit elongated and/or hyperflagellated morphologies during swarming, while other swarming species show no evidence of an elongated cell phenotype.
FIG 1.
Bacterial surface motility modes. Swarming is flagellum-mediated motility assayed on top of semisolid-agar assays. Twitching is TFP-mediated motility assayed in the interstitial space between the agar-dish interface. Gliding is cell protein-associated motility assayed on top of semisolid-agar assays. Sliding is spreading of bacteria independent of appendages assayed on top of semisolid-agar assays. The arrow indicates the point of assay inoculation, and the red zone indicates the region of bacterial spreading.
During swarming, water must be drawn to the surface to allow flagellar rotation, and bacteria must be able to overcome frictional forces due to viscosity in the thin liquid layer through which they move as a group. The mechanisms by which bacteria promote their swarming are as varied as the bacteria that use them (5, 9–11). Swarming bacteria can be further divided into two general groups, robust swarmers and temperate swarmers (12). Robust swarmers can move on hard surfaces, displaying a drastically changed morphology with hyperflagellation and elongation. Robust swarmers Proteus mirabilis and Vibrio parahaemolyticus can show motility on agar with a concentration as high as 2% (13). For P. mirabilis, hyperflagellation greatly improves swarming, allowing movement in more viscous environments (14). Temperate swarmers, like Bacillus subtilis, Pseudomonas aeruginosa, or Escherichia coli, often require softer surfaces (nearer 0.5% [wt/vol] agar concentrations) to promote swarming (12, 15). Though classified as a temperate swarmer, undomesticated strains of B. subtilis can swarm easily on 0.7% agar (16). E. coli, however, swarms “reluctantly” under soft-agar conditions, highlighting the range of swarming in this general group (17, 18). Temperate swarmers may not show markedly different cell morphology during swarming, though it is still common for cells to hyperflagellate and elongate to a lesser extent. Temperate swarmers often produce surfactants to lower surface tension in the thin liquid layer, and many studies have reported that B. subtilis surfactin and P. aeruginosa rhamnolipid are required for swarming (12, 16, 19–24). Interestingly, some studies for P. aeruginosa have now shown that flagellum-mediated swarming can still be achieved in the absence of its surfactant rhamnolipid (e.g., for a ΔrhlAB mutant strain) with certain combinations of nutrients and/or agar conditions (20, 25). Further delineation of these chemical influences upon regulation and swarming motility will significantly improve our understanding of the motility that we define operationally as swarming.
Twitching.
Type IV pili (TFP) are common bacterial surface appendages used for attachment, DNA transfer, pathogenesis, and motility. TFP can be found in a wide variety of bacteria, including species of Acinetobacter, Bacteroides, Caulobacter, Escherichia, Geobacter, Kingella, Myxococcus, Neisseria, Pseudomonas, Vibrio, and Xylella. The structures of these appendages vary depending on the species, but their ability to attach to surfaces seems global. Attaching to various surfaces, including glass and human cells, can be helpful in microcolony formation and virulence. The three subgroups of TFP, type IVa, IVb, and IVb-tight adherence pili, have a conserved hydrophobic alpha-helical N terminus involved in mediating pilus assembly, though the lengths of pilin-subunit chains vary (26, 27).
Early definitions of twitching did not specify a mechanism but noted that translocation occurred in flagellated and nonflagellated bacteria. Single cells were originally described to move in a “jerky” manner, not following the long axis of the cell (2). We now know that twitching is one form of TFP-mediated surface motility, during which TFP are extended, attach to a surface, and are retracted to move the cell (28–30). Furthermore, many twitching bacteria twitch quite smoothly in groups. Like swarming, twitching is a way of exploring surfaces for nutrients or toxins, allowing bacteria to move toward more favorable environments or escape potential dangers (26).
Gliding.
Gliding is functionally defined as surface motility by an active process, following the long axis of the cell and without the use of flagella or other propulsive appendages (31, 32). Common in the deltaproteobacteria, cyanobacteria, and flavobacteria, gliding is often considered a rarer form of bacterial motility. At present, bacterial gliding is best characterized in three bacterial clusters: the order Myxococcales, the phylum Bacteroidetes, and the genus Mycoplasma (5, 33, 34). The hallmark gliding organisms of these three clusters are Myxococcus xanthus, Flavobacterium johnsoniae, and Mycoplasma mobile, respectively. These gliding bacteria do not utilize machinery that the field currently perceives as appendages (Fig. 1). Rather, these actively moving bacteria utilize surface protein complexes that serve to move the cell. Unlike the strong homology that has been determined for flagellar or type IV pilus components of swimming, swarming, and twitching bacteria, the motility machineries used by gliding bacteria all appear to be quite distinct. The regulatory elements and biochemistry that have been discerned for M. xanthus, F. johnsoniae, M. mobile, and otherwise share no common homology outside their respective clusters. It is likely that gliding evolved independently in several bacterial classes (35), but each of these species exhibits characteristics consistent with tank tread locomotion mediated by membrane-spanning complexes (36–38).
Sliding.
There are numerous species that have been shown to move on surfaces without the aid of any appendage or known cellular component. In some cases, this is specifically labeled as sliding, termed by Henrichsen as “…a kind of surface translocation produced by the expansive forces in a growing culture in combination with special surface properties of the cells resulting in reduced friction between cell and substrate” (2). Operationally, if a bacterium exhibits a spreading phenotype with no attribution to an active motility apparatus, we can reasonably characterize such behavior as sliding (Fig. 1). Escherichia coli and species of Bacillus, Legionella, Mycobacterium, Pseudomonas, Sinorhizobium, Staphylococcus, Streptomyces, and Vibrio all exhibit sliding motility. The key component for these sliding bacteria appears to be their utilization of self-secreted products to aid their spreading. Many, but not all, of these sliding bacteria produce a surfactant (39). For example, Mycobacterium smegmatis exhibits a sliding phenotype that can be attributed to its production of glycolipids (40). Sliding of Staphylococcus aureus has been attributed to the agr quorum sensing system and production of phenol-soluble modulins (PSMs), which are strongly associated with S. aureus pathogenesis (41).
Assessment of the sliding of some bacteria can be confusing, as even well-known flagellum-motile bacteria have been shown to slide. Much of this characterization has been completed investigating flagellum-deficient or -impaired strains of these bacteria. For example, a flagellum-deficient hag mutant of B. subtilis will slide sufficiently to cover an entire plate assay in the presence of sufficient potassium, iron, and magnesium on select agarose concentrations (42). Pseudomonas syringae pv. tomato has also been shown to slide as an alternative plant colonization method to swarming, though this was not observed without its surfactant, syringafactin (43). Flagellum- and TFP-deficient mutants of P. aeruginosa also slide (44). Salmonella enterica serovar Typhimurium slides in the absence of flagella with the aid of a cell surface protein where low magnesium concentrations, but not wetting agents, are required (45). It is clear that surface motility improves survival and surface colonization, promoting the evolution of novel modes of translocation. While sometimes not as dynamic as other forms of motility, there is much to learn when it comes to these and potentially undiscovered modes of appendage-free surface motility.
PHYSICAL INFLUENCES ON SURFACE MOTILITY
Agar concentration.
Surface conditions greatly affect surface motility. In laboratory settings, this translates to surface hardness and moisture content of semisolid assay plates, which are most commonly controlled by agar type and concentration.
Various agars and agar substitutes can be used in bacterial motility assays. Agar, which has been used in microbiology since the 1880s, is a purified phycocolloid (containing both agarose and agaropectin fractions) collected from red-purple marine seaweed. The Difco & BBL manual highlights some differences found just throughout agars available from that company, including calcium and magnesium concentrations (46). Hard-agar plates usually contain 1 to 2% (wt/vol) agar, while motility assays might use 0.2% to 0.8%, depending on the motility of interest (3, 47). Gellan gum (found under names, like Gelzan, Gelrite, Gel-Gro, or Phytagel) is an anionic polysaccharide produced by Sphingomonas elodea (previously Pseudomonas elodea) that has been used as an alternative to agar since the 1980s (25, 48–51). Gellan gum generally has better optical clarity than agar and higher thermal stability; both of these properties provide some advantages for microscopy (48, 52–54). Different gelling agents can have different losses on drying, influencing wettability. Eiken agar, for example, has been called “more wettable” than Bacto agar, because swarm motility was observed for reluctant swarmers, like E. coli K-12, when cultured on Eiken agar (17).
