ABSTRACT
Experimental studies of transcriptional regulation in bacteria require the ability to precisely measure changes in gene expression, often accomplished through the use of reporter genes. However, the boundaries of promoter sequences required for transcription are often unknown, thus complicating the construction of reporters and genetic analysis of transcriptional regulation. Here, we analyze reporter libraries to define the promoter boundaries of the luxCDABE bioluminescence operon and the betIBA-proXWV osmotic stress operon in Vibrio harveyi. We describe a new method called rapid arbitrary PCR insertion libraries (RAIL) that combines the power of arbitrary PCR and isothermal DNA assembly to rapidly clone promoter fragments of various lengths upstream of reporter genes to generate large libraries. To demonstrate the versatility and efficiency of RAIL, we analyzed the promoters driving expression of the luxCDABE and betIBA-proXWV operons and created libraries of DNA fragments from these loci fused to fluorescent reporters. Using flow cytometry sorting and deep sequencing, we identified the DNA regions necessary and sufficient for maximum gene expression for each promoter. These analyses uncovered previously unknown regulatory sequences and validated known transcription factor binding sites. We applied this high-throughput method to gfp, mCherry, and lacZ reporters and multiple promoters in V. harveyi. We anticipate that the RAIL method will be easily applicable to other model systems for genetic, molecular, and cell biological applications.
IMPORTANCE Gene reporter constructs have long been essential tools for studying gene regulation in bacteria, particularly following the recent advent of fluorescent gene reporters. We developed a new method that enables efficient construction of promoter fusions to reporter genes to study gene regulation. We demonstrate the versatility of this technique in the model bacterium Vibrio harveyi by constructing promoter libraries for three bacterial promoters using three reporter genes. These libraries can be used to determine the DNA sequences required for gene expression, revealing regulatory elements in promoters. This method is applicable to various model systems and reporter genes for assaying gene expression.
KEYWORDS: reporter fusion, isothermal DNA assembly, arbitrary PCR, Vibrio harveyi, quorum sensing, bioluminescence, gene reporters
INTRODUCTION
Central to the study of bacterial physiology and development is the ability to monitor and quantify gene expression. Monitoring of gene expression is greatly aided by the use of gene reporter fusions. Transcriptional and translational fusion constructs facilitate single-cell and population-wide gene expression investigations to study the influence of regulatory factors, perform genetic screens, and visualize protein localization patterns. Typically, such reporters are cloned downstream of regulatory promoters or genes of interest and introduced into a model bacterial system, either on replicating plasmids or integrated into the genome. Numerous reporter genes have traditionally been used to assay the expression of genes such as lux (bacterial luciferase), lacZ (β-galactosidase), phoA (alkaline phosphatase), bla (β-lactamase), and cat (chloramphenicol acetyltransferase) (1, 2). However, the advent of more modern techniques has allowed for the use of fluorescent proteins such as green fluorescent protein (GFP) for these studies without the need for substrates or specialized medium (1–4).
To adequately and efficiently study the expression pattern of a particular gene, the defined regulatory region controlling promoter activity must be known. The region upstream of the promoter driving luxCDABE transcription in Vibrio harveyi is an example of a locus with a large and undefined regulatory region, which has limited studies of gene regulation. This particular promoter is of interest because it drives expression of the bioluminescence genes with a >100-fold increase in transcription and a >1,000-fold increase in bioluminescence production under activating conditions (i.e., quorum sensing) (5–10). It was previously suggested that the lux promoter requires ∼400 bp upstream of the translation start site and ∼60 bp downstream of the start codon for full activation of the cat reporter gene (9). The requirement for a large promoter region is due in part to the presence of seven binding sites for the transcription factor LuxR upstream of the primary transcription start site, each of which is necessary for maximal activation of the promoter (5, 6, 9). The ∼400-bp region of PluxCDABE is relatively large compared to some bacterial regulatory promoters (e.g., the lac promoter) but comparable in size to other promoters with evidence of cooperative binding between transcription factors and DNA looping (e.g., the araBAD promoter) (5, 11–13). Indeed, full activation of the luxCDABE promoter requires the transcription factor LuxR and nucleoid-associated protein integration host factor (IHF), and DNA looping by IHF is proposed to drive interactions between LuxR and RNA polymerase (RNAP) for transcription activation (6).
Another V. harveyi operon that has an unknown promoter region is betIBA-proXWV. The betIBA-proXWV osmoregulation genes encode proteins required for the synthesis and transport of the osmoprotectant glycine betaine (14). These genes are autoregulated by the BetI repressor and activated 3- to 10-fold by LuxR (14). There are two sites in the betIBA-proXWV promoter that have been shown to be bound by LuxR in vitro and in vivo, though the role of these sites in transcriptional regulation has not yet been tested (5, 14). For both the luxCDABE and betIBA-proXWV operons, the boundaries of the promoters are not defined, and thus, mechanistic studies of transcriptional regulation of these operons is limited.
Here, we describe a new method for rapidly generating reporter plasmids that we used to define promoter regions. The rapid arbitrary PCR insertion libraries (RAIL) method exploits the power of arbitrary PCR and isothermal DNA assembly (IDA) to insert semirandomized fragments of promoter DNA into reporter plasmids (15–17). Using RAIL, we generated libraries containing fragments of various lengths of the region upstream of the luxCDABE operon transcriptionally fused to gfp. We used flow cytometry sorting to screen the library of promoter fragments for reporter expression and next-generation sequencing to map the 3′ boundary of the luxCDABE promoter required for full activation. We also applied this method to two additional promoter regions in V. harveyi (betIBA-proXWV and VIBHAR_06912), and we demonstrated the versatility of the system by using two additional reporters, mCherry and β-galactosidase. This approach enabled us to identify the regions required for gene expression for multiple promoters and simultaneously produce usable gene reporter constructs. Our method should be widely applicable to any system for which gene reporters have been established and represents a simple and efficient technique to construct reporter fusions for molecular, genetic, and cell biology studies.
RESULTS
Measurement of transcription activation from the V. harveyi luxCDABE promoter using fluorescent reporter fusions.
