ABSTRACT
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen with distinct acute and chronic virulence phenotypes. Whereas acute virulence is typically associated with expression of a type III secretion system (T3SS), chronic virulence is characterized by biofilm formation. Many of the phenotypes associated with acute and chronic virulence are inversely regulated by RsmA and RsmF. RsmA and RsmF are both members of the CsrA family of RNA-binding proteins and regulate protein synthesis at the posttranscriptional level. RsmA activity is controlled by two small noncoding regulatory RNAs (RsmY and RsmZ). Bioinformatic analyses suggest that RsmY and RsmZ each have 3 or 4 putative RsmA binding sites. Each predicted binding site contains a GGA sequence presented in the loop portion of a stem-loop structure. RsmY and RsmZ regulate RsmA, and possibly RsmF, by sequestering these proteins from target mRNAs. In this study, we used selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) chemistry to determine the secondary structures of RsmY and RsmZ and functional assays to characterize the contribution of each GGA site to RsmY/RsmZ activity. Our data indicate that RsmA has two preferential binding sites on RsmY and RsmZ, while RsmF has one preferential binding site on RsmY and two sites on RsmZ. Despite RsmF and RsmA sharing a common consensus site, RsmF binding properties are more restrictive than those of RsmA.
IMPORTANCE CsrA homologs are present in many bacteria. The opportunistic pathogen Pseudomonas aeruginosa uses RsmA and RsmF to inversely regulate factors associated with acute and chronic virulence phenotypes. RsmA has an affinity for RsmY and RsmZ higher than that of RsmF. The goal of this study was to understand the differential binding properties of RsmA and RsmF by using the RsmY and RsmZ regulatory small RNAs (sRNAs) as a model. Mutagenesis of the predicted RsmA/RsmF binding sites on RsmY and RsmZ revealed similarities in the sites required to control RsmA and RsmF activity in vivo. Whereas binding by RsmA was relatively tolerant of binding site mutations, RsmF was sensitive to disruption to all but two of the sites, further demonstrating that the requirements for RsmF binding activity in vivo and in vitro are more stringent than those for RsmA.
KEYWORDS: Pseudomonas aeruginosa, RsmA, RsmF, CsrA, RsmY, RsmZ, sRNA, type III secretion
INTRODUCTION
The survival of opportunistic bacterial pathogens is dependent upon the ability to sense and adapt to changing conditions within the host and environment. The most common adaptive response is to alter gene expression and/or protein synthesis patterns. Large-scale alterations to the transcriptome/proteome often involve global regulators (1). Global regulation can occur at the transcriptional, posttranscriptional, and/or posttranslational level. A well-studied posttranscriptional regulator is Escherichia coli CsrA (carbon storage regulator), which plays a central role in carbon metabolism (2, 3). CsrA is a homodimeric RNA-binding protein that can have both negative and positive effects on protein synthesis (4). The general mechanism of repression by CsrA is to physically interfere with translation initiation by binding to a site that overlaps the ribosome binding site on target mRNAs (5–7). The CsrA consensus binding site consists of a core GGA sequence located in the single-stranded region of an RNA stem-loop structure, with the highest-affinity sites possessing pentaloop structures (8). Since each CsrA dimer contains two RNA-binding domains, full regulatory control often involves binding to an additional site on the target RNA (9). Although CsrA usually inhibits translation, examples of positive regulation have been described, the mechanisms of which include protection of the target mRNA from degradation and an increased rate of translation initiation (6, 10).
CsrA orthologs are found in a wide range of bacteria (7). The opportunistic pathogen Pseudomonas aeruginosa has two CsrA orthologs: RsmA (regulator of secondary metabolites) and RsmF (described below). RsmA was first shown to play a role in controlling the production of virulence factors, including elastase, LasA protease, hydrogen cyanide, and pyocyanin (11). The RsmA regulon is now known to consist of >500 target genes (12, 13). Notably, RsmA inhibits the synthesis of several gene products thought to play a role in persistent colonization of the airways in cystic fibrosis patients. Those products include components of a type VI secretion system (T6SS) and factors that promote biofilm formation (14, 15). Conversely, RsmA positively regulates genes associated with acute infection, including the type III secretion system (T3SS) and type IV pili (12, 16). RsmA activity is regulated by two small noncoding RNAs, designated RsmY and RsmZ. RsmY and RsmZ possess multiple RsmA binding sites that function to sequester RsmA from target mRNAs (17). Expression of rsmY and rsmZ is regulated at the transcriptional level by the GacS/A two-component system and two orphan sensor kinases, LadS and RetS (18, 19). The environmental signals resulting in altered RsmY and RsmZ synthesis are poorly understood. Nevertheless, sequestration of RsmA by RsmY/RsmZ results in reduced RsmA availability and preferential expression of virulence factors associated with chronic colonization. Conversely, high RsmA availability favors expression of factors associated with acute virulence (20). Because of this inverse relationship, RsmA is thought to play a pivotal role in controlling the transition from acute to chronic colonization (12, 13, 21).
Several of the pseudomonads, including P. fluorescens, P. protegens, P. putida, and P. aeruginosa, have multiple CsrA proteins (22–25). In P. fluorescens, RsmA and RsmE are structurally similar and appear to be redundant in function (23). P. putida RsmA, RsmE, and RsmI are also homologous, but distinct roles for each have been described (26). In contrast, P. aeruginosa RsmA and RsmF (also called RsmN) are structurally distinct and share some, but not all, regulatory targets (24, 25, 27). Both RsmA and RsmF are homodimers, with similar RNA-binding surfaces composed of five β-sheets, but they differ in how the RNA-binding surface is formed. The RNA-binding surface in RsmF is formed by β1, β3, and β4 from one polypeptide and β2 and β5 from the second polypeptide (24, 25). In other CsrA proteins, including RsmA, the RNA-binding surface consists of β2, β3, and β4 from one polypeptide and β1 and β5 from the second polypeptide (28–30). Despite the significant difference in structure, both proteins possess a similarly positioned arginine residue (R44 in RsmA and R62 in RsmF) that is critical for RNA-binding activity (24, 25, 30). The RNA-binding activity of both proteins is also dependent on the core GGA motif, since RNA targets carrying CCU substituted for GGA are poorly bound (24). Finally, data from systematic evolution of ligands by exponential enrichment experiments (SELEX) found that RsmA and RsmF share a consensus binding sequence (CAnGGAyG), with 100% conservation of the core GGA sequence (underlined) (27).
Although RsmA and RsmF share several binding targets, including RsmY and RsmZ, there are significant differences in the interactions with those targets. Data from electrophoretic mobility shift assays (EMSAs) suggest that three RsmA dimers bind to RsmZ with affinities in the low nanomolar range (24). RsmA binding to RsmY is also in the low nanomolar range. In contrast, only one RsmF dimer appears to bind to RsmZ, and the affinities for both RsmY and RsmZ are reduced nearly 50-fold relative to those of RsmA (24). In this study, we report the preferences of RsmA and RsmF for the predicted binding sites in RsmY and RsmZ as a means to better understand the basis for differential binding of target RNAs.