In general, these motility assays utilize a narrow range of agar concentrations. At lower or higher agar concentrations, these bacteria may exhibit an alternate motility phenotype or just stop movement altogether. For example, swarming motility in Rhizobium leguminosarum was inhibited on the hard surface of 1.3% (wt/vol) agar but was optimal at 0.7%, with swimming occurring at the softer 0.5% concentration (55). Swarming in Clostridium and Proteus species was impaired on agar concentrations between 3% and 6% (6). P. aeruginosa swarming is studied using 0.4% to 0.7% agar, while TFP twitching is studied using 1.0% agar (26, 56).
Wettability.
All bacterial surface motility involves some form of a hydrated surface. Limited work has been done to characterize the surfaces of bacterial habitats, in laboratory assays or otherwise, but we can use the physics definitions to gain some insight into what parameters might be important. One measure of the liquid present is “wettability.” Wettability refers to how much a liquid interacts with a surface; macroscopically, this is measured by the equilibrium or static advancing contact angle between the edge of the droplet and the surface. Partial wetting occurs when the contact angle (θ) is between 0° and 90°; complete wetting is a state where only the dynamic contact angle can be measured as a drop spreads over a surface (57). Young's equation (equation 1) (58), which relates contact angle and interfacial tensions, is often used to describe wettability of a surface, where γ is the interfacial surface tension between gas (g), liquid (l), and/or the solid surface (s).
| (1) |
A spreading parameter (S) can also be used to describe the wettability of a surface. S, like θ, can also describe partial or complete wetting of a surface, where a negative value indicates partial wetting, while a positive value indicates complete wetting. S can be described by the traditional 3-phase interfacial surface tensions (equation 2) (58, 59) or, in the presence of a surfactant, in terms of the surface tension of the surfactant σm and the thin liquid film σF (equation 3) (58).
| (2) |
| (3) |
Surfactants or other wetting agents can also cause shear stresses at the liquid-vapor interface, resulting in Marangoni gradients that cause differences in the thickness of the liquid layer; these gradients bear a striking resemblance to the characteristic branched patterns seen in many species of swarm bacteria (57, 60). Indeed, some have modeled swarming as a Marangoni flow, using shear stress, viscosity of the thin liquid layer, and the spreading velocity and height of a nongrowing colony (4).
These parameters describing a surface could make all the difference in a motility assay. For example, swarming and sliding patterns can look very similar at the macro level; knowing if the motility assay is completely or partially wetted would help distinguish the motility at play. The wettability can be affected by how long the plates are allowed to dry (a control on surface tension at the liquid-gas interface) and how much moisture evaporates from the agar matrix (a control on overall water availability, affecting how much water could be pulled to the surface during swarming). Observing the spread of waterproof India ink mixed with bacterial inoculum can give a good indication of whether the surface of a plate will allow passive spreading or if moving bacteria will require the adaptations of swarming mentioned earlier (56, 61). Few reports have measured these physical parameters, with most studies relying on the chemical properties of surfactants to control surface tension. Detailed characterization of the wettability and other properties for hydrated surfaces of relevance (62, 63) has not yet been conducted for more applied surfaces, such as infected host cells or soil particles in the subsurface. Thus, the influence of surfaces relevant to pathogenic and environmental bacteria alike is largely unexplored.
Overcoming drag with production of surfactants and osmolytes.
Though surface moisture is critical to swarming motility and organism survival, it also presents physical interactions between the agar surface, the liquid layer, the bacterial cell body, and bacterial appendages in the form of different forces that must be overcome for individual cells to move and groups to spread (11, 64, 65). There are no “dry” surfaces within the context of surface-motile bacteria. The strength and distribution of these forces can change how bacteria deploy flagella or TFP (65). Bacteria have evolved many ways to deal with these challenges, but generally, two methods prevail.
To overcome hydrodynamic forces between the bacteria and the agar, as well as viscous forces within the liquid layer, many bacteria produce surfactants. These surfactants are amphipathic compounds that act as lubricants, lowering surface tension by straddling hydrophilic and hydrophobic surfaces to allow easier movement (66, 67). (Detergents such as Tween and SDS are common synthetic surfactants.) Bacillus subtilis produces the lipopeptide surfactin (3, 12). Paenibacillus species produce the surfactant CmoA (68). P. aeruginosa, as well as other Pseudomonas species, produces rhamnolipid, the production of which is tightly controlled by quorum sensing intercellular signaling (66, 67). Rhamnolipid is known to influence P. aeruginosa virulence, and its production is controlled via the rhl quorum sensing regulon (69). This quorum sensing dependence reinforces the idea that swarming is a community motility, as in some species it requires sufficient numbers to produce required compounds. Other surfactants include serrawettin in Serratia marcescens, serrawettin W2 in Serratia liquefaciens, and rubiwettins in Serratia rubidaea (13, 70). These surfactants and osmolytes aiding surface motility can also be derived from cellular components, such as lipopolysaccharide in Gram-negative bacteria (71). It is important to note, however, that surfactants and other tension-active compounds are not required for swarming for all bacteria. E. coli and Salmonella spp. are not known to produce any surfactant, yet they swarm under the proper conditions. While surfactant-deficient mutants of S. marcescens, B. subtilis, and other species have been reported to swarm on softer or alternate agar surfaces (12, 19), as discussed above, it is common that the base swarm plate assay does not readily distinguish between swarming and sliding.
When there is not sufficient water at the surface, some bacteria produce nonsurfactant osmotic agents to pull water from the matrix. The production of such an osmolyte was first shown for S. Typhimurium (72). In P. mirabilis, the produced osmolyte includes polysaccharides and glycine betaine in the extracellular matrix (12). Other biologically relevant osmolytes broadly fall into the categories of amino acids, sugars, and salts (11).
Surface conditions and initiation of motility.
As confirmation of the importance of surface conditions, many bacteria can “sense” the surface properties to initiate differentiation from swimming cells to swarmer cells (73). The regulation of this differentiation and other physical “mechanosensing” behavior is largely uncharacterized for most motile bacteria. It is now clear that proteins associated with both TFP and flagella are involved in some form of surface sensing. Much of this discovery has been conducted by discerning how TFP and flagella, in species such as C. crescentus, interact with hard surfaces and how secondary signaling cascades regulate subsequent surface colonization (74, 75). Work with P. aeruginosa has shown that PilY1 is a dual regulator that distinctly influences both polysaccharide formation and motility (76). Regulation of surface sensing distinctly tied to surface motility is most apparent by studies with species that differentiate into hyperflagellated and elongated cells upon surface contact, such as those of Proteus mirabilis (77) and Vibrio parahaemolyticus (78). Numerous genes have been shown to be upregulated during this differentiation, which have been attributed to both surface and environmental cues (5, 13). Azospirillum and Vibrio species, for example, induce multiple flagella upon surface contact. Other sensing systems are also possible, like the cell envelope sensing of P. mirabilis, which uses Umo proteins upregulated during swarming (12).
Surface conditions affecting twitching and other TFP-mediated motilities have not been as thoroughly investigated as swarming. The attachment of TFP to surfaces requires specific proteins that serve as adhesins, though exactly how the cells regulate attachment and detachment is still largely unknown. It is commonly agreed that some wetness is required. Liu et al. (79) showed, for example, that Ralstonia solanacearum twitching was enhanced under high-humidity conditions. Differences in TFP-mediated motilities have also been observed with different types of gelling agents in certain species. R. solanacearum twitching motility, in that same study, was observed to be better on BG agar than on agarose plates (79), and in Acinetobacter baumannii, robust twitching motility has been observed, but the patterns differ dramatically when grown on Difco agar versus Eiken agar, with Difco agar exhibiting branching patterns and Eiken agar forming more of a lawn (80). A. baumannii TFP-mediated motility has also been shown to be enhanced on agarose, rather than Noble or granulated agar (81).