To study the mechanism of LuxR regulation of the luxCDABE promoter, we constructed four reporter plasmids containing various fragments of the luxCDABE locus transcriptionally fused to gfp by traditional cloning methods (Fig. 1A). Each plasmid contains the same 5′ end (∼400 bp upstream of the luxC open reading frame [ORF]), and the 3′ ends vary as follows: (i) 2 bp after the transcription start site at −26 (pJV369), (ii) at the LuxC translation start site (pJV367), (iii) 36 bp into the luxC ORF (pSO04), and (iv) 407 bp into the luxC ORF (pJV365) (Fig. 1A). All plasmid constructs contained the seven LuxR binding sites previously found to be essential for transcriptional activation. Only plasmid pJV365 contained LuxR site H, which has previously been shown to be nonessential for activation. We chose the lengths of these fragments to investigate the requirement for the 5′ untranslated region (UTR), site H, and various lengths of the luxC ORF. We first tested LuxR activation of these reporter plasmids in Escherichia coli because expression of luxR in E. coli is sufficient to drive high levels of transcription of the luxCDABE operon (5, 10, 18), and the use of E. coli is more efficient for transformation. Activation of luxCDABE promoter transcription was assayed in E. coli strains containing either a second plasmid constitutively expressing luxR (pKM699) or an empty vector (pLAFR2). The plasmid containing the 3′ boundary 36 bp into the luxC ORF was highly expressed (Fig. 1B), which is consistent with a previous study using a nearly identical promoter fragment (see Fig. S1A in the supplemental material) (5). The strain containing pJV369 with the DNA fragment up to and including the transcription start site also displayed high levels of GFP. Activation was appreciably decreased (∼7-fold) for the pJV367 strain containing the 5′ UTR but ending at the LuxC translation start site compared to the pSO04 strain (Fig. 1B). Also, the strain containing pJV365 with 407 bp of the luxC ORF was not activated >2-fold (Fig. 1B). A similar trend was obtained when these constructs were conjugated into V. harveyi strains and the GFP expression levels in the wild-type and ΔluxR mutant strains were compared (Fig. S1B). From these data, we conclude that a promoter fragment ending 2 bp past the transcription start site is sufficient for activation. Further, we revealed that various lengths of 3′ constructs fused to gfp produce unexpected changes in gene expression across the luxCDABE promoter.
FIG 1.
Promoter fusion plasmids for the luxCDABE genes. (A) Diagram of the regions of the luxCDABE promoter present in the plasmids listed. LuxR binding sites (BS) are shown as gray boxes, and the letters correspond to LuxR binding sites A through H. Transcription start sites are indicated by black arrows. The LuxC translation start site is shown as +1. Lengths of constructs are shown relative to the LuxC translation start site. (B) Relative GFP expression per unit of OD600 (GFP/OD600) for E. coli strains containing plasmids with various luxCDABE promoter fragments fused to gfp as indicated in panel A. The strains also contain either a plasmid constitutively expressing LuxR (pKM699) or an empty vector (pLAFR2). Relative expression was calculated by dividing the values for the pKM699-containing strain by the pLAFR2-containing strain. Different letters (a, b, and c) indicate significant differences (P < 0.05; one-way analysis of variance [ANOVA], followed by Tukey's multiple-comparison test of log-transformed data; n = 3).
The RAIL method.
Our observation that various 3′ ends of the luxCDABE promoter greatly affected gene expression led us to expand our analysis of the expression profile of promoter fusions across the entire locus. Therefore, we needed to construct numerous promoter fragments transcriptionally fused to a fluorescent reporter. Instead of constructing each of these plasmids individually, we designed a cloning technique combining the power of arbitrary PCR and IDA (also called Gibson assembly) (15–17). This method enabled us to simultaneously amplify fragments of various lengths and clone them into a vector backbone to create a library in four simple steps (Fig. 2). First, arbitrary primers were used in a preliminary round of PCR in conjunction with a primer that specifically anneals to the promoter (Fig. 2, primer 1F). Four arbitrary primers were synthesized with eight sequential random nucleotides anchored at the 3′ end with two specific nucleotides: AA, TT, AT, or TA (Table S1). We chose to use A-T pairs to anchor the primer because of the low G+C content of V. harveyi. Each of these four primers also contains a linker at the 5′ end (Fig. 2, primer 1R). The first round of PCR produced a range of products that varied in length from 100 to >3,000 bp and appeared as faint smears of products, as expected for random priming (Fig. 2). For some loci, no smear could be visualized by gel electrophoresis after the first round of PCR, but this did not impact the success of the second round of amplification.
FIG 2.
Schematic of the RAIL method for constructing libraries of promoter fusions. In PCR round 1, primers 1F and 1R were used to amplify a range of products specific to the luxC promoter. The 1R primers have eight random nucleotides (N) incorporated, are anchored by two nucleotides (AA, AT, TA, or TT), and contain a linker (shown in blue). In PCR round 2, the 2F and 2R primers were used to further amplify and add a linker to the products. Primer 2F anneals just downstream of 1F and contains a linker (shown in green). Primer 2R anneals to the linker region of primer 1R. The linear plasmid backbone was prepared by either restriction digestion or PCR. The library of products was inserted into the linear plasmid backbone via IDA with the homologous sequences present in the two linker regions (green and blue). The final plasmids contain fragments of the luxC promoter fused to the reporter. The gel images shown are examples of products from PCR rounds 1 and 2 for the luxCDABE locus with arbitrary R1 primers JCV1135-1138 and F1 primer SO71 in round 1 (Table S1). The products of the round 1 reactions were used in round 2 with primers SO72 and JCV1139 (Table S1).
In the second step, the products from round 1 were further amplified with a nested primer (primer 2F) and a linker with homology to the plasmid backbone was added (Fig. 2). Primer 2R anneals to the linker on primer 1R. The second round of PCR using these primers was performed with the products from round 1 as templates. This second step served to increase the amount of DNA product and to add a linker to the 5′ end. Each reaction in round 2 produced a smear of products that contained homology to the plasmid backbone at their 5′ and 3′ ends (Fig. 2). The smear of products can also be gel extracted to the desired size. In the third step, the plasmid backbone was PCR amplified or digested by specific restriction enzymes to form a linear product (Fig. 2). In the fourth and final step, IDA was performed to clone the promoter fragments into the plasmid backbone and the mixture was transformed into E. coli to obtain isolated clones (Fig. 2).
Defining the 3′ boundary of the luxCDABE operon by RAIL.