RESULTS
Structural analyses of RsmY and RsmZ.
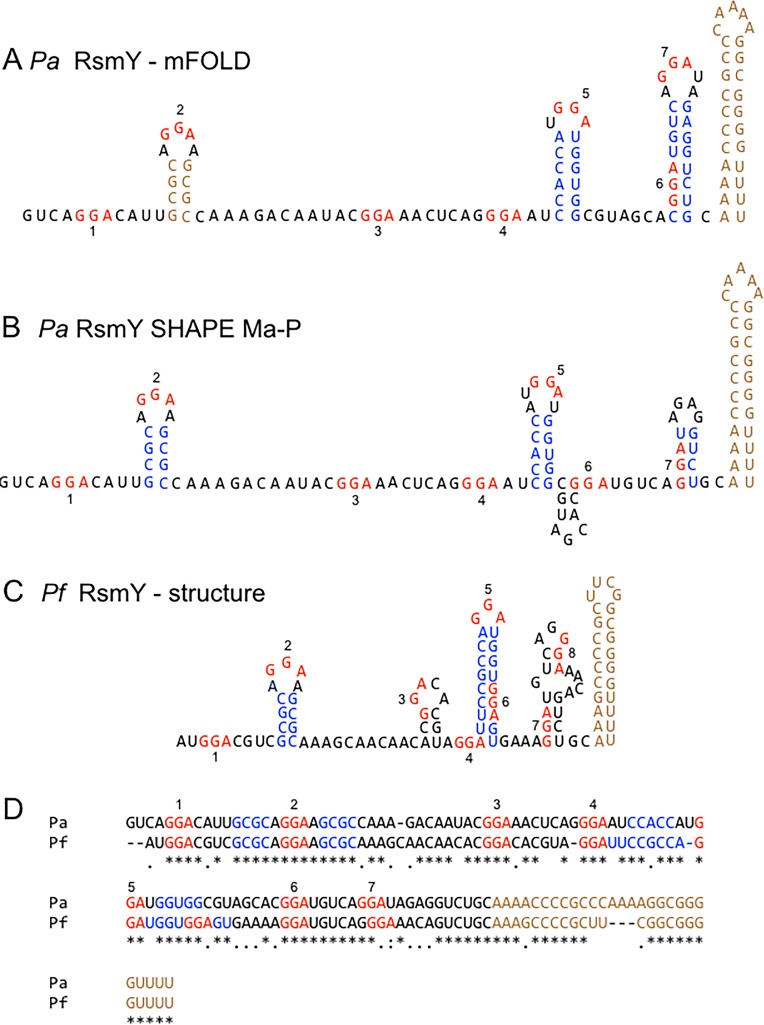
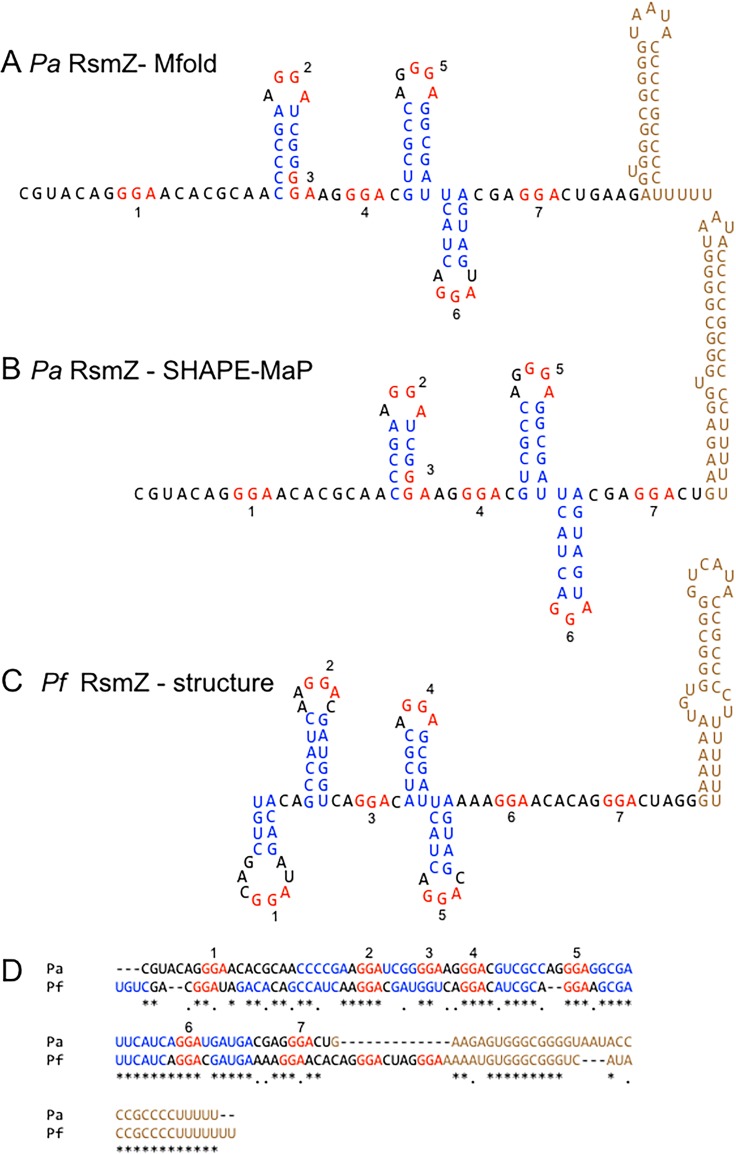
We previously reported that RsmF binds to the RsmY and RsmZ RNAs with significantly lower affinities than those of RsmA (24). To gain further insight into the differential binding properties of RsmA and RsmF, we sought to characterize RsmA/RsmF binding sites on RsmY and RsmZ. The consensus binding sites determined by SELEX for both RsmA and RsmF contain a critical GGA sequence (CAnGGAyG) that is presented in the loop portion of a stem-loop structure (4, 27). RsmY and RsmZ each contain seven GGA sequences, designated GGA1 to GGA7 starting from the 5′ end of the RNAs (Fig. 1A and 2A). Comparison of each GGA sequence to the full RsmA/RsmF consensus binding site (CAnGGAyG) shows matches that range from 4 to 7 positions (see Fig. S1 in the supplemental material). Mfold predictions suggest that three of the RsmY GGA sequences (sites 2, 5, and 7) and three of the RsmZ GGA sequences (sites 2, 5, and 6) are presented in the loop portion of stem-loop structures (Fig. 1A and 2A) (31). In some instances, CsrA proteins also bind unpaired GGA sequences that are not presented in stem-loop structures. The Mfold predictions show that RsmY GGA sequences 1, 3, and 4 and RsmZ GGA sites 1, 4, and 7 are unpaired, making each potentially available for RsmA/RsmF interactions (Fig. 1A and 2A).