The characteristics of a surface are especially important when bacteria are looking to switch from a motile state to a sessile state, attaching to host cells or other surfaces. Mucin, readily prevalent in the lung, promotes P. aeruginosa swarming (82). Surface motility is also affected by the surface charge of abiotic surfaces, like glass or polymer brushes, affecting later biofilm formation (83). Work with P. aeruginosa also indicates that TFP bind preferentially to the apical surface, while flagella bind to the basal lateral surface of lung epithelia (84). TFP of Neisseria gonorrhoeae bind to host cell lipids (85), while TFP of Neisseria meningitidis bind to at least one host glycoprotein, CD147 (86). Ex vivo work has made the impact of TFP cues more clear. Phospholipids have also been shown to serve as attractants for M. xanthus and P. aeruginosa TFP (87–90). Gliding has often been associated with a conditioned surface as well. For example, M. xanthus has been shown to deposit slime using motility complexes, modifying the surface to reinforce adhesion during gliding (91).
Temperature.
Temperature can affect swarming motility in a manner distinct from promoting or limiting growth. Some bacteria, like Bradyrhizobium spp. (92) and S. marcescens (93), swarm better at higher temperatures (i.e., 30°C versus 23°C and 37°C versus 30°C, respectively). Others, like isolates from Lake Baikal in Siberia (94) and R. leguminosarum (55), show better swarm motility at lower temperatures (under 30°C). This could be the result of a physiological change altering excretions needed for swarming, like secretion of biosurfactants. Additionally, temperature can have an indirect effect, changing other physical properties already discussed. Surface wetness, particularly, will change, as warmer temperatures create a drier surface over time.
NUTRIENT INFLUENCES ON SURFACE MOTILITY
Carbon.
When surface conditions provide the optimal physical environment for swarming, available nutrients have significant effects, such as inhibiting swarming or changing phenotypic responses (Table 2). Changing the carbon source available, for example, can foster phenotypic changes in P. aeruginosa swarming. Köhler et al. showed that P. aeruginosa forms a dendritic pattern when grown on glucose in minimal medium, while glycerol impairs swarming (95). Additional phenotypic changes are observed when succinate or glutamate is used as the sole source of carbon (20). Significant differences in P. aeruginosa swarm phenotype and associated regulation are readily apparent in comparisons of swarm assays utilizing rich media, minimal media containing amino acids, or minimal media containing glucose (25). Changes in swarming pattern can also be seen in R. leguminosarum, as it swarms on glycerol and erythritol as sole carbon sources (55). Salmonella Typhimurium is also characterized as requiring glucose (or other select six-carbon sugars) to exhibit a swarming phenotype (18). E. coli K-12 has similarly been observed to require glucose for swarming motility; the reason for this is not known (96).
Available carbon also has an effect on twitching motility. Nutrient-supplemented conditions, achieved upon adding tryptone or bovine serum albumin (BSA) to minimal media, have been shown to reduce twitching motility in P. aeruginosa by inducing a switch in how the bacteria deploy TFP (97). Neisseria gonorrhoeae twitching motility requires both l-glutamine and pyruvate (30).
Though required for surface motility in some species, as mentioned above, glucose inhibits surface motility in other species. Once thought to be nonmotile, it is now known that Clostridium perfringens produces TFP. C. perfringens TFP motility is inhibited in the presence of glucose and other readily metabolized sugars, as it was shown that pilT and pilD genes required for TFP production are downregulating via catabolite repression (98, 99) and possibly other factors. Exploratory sliding of Streptomyces venezuelae specifically requires the presence of trimethylamine (and low glucose) (100), which is obtained when utilizing yeast and fungi as food.
Nitrogen.
Nitrogen source can induce an effect on swarming. In P. aeruginosa, nitrogen metabolism is linked with biosurfactant production. Nitrogen exhaustion activates rhamnolipid production, though it does not favor production down the line. Rhamnolipid production is inhibited by ammonia, glutamine, asparagine, and arginine as nitrogen sources; nitrate, glutamate, and aspartate all serve to promote production (66, 101), indicating that these nitrogen sources would promote swarming. A nitrogen-related small RNA (sRNA) (NrsZ) induced under nitrogen limitation has also been shown to regulate swarming motility in P. aeruginosa through posttranscriptional control of rhlA (102), further connecting nitrogen availability and swarming in that species. Armbruster et al. (103) identified the following five swarming cues for P. mirabilis that were also found in human urine: l-arginine, dl-histidine, l-glutamine, malate, and dl-ornithine. When added to basal medium, these nitrogen-associated amino acid cues resulted in various swarm diameters.
Phosphorus.
Phosphorus is another essential nutrient for bacteria used in many metabolic processes and energy storage. Phosphorus as an inorganic polyphosphate has been shown to be instrumental in the motility of most bacterial pathogens. These pathogens include P. aeruginosa, Klebsiella pneumoniae, Vibrio cholerae, and Salmonella Typhimurium, all of which, without the ppk gene that encodes the polyphosphate kinase that allows polyphosphate synthesis from ATP, are swarming deficient due to the interruption of flagellar rotation (104). Phosphorus is also important for swarming of S. liquefaciens, as the expression of flagella and phospholipase is controlled by the same operon (70). P. aeruginosa swarming is linked to phosphate-deficient conditions. Swarming decreases when phosphate concentrations transition from high to sufficient but increases when transitioning from sufficient to insufficient (105). Phosphate uptake in P. aeruginosa has also been connected to quorum sensing (QS) signaling, which is necessary for swarming motility; hydroxyl-alkyl-quinolone signal production is enhanced under low-phosphate conditions, and production is halted altogether when the phoB gene is deleted (106).
Twitching motility can also be dependent on the presence of phosphate, particularly inorganic polyphosphate, as mentioned previously in connection with swarming and flagellar motion. A study by Rashid and Kornberg (107) found that P. aeruginosa mutants deficient in polyphosphate kinase were also deficient in twitching. The mutants, however, still formed apparently normal TFP when examined by electron microscopy and were motile, indicating that the ability to use inorganic polyphosphate is more important for controlling pilus extension and retraction than for formation. P. aeruginosa has also been shown to deploy TFP in such a way as to enhance twitching motility when medium is depleted of phosphorus (97).
Oxygen.
The effect of oxygen on bacterial swimming and chemotaxis is well established (108, 109), but its effects on surface motility have been less well explored. This is somewhat surprising, given that many pathogens thrive in low-oxygen environments. Oxygen is not even always required; Clostridium species are obligate anaerobes and yet still exhibit swarming and twitching motilities. P. aeruginosa, for example, thrives in the cystic fibrosis lung, where oxygen levels are closer to 13% (compared to 21% atmospheric), and carbon dioxide levels can reach up to 6% (compared to 0.03% atmospheric) when the airway is clogged with mucus (110). In vitro, P. aeruginosa swarming is impaired when the environment is depleted of oxygen; however, enrichment with CO2 allows P. aeruginosa to spread on high agar concentrations (23). This spreading did not require flagella, pili, or morphological changes but was dependent upon the presence of a surfactant, indicating that it might be a type of sliding, based on the definitions used in this review.
Trace nutrients.
Various micronutrients are essential for cell development and can have major influences on surface motility. Trace nutrients mostly affect surface motility via metabolic or signaling processes, rather than intrinsic properties of the element. Here, we describe the various responses bacteria can have to select trace nutrients and how those can affect surface motility.
Iron.
One of the most important micronutrients is iron. It is often used in enzymes and metabolic processes but is most abundant in the poorly soluble ferric form. Bacteria need specialized systems for binding and acquiring iron; these often take the form of siderophores, which are small high-affinity iron-chelating compounds that increase ferric solubility for the cell. Bacteria are known to recognize their own siderophores, as well as those of other bacterial species (xenosiderophores), so they can steal iron from surrounding competitors (111, 112).