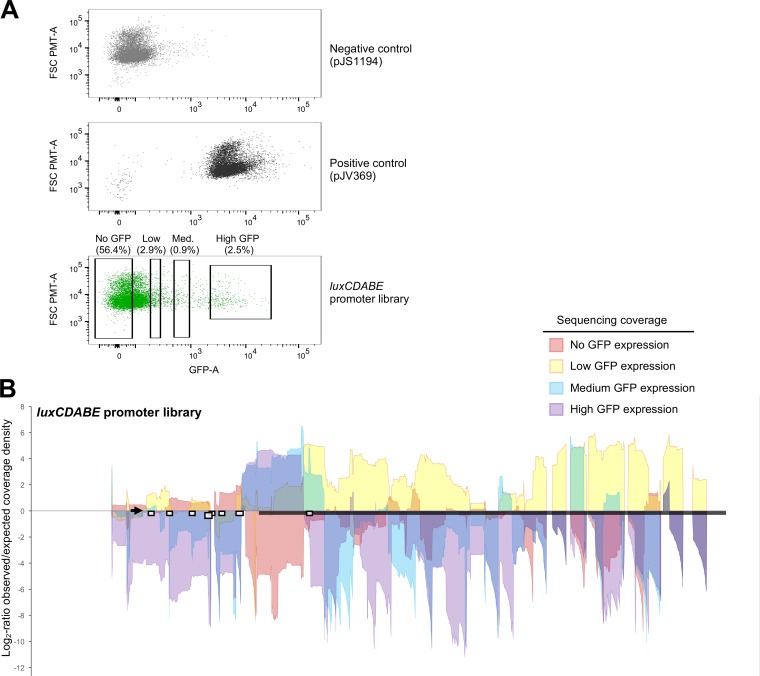
We used the RAIL method to generate a large library of plasmids with promoter fragments fused to gfp. This library had fixed 5′ ends and various 3′ ends generated by combining PCR products from four arbitrary primers, as shown in Fig. 2, and inserts ranging from ∼50 to >1,000 bp. We screened for gfp activation by fluorescence-activated cell sorting (FACS). The libraries were sorted by FACS into four groups with no, low, medium, and high GFP expression (Fig. 3A). The no-GFP pool contained cells expressing fluorescence similar to or lower than that of the negative-control strain. The high-GFP pool contained cells with fluorescence expression similar to or higher than that of the positive-control strain. The low- and medium-GFP pools were arbitrarily chosen to collect cells in the intermediate region between no GFP and high GFP without any overlap among the four bins (Fig. 3A). Illumina sequencing of the plasmid DNA from these pools enabled us to visualize the 3′-terminal end of the region cloned into the plasmid by graphing the location of the sequencing coverage (42 bp) and the 3′-terminal nucleotides (Fig. 3B; Fig. S2A). From these graphs, we pinpointed the boundary in the luxCDABE promoter required for maximum expression and showed the expression profile for promoter fragments across the entire locus (Fig. 3B; Fig. S2A). The plasmids containing promoter fragments that terminated at nucleotide +129 (relative to +1, the start of the luxC ORF) were highly enriched in the high-expression pool, and plasmids in the no-expression pool were specifically de-enriched in this same location (Fig. 3B; Fig. S2A). The high-expression pool had a clear 3′ boundary at +129, which is 16 bp upstream of LuxR site H. Thus, a DNA fragment that terminates at +129 includes LuxR sites A, B, C, D, E, F, and G (6). The observation that LuxR site H was not included in this region of high expression is consistent with previous findings that site H is nonessential for transcription activation at high cell density in V. harveyi (6). We conclude from these data that fragments with 3′ ends longer than +129 were decreased in reporter gene expression. There is also a clear edge where sequencing coverage drops off for the high-expression pool at −55 (Fig. 3B). However, the exact minimum boundary cannot be determined because we did not use every combination of anchor nucleotides in the arbitrary primers. Also, within the high-expression pool, we noted a peak of sequencing coverage that started at +36, suggesting that this is the minimum length promoter sufficient for high GFP expression in this library (Fig. S2A). Thus, promoter fragments with ends ranging from +36 through +129 yield maximum expression levels without being detrimental. The medium-expression pool contained sequences that terminated at +199, which is located 32 bp beyond LuxR site H (Fig. 3B). Plasmids with promoter fragments that extended throughout the luxC ORF past +199 had low levels of expression, whereas plasmids without GFP expression were limited to promoter regions upstream of −55 (Fig. 3B). We conclude that long promoter fragments decrease GFP expression and are not suitable reporter plasmids. We also conclude that plasmids containing promoter fragments shorter than −55 are not sufficient to activate transcription. Collectively, by analyzing the sequencing data of RAIL libraries, we located the DNA region that is sufficient for maximal transcription activation (−393 to +36), validated previous findings that LuxR sites A through G are required for activation of luxCDABE (6), and defined the expression profile for the luxCDABE locus in the context of a transcriptional fusion to gfp.
FIG 3.
Flow cytometry sorting and next-generation sequencing of the PluxCDABE-gfp library. (A) FACS GFP expression data from three E. coli cultures, (i) a negative-control strain containing an empty-vector control (pJS1194) and a plasmid expressing LuxR (pKM699), (ii) a positive-control strain containing a PluxCDABE-gfp reporter plasmid (pJV369) and pKM699, and (iii) the PluxCDABE library of plasmids in E. coli containing pKM699. Gates (boxes) indicate the cells sorted (percentage of the total population) into four bins with no, low, medium, and high GFP expression. Data were obtained with FlowJo software. (B) Genomic regions associated with differential expression of the luxCDABE locus. The nucleotide coverage of the reads (42 bp) is shown for different populations of cells with distinct levels of GFP reporter expression as indicated in the color key. Data are graphed as the nucleotide position (x axis) versus the log2 ratio of observed coverage density divided by the expected coverage density (as determined by the read counts observed in the total library). Areas with positive values in log2 observed/expected coverage densities indicate an enrichment of sequence reads in that region, and areas with negative values indicate lower-than-expected-frequency reads. The thick black bar indicates the location of the luxC ORF. The locations of LuxR binding sites are indicated by white boxes. The locus to which the 2F primer anneals is indicated by the arrow.
Defining the 3′ boundary of the betIBA-proXWV operon by RAIL.
We next used the RAIL strategy to construct reporter clones for the betIBA-proXWV operon with a different fluorescent reporter, mCherry. We screened the promoter clones individually before using the high-throughput flow cytometry method to analyze the library to test whether we could identify useful promoter clones via small-scale screening. Approximately 40 plasmids were screened by restriction digestion for inserts of various sizes, and the inserts were sequenced to determine the size of the inserted region. We observed that plasmids containing regions shorter than the predicted transcription start sites did not show any activation compared to the empty-vector control strain (Fig. 4B, pCH28 as an example). However, plasmids with larger regions that extended into the betI ORF were activated by LuxR, such as pCH50 and pCH72 (Fig. 4B). Plasmids containing the entire betI gene did not display activation (Fig. 4B, pCH75). These data show that the RAIL method can be used for small-scale screening for promoter clones by individually assaying plasmids.
FIG 4.