FIG 1.
Predicted and determined structures of RsmY. (A) Predicted P. aeruginosa (Pa) RsmY secondary structure determined by Mfold modeling. (B) SHAPE-MaP structure for P. aeruginosa RsmY. (C) P. fluorescens (Pf) RsmY secondary structure predicted by Mfold. (D) Alignment of P. aeruginosa and P. fluorescens rsmY sequences. GGA motifs are numbered 1 to 7 and highlighted in red. Stem structures are highlighted in blue, and terminator regions are shown in brown.
FIG 2.
Predicted and determined structures of RsmZ. (A) Predicted Mfold structure for P. aeruginosa RsmZ. (B) SHAPE-MaP structure for P. aeruginosa RsmZ. (C) P. fluorescens RsmZ NMR structure. (D) Alignment of P. aeruginosa and P. fluorescens rsmZ sequences. GGA motifs are numbered 1 to 7 and highlighted in red. Stem structures are highlighted in blue, and terminator regions are shown in brown.
To experimentally verify the RsmY and RsmZ secondary structure, we performed selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) (32–34). SHAPE-MaP determines the reactivity of the 1-methyl-7-nitroisatoic anhydride (1M7) chemical probe with each nucleotide in an RNA. Higher reactivity indicates more flexibility, which is generally indicative of unpaired nucleotides. The free energy of RNA folding incorporated into the SHAPE-MaP data predicts minimum free energy structures that are different from the Mfold predictions. Both Mfold and SHAPE-MaP are consistent in predicting that RsmY GGA sequences 2 and 5 are presented in the loop portion of stem-loop structures and that GGA sequences 1, 3, and 4 are unpaired (Fig. 1A and B). The only major discrepancy between the Mfold and SHAPE predictions for RsmY is whether GGA site 7 is presented in the loop portion of a stem-loop structure, as predicted by Mfold, or remains base paired and inaccessible for RsmA/RsmF binding, as predicted by SHAPE-MaP. An in vivo cleavage structure for P. fluorescens RsmY found that GGA sites 2 and 5 are presented in the loop portion of stem-loop structures (Fig. 1C) (35). Alignment of RsmY sequences from both organisms shows strong conservation of the nucleotides that comprise the stem and loop regions for GGA sites 2 and 5 (Fig. 1D). Consistent with this, the P. aeruginosa RsmY SHAPE-MaP and P. fluorescens RsmY structures for GGA sites 2 and 5 bear a striking resemblance to one another. These combined data suggest that P. aeruginosa RsmY GGA site 7 may not be accessible for RsmA/RsmF.
The predicted SHAPE-MaP structure for RsmZ has three GGA sequences presented in stem-loop structures (sites 2, 5, and 6) and three GGA sequences that are unpaired (sites 1, 4, and 7) (Fig. 2B). The Mfold and SHAPE-MaP data are very similar and differ only in the terminator region (Fig. 2A and B). A recent nuclear magnetic resonance (NMR) structure for P. fluorescens RsmZ found that GGA sites 1, 2, 5, and 6 are presented in the loop portion of stem-loop structures (36) (Fig. 1C). Alignment of the RsmZ sequences from both organisms shows strong conservation of the nucleotides that comprise the stem and loop regions for GGA sites 5 and 6, weak conservation around site 2, and minimal conservation around site 1 (Fig. 1D). Consistent with this, the P. aeruginosa RsmZ SHAPE-MaP and P. fluorescens RsmZ NMR structures for GGA sites 2, 5, and 6 resemble one another. These combined data suggest that P. aeruginosa RsmZ GGA site 1 may not be presented in the context of a stem-loop structure.
Roles of RsmY and RsmZ GGA sequences in vivo.
To determine which GGA sites are important for RsmY and RsmZ regulatory activity, each of the GGA sequences within stem-loop structures predicted by either Mfold or SHAPE-MaP was changed to CCU. Select double, triple, and quadruple substitution mutants were also constructed as described below. Each of the RNAs was expressed from a plasmid under the control of an arabinose-inducible promoter and tested for regulatory activity in vivo. Regulatory activity was measured using a previously described PtssA1′-′lacZ translational reporter (24) integrated at the chromosomal ΦCTX phage attachment site of P. aeruginosa strain PA103. The tssA1 operon encodes components of T6SS-1, and tssA1 translation is repressed by the direct binding of RsmA to the tssA1 mRNA leader region, as seen for wild-type cells and mutants lacking rsmF, rsmYZ, or rsmFYZ (Fig. 3) (12, 24). Conversely, reporter activity is derepressed when rsmA is deleted from the wild-type and rsmYZ backgrounds. Although deletion of rsmF alone has no effect on PtssA1′-′lacZ reporter activity, mutants lacking both rsmA and rsmF (i.e., rsmAF and rsmAFYZ mutants) have much higher levels of reporter activity than those of the single rsmA mutant and the rsmAYZ mutant (Fig. 3).
FIG 3.
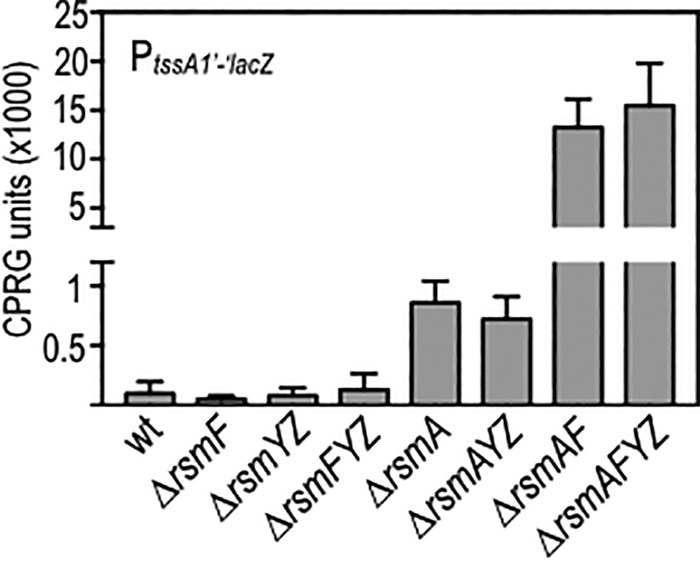
Control of tssA1′-′lacZ translational reporter activity by the RsmAFYZ system. The indicated mutants carrying the tssA1′-′lacZ translational reporter were cultured to mid-exponential phase and assayed for β-galactosidase activity by use of CPRG as a substrate.