Iron does not seem to influence swarming species in one particular way. Lin et al. (113) found that ferric iron availability determines when swarming initiates in S. marcescens CH-1, though it had no effect on swarm expansion rate. High concentrations of Fe3+ delayed swarm initiation, while low levels reduced the lag phase. Hao et al. (114) showed that Salmonella enterica requires siderophores to colonize alfalfa roots. Mutants deficient in siderophore production were unable to swarm and thereby unable to colonize the host plant surface. A similar effect was shown in P. aeruginosa PA14, where interruptions in the transport of siderophores, pyoverdin, and pyochelin significantly reduced swarming (115). Böttcher and Clardy (111) showed that the siderophore avaroferrin, produced by marine bacterium Shewanella algae, inhibits the swarming motility of Vibrio alginolyticus. However, when applied with excess ferric iron, the inhibitory action was eliminated, indicating that avaroferrin either limited V. alginolyticus access to iron necessary for swarming or could not block swarming in its iron-bound form. Another organism of the genus, Vibrio parahaemolyticus, uses iron regulation in its swarming as well but in a different form. For V. parahaemolyticus, iron-limiting conditions enhance swarming motility, at least in part, by signaling the synthesis of lateral flagella (9). Lateral flagellar expression is also connected to another micronutrient, calcium, which increases swarming when present in high concentrations (116).
Iron levels can also affect twitching motility. Iron-limiting conditions can induce twitching, as seen with P. aeruginosa (26, 97, 117, 118). Interestingly, iron-limited conditions are also connected to the production of rhamnolipid (discussed above, with other swarming surfactants). Generally, surfactants are not understood to be required for twitching motility (and may even inhibit adhesion), but a rhamnolipid-deficient mutant under iron-limited conditions was limited for twitching until complemented with a plasmid containing the rhlAB operon (119). This connection to surfactant could be explained by a quorum sensing cascade. P. aeruginosa uses a dual-cascade QS signaling system involving the las and rhl systems, and in low-iron environments, it has been reported that twitching requires rhlI, which encodes the butyryl homoserine lactone QS signal synthase (118). However, a detailed mechanism between the P. aeruginosa rhl system and twitching motility has not yet been established.
The effects of iron on motility have also been observed in the nonflagellated opportunistic pathogen Acinetobacter baumannii. A. baumannii expresses multiple iron acquisition systems, making it adept at sequestering iron from its environment and allowing it to thrive in low-iron environments (like a human host) (120). In this species, however, twitching motility is increased with increased extracellular iron (81, 121). In Xylella fastidiosa, iron regulates the transcription of pilUT and pilNOP, among others, which are involved in TFP function (122). Through these examples, we see that surface motility is closely associated with the availability of this important nutrient.
Calcium.
Calcium has a major effect on twitching in multiple species. Womack et al. found that M. xanthus surface motility (both A and S types) is dependent upon calcium concentrations between 0.1 and 0.3 mM (123). Cyanobacterial surface motility and phototaxis have also been linked with calcium availability (124), where “thick” pili are analogous to TFP and are required to maintain motility (125). P. aeruginosa does not exhibit normal pilus function when it loses its calcium-binding site in the C-terminal domain PilY1 protein, indicating that calcium binding and release are essential for forming functional pili. The same study also concluded that when calcium is bound to pili, pilus retraction is inhibited, revealing calcium as a twitching regulator (28). Similar binding requirements have been seen in Kingella kingae, an important pathogen in young children. K. kingae produces two PilC-like proteins, PilC1 and PilC2, which act as adhesins at the end of TFP. Both of these proteins bind calcium, but the binding site on PilC1 is required for twitching motility (126).
As many human fluids have high calcium concentrations and twitching has been shown to help in host colonization, calcium contributes to the virulence of bacteria and human infection. Calcium can also be important for plant pathogens, like Xylella fastidiosa, that forms biofilms in the xylem vessels of plants, cutting off water supply to specific parts of the plant. Cruz et al. saw that in high-calcium environments, X. fastidiosa cells had significant increases in colony fringe size and moved at higher speeds than those cells where the calcium chelator EGTA was present. They also saw that calcium increased attachment strength to both surfaces and other cells (127). Without a PilY1 homolog containing a calcium binding motif, this twitching enhancement with calcium was abolished. Interestingly, this study also found that two other divalent cations, zinc and copper, significantly decreased twitching motility (128).
OPEN QUESTIONS
Role of chemotaxis in directing swarming.
Chemotaxis was originally found to be a behavior in which flagellum-mediated swimming cells sense a chemical gradient and move toward environments that favor growth. This behavior is well described for E. coli, which alternates the direction of flagellar rotation, leading to oscillating rounds of straight swimming and tumbling as flagella rotate in counterclockwise (CCW) and clockwise (CW) directions, respectively (129).
Surface-motile cells must sense the surface, respond to changes in viscosity, and retain the ability to move toward or away from environmental cues. Efficient surface motility couples external physical and chemical signals to internal pathways using chemotaxis and chemotaxis-like pathways that help microbes navigate life on a surface. “Chemotactic-like” behavior has been observed for several bacterial species, but the regulation of a complete system homologous to swimming chemotaxis has yet to be elucidated. Currently, chemotaxis pathways have been found to play some role in swarming and twitching but not gliding or sliding, which do not require pili or flagella (42, 44, 130, 131). However, it is not currently clear whether canonical chemotaxis regulation actually influences swarming, as this motility still occurs when CheY is uncoupled from the flagellar machinery (132). Links between the Wsp (133, 134), CheRI (135), and Sad (136) protein systems in P. aeruginosa all point to the possibility of true swarming regulation via signal transduction. Continued investigation of mechanosensing (137–139) cues and regulation in the context of regulating surface motility promises to greatly inform our understanding.
The che mutants further highlight important changes in flagella. Such che mutants of S. Typhimurium have shorter and fewer flagella due to the downregulation of “late” motility genes (18). Flagella, which function downstream in classic chemotaxis, in swarming possess the ability to sense hydration outside the bacterial cell. When hydration is limited, flagella prematurely stop their inherent growth by blocking flagellar subunit synthesis and secreting a transcription inhibitor (73). This flagellar adaptation keeps bacteria from attempting to move on a surface that is too dry, which could damage the bacterial envelope. Additionally, some flagella have developed the ability to reverse motor direction through a chemotactic-like response, allowing increased motility on drier surfaces (132).
Impact of chemotactic regulation on twitching.
The Pil-Chp pathways are required for proper pilus assembly and function, as they are necessary for twitching motility (54, 140). Growth of P. aeruginosa on a surface increases levels of the secondary messenger, cAMP, dependent on pili and the Pil-Chp pathways (141). The cAMP messenger, interacting with the protein receptor, Vfr, in turn upregulates virulence factors (142) and increases nonreversible surface attachment (143). Activation of the Pil-Chp pathways occurs with surface exposure, without the addition of any chemical signals. Activation requires both pilus extension and retraction, exhibiting increased activation with higher percentages of agarose (144). Once twitching has been initiated, this surface motility is known to increase a phosphatidylethanolamine (PE) gradient (87). Oliveira et al., using single-cell tracking in microfluidic devices, showed that cells are capable of a chemotactic response that in appearance is reminiscent of classic chemotaxis. Twitching cells use pili located at the two poles to allow movement in different directions. In the presence of a chemoattractant, cells preferentially move toward the attractant, while a pilG mutant does not exhibit this bias (145).
INTERCONNECTED MOTILITY MODES AND CONDITIONAL PRODUCTION OF SLIDING AGENTS
M. xanthus TFP and gliding surface motilities generally occur at the same time. P. aeruginosa swarming and twitching are not known to coincide. Numerous twitching, swarming, and gliding motilities must be influenced by sliding that has been discerned for impaired mutants of the same species. Given the many exogenous nutrient and surface cues that have been identified for select instances and species, much remains to be placed in context to understand how bacteria utilize surface motility to colonize their native environments.
Collective motion and switching between motility modes.