Expression data from the PbetIBA-proXWV-mCherry library. (A) Diagram of the regions of the betIBA-proXWV promoter present in various plasmids. LuxR binding sites (BS) and the BetI binding site are indicated. Putative transcription start sites are indicated by black arrows. The BetI translation start site is shown as +1. Lengths of constructs are shown relative to the BetI translation start site. (B) Relative mCherry expression per unit of OD600 (mCherry/OD600) for E. coli strains as calculated by dividing the values for the pKM699-carrying strain by those for the pLAFR2-carrying strain. The strains contained plasmids with various betIBA-proXWV promoter fragments fused to mCherry, as indicated in panel A. Different letters (a, b, c, and d) indicate significant differences (P < 0.05; one-way ANOVA, followed by Dunnett's multiple-comparison test; n = 3). (C) FACS data showing mCherry expression for three E. coli cultures, (i) a negative-control strain containing pCH76 and pKM699, (ii) a positive-control strain containing pCH50 and pKM699, and (iii) the PbetIBA-proXWV library in E. coli containing pKM699. Gates indicate the cells sorted (percentage of the total population) into two bins with no and high mCherry expression. (D) Genomic regions associated with differential expression of the betIBA-proXWV locus. The nucleotide coverage of the reads (42 bp) is shown for different populations of cells with distinct levels of mCherry reporter expression as indicated in the color key. Data are graphed as the nucleotide position (x axis) versus the log2 ratio of observed coverage density divided by the expected coverage density (as determined by the read counts observed in the total library). Areas with positive values in log2 observed/expected coverage densities indicate an enrichment of sequence reads in that region, and areas with negative values indicate lower-than-expected-frequency reads. The thick black bar indicates the location of the betI ORF. The locations of LuxR binding sites are indicated by white boxes. The locus to which the 2F primer anneals is indicated by the arrow.
We next synthesized a large library of betIBA-proXWV promoter fusions to mCherry by using RAIL. It is important to note that only one arbitrary primer was used to generate this library, which limited the range of PCR products across the locus. This library of clones was sorted by FACS for those that maximally expressed mCherry (Fig. 4C). The dynamic range of the betIBA-proXWV promoter is substantially smaller than that of luxCDABE, resulting in an approximately 3-fold difference in expression in the averages of the positive and negative controls. Thus, we chose to sort cells with fluorescence levels above that of the negative-control strain (Fig. 4C). In doing so, we lost cells that exhibited intermediate fluorescence levels but could therefore be assured that all of the cells we collected were expressing high levels of fluorescence. The Illumina sequencing coverage and 3′-terminal nucleotides of the DNA in the two pools was graphed (Fig. 4D; Fig. S2B). Sequencing analyses revealed the minimum 3′ boundary of the betIBA-proXWV promoter to be at −13 (Fig. 4D; Fig. S2B) relative to +1, the start of the betI ORF, suggesting that the −46 transcription start site is the primary site for this locus. The high-expression pool contained plasmids with DNA fragments up through the first portion of the betI ORF at +25, which then tapered off (Fig. 4D; Fig. S2B). Plasmids with fragments that extended more than halfway through the betI gene displayed low or no expression. Collectively, these data showed that similar to the luxCDABE locus, transcription reporters were functional if they contained DNA fragments past the 3′ boundary near the transcription start site. However, longer fragments extending into the ORF decreased reporter gene expression.
Versatility of the RAIL method for cloning with other promoters and reporter genes.
We also successfully used the RAIL technique to generate a promoter library by using the lacZ reporter for another V. harveyi gene, VIBHAR_06912, which encodes a transcription factor. VIBHAR_06912 expression is repressed by LuxR (19), and this is likely indirect repression because there are no detectable LuxR binding sites in this region (5). Using RAIL, multiple clones with various promoter lengths were generated as transcriptional fusions to lacZ, and V. harveyi strains were assayed for β-galactosidase activity (Fig. 5A). All of the plasmids with long promoters were repressed by LuxR in the wild-type strain compared to the ΔluxR mutant strain (Fig. 5B). Conversely, a plasmid with a short fragment (pJV342) showed the same level of β-galactosidase activity in the wild-type strain as in the ΔluxR mutant strain (Fig. 5B). We note that each construct promoted transcription to a different level, even in the absence of LuxR repression. This suggests that other regulatory elements in addition to LuxR affect transcription at this locus. Thus, we conclude that we again generated functional promoter fusion plasmids for this promoter for future studies of gene expression and regulation of VIBHAR_06912.
FIG 5.
VIBHAR_06912 promoter-reporter fusion plasmids. (A) Diagram of the regions of the VIBHAR_06912 promoter present in various plasmids. The putative transcription start site is indicated by a black arrow. The VIBHAR_06912 translation start site is shown as +1. Lengths of constructs are shown relative to the VIBHAR_06912 translation start site. (B) Modified Miller units are shown for the wild-type (BB120) and ΔluxR mutant (KM669) strains containing various plasmids as indicated in panel A. Asterisks indicate significant differences between the wild-type and ΔluxR mutant strains with the various plasmids (P < 0.05; ns, not significant; n = 3; two-way ANOVA, followed by Sidak's multiple-comparison test of log-transformed data).
Reporter gene affects measurement of gene expression.
We noted that for each of the three promoters we studied, plasmid constructs that contained promoter regions that extended into the ORF of the first gene had variable levels of expression. For example, the pJV365 plasmid that included 407 bp of the luxC gene only expressed GFP ∼2-fold more in the presence of LuxR than in its absence (Fig. 1B). This is in contrast to plasmid pMGM115 from the study of Miyamoto et al., which contains the full luxC ORF and displays maximal activation of the cat gene (Fig. S1A) (∼50-fold more than truncated promoters) (9). To examine these contradictory results further, we constructed plasmids containing the entire luxC gene and its promoter region driving gfp, lacZ, or mCherry expression (Fig. 6A and B). These constructs contained the intragenic region between luxC and luxD (15 bp), and the reporter gene was cloned in place of the luxD ORF (Fig. 6B). We observed that the lacZ and mCherry plasmids were activated 16- to 20-fold, whereas the gfp construct was activated only 1.6-fold by LuxR in E. coli (Fig. 6C). The gfp (pSO05) and mCherry (pSO11) plasmids had similar levels of activation when they were introduced into wild-type V. harveyi, though neither was expressed maximally (Fig. S1B and C).
FIG 6.