To specifically examine the regulation of RsmA and RsmF activity, the following experiments were performed in the rsmFYZ and rsmAYZ mutant backgrounds, respectively. Whereas rsmFYZ and rsmAYZ strains carrying a vector control (pJN105) have low levels of PtssA1′-′lacZ reporter activity, plasmid-expressed RsmY or RsmZ sequester RsmA or RsmF, resulting in significant activation of reporter activity (Fig. 4A to D). Examination of the RsmY GGA mutant panel expressed in the rsmFYZ background revealed that the site 2 and site 5 GGA mutants had significantly reduced PtssA1′-′lacZ reporter activity, while the site 7 GGA mutant had activity levels similar to those of wild-type RsmY (Fig. 4A). The GGA27 and GGA57 double mutants (with mutations of GGA sites 2 and 7 and GGA sites 5 and 7, respectively) had activities similar to those of the single GGA2 and GGA5 mutants, further demonstrating that GGA7 is dispensable for RsmY activity. The GGA25 double mutant and GGA257 triple mutant also had significant reductions in activity compared to the activity of wild-type RsmY, indicating that sites 2 and 5 are necessary for maximal sequestration of RsmA.
FIG 4.
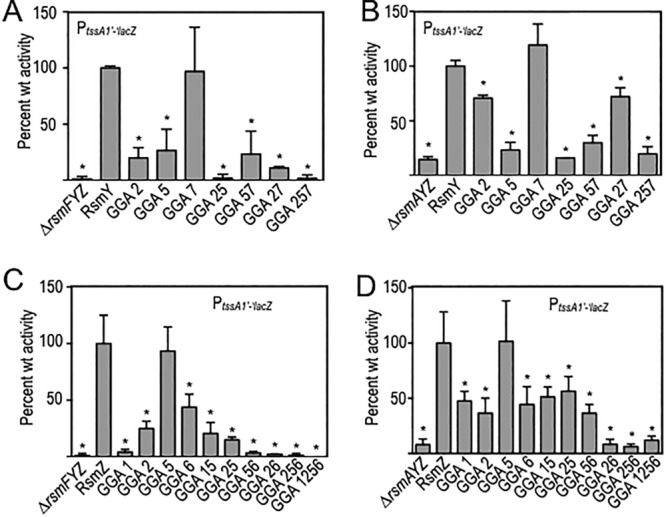
Regulation of RsmA and RsmF activities by mutant RsmY and RsmZ regulatory RNAs. The ΔrsmFYZ (A and C) and ΔrsmAYZ (B and D) mutants bearing the tssA1′-′lacZ translational reporter were transformed with the indicated RsmY (A and B) or RsmZ (C and D) expression plasmids. Transformants were cultured to mid-exponential phase in the presence of 0.1% arabinose to induce expression of the respective RNAs and then assayed for β-galactosidase activity. Reported values represent the averages for at least three experiments, with standard errors indicated by error bars. The indicated statistically significant differences (*, P < 0.05) are relative to strains expressing either native RsmY (A and B) or RsmZ (C and D), normalized to 100% activity.
Using the rsmAYZ background to examine RsmF activity, the RsmY GGA site 5 mutant had a significant defect in regulation of RsmF activity, the site 2 mutant had a modest but still significant defect, and the site 7 mutant had activity levels to similar to those of native RsmY (Fig. 4B). As observed above for RsmA, the GGA27 and GGA57 double mutants had activities similar to those of the single GGA2 and GGA5 mutants, indicating that site 7 does not significantly contribute to regulation of RsmA or RsmF activity (Fig. 4B). These combined findings suggest that GGA sites 2 and 5 are the primary sites on RsmY for RsmF binding.
Using the same strategy, we next examined which GGA sites in RsmZ are important for controlling RsmA and RsmF activity by creating single, double, triple, and quadruple mutants. Examination of the RsmZ mutant panel in the rsmFYZ background revealed that the site 1, 2, and 6 mutants had reduced activity relative to that of native RsmZ and that sites 1, 2, and 6 are thus required for maximal activity (Fig. 4C). The site 1 mutant in particular was completely devoid of activity and had the strongest phenotype of all of the single mutants examined. The site 5 GGA mutant had activity levels similar to that of native RsmZ. A role for GGA site 5 cannot be excluded, though, because the activity of the GGA56 double mutant was significantly lower than that of the site 6 GGA mutant alone.
We saw a similar trend for RsmF in that RsmZ sites 1, 2, and 6 were required for maximal activity (Fig. 4D). The phenotype of the site 1 mutant, however, was not nearly as pronounced in the rsmAYZ mutant. Similar to our findings for RsmA, RsmZ site 5 was also largely dispensable for regulation of RsmF activity.
Decreased stability of mutant RsmY and RsmZ RNAs does not account for defects in regulatory activity in vivo.
The simplest interpretation of the above data is that the decreased activity of the mutant RsmY and RsmZ RNAs correlates with a reduced capacity to bind and sequester RsmA/F. An alternative explanation is that the GGA-to-CCU substitutions reduced the stability of the RNAs under steady-state conditions. Mfold predictions for the mutant RNAs, however, indicate that the single CCU substitutions did not alter the secondary structures of the RNAs. The CCU substitutions, however, might create or destroy RNase cleavage sites. To directly examine RsmY and RsmZ stability, we performed quantitative reverse transcription-PCR (qRT-PCR). RNA samples were isolated from each RsmY- or RsmZ-expressing strain (grown to mid-exponential phase). The expression level of each RNA was normalized to that of rimM, a housekeeping gene encoding a ribosomal protein (37), and expression levels are reported relative to those of wild-type RsmY or RsmZ. Only two of the mutant RNAs, RsmY GGA57 and RsmZ GGA1256, had >2-fold reductions in expression, though these were not significant based on analysis of variance (ANOVA) (Fig. 5A and B). The lower steady-state level of RsmZ GGA1256 may have contributed to the significant loss of activity observed for RsmA (Fig. 4C). The remaining RsmY and RsmZ mutants had expression levels similar to or greater than those of wild-type RsmY and RsmZ, with RsmY GGA27, RsmZ GGA6, and RsmZ GGA256 levels being increased >3-fold, for reasons that are unclear (Fig. 5A and B). These findings indicate that the reduced stability of the single GGA mutant RsmY and RsmZ RNAs did not significantly contribute to the observed changes in regulation of RsmA and RsmF activity.
FIG 5.