In most circumstances, these surface-motile bacteria are moving not just in groups, but as a group. Such behavior is described as collective motion. Some research has worked to define the behavior of single cells within motile groups (146–148), but much remains to be learned about the regulation of single cells and group behaviors that control bacterial surface motility and colonization of new surfaces. For example, it is not at all clear if the transcriptional and translational profiles of single cells within motile groups are equivalent. Additionally, for those bacteria that exhibit multiple surface motility modes, the cues and regulation that promote one motility mode versus another are also very unclear.
OUTLOOK
Mechanisms to effectively identify and colonize favorable surface growth environments are a great advantage to bacteria. Surface-motile bacteria explore environments and spread over and colonize surfaces to find nutrients and escape toxins. The machinery involved can have multiple uses, can simultaneously influence motility, and could offer different advantages in different environments (148, 149). We have only recently started to understand the sensitivity of these motility responses to the surrounding environment. Surface firmness, wetness, temperature, nutrients, and cell densities can all play a role in the surface motilities of bacteria and are all interconnected within an environment. The two-dimensional (2D) and three-dimensional (3D) scales of bacterial surface motility are largely neglected and require further study.
Also, many of the environmental factors discussed here are interconnected. Temperature correlates to humidity, which in turn controls surface wetness; surface wetness can also be determined by the firmness of the surface, as high agar concentrations retain water better. Oxygen effects are likely connected with iron or other trace metals for many aerobic bacteria. Most research addressing surface motility phenotypes and regulation would benefit from a more rigorous examination of the base lab assay we utilize for such research. Consideration of well-defined physics and chemistry concepts (e.g., Raoult's law, Marangoni effect, and relative humidity) to the design, use, and interpretation of these assays will significantly improve our potential to understand bacterial surface motility.
The nutrients available to the bacteria, be it nitrogen, calcium, iron, or carbon, can enhance or inhibit movement, as the metabolic cycles for these nutrients are often linked to cell machinery and excretions required for motility. Excretions that facilitate motility are also often connected to quorum sensing, meaning that production is dependent on cell concentrations. Careful continued study of bacterial motility in response to changes in interconnected surface cues is needed to further understand the forces that guide these motilities in order to gain important insight concerning microbial motility responses in relevant but complex environments, such as sand grains, root surfaces, or eukaryotic host cells.
ACKNOWLEDGMENTS
Support for this work was provided by NIH (NIAID) under grants R21AI109417 and R01AI113219 (to J.D.S.) and with support from the Indiana CTSI (to A.A.W.), funded in part by award KL2TR001106 from the NIH/NCATS Clinical and Translational Sciences Award.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Biographies

Anne E. Mattingly is currently a Ph.D. candidate in environmental engineering at the University of Notre Dame. She earned her B.S. in chemical and biological engineering from Colorado State University in 2013 and shortly thereafter began her Ph.D. work in the Shrout lab at Notre Dame. Her research focuses on the surface motility of the opportunistic pathogen Pseudomonas aeruginosa and how it responds to various environmental cues in the absence of key quorum sensing molecules.

Abigail A. Weaver received her Ph.D. in biochemistry from the University of Notre Dame. She is currently conducting postdoctoral research in the Shrout lab at the University of Notre Dame. Her past research was in the development of paper-based sensors using chemical and microbial responses to identify counterfeit and low-quality pharmaceuticals. Continuing her work with microbes, she is now studying the microbial communities of prosthetic joint infections and the impact of polymicrobial interactions on bacterial movement.

Aleksandar Dimkovikj received a B.S. in marine science and a B.S. in biochemistry from Coastal Carolina University in the fall of 2014. After those studies, he was employed as a temporary field and laboratory technician by the Environmental Quality Laboratory at Coastal Carolina University examining Enterobacter spp. in river samples and remediated wastewater. In the summer of 2015, he began a Ph.D. program at the University of Notre Dame with a focus in environmental microbiology. His recent research in the Shrout lab has primarily targeted the behavior of Pseudomonas aeruginosa under various nutrient conditions under different stressors.

Joshua D. Shrout received his Ph.D. in environmental engineering from the University of Iowa and then conducted postdoctoral research with Matthew Parsek in microbiology at the University of Washington. He is currently an associate professor in the Department of Civil and Environmental Engineering and Earth Sciences and the Department of Biological Sciences at the University of Notre Dame. Since 2003, his research has investigated the behavior and regulation of bacteria on surfaces in response to chemical and physical cues of the surrounding environment and neighboring organisms.
REFERENCES
- 1.Wei Y, Wang XL, Liu JF, Nememan I, Singh AH, Weiss H, Levin BR. 2011. The population dynamics of bacteria in physically structured habitats and the adaptive virtue of random motility. Proc Natl Acad Sci U S A 108:4047–4052. doi: 10.1073/pnas.1013499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrichsen J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol Rev 36:478–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fauvart M, Phillips P, Bachaspatimayum D, Verstraeten N, Fransaer J, Michiels J, Vermant J. 2012. Surface tension gradient control of bacterial swarming in colonies of Pseudomonas aeruginosa. Soft Matter 8:70–76. doi: 10.1039/C1SM06002C. [DOI] [Google Scholar]
- 5.Jarrell K, McBride M. 2008. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 6.Hayward NJ, Incledon GM, Spragg JE. 1978. Effect of firm agar on the swarming of Proteus and Clostridium species and on the colonies of clinically important bacteria (plates IX–XI). J Med Microbiol 11:155–164. doi: 10.1099/00222615-11-2-155. [DOI] [PubMed] [Google Scholar]
- 7.Ingham CJ, Ben Jacob E. 2008. Swarming and complex pattern formation in Paenibacillus vortex studied by imaging and tracking cells. BMC Microbiol 8:36. doi: 10.1186/1471-2180-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Be'er A, Smith RS, Zhang HP, Florin E-L, Payne SM, Swinney HL. 2009. Paenibacillus dendritiformis bacterial colony growth depends on surfactant but not on bacterial motion. J Bacteriol 191:5758–5764. doi: 10.1128/JB.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarter L, Silverman M. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol 4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 10.Gygi D, Rahman MM, Lai HC, Carlson R, Guard-Petter J, Hughes C. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol Microbiol 17:1167–1175. doi: 10.1111/j.1365-2958.1995.mmi_17061167.x. [DOI] [PubMed] [Google Scholar]
- 11.Partridge J, Harshey R. 2013. More than motility: Salmonella flagella contribute to overriding friction and facilitating colony hydration during swarming. J Bacteriol 195:919–929. doi: 10.1128/JB.02064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge JD, Harshey RM. 2013. Swarming: flexible roaming plans. J Bacteriol 195:909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberl L, Molin S, Givskov M. 1999. Surface motility of Serratia liquefaciens MG1. J Bacteriol 181:1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuson HH, Copeland MF, Carey S, Sacotte R, Weibel DB. 2013. Flagellum density regulates Proteus mirabilis swarmer cell motility in viscous environments. J Bacteriol 195:368–377. doi: 10.1128/JB.01537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamatkar N, Shrout J. 2011. Surface hardness impairment of quorum sensing and swarming for Pseudomonas aeruginosa. PLoS One 6:e20888. doi: 10.1371/journal.pone.0020888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 17.Berg H. 2005. Swarming motility: it better be wet. Curr Biol 15:R599–R600. doi: 10.1016/j.cub.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Harshey RM, Matsuyama T. 1994. Dimorphic transition in Escherichia coli and Salmonella Typhimurium surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci U S A 91:8631–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 20.Shrout J, Chopp D, Just C, Hentzer M, Givskov M, Parsek M. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 21.Caiazza NC, Shanks RMQ, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Déziel E, Lépine F, Milot S, Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- 23.Nozawa T, Tanikawa T, Hasegawa H, Takahashi C, Ando Y, Matsushita M, Nakagawa Y, Matsuyama T. 2007. Rhamnolipid-dependent spreading growth of Pseudomonas aeruginosa on a high-agar medium: marked enhancement under CO2-rich anaerobic conditions. Microbiol Immunol 51:703–712. doi: 10.1111/j.1348-0421.2007.tb03959.x. [DOI] [PubMed] [Google Scholar]