Measuring transcription of plasmids with long promoter regions fused to reporters. (A) Diagram of the regions of the luxCDABE promoter present in various constructs. LuxR binding sites (BS) are indicated. Transcription start sites are indicated by black arrows. The LuxC translation start site is shown as +1. Lengths of constructs are shown relative to the LuxC translation start site. Plasmids containing mCherry, gfp, or lacZ fusions are indicated. (B) Diagram of plasmids containing luxCDABE promoters and the luxC ORF fused to gfp (pSO05), lacZ (pSO10), or mCherry (pSO11). Each construct contains the 15-bp sequence between luxC and luxD as shown. (C) Relative expression of reporters (gfp, mCherry, and lacZ) is shown for E. coli strains containing either a plasmid constitutively expressing LuxR (pKM699) or an empty vector (pLAFR2). Relative expression was calculated by dividing the values for the pKM699-containing strain by the pLAFR2-containing strain. LacZ expression was determined by modified Miller assays, and gfp and mCherry expression was assayed with a plate reader. Different letters (a, b) indicate significant differences (P < 0.05; one-way ANOVA, followed by Tukey's multiple-comparison test of log-transformed data; n = 3). (D) Relative expression of mCherry (mCherry/OD600) in E. coli strains containing either pKM699 or pLAFR2 calculated as described for panel C. Different letters (a, b, and c) indicate significant differences (P < 0.05; one-way ANOVA, followed by Tukey's multiple-comparison test of log-transformed data; n = 3). (E) Relative gfp transcript levels determined by qRT-PCR in E. coli strains containing either pKM699 or pLAFR2 calculated as described for panel C. Different letters (a and b) indicate significant differences (P < 0.05; one-way ANOVA, followed by Tukey's multiple-comparison test of log-transformed data; n = 6).
We hypothesized that the observed decrease in activation with longer fragments might be due to instability of the transcript when the luxC ORF is present upstream of the gfp reporter. Thus, we constructed mCherry reporter plasmids containing the same four luxCDABE promoter fragments that were fused to gfp in Fig. 1A and assayed these in E. coli (Fig. 6A). We verified that the shortest region tested (2 bp past the primary transcription start site) was sufficient for activation, and there was no significant difference in expression with a construct containing a slightly longer promoter fragment (Fig. 6D), and the three shortest fragments were activated >50-fold (Fig. 6D). However, as seen with GFP, the plasmid with the longest promoter fragment (e.g., pJV366 with 407 bp of the luxC ORF) yielded a significantly lower level of mCherry expression (Fig. 6D) (activated 17-fold), which was similar to the construct containing the entire luxC ORF (Fig. 6C, pSO11) (activated 17-fold). A similar trend was observed in V. harveyi with these mCherry plasmids (Fig. S1C). Thus, we conclude that constructs containing long fragments indeed decrease the expression of downstream reporters, and with some of these, large decreases occur (i.e., gfp). This result is not observed with expression of the luxCDABE operon in vivo; the expression levels of the five genes in the operon are similar and do not differ by more than 2-fold from one another, as determined by microarray analysis (19).
To examine these results, we measured gfp transcript levels for several PluxC reporter plasmids in E. coli. The relative gfp transcript levels from quantitative reverse transcription (qRT)-PCR measurements were high for the three plasmids containing short regions of the luxCDABE promoter, but as seen with GFP expression measurements, gfp transcript levels were significantly ∼43-fold lower in a strain containing the pJV365 plasmid containing 407 bp into the luxC ORF than in a strain containing pJV369 (Fig. 6E). The gfp transcript levels were significantly lower in pJV365 than in all of the other plasmids with shorter promoter fragments tested. Thus, we conclude that the decrease in GFP expression in the pJV365-containing strain is due to a decrease in transcript levels, which may be caused by either transcript instability or a decrease in transcription initiation or elongation in plasmids with long promoter fragments. We did not observe a significant decrease in gfp transcript levels with pJV367 as observed with GFP expression (Fig. 1B), suggesting that the decrease in GFP expression may be due to constraints at the posttranscriptional or translational level. These results indicate that testing of multiple promoter fusions is beneficial for identifying a promoter-reporter fusion that functions in vivo to mimic expression from the native locus.
DISCUSSION
We have developed the RAIL method for rapid construction of promoter fusion plasmids and demonstrated that this approach can be applied to multiple promoters and reporter genes. The RAIL strategy can be used to quickly generate a few reporters or to create large libraries of promoter fusions for high-throughput analysis of the regions that drive transcription activation. The method requires simple cloning steps, and once the system is designed for a particular plasmid backbone, only two locus-specific primers are needed. For our plasmid backbone, we designed arbitrary primer sets for creating fusions to gfp, mCherry, and lacZ that can be used with any gene locus (Table S1), and these primers can be easily modified for use in any plasmid with a reporter gene.
Our library sets revealed several important findings with regard to the expression profiles of the luxCDABE and betIBA-proXWV promoters. First, we validated previous work describing the requirement for LuxR binding sites in these promoters (5, 6, 9, 14). Second, we identified the promoter region that is required for high levels of transcription activation for these two promoters. We did not resolve the 3′ boundary to a specific nucleotide locus in these experiments because we did not use every combination of anchor nucleotides in the arbitrary primers and restricted our analysis to combinations of A and T pairs. To acquire complete coverage, a full set of random primers with every combination of nucleotides as anchors should be used. However, with this resolution, we clearly found a marked difference in plasmids containing various fragments of the promoters such that we could identify the region sufficient for maximum gene expression. Even when only one arbitrary primer was used, as in the case of the betI library, we were still able to determine regions of maximal regulation, but with less coverage. Therefore, we have shown that the RAIL method is applicable for small-scale studies in which perhaps only one primer is used for quicker analysis or large-scale studies where a combination of arbitrary primers will result in greater coverage of the promoter region and produce higher resolution. Further, arbitrary primer design in these studies was limited to terminal A-T combinations because of low G+C content in V. harveyi (∼45%). In GC-rich organisms, we propose that arbitrary primers should instead be designed with terminal G-C combinations for more precise anchoring. Smaller fragments may be sufficient to drive the same level of gene expression, which can be tested with the full series of anchor nucleotides in the arbitrary primers. Further, the minimum 5′ ends of the promoters in this study are not known, so 5′ ends were chosen several hundred base pairs upstream of the ORF (∼400 to 1,000 bp). Future studies could use the same approach to map the 5′ boundary of these two promoters, which is a separate but intriguing question.
Third, our data conclusively demonstrate that there is no requirement for the region downstream of the transcription start site for full activation of the luxCDABE promoter. This finding is important because a previous study by Miyamoto et al. also tested promoter regions for luxCDABE via a cat promoter (9). Among the various constructs tested in that study, the pMGM127 plasmid contains a region truncated slightly upstream of the −26 transcription start site (the specific 3′ end is undefined in the article) and the pMGM116 plasmid includes a 3′ end at +61 relative to the luxC start codon (Fig. S1A). The shorter promoter in pMGM127 shows no transcription activation, whereas the longer promoter in pMGM116 had full activation of the cat reporter (9). These data and other observations have led to an anecdotal hypothesis in the field that there is an element downstream of the transcription start site that is required for full activation of the luxCDABE promoter. Our data refute this hypothesis because the pJV369 plasmid does not include the 5′ UTR and is maximally activated in both E. coli and V. harveyi.