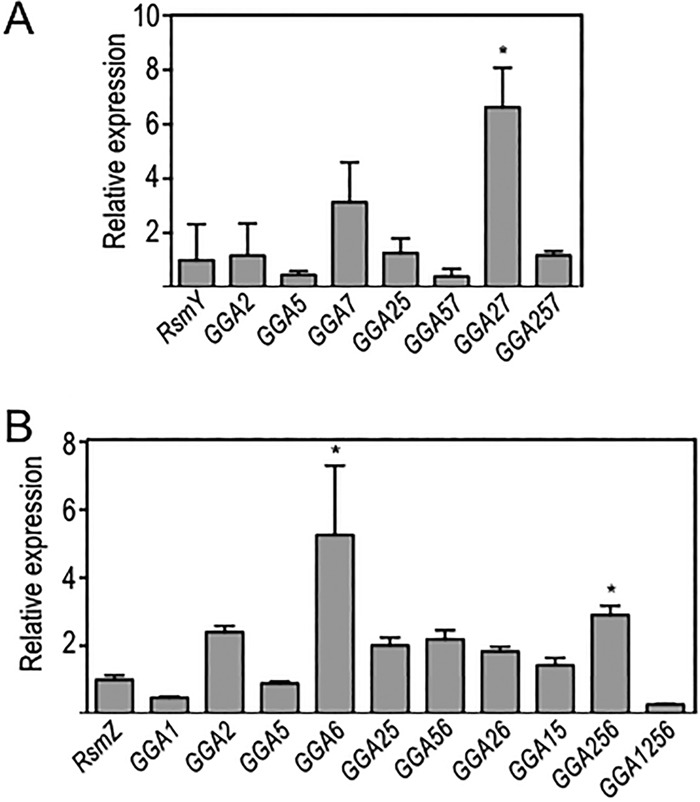
RsmY and RsmZ steady-state expression levels. Cells were cultured to mid-exponential phase in the presence of 0.1% arabinose to induce expression of the respective RNAs, at which time samples were harvested for RNA preparation and quantitation by qRT-PCR. Levels of mutant RsmY (A) or RsmZ (B) RNAs relative to that of native RsmY (A) or RsmZ (B) (normalized to 1.0) are shown. Each RNA sample was normalized to rimM. The reported values represent the averages for at least three experiments, with standard errors indicated by error bars. The indicated statistically significant differences (*, P < 0.05) are relative to strains expressing either native RsmY (A) or RsmZ (B).
Effects of RsmY GGA substitutions on RsmA and RsmF binding in vitro.
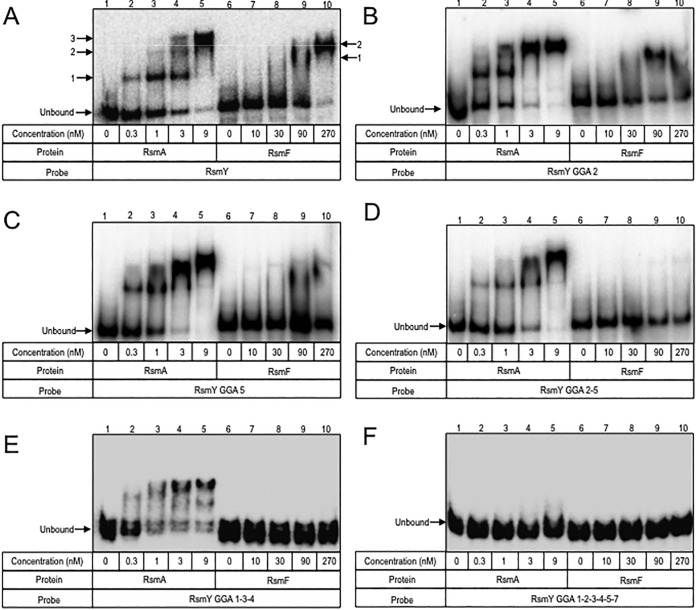
To further investigate the interactions between RsmA/RsmF and RsmY/RsmZ, we measured in vitro binding by using EMSAs. Full-length RsmY and RsmZ and mutant RNAs were synthesized in vitro, radiolabeled at the 5′ end, and incubated with purified RsmAHis or RsmFHis prior to electrophoresis in nondenaturing gels. As previously reported (24), RsmAHis binds RsmY with a high affinity, resulting in the formation of three distinct complexes (Fig. 6A). These complexes likely reflect binding by multiple RsmAHis dimers and/or RsmAHis interactions with different sites on RsmY that alter migration of the complexes. Disruption of RsmY site GGA2, GGA5, or both (GGA25) had no significant effect on RsmAHis affinity or complex formation (Fig. 6B to D). The GGA57 double mutant also had no effect on RsmA affinity, but only two distinct complexes were observed (Fig. S2A). Curiously, disruption of the GGA7 and GGA27 sites resulted in an ∼10-fold higher affinity of RsmAHis (Fig. S1B and C). Although three distinct complexes were seen with the GGA7 mutant, only two complexes were evident with the GGA27 mutant. A significant reduction in RsmAHis affinity was observed with the GGA257 triple mutant, but complex formation was still observed at higher RsmAHis concentrations (Fig. S2D). To determine whether lower-affinity interactions at suboptimal GGA sites account for residual binding to the GGA257 mutant, mutations were introduced into GGA sites 1, 3, and 4. Whereas RsmA still bound the GGA134 mutant with high affinity (Fig. 6E), binding to the GGA123457 mutant was entirely ablated (Fig. 6F). These findings suggest that GGA sites 2, 5, and 7 are the primary determinants for RsmAHis-RsmY complex formation in vitro and that sites 1, 3, and 4 make minor contributions to binding.
FIG 6.
RsmA and RsmF binding to mutant RsmY RNAs. (A to F) The indicated RNAs were synthesized in vitro, radiolabeled, incubated with RsmA (lanes 1 to 5) or RsmF (lanes 6 to 10), and analyzed by nondenaturing polyacrylamide gel electrophoresis. Complexes formed upon RsmA or RsmF binding to native RsmY are indicated by arrows and numbered in panel A. Unbound RNA is indicated by an arrow in each panel.
The binding experiments with RsmF were performed with higher protein concentrations than those used in the RsmA assays because the affinity of RsmF for RsmY is ∼10-fold lower than that of RsmA (Fig. 6A). In general, the RNA-binding activity of RsmF was more sensitive to the GGA substitutions than that of RsmA. RsmFHis binding was detectable only for the GGA2 and GGA7 single mutants (Fig. 6B; Fig. S2B). None of the remaining RsmY mutant probes bound to RsmFHis, even at high protein concentrations (Fig. 6; Fig. S2).
Effects of RsmZ GGA substitutions on RsmA and RsmF binding.