- 24.Ghelardi E, Salvetti S, Ceragioli M, Gueye SA, Celandroni F, Senesi S. 2012. Contribution of surfactin and SwrA to flagellin expression, swimming, and surface motility in Bacillus subtilis. Appl Environ Microbiol 78:6540–6544. doi: 10.1128/AEM.01341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattingly AE, Kamatkar N, Borlee BR, Shrout JD. 2018. Multiple environmental factors influence the importance of the phosphodiesterase DipA upon Pseudomonas aeruginosa swarming. Appl Environ Microbiol doi: 10.1128/aem.02847-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burrows L. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 27.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 28.Orans J, Johnson M, Coggan K, Sperlazza J, Heiniger R, Wolfgang M, Redinbo M. 2010. Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc Natl Acad Sci U S A 107:1065–1070. doi: 10.1073/pnas.0911616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skerker JM, Berg HC. 2001. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A 98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 31.McBride MJ. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol 55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 32.Nan B. 2017. Bacterial gliding motility: rolling out a consensus model. Curr Biol 27:R154–R156. doi: 10.1016/j.cub.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Nan B, Zusman DR. 2011. Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet 45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyata M. 2010. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol 64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 35.Mignot T. 2007. The elusive engine in Myxococcus xanthus gliding motility. Cell Mol Life Sci 64:2733–2745. doi: 10.1007/s00018-007-7176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faure LM, Fiche JB, Espinosa L, Ducret A, Anantharaman V, Luciano J, Lhospice S, Islam ST, Treguier J, Sotes M, Kuru E, Van Nieuwenhze MS, Brun YV, Theodoly O, Aravind L, Nollmann M, Mignot T. 2016. The mechanism of force transmission at bacterial focal adhesion complexes. Nature 539:530–535. doi: 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nan BY, Zusman DR. 2016. Novel mechanisms power bacterial gliding motility. Mol Microbiol 101:186–193. doi: 10.1111/mmi.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shrivastava A, Roland T, Berg Howard C. 2016. The screw-like movement of a gliding bacterium is powered by spiral motion of cell-surface adhesins. Biophys J 111:1008–1013. doi: 10.1016/j.bpj.2016.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hölscher T, Kovacs AT. 2017. Sliding on the surface: bacterial spreading without an active motor. Environ Microbiol 19:2537–2545. doi: 10.1111/1462-2920.13741. [DOI] [PubMed] [Google Scholar]
- 40.Gupta KR, Kasetty S, Chatterji D. 2015. Novel functions of (p)ppGpp and cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl Environ Microbiol 81:2571–2578. doi: 10.1128/AEM.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peschel A, Otto M. 2013. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol 11:667. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fall R, Kearns DB, Nguyen T. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol 6:31. doi: 10.1186/1471-2180-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogales J, Vargas P, Farias GA, Olmedilla A, Juan SJ, Gallegos MT. 2015. FleQ coordinates flagellum-dependent and -independent motilities in Pseudomonas syringae pv. tomato DC3000. Appl Environ Microbiol 81:7533–7545. doi: 10.1128/AEM.01798-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray TS, Kazmierczak BI. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190:2700–2708. doi: 10.1128/JB.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park SY, Pontes MH, Groisman EA. 2015. Flagella-independent surface motility in Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 112:1850–1855. doi: 10.1073/pnas.1422938112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Difco. 2009. Difco & BBL manual: manual of microbiological culture media, 2nd ed Becton, Dickinson and Company, Sparks, MD. [Google Scholar]
- 47.Morales-Soto N, Anyan ME, Mattingly AE, Madukoma CS, Harvey CW, Alber M, Déziel E, Kearns DB, Shrout JD. 2015. Preparation, imaging, and quantification of bacterial surface motility assays. J Vis Exp doi: 10.3791/52338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin CC, Casida LE Jr.. 1984. Gelrite as a gelling agent in media for the growth of thermophilic microorganisms. Appl Environ Microbiol 47:427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansson PE, Lindberg B, Sandford PA. 1983. Structural studies of gellan gum, an extracellular polysaccharide elaborated by Pseudomonas elodea. Carbohydr Res 124:135–139. doi: 10.1016/0008-6215(83)88361-X. [DOI] [Google Scholar]
- 50.Kang KS, Veeder GT, Mirrasoul PJ, Kaneko T, Cottrell IW. 1982. Agar-like polysaccharide produced by a Pseudomonas species–production and basic properties. Appl Environ Microbiol 43:1086–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Neill MA, Selvendran RR, Morris VJ. 1983. Structure of the acidic extracellular gelling polysaccharide produced by Pseudomonas-Elodea. Carbohydr Res 124:123–133. doi: 10.1016/0008-6215(83)88360-8. [DOI] [PubMed] [Google Scholar]
- 52.Shungu D, Valiant M, Tutlane V, Weinberg E, Weissberger B, Koupal L, Gadebusch H, Stapley E. 1983. Gelrite as an agar substitute in bacteriological media. Appl Environ Microbiol 46:840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Semmler AB, Whitchurch CB, Mattick JS. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 54.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 55.Tambalo DD, Yost CK, Hynes MF. 2010. Characterization of swarming motility in Rhizobium leguminosarum bv. viciae. FEMS Microbiol Lett 307:165–174. doi: 10.1111/j.1574-6968.2010.01982.x. [DOI] [PubMed] [Google Scholar]
- 56.Morales-Soto N, Anyan ME, Mattingly AE, Madukoma CS, Harvey CW, Alber M, Deziel E, Kearns DB, Shrout JD. 2015. Preparation, imaging, and quantification of bacterial surface motility assays. J Vis Exp doi: 10.3791/52338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee KS, Ivanova N, Starov VM, Hilal N, Dutschk V. 2008. Kinetics of wetting and spreading by aqueous surfactant solutions. Adv Colloid Interface 144:54–65. doi: 10.1016/j.cis.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Afsar-Siddiqui AB, Luckham PF, Matar OK. 2003. The spreading of surfactant solutions on thin liquid films. Adv Colloid Interface Sci 106:183–236. doi: 10.1016/S0001-8686(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 59.Härth M, Schubert DW. 2012. Simple approach for spreading dynamics of polymeric fluids. Macromol Chem Phys 213:654–665. doi: 10.1002/macp.201100631. [DOI] [Google Scholar]
- 60.Troian SM, Wu XL, Safran SA. 1989. Fingering instability in thin wetting films. Phys Rev Lett 62:1496–1499. doi: 10.1103/PhysRevLett.62.1496. [DOI] [PubMed] [Google Scholar]
- 61.Patrick JE, Kearns DB. 2009. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J Bacteriol 191:7129–7133. doi: 10.1128/JB.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tecon R, Or D. 2016. Bacterial flagellar motility on hydrated rough surfaces controlled by aqueous film thickness and connectedness. Sci Rep 6:19409. doi: 10.1038/srep19409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dechesne A, Wang G, Gülez G, Or D, Smets BF. 2010. Hydration-controlled bacterial motility and dispersal on surfaces. Proc Natl Acad Sci U S A 107:14369–14372. doi: 10.1073/pnas.1008392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purcell EM. 1977. Life at low Reynolds number. Am J Phys 45:3–11. doi: 10.1119/1.10903. [DOI] [Google Scholar]