Finally, our analysis of various promoter-reporter fusion plasmids demonstrated that not all reporter fusions are created equal and suggests that testing various reporter constructs for each gene of interest is beneficial to finding the optimal reporter for downstream assays. We noted that plasmid constructs with long fragments of the luxCDABE and betIBA-proXWV promoters that included sections of the first ORF in the operon were substantially decreased in expression, and we showed that this is effective at the transcript level for luxC-gfp fusions (Fig. 6E). However, we also noted that the strains containing the pJV367 plasmid that had a decrease in GFP fluorescence did not exhibit a decrease in gfp transcript levels (Fig. 6E). This result implies that the 7-fold decrease in GFP fluorescence is due to posttranscriptional or translational effects such as mRNA secondary structure that may block translation initiation.
We have focused attention on our results for the multiple plasmids with long luxCDABE promoter fragments that show significantly decreased levels of reporter expression (pJV365, pJV366, pSO05, pSO10, and pSO11). Our qRT-PCR analysis showed that transcript levels were significantly lower for pJV365 than for its counterpart plasmids with shorter promoter fragments. These data suggest that decreased reporter expression for all of the other long-promoter plasmids may also be due to decreased transcript levels. There are at least two possible reasons why the pJV365 plasmid has decreased transcript levels. One possibility is that transcripts generated with fragments of the luxCDABE operon fused to the gfp gene fold into unstable secondary structure and be subject to degradation. However, we suspect that this explanation is unlikely to be the cause of low expression for every plasmid with a fragment longer than +129, as we would predict that at least some would be stable. A second possibility is that LuxR binding to site H is acting as a roadblock to transcription elongation, which results in the abrupt drop in GFP expression for plasmids containing promoter regions that terminate after site H. Previously, we showed that scrambling of site H does not decrease LuxR activation of β-galactosidase expression in a luxC-lacZ reporter plasmid under conditions in which LuxR is maximally expressed at high cell densities in V. harveyi (6). However, the results of our expression profiling experiment with E. coli with the luxCDABE promoter library suggest that plasmids that contain LuxR site H have decreased levels of transcription activation and are strictly in the low GFP expression pool (Fig. 3B). LuxR has an extremely high affinity for site H, with a Kd of 0.6 nM, one of the tightest LuxR binding affinities in the genome (5). Thus, it is curious that LuxR binds at this locus with no apparent activation defect when tested at high cell density in V. harveyi.
Protein roadblocks have been described in bacteria and eukaryotes that hinder transcription, and elongation factors aid in transcription elongation through these roadblocks by various mechanisms (e.g., Mfd in E. coli) (20). In addition, when multiple RNAP molecules are initiated from the same promoter, these trailing RNAP complexes can “push” a stalled RNAP through a roadblock (21). Thus, it is possible that higher levels of transcription initiation of the luxCDABE promoter in V. harveyi at high cell densities drive transcription elongation through site H, whereas lower levels of LuxR at low cell densities in V. harveyi or in our synthetic E. coli system are not sufficient to push through the LuxR site H roadblock. LuxR concentrations are low in the cell at low cell densities, and thus, the relatively few LuxR molecules likely bind to the highest-affinity sites, such as site H in the luxC ORF. As cells grow to high densities, LuxR accumulates (19, 22, 23) and enables LuxR binding to other sites, which drives high levels of transcription initiation and may relieve binding of LuxR to site H to allow RNAP elongation. Alternatively, the roadblock might be relieved by restructuring of the DNA architecture at the locus. Because we have already shown that IHF binds to multiple places in the luxCDABE region and its binding is positively cooperative with LuxR, this DNA bending may play a role in the removal of transcription roadblocks. We also observed a sharp difference between the medium- and low-expression pools just downstream of the LuxR site H (Fig. 3B), suggesting that there may be yet another roadblock in this region. Future studies should elucidate the role of LuxR binding sites within ORFs in V. harveyi, which are observed throughout the genome (5).
In conclusion, the RAIL method offers a rapid and efficient method to obtain libraries of reporter fusions that can be used for various studies of gene expression and regulation. Often in bacterial genetics, researchers attempt to create promoter fusions by cloning a reporter gene in place of the translation start site, and this would have yielded suboptimal reporters for the luxCDABE promoter. Anecdotally, and as we experienced with the betIBA-proXWV and luxCDABE promoters, one often needs to construct multiple reporter fusions to identify a promoter region that drives gene expression mimicking native locus gene expression. Thus, our method is more efficient by generating numerous clones in a single cloning experiment. We envision the use of the RAIL method for numerous other purposes, such as creating functional GFP fusions for studying protein localization, identifying cis-regulatory sequences in promoters (e.g., protein binding sequences), inserting affinity tags for purification strategies, and identifying highly expressed soluble constructs for protein purification. Finally, this method should be applicable to any model organism for which genetic cloning techniques have been established.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli strains S17-1λpir, DH10B, and derivatives (see Table S2 in the supplemental material) were used for cloning and in vivo assays. E. coli strains were grown with shaking at 275 rpm at 30°C in lysogeny broth (LB) augmented with 10 μg/ml chloramphenicol and 10 μg/ml tetracycline when required. The V. harveyi BB120 is strain ATCC BAA-1116, which was recently reassigned to Vibrio campbellii (24). It is referred to as V. harveyi here for consistency with previous literature. BB120 and derivatives (Table S2) were grown at 30°C with shaking at 275 rpm in LB marine (LM) medium supplemented with 10 μg/ml chloramphenicol when required. LM medium is prepared similarly to LB (10 g of tryptone, 5 g of yeast extract) but with 20 g of NaCl instead of the 10 g used for LB.
Molecular methods.