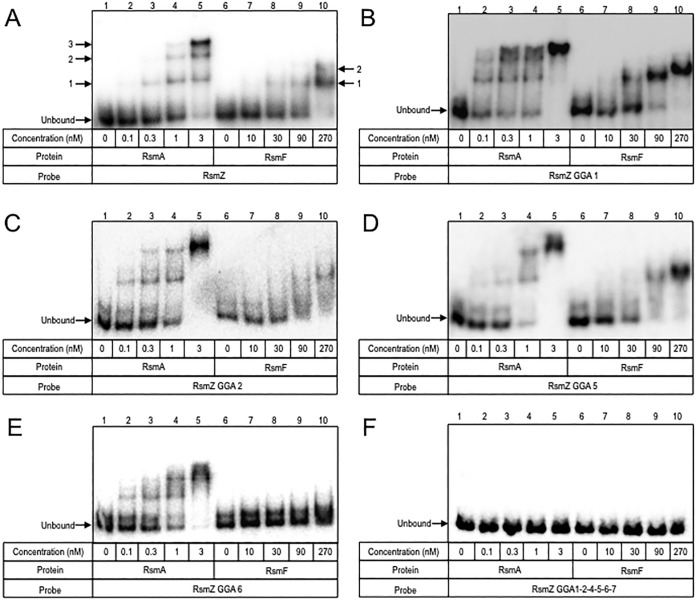
RsmAHis binds RsmZ with a high affinity and forms three distinct complexes (Fig. 7A) (24). Disruption of GGA site 1, 2, 5, or 6 had a significant effect on RsmAHis binding, with a 12- to 40-fold affinity reduction. Despite the reduction in affinity, RsmA binding to the GGA1, GGA2, GGA5, and GGA6 mutants still resulted in the formation of three distinct complexes (Fig. 7B to E). In contrast, the GGA26, GGA15, and GGA56 double mutants demonstrated differences in complex formation, with the most affected mutant being GGA15, for which only one complex was detected (Fig. 7B and C). RsmA still bound mutant RsmZ probes lacking either three (sites 2, 5, and 6) or four (sites 1, 2, 5, and 6) of the GGA sequences presented in stem-loop structures (Fig. S3E and F). To determine if lower-affinity interactions could account for binding to the RsmZ GGA1256 mutant, mutations were introduced at GGA sites 4 and 7. Although the RsmAHis affinity was not reduced for RsmZ GGA147 (Fig. S3G), disruption of GGA sites 1, 2, and 4 to 7 resulted in a complete loss of RsmAHis binding (Fig. 7F).
FIG 7.
RsmA and RsmF binding to mutant RsmZ RNAs. (A to F) Labeled RNAs were incubated with RsmA (lanes 1 to 5) or RsmF (lanes 6 to 10) and analyzed by nondenaturing polyacrylamide gel electrophoresis. Complexes formed upon RsmA or RsmF binding to native RsmZ are indicated by arrows and numbered in panel A. Unbound RNA is indicated by an arrow in each panel.
Compared to that of RsmA, RsmF has a reduced affinity for wild-type RsmZ, and only 1 or 2 shift products are formed (Fig. 7A). Whereas disruption of GGA site 2 or 6 resulted in a significant reduction in RsmFHis affinity (>270 nM) (Fig. 7C and E), GGA substitutions at sites 1 and 5 had no significant effect on affinity or complex formation (Fig. 7B and D). Consistent with the former finding, RsmF was unable to bind to the double mutants lacking GGA sites 2 and 6 or 5 and 6 (Fig. S3B and C), and binding was unaffected in the GGA15 mutant (Fig. S3D). The one anomalous finding was that the GGA25 mutant still supported RsmF binding (Fig. S3A), which contrasts with our finding that GGA site 2 is critical for binding (Fig. 7C). The addition of further GGA substitutions in RsmZ completely blocked RsmF binding (Fig. 7F; Fig. S3E to G).
The discrepancy between the in vivo and in vitro findings for RsmA likely reflects limitations of the in vitro binding assay. One possibility is that another RNA-binding protein interacts with RsmY and/or RsmZ and blocks RsmA binding at suboptimal GGA sites in vivo. One candidate protein is the RNA chaperone Hfq. Hfq binds to and stabilizes RsmY (38). We find that Hfq also binds to RsmZ, with an affinity in the low nanomolar range (Fig. S4A). To test whether Hfq prevents RsmA from binding to some GGA sites in vivo, wild-type RsmY or the RsmY GGA25 double mutant was expressed in wild-type PA103 or an hfq mutant. RsmA availability was measured using a previously described PexsD-lacZ transcriptional reporter (39). The PexsD-lacZ reporter measures type III secretion gene expression and is dependent upon RsmA. Whereas overexpression of wild-type RsmY significantly reduced PexsD-lacZ reporter activity by sequestering RsmA, the RsmY GGA25 mutant lacked this activity in both the wild-type and Δhfq backgrounds (Fig. S4B). Thus, even in the absence of Hfq, the RsmY GGA25 mutant was unable to effectively sequester RsmA.
DISCUSSION
The previous observation that RsmA and RsmF bind RsmY and RsmZ with different affinities led us to investigate the binding interactions with the RNAs. RsmY and RsmZ both have 7 GGA motifs within their sequence. Mfold predictions indicate that RsmY GGA sites 2, 5, and 7 are presented in the loop portion of stem-loop structures. Functional analyses of those sites identified roles for GGA sites 2 and 5 in the sequestration of both RsmA and RsmF. A role for GGA7 could not be demonstrated, consistent with SHAPE-MaP findings showing that GGA7 is base paired in a stem rather than being presented in a stem-loop structure. This conclusion is further supported by findings for the P. fluorescens RsmY homolog, wherein the sites analogous to P. aeruginosa GGA2 and GGA5 serve as the primary determinants for activity in vivo (35).
Mfold and SHAPE-MaP data for RsmZ are in agreement that GGA sites 2, 5, and 6 are presented in stem-loop structures. The NMR structure of P. fluorescens RsmZ also shows that the sites analogous to P. aeruginosa GGA sites 2, 5, and 6 are presented in the same manner. Functional assays, however, identified primary roles for GGA sites 1, 2, and 6 in the sequestration of both RsmA and RsmF. A minor role for GGA site 5 was also evident in the GGA25 and GGA56 double mutants. The finding that in vivo activity requires GGA site 1 was unexpected, as the Mfold and SHAPE-MaP data indicated that site 1 is not presented in a stem-loop structure. We offer three potential interpretations of this observation. First, the GGA-to-CCU substitution in site 1 might significantly alter RsmY folding and render the RNA nonfunctional. Neither Mfold predictions for the site 1 mutant nor qRT-PCR measurements of the RsmZ GGA1 half-life, however, suggest this to be the case. The second possibility is that RsmA/RsmF binding to site 1 is not dependent upon GGA being presented in a stem-loop structure. A similar situation has been demonstrated for several CsrA targets (40, 41). The final possibility is that the Mfold and SHAPE-MaP data are incorrect and that GGA site 1 is presented in a stem-loop structure. This seems unlikely, however, as the sequence surrounding site 1 bears little resemblance to P. fluorescens GGA1 and the best potential stem is only 2 bp long, and likely unstable.