- 65.Conrad JC. 2012. Physics of bacterial near-surface motility using flagella and type IV pili: implications for biofilm formation. Res Microbiol 163:619–629. doi: 10.1016/j.resmic.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 66.Soberón-Chávez G, Lépine F, Déziel E. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 67.Tremblay J, Richardson A-P, Lépine F, Déziel E. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 9:2622–2630. doi: 10.1111/j.1462-2920.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 68.Kobayashi K, Kanesaki Y, Yoshikawa H. 2016. Genetic analysis of collective motility of Paenibacillus sp. NAIST15-1. PLoS Genet 12:e1006387. doi: 10.1371/journal.pgen.1006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Delden C, Iglewski BH. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis 4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Givskov M, Eberl L, Molin S. 1997. Control of exoenzyme production, motility and cell differentiation in Serratia liquefaciens. FEMS Microbiol Lett 148:115–122. doi: 10.1111/j.1574-6968.1997.tb10276.x. [DOI] [Google Scholar]
- 71.Toguchi A, Siano M, Burkart M, Harshey RM. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol 182:6308–6321. doi: 10.1128/JB.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen B, Turner L, Berg H. 2007. The wetting agent required for swarming in Salmonella enterica serovar Typhimurium is not a surfactant. J Bacteriol 189:8750–8753. doi: 10.1128/JB.01109-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey R. 2005. Sensing wetness: a new role for the bacterial flagellum. EMBO J 24:2034–2042. doi: 10.1038/sj.emboj.7600668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellison CK, Kan JB, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke ZL, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hug I, Deshpande S, Sprecher KS, Pfohl T, Jenal U. 2017. Second messenger-mediated tactile response by a bacterial rotary motor. Science 358:531–534. doi: 10.1126/science.aan5353. [DOI] [PubMed] [Google Scholar]
- 76.O'Toole GA, Wong GCL. 2016. Sensational biofilms: surface sensing in bacteria. Curr Opin Microbiol 30:139–146. doi: 10.1016/j.mib.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cusick K, Lee Y-Y, Youchak B, Belas R. 2012. Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation. J Bacteriol 194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCarter LL. 2004. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 79.Liu H, Kang Y, Genin S, Schell MA, Denny TP. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215–3229. doi: 10.1099/00221287-147-12-3215. [DOI] [PubMed] [Google Scholar]
- 80.Clemmer KM, Bonomo RA, Rather PN. 2011. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McQueary C, Kirkup B, Si Y, Barlow M, Actis L, Craft D, Zurawski D. 2012. Extracellular stress and lipopolysaccharide modulate Acinetobacter baumannii surface-associated motility. J Microbiol 50:434–443. doi: 10.1007/s12275-012-1555-1. [DOI] [PubMed] [Google Scholar]
- 82.Yeung A, Parayno A, Hancock R. 2012. Mucin promotes rapid surface motility in Pseudomonas aeruginosa. mBio 3:e00073-. doi: 10.1128/mBio.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rzhepishevska O, Hakobyan S, Ruhal R, Gautrot J, Barbero D, Ramstedt M. 2013. The surface charge of anti-bacterial coatings alters motility and biofilm architecture. Biomater Sci 1:589–602. doi: 10.1039/c3bm00197k. [DOI] [PubMed] [Google Scholar]
- 84.Bucior I, Pielage JF, Engel JN. 2012. Pseudomonas aeruginosa pili and flagella mediate distinct binding and signaling events at the apical and basolateral surface of airway epithelium. PLoS Pathog 8:e1002616. doi: 10.1371/journal.ppat.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee SW, Higashi DL, Snyder A, Merz AJ, Potter L, So M. 2005. PilT is required for PI(3,4,5)P3-mediated crosstalk between Neisseria gonorrhoeae and epithelial cells. Cell Microbiol 7:1271–1284. doi: 10.1111/j.1462-5822.2005.00551.x. [DOI] [PubMed] [Google Scholar]
- 86.Bernard SC, Simpson N, Join-Lambert O, Federici C, Laran-Chich M-P, Maïssa N, Bouzinba-Ségard H, Morand PC, Chretien F, Taouji S, Chevet E, Janel S, Lafont F, Coureuil M, Segura A, Niedergang F, Marullo S, Couraud P-O, Nassif X, Bourdoulous S. 2014. Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization. Nat Med 20:725. doi: 10.1038/nm.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kearns DB, Robinson J, Shimkets LJ. 2001. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol 183:763–767. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kearns DB, Shimkets LJ. 1998. Chemotaxis in a gliding bacterium. Proc Natl Acad Sci U S A 95:11957–11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller RM, Tomaras AP, Barker AP, Voelker DR, Chan ED, Vasil AI, Vasil ML. 2008. Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids: specificity and metabolic requirements. J Bacteriol 190:4038–4049. doi: 10.1128/JB.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kearns DB, Shimkets LJ. 2001. Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol 9:126–129. doi: 10.1016/S0966-842X(01)01948-5. [DOI] [PubMed] [Google Scholar]
- 91.Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O. 2012. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Natl Acad Sci U S A 109:10036–10041. doi: 10.1073/pnas.1120979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vicario JC, Dardanelli MS, Giordano W. 2015. Swimming and swarming motility properties of peanut-nodulating rhizobia. FEMS Microbiol Lett 362:6. doi: 10.1093/femsle/fnu038. [DOI] [PubMed] [Google Scholar]
- 93.Lai HC, Soo PC, Wei JR, Yi WC, Liaw SJ, Horng YT, Lin SM, Ho SW, Swift S, Williams P. 2005. The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J Bacteriol 187:3407–3414. doi: 10.1128/JB.187.10.3407-3414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soutourina OA, Semenova EA, Parfenova VV, Danchin A, Bertin P. 2001. Control of bacterial motility by environmental factors in polarly flagellated and peritrichous bacteria isolated from Lake Baikal. Appl Environ Microbiol 67:3852–3859. doi: 10.1128/AEM.67.9.3852-3859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Köhler T, Curty L, Barja F, van Delden C, Pechère J. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. 2007. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol 189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ni L, Yang S, Zhang R, Jin Z, Chen H, Conrad JC, Jin F. 2016. Bacteria differently deploy type-IV pili on surfaces to adapt to nutrient availability. NPJ Biofilms Microbiomes 2:15029. doi: 10.1038/npjbiofilms.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mendez M, Huang IH, Ohtani K, Grau R, Shimizu T, Sarker M. 2008. Carbon catabolite repression of type IV pilus-dependent gliding motility in the anaerobic pathogen Clostridium perfringens. J Bacteriol 190:48–60. doi: 10.1128/JB.01407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu H, Bouillaut L, Sonenshein A, Melville S. 2013. Use of a mariner-based transposon mutagenesis system to isolate Clostridium perfringens mutants deficient in gliding motility. J Bacteriol 195:629–636. doi: 10.1128/JB.01288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones SE, Ho L, Rees CA, Hill JE, Nodwell JR, Elliot MA. 2017. Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife 6:e21738. doi: 10.7554/eLife.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mulligan C, Gibbs B. 1989. Correlation of nitrogen metabolism with biosurfactant production by Pseudomonas aeruginosa. Appl Environ Microbiol 55:3016–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wenner N, Maes A, Cotado-Sampayo M, Lapouge K. 2014. NrsZ: a novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ Microbiol 16:1053–1068. doi: 10.1111/1462-2920.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Armbruster C, Hodges S, Mobley H. 2013. Initiation of swarming motility by Proteus mirabilis occurs in response to specific cues present in urine and requires excess l-glutamine. J Bacteriol 195:1305–1319. doi: 10.1128/JB.02136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rashid MH, Rao NN, Kornberg A. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol 182:225–227. doi: 10.1128/JB.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bains M, Fernandez L, Hancock REW. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ Microbiol 78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jensen V, Löns D, Zaoui C, Bredenbruch F, Meissner A, Dieterich G, Münch R, Häussler S. 2006. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J Bacteriol 188:8601–8606. doi: 10.1128/JB.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Adler J. 1966. Chemotaxis in bacteria. Science 153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 109.Armitage JP. 1997. Behavioural responses of bacteria to light and oxygen. Arch Microbiol 168:249–261. doi: 10.1007/s002030050496. [DOI] [PubMed] [Google Scholar]
- 110.Høiby N. 2006. P. aeruginosa in cystic fibrosis patients resists host defenses, antibiotics, vol 1 American Society for Microbiology, Washington, DC. [Google Scholar]