Oligonucleotides (Table S1) were purchased from Integrated DNA Technologies. All PCRs were performed with Phusion HF polymerase (New England BioLabs) or iProof polymerase (Bio-Rad). Restriction enzymes, enzymes for IDA (15), and deoxynucleoside triphosphates (dNTPs) were obtained from New England BioLabs. DNA samples were visualized on 1% agarose gels. Standard cloning methods and primers for the single plasmid constructs listed in Table S3 are available upon request. Standard sequencing of single plasmid constructs was conducted by ACGT, Inc., and Eurofins Genomics. To measure the expression levels of fluorophore reporter plasmids, E. coli and V. harveyi strains were grown overnight at 30°C with shaking at 275 rpm. Strains were diluted 100-fold in growth medium and selective antibiotics in 96-well plates (black with a clear bottom), covered with microporous sealing tape (USA Scientific), and incubated at 30°C with shaking at 275 rpm for 16 to 18 h. Fluorescence and optical density at 600 nm (OD600) of strains expressing mCherry and gfp were measured with either a BioTek Synergy H1 or a Cytation plate reader. Miller assays were conducted as previously described (6). RNA extraction and qRT-PCR were performed and analyzed as described previously (14) with primers listed in Table S1 on a StepOne Plus real-time PCR machine (Applied Biosystems). Transcript levels were normalized to the level of expression of the internal standard recA, and the standard-curve method was used for data analysis. The error bars on graphs represent the standard deviations of measurements of at least three biological samples. Statistical analysis was performed with GraphPad Prism version 7.0c. Additional information about statistical analyses pertinent to each result set is included in the figure legends.
RAIL: construction of promoter libraries by arbitrary PCR.
The arbitrary PCR method was adapted from a study by Schmidt et al. (25), with several modifications. The first round of PCR was conducted with two primers, a forward primer specific to the promoter of interest and a reverse primer for random DNA amplification (Fig. 2, primers 1F and 1R, respectively). The 1R primer includes a priming sequence followed by eight random nucleotides (′N′) and terminating in two defined nucleotide anchors, AT, TA, TT, or AA (Table S1). For PCR round 1, ∼10 to 100 ng/μl genomic DNA from V. harveyi BB120 or a plasmid containing the region of interest was added as the template. The reaction mixture included 200 μM dNTPs, 250 μM primers, 5% dimethyl sulfoxide (DMSO), 0.5 μl of Phusion polymerase, and 1× Phusion buffer. Cycling parameters were as follows and as previously published (25): an initial denaturation at 95°C for 5 min; 5 cycles of 95°C for 30 s, 25°C for 30 s, and 72°C for 2.5 min; 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 2.5 min; and a final extension step of 72°C for 10 min. PCRs were purified with the GeneJet PCR purification kit (Thermo Scientific) and eluted in 30 to 50 μl of elution buffer. The second round of PCR used primers 2F and 2R (Fig. 2). The forward primer (2F) included 30 nucleotides of homology to the plasmid backbone for IDA and a sequence specific to the promoter of interest that is nested downstream of the 1F primer. The reverse primer (2R) included the priming sequence that is identical to that of primer 1R. To perform PCR round 2, 5 μl of the purified DNA from round 1 was used as the template. These reaction mixtures also included 200 μM dNTPs, 250 μM primers, 5% DMSO, 0.5 μl of Phusion polymerase, and 1× Phusion buffer. The cycling parameters were as follows: an initial denaturation at 95°C for 5 min; 35 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 2.5 min; and a final extension step of 72°C for 10 min. PCR products were separated by agarose gel electrophoresis and visualized by UV transillumination, and the products were gel extracted to the desired target size with a GeneJet gel extraction kit (Thermo Scientific).
Cloning of arbitrary PCR inserts into the plasmid backbone was performed by IDA as described previously (15). Library inserts were incubated in IDA reaction mixtures with 100 ng of plasmid backbone, and these reaction mixtures were transformed into electrocompetent E. coli ElectroMAX DH10B cells (ThermoFisher) and plated on medium with selective antibiotics. DNA from individual colonies was first screened by restriction digestion and sequenced to confirm that inserts of the desired size were incorporated. For generation of libraries for sorting, >50,000 colonies were collected from plates and mixed in LB selective medium and the culture was stored at −80°C. DNA extracted from this library was transformed into electrocompetent E. coli S17-1λpir cells containing a plasmid expressing luxR (pKM699). After this second transformation step, >50,000 colonies were collected from plates and mixed in LB selective medium and the culture was stored at −80°C.
Flow cytometry sorting.
A BD FACSAria II was used to sort E. coli cells on the basis of expression of the fluorescent reporter. In preparation for sorting, the library culture from the frozen stock was grown in 50 ml of selective medium at 30°C with shaking to an OD600 of 0.5. DNA was extracted from this culture as the input DNA sample for sequencing. Control cultures were used to set the sorting gates. The positive controls for maximal expression were strains containing pKM699 (expressing luxR) and a plasmid construct that demonstrated high levels of expression for PluxC (pJV369, PluxC-gfp positive-control plasmid) or PbetI (pCH50, PbetI-mCherry positive-control plasmid). The negative controls were strains containing pKM699 and pJS1194 (gfp negative-control plasmid) or pCH76 (mCherry negative-control plasmid), which lack promoters in front of gfp or mCherry, respectively. The library culture was sorted into bins with >10,000 cells per bin. For PluxC sorting, there were four bins with no, low, medium, or high GFP expression. For PbetI sorting, there were two bins with high or no mCherry expression. The sorted cell cultures were incubated in 5 ml of selective medium at 30°C with shaking at 275 rpm and grown to stationary phase. Cultures were stored at −80°C, and the DNA was extracted for sequencing with the GeneJet Miniprep kit (Thermo Scientific).
Illumina library preparation and sequencing.
DNA extracted from all sorted samples and input controls was purified over a Performa DTR gel filtration cartridge (Edge Biosystems). Next, the DNA was sheared with a Covaris S220 in 6- by 16-mm microtubes to an average size of 400 bp and analyzed on an Agilent 2200 TapeStation with D1000 ScreenTape. The sheared DNA samples (1 μg) were each treated with terminal deoxynucleotidyl transferase (TdT) in a tailing reaction mixture using TdT (Promega) and a mixture of dCTP and ddCTP (475 and 25 μM final concentrations, respectively) to generate a poly(C) tail. The reaction mixtures were incubated at 37°C for 1 h, heat inactivated at 75°C for 20 min, and cleaned over a Performa DTR gel filtration cartridge. Next, two rounds of PCR were performed to attach sequences for Illumina sequencing (Table S1). In round 1, a C tail-specific primer (olj376) and a gene-specific primer (CH060 for the gfp library and CH063 for the mCherry library) were used to amplify the promoter fragments that were cloned into the plasmid by the RAIL method. This PCR used Taq polymerase (NEB) under the following cycling conditions: an initial denaturation at 95°C for 2 min; 24 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 2 min; and a final extension step of 72°C for 2 min. The round 2 PCR used a nested gene-specific primer (CH061 for the gfp library and CH064 for the mCherry library) to provide added specificity and also to append the linker sequence needed for Illumina sequencing. The second primer in the reaction mixture contained different barcodes for each sample to enable the libraries to be pooled and sequenced simultaneously (Table S1; BC37-44). The round 2 PCRs were performed with Taq polymerase and cycling as follows: an initial denaturation at 95°C for 2 min; 12 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 2 min; and a final extension step of 72°C for 2 min. These final PCR products were examined by DNA gel electrophoresis, at which point a smear of products was visible on the gel.