Assays using the tssA1 translational reporter defined clear roles for RsmY GGA sites 2 and 5 and RsmZ GGA sites 1, 2, and 6 in the sequestration of RsmA/RsmF in vivo. RNA probes bearing the same substitutions, however, had only modest effects on RsmA binding activity in vitro. The discrepancy between the in vivo and in vitro findings likely reflects limitations of the in vitro binding assay. Interaction partners of RsmY and RsmZ are not limited to RsmA and RsmF. While our data suggest that Hfq does not account for the discrepancy, there are other proteins that interact with RsmY and RsmZ. Polynucleotide phosphorylase (PNPase), a protein involved in RNA processing and a component of the RNA degradosome, binds to and promotes RsmY and RsmZ turnover (42). A role for PNPase in differential processing of the mutant RNAs, however, is not supported by the qRT-PCR data, which show no significant differences in the half-lives of the mutant RNAs. The ability of RsmA to bind to RsmY GGA25 in vitro may also be due to differences under conditions (ion homeostasis) that contribute to RsmY's ability to fold in vivo versus in vitro conditions.
Analyses of the mutant RsmY and RsmZ RNAs showed that RsmF has more stringent binding requirements than those of RsmA. As best illustrated in Table 1, RsmA demonstrated high-affinity binding to 18 of the 21 RsmY and RsmZ mutant probes. In contrast, RsmF bound only 7 of the same probes, with significantly lower affinities. These findings are in agreement with the previous SELEX data (27) and further support the hypothesis that RsmF requires 2 or more optimal binding sites for high-affinity binding to target RNAs. It is unclear why RsmF binding activity is restricted in this manner or how that might affect the biology of the regulatory system. An answer to the latter question is complicated by the fact that a role for RsmY and RsmZ in the sequestration of RsmF has not been demonstrated under physiologically relevant conditions. The challenges lie in the fact that RsmA activity is dominant over that of RsmF, thus necessitating a requirement to remove RsmA from the system to assess RsmY/RsmZ effects on RsmF function, and that RsmYZ transcription is entirely RsmA dependent (i.e., in the absence of rsmA, no RsmY/RsmZ is produced). Experiments to date have therefore relied upon RsmY/RsmZ overexpression to demonstrate effects on RsmF function, but the results may be misleading. Another possibility is that biologically relevant targets, including primary sRNAs that sequester RsmF and mRNA targets subject to RsmF regulatory control, remain to be identified.
TABLE 1.
RsmA and RsmF affinities for RsmY and RsmZ sRNAs
| RNA probe | Affinity (nM) |
|
|---|---|---|
| RsmA | RsmF | |
| RsmY | 4 | 41 |
| RsmY GGA2 | 2 | 13 |
| RsmY GGA5 | 2 | >270 |
| RsmY GGA7 | 0.2 | 99 |
| RsmY GGA25 | 3 | >270 |
| RsmY GGA57 | 2 | >270 |
| RsmY GGA27 | 0.3 | >270 |
| RsmY GGA134 | 0.5 | >270 |
| RsmY GGA257 | >9 | >270 |
| RsmY123457 | >9 | >270 |
| RsmZ | 0.1 | 84 |
| RsmZ GGA1 | 0.2 | 79 |
| RsmZ GGA2 | 2 | >270 |
| RsmZ GGA5 | 1 | 34 |
| RsmZ GGA6 | 4 | >270 |
| RsmZ GGA25 | 0.1 | 16 |
| RsmZ GGA56 | 4 | >270 |
| RsmZ GGA26 | 0.2 | >270 |
| RsmZ GGA15 | 0.6 | 63 |
| RsmZ GGA147 | 0.1 | >270 |
| RsmZ GGA256 | 0.4 | >270 |
| RsmZ GGA1256 | 1 | >270 |
| RsmZ GGA124567 | >3 | >270 |
RsmA and RsmF are important global regulators contributing to the transition from the acute to the chronic infection phenotype. It remains to be determined if RsmY and RsmZ are the only sRNAs controlling RsmA and RsmF activity. RsmW, an sRNA upregulated during biofilm growth, was recently found to bind RsmA (43), adding another facet to RsmA regulation. Another factor to consider for the Rsm system is the temporal expression of RsmA and RsmF in comparison to that of the sRNAs, as observed in P. fluorescens. RsmX, RsmY, and RsmZ in P. fluorescens each have expression peaks at different stages of growth (44). The different affinities of RsmA and RsmF for RsmY and RsmZ may provide a mechanism for fine-tuning the expression of genes under the control of the Rsm system.
MATERIALS AND METHODS
Strain and plasmid construction.
Routine cloning was performed with E. coli DH5α cultured in LB-Lennox medium with gentamicin (15 μg/ml) as required. P. aeruginosa ΔrsmA, ΔrsmF, ΔrsmAF, ΔrsmYZ, and ΔrsmAYZ mutants in the strain PA103 background were reported previously (see Table S1 in the supplemental material) (24). The ΔrsmFYZ mutant was constructed by introducing the previously described pEX18G2ΔrsmF allelic exchange vector (45) into the ΔrsmYZ mutant. Merodiploids were resolved by sucrose counterselection as previously described (46). The RsmY expression plasmid was constructed by positioning the rsmY transcription start site immediately downstream of the PBAD promoter start site, using the Gibson assembly method (New England BioLabs). Briefly, the PBAD promoter region from pJN105 (primers 117830775 and 117830776) and rsmY (primers 118617707 and 118617708) were each amplified by PCR and then assembled into the MluI and SacI restriction sites of pJN105 (47). pRsmY and pRsmZ vectors bearing single GGA-to-CCT substitutions or various combinations thereof were assembled from two overlapping PCR products by use of the Gibson method and then introduced into the NruI and PvuI sites of pJN105 as outlined in Table S2.
β-Galactosidase assays.
PA103 strains (Table S1) were grown overnight at 37°C on Vogel-Bonner medium (VBM) containing gentamicin (80 μg/ml) as required. The next day, cell suspensions were prepared in Trypticase soy broth (TSB) and diluted to an A600 of 0.1 in TSB supplemented with 100 mM monosodium glutamate, 1% glycerol, and 1 mM EGTA. Arabinose (0.4%) was also added to induce rsmY or rsmZ expression from the PBAD promoter. The cultures were incubated at 37°C and harvested when the A600 reached 1.0. β-Galactosidase activity was measured using the Miller method as previously described (39), using the substrate chlorophenol red-β-d-galactopyranoside (CPRG). CPRG activity was determined by measuring product formation at 578 nm and using an adaptation of the Miller equation, as follows: CPRG units = [A578/culture A600/time (min)/culture volume (ml)] × 1.00.
Electrophoretic mobility shift assays.