- 111.Böttcher T, Clardy J. 2014. A chimeric siderophore halts swarming Vibrio. Angew Chem Int Ed Engl 53:3510–3513. doi: 10.1002/anie.201310729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol 17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin CS, Tsai YH, Chang CJ, Tseng SF, Wu TR, Lu CC, Wu TS, Lu JJ, Horng JT, Martel J, Ojcius DM, Lai HC, Young JD. 2016. An iron detection system determines bacterial swarming initiation and biofilm formation. Sci Rep 6:36747. doi: 10.1038/srep36747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hao L-Y, Willis D, Andrews-Polymenis H, McClelland M, Barak J. 2012. Requirement of siderophore biosynthesis for plant colonization by Salmonella enterica. Appl Environ Microbiol 78:4561–4570. doi: 10.1128/AEM.07867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Strehmel J, Neidig A, Nusser M, Geffers R, Brenner-Weiss G, Overhage J. 2015. Sensor kinase PA4398 modulates swarming motility and biofilm formation in Pseudomonas aeruginosa PA14. Appl Environ Microbiol 81:1274–1285. doi: 10.1128/AEM.02832-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gode-Potratz C, Chodur DM, McCarter LL. 2010. Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J Bacteriol 192:6025–6038. doi: 10.1128/JB.00654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 118.Patriquin G, Banin E, Gilmour C, Tuchman R, Greenberg E, Poole K. 2008. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 190:662–671. doi: 10.1128/JB.01473-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Glick R, Gilmour C, Tremblay J, Satanower S, Avidan O, Déziel E, Greenberg E, Poole K, Banin E. 2010. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 192:2973–2980. doi: 10.1128/JB.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zimbler D, Penwell W, Gaddy J, Menke S, Tomaras A, Connerly P, Actis L. 2009. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 22:23–32. doi: 10.1007/s10534-008-9202-3. [DOI] [PubMed] [Google Scholar]
- 121.Harding C, Tracy E, Carruthers M, Rather P, Actis L, Munson R Jr.. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zaini PA, Fogaca AC, Lupo FGN, Nakaya HI, Vencio RZN, da Silva AM. 2008. The iron stimulon of Xylella fastidiosa includes genes for type IV pilus and colicin V-like bacteriocins. J Bacteriol 190:2368–2378. doi: 10.1128/JB.01495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Womack B, Gilmore D, White D. 1989. Calcium requirement for gliding motility in myxobacteria. J Bacteriol 171:6093–6096. doi: 10.1128/jb.171.11.6093-6096.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhaya D, Takahashi A, Grossman A. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci U S A 98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Devaki B, Nicole RB, Donald B, Arthur G. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol Microbiol 37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 126.Porsch E, Johnson M, Broadnax A, Garrett C, Redinbo M, St. Geme J III. 2013. Calcium binding properties of the Kingella kingae PilC1 and PilC2 proteins have differential effects on type IV pilus-mediated adherence and twitching motility. J Bacteriol 195:886–895. doi: 10.1128/JB.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cruz L, Cobine P, De La Fuente L. 2012. Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl Environ Microbiol 78:1321–1331. doi: 10.1128/AEM.06501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cruz LF, Parker JK, Cobine PA, De la Fuente L. 2014. Calcium-enhanced twitching motility in Xylella fastidiosa is linked to a single PilY1 homolog. Appl Environ Microbiol 80:7176–7185. doi: 10.1128/AEM.02153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 130.Stewart CR, Rossier O, Cianciotto NP. 2009. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J Bacteriol 191:1537–1546. doi: 10.1128/JB.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nogales J, Bernabeu-Roda L, Cuellar V, Soto MJ. 2012. ExpR is not required for swarming but promotes sliding in Sinorhizobium meliloti. J Bacteriol 194:2027–2035. doi: 10.1128/JB.06524-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mariconda S, Wang Q, Harshey RM. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol Microbiol 60:1590–1602. doi: 10.1111/j.1365-2958.2006.05208.x. [DOI] [PubMed] [Google Scholar]
- 133.Huangyutitham V, Guvener ZT, Harwood CS. 2013. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio 4:e00242-. doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schmidt J, Musken M, Becker T, Magnowska Z, Bertinetti D, Moller S, Zimmermann B, Herberg FW, Jansch L, Haussler S. 2011. The Pseudomonas aeruginosa chemotaxis methyltransferase CheR1 impacts on bacterial surface sampling. PLoS One 6:e18184. doi: 10.1371/journal.pone.0018184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chaban B, Hughes HV, Beeby M. 2015. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol 46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 138.Ellison C, Brun YV. 2015. Mechanosensing: a regulation sensation. Curr Biol 25:R113–R115. doi: 10.1016/j.cub.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 140.Bertrand JJ, West JT, Engel JN. 2010. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J Bacteriol 192:994–1010. doi: 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4:253–263. doi: 10.1016/S1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 143.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GC, O'Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oliveira NM, Foster KR, Durham WM. 2016. Single-cell twitching chemotaxis in developing biofilms. Proc Natl Acad Sci U S A 113:6532–6537. doi: 10.1073/pnas.1600760113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Be'er A, Harshey RM. 2011. Collective motion of surfactant-producing bacteria imparts superdiffusivity to their upper surface. Biophys J 101:1017–1024. doi: 10.1016/j.bpj.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang A, Tang WS, Si TY, Tang JX. 2017. Influence of physical effects on the swarming motility of Pseudomonas aeruginosa. Biophys J 112:1462–1471. doi: 10.1016/j.bpj.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Anyan ME, Amiri A, Harvey CW, Tierra G, Morales-Soto N, Driscoll CM, Alber MS, Shrout JD. 2014. Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 111:18013–18018. doi: 10.1073/pnas.1414661111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shi W, Zusman DR. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Natl Acad Sci U S A 90:3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shemesh M, Pasvolsky R, Zakin V. 2014. External pH is a cue for the behavioral switch that determines surface motility and biofilm formation of Alicyclobacillus acidoterrestris. J Food Prot 77:1418–1423. doi: 10.4315/0362-028X.JFP-13-425. [DOI] [PubMed] [Google Scholar]
- 151.Mauriello EM, Mignot T, Yang Z, Zusman DR. 2010. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol Mol Biol Rev 74:229–249. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Barrionuevo MR, Vullo DL. 2012. Bacterial swimming, swarming and chemotactic response to heavy metal presence: which could be the influence on wastewater biotreatment efficiency? World J Microbiol Biotechnol 28:2813–2825. doi: 10.1007/s11274-012-1091-5. [DOI] [PubMed] [Google Scholar]
- 153.Porsch EA, Kehl-Fie TE, St. Geme JW III. 2012. Modulation of Kingella kingae adherence to human epithelial cells by type IV pili, capsule, and a novel trimeric autotransporter. mBio 3:e00372-12. doi: 10.1128/mBio.00372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun H, Zusman D, Shi W. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol 10:1143–1146. doi: 10.1016/S0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 155.Huang BX, Whitchurch CB, Mattick JS. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J Bacteriol 185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Braeken K, Daniels R, Vos K, Fauvart M, Bachaspatimayum D, Vanderleyden J, Michiels J. 2008. Genetic determinants of swarming in Rhizobium etli. Microb Ecol 55:54–64. doi: 10.1007/s00248-007-9250-1. [DOI] [PubMed] [Google Scholar]
- 157.Belas M, Colwell R. 1982. Adsorption kinetics of laterally and polarly flagellated Vibrio. J Bacteriol 151:1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kojima M, Kubo R, Yakushi T, Homma M, Kawagishi I. 2007. The bidirectional polar and unidirectional lateral flagellar motors of Vibrio alginolyticus are controlled by a single CheY species. Mol Microbiol 64:57–67. doi: 10.1111/j.1365-2958.2007.05623.x. [DOI] [PubMed] [Google Scholar]
- 159.Kurre R, Maier B. 2012. Oxygen depletion triggers switching between discrete speed modes of gonococcal type IV pili. Biophys J 102:2556–2563. doi: 10.1016/j.bpj.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gu Y, Hou YG, Huang DP, Hao ZX, Wang XF, Wei Z, Jousset A, Tan SY, Xu DB, Shen QR, Xu YC, Friman VP. 2017. Application of biochar reduces Ralstonia solanacearum infection via effects on pathogen chemotaxis, swarming motility, and root exudate adsorption. Plant Soil 415:269–281. doi: 10.1007/s11104-016-3159-8. [DOI] [Google Scholar]