Sequencing was performed on a NextSeq 500 with a NextSeq 75 reagent kit, 42- by 42-bp paired-end run parameters, and gene-specific primers (CH062 for the gfp library and CH065 for the mCherry library). Reads were checked with FastQC (v0.11.5; available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc) to ensure that the quality of the data was high and that there were no noteworthy artifacts associated with the reads. The R1 read from paired-end Illumina NextSeq reads were quality and adapter trimmed with Trimmomatic (v0.33) (26) by using the following parameters: ILLUMINACLIP:<adapters>:3:20:6 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:25. Reads were high quality, with 80 to 85% of them surviving trimming. Reads were mapped against the V. campbellii ATCC BAA-1116 genome (including molecules NC_009777, NC_009783, and NC_009784) with Bowtie 2.3.2 (27) and visualized with JBrowse (version 1.10.12) to analyze the alignment with the betI and luxC gene regions. Analysis of the promoter-seq data hinged on contrasting high-, medium-, low-, and no-expression cells with the null distribution. The null distribution was determined by sequencing cells collected without any sorting applied. The reads associated with the null and high-, medium-, low-, and no-expression data sets for the luxC library (or null and high- and no-expression data sets for the betI library) were normalized by transforming the data into the fraction of bases covered, which was defined as the depth of coverage at a particular base divided by the total depth of coverage over the particular promoter region (betI, bases 1361141 to 1362440 in NC_009784; luxC, bases 1424774 to 1426907 in NC_009784). Analysis was performed by plotting the log2 ratio of the observed fraction of bases covered (for the high-, medium-, low-, or no-expression cell collection) over the expected (null) distribution.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ankur Dalia for assistance with the design of the Illumina sequencing protocol and helpful discussions. We also thank James Ford of the Indiana University Center for Genomics and Bioinformatics for Illumina sequencing experiments, bioinformatics analysis, and assistance with graphing of the results. We also acknowledge Christiane Hassel of the Indiana University Flow Cytometry Core Facility for assistance with FACSAria II analysis and sorting. We thank Ryan Fritts for assistance with statistical analyses. We appreciate the use of Jason Tennessen's BioTek Cytation plate reader, and we thank Ryan Chaparian and Blake Petersen for assistance with experiments.
This work was supported by National Institutes of Health (NIH) grant 1R35GM124698-01 to J.C.V.K. and startup funds from Indiana University to J.C.V.K. and M.L.B.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00724-17.
For a commentary on this article, see https://doi.org/10.1128/JB.00039-18.
REFERENCES
- 1.Rosochacki SJ, Matejczyk M. 2002. Green fluorescent protein as a molecular marker in microbiology. Acta Microbiol Pol 51:205–216. [PubMed] [Google Scholar]
- 2.Jiang T, Xing B, Rao J. 2008. Recent developments of biological reporter technology for detecting gene expression. Biotechnol Genet Eng Rev 25:41–75. doi: 10.5661/bger-25-41. [DOI] [PubMed] [Google Scholar]
- 3.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnone MI, Dmochowski IJ, Gache C. 2004. Using reporter genes to study cis-regulatory elements. Methods Cell Biol 74:621–652. doi: 10.1016/S0091-679X(04)74025-X. [DOI] [PubMed] [Google Scholar]
- 5.van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. 2013. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. mBio 4:e00278-13. doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaparian RR, Olney SG, Hustmyer CM, Rowe-Magnus DA, van Kessel JC. 2016. Integration host factor and LuxR synergistically bind DNA to coactivate quorum-sensing genes in Vibrio harveyi. Mol Microbiol 101:823–840. doi: 10.1111/mmi.13425. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee J, Miyamoto CM, Zouzoulas A, Lang BF, Skouris N, Meighen EA. 2002. MetR and CRP bind to the Vibrio harveyi lux promoters and regulate luminescence. Mol Microbiol 46:101–111. doi: 10.1046/j.1365-2958.2002.03128.x. [DOI] [PubMed] [Google Scholar]
- 8.Meighen EA. 1991. Molecular biology of bacterial bioluminescence. Microbiol Rev 55:123–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto CM, Smith EE, Swartzman E, Cao JG, Graham AF, Meighen EA. 1994. Proximal and distal sites bind LuxR independently and activate expression of the Vibrio harveyi lux operon. Mol Microbiol 14:255–262. doi: 10.1111/j.1365-2958.1994.tb01286.x. [DOI] [PubMed] [Google Scholar]
- 10.Swartzman E, Silverman M, Meighen EA. 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol 174:7490–7493. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reznikoff WS. 1992. The lactose operon-controlling elements: a complex paradigm. Mol Microbiol 6:2419–2422. doi: 10.1111/j.1365-2958.1992.tb01416.x. [DOI] [PubMed] [Google Scholar]
- 12.Dunn TM, Hahn S, Ogden S, Schleif RF. 1984. An operator at −280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A 81:5017–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galagan J, Lyubetskaya A, Gomes A. 2013. ChIP-Seq and the complexity of bacterial transcriptional regulation. Curr Top Microbiol Immunol 363:43–68. [DOI] [PubMed] [Google Scholar]
- 14.van Kessel JC, Rutherford ST, Cong JP, Quinodoz S, Healy J, Bassler BL. 2015. Quorum sensing regulates the osmotic stress response in Vibrio harveyi. J Bacteriol 197:73–80. doi: 10.1128/JB.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 16.Welsh J, McClelland M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Showalter RE, Martin MO, Silverman MR. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J Bacteriol 172:2946–2954. doi: 10.1128/jb.172.6.2946-2954.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. 2013. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol 195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JS, Marr MT, Roberts JW. 2002. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109:757–767. doi: 10.1016/S0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 21.Epshtein V, Toulme F, Rahmouni AR, Borukhov S, Nudler E. 2003. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J 22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters CM, Bassler BL. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng SW, Wang Y, Tu KC, Long T, Mehta P, Wingreen NS, Bassler BL, Ong NP. 2010. Measurement of the copy number of the master quorum-sensing regulator of a bacterial cell. Biophys J 98:2024–2031. doi: 10.1016/j.bpj.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin B, Wang Z, Malanoski AP, O'Grady EA, Wimpee CF, Vuddhakul V, Alves N Jr, Thompson FL, Gomez-Gil B, Vora GJ. 2010. Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep 2:81–89. doi: 10.1111/j.1758-2229.2009.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt KH, Pennaneach V, Putnam CD, Kolodner RD. 2006. Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol 409:462–476. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.