DNA templates carrying rsmY, rsmZ, or rsmY or -Z bearing point mutations within the GGA sequences were PCR amplified and end labeled with [γ-32P]ATP as previously described (24). RsmAHis or RsmFHis was purified as previously described (24). RsmAHis or RsmFHis at the concentrations indicated in the figures was incubated with the RNA probes in 1× binding buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 100 mM KCl) with 3.25 ng/μl total yeast tRNA (Life Technologies), 10 mM dithiothreitol (DTT), 5% (vol/vol) glycerol, and 0.1 U RNase Out (Life Technologies). Reaction mixtures were incubated at 37°C for 30 min, mixed with 2 μl of gel loading buffer II (Life Technologies), and immediately subjected to electrophoresis on 7.5 or 10% native polyacrylamide glycine gels (10 mM Tris-HCl, pH 7.5, 380 mM glycine, 1 mM EDTA) at 4°C. Imaging was performed using an FLA-7000 phosphorimager (Fujifilm) and analyzed using MultiGauge v3.0 software.
qRT-PCR.
PA103 cells grown to an A600 of 1.0 were added to 2 volumes of RNAprotect (Qiagen Sciences, Germantown, MD) prior to RNA collection. RNA was extracted using an RNeasy minikit (Qiagen Sciences, Germantown, MD) per the manufacturer's instructions. cDNA was generated by use of the specific primers listed in Table S3, using reaction mixtures containing 100 ng of RNA, 9 μl RNase-free water, 1 μl oligonucleotide mix (2 pmol/μl of specific primer), and 1 μl (10 mM) deoxynucleoside triphosphate (dNTP) mix. Reaction mixtures were heated at 65°C for 5 min and then placed on ice for 1 min. Four microliters of 5× first-strand buffer, 1 μl of 0.1 M DTT, 1 U of RNase Out, and 1 μl of Superscript III reverse transcriptase (Life Technologies, Grand Island, NY) were added to each reaction mixture and incubated at 50°C for 1 h. Each reaction mixture was heat inactivated at 70°C for 15 min. cDNA (2 ng) and 1.8 μl (each) of forward and reverse primers (Table S3) were then added to Power SYBR green PCR master mix (Life Technologies, Grand Island, NY). qPCRs were conducted by the University of Iowa IIHG core.
SHAPE-MaP analysis.
RNA structure cassettes containing rsmY or rsmZ (RsmY_SHAPE_gBlock and RsmZ_SHAPE_gBlock) were synthesized as gBlock DNA templates (Table S3) for in vitro transcription. Each cassette carries a 5′ T7 RNA polymerase promoter (TAATACGACTCACTATAGGG) and rsmY or rsmZ flanked by a stable UUCG tetraloop (GGCCTTCGGGCCAA) at the 5′ end and two tandem UUCG tetraloops (TCGATCCGGTTCGCCGGATCCAAATCGGGCTTCGGTCCGGTTC) at the 3′ end, as previously described (32). In vitro RNA synthesis and structural probing with 1-methyl-7-nitroisatoic anhydride (1M7) were performed as previously described (27). Sequencing was performed on a MiSeq desktop sequencer (Illumina) by use of a MiSeq reagent kit v3 (600 cycles). SHAPE reactivity values for each nucleotide were determined using the ShapeMapper pipeline (33). Structures were determined using RNAstructure to incorporate SHAPE reactivities into the folding free energy model to identify the structure that best agrees with the experimental data (48).
Statistical analysis.
One-way ANOVA was performed using Prism 6.0 (GraphPad Software, Inc., La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (AI097264 to M.C.W. and T.L.Y.). K.H.S. was supported by NIH training grants T32 GM082729 and T32 AI07511.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00736-17.
REFERENCES
- 1.Gottesman S. 1984. Bacterial regulation: global regulatory networks. Annu Rev Genet 18:415–441. doi: 10.1146/annurev.ge.18.120184.002215. [DOI] [PubMed] [Google Scholar]
- 2.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romeo T, Gong M. 1993. Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J Bacteriol 175:5740–5741. doi: 10.1128/jb.175.17.5740-5741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P, Romeo T. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 7.Timmermans J, Van Melderen L. 2010. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapouge K, Perozzo R, Iwaszkiewicz J, Bertelli C, Zoete V, Michielin O, Scapozza L, Haas D. 2013. RNA pentaloop structures as effective targets of regulators belonging to the RsmA/CsrA protein family. RNA Biol 10:1031–1041. doi: 10.4161/rna.24771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. 2009. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol 392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P. 2013. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 87:851–866. doi: 10.1111/mmi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Camara M, Haas D, Williams P. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol 183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burrowes E, Baysse C, Adams C, O'Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 14.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulcahy H, O'Callaghan J, O'Grady EP, Adams C, O'Gara F. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect Immun 74:3012–3015. doi: 10.1128/IAI.74.5.3012-3015.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papenfort K, Vogel J. 2010. Regulatory RNA in bacterial pathogens. Cell Host Microbe 8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. 2014. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 196:357–366. doi: 10.1128/JB.01199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulcahy H, O'Callaghan J, O'Grady EP, Macia MD, Borrell N, Gomez C, Casey PG, Hill C, Adams C, Gahan CG, Oliver A, O'Gara F. 2008. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect Immun 76:632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zha D, Xu L, Zhang H, Yan Y. 2014. The two-component GacS-GacA system activates lipA translation by RsmE but not RsmA in Pseudomonas protegens Pf-5. Appl Environ Microbiol 80:6627–6637. doi: 10.1128/AEM.02184-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimmann C, Valverde C, Kay E, Haas D. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol 187:276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC. 2013. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris ER, Hall G, Li C, Heeb S, Kulkarni RV, Lovelock L, Silistre H, Messina M, Camara M, Emsley J, Williams P, Searle MS. 2013. Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21:1659–1671. doi: 10.1016/j.str.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huertas-Rosales O, Ramos-Gonzalez MI, Espinosa-Urgel M. 2016. Self-regulation and interplay of Rsm family proteins modulate the lifestyle of Pseudomonas putida. Appl Environ Microbiol 82:5673–5686. doi: 10.1128/AEM.01724-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulmeyer KH, Diaz MR, Bair TB, Sanders W, Gode CJ, Laederach A, Wolfgang MC, Yahr TL. 2016. Primary and secondary sequence structure requirements for recognition and discrimination of target RNAs by Pseudomonas aeruginosa RsmA and RsmF. J Bacteriol 198:2458–2469. doi: 10.1128/JB.00343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rife C, Schwarzenbacher R, McMullan D, Abdubek P, Ambing E, Axelrod H, Biorac T, Canaves JM, Chiu HJ, Deacon AM, DiDonato M, Elsliger MA, Godzik A, Grittini C, Grzechnik SK, Hale J, Hampton E, Han GW, Haugen J, Hornsby M, Jaroszewski L, Klock HE, Koesema E, Kreusch A, Kuhn P, Lesley SA, Miller MD, Moy K, Nigoghossian E, Paulsen J, Quijano K, Reyes R, Sims E, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, White A, Wolf G, Xu Q, Hodgson KO, Wooley J, Wilson IA. 2005. Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 A resolution reveals a new fold. Proteins 61:449–453. doi: 10.1002/prot.20502. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. 2005. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol 187:3496–3501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heeb S, Kuehne SA, Bycroft M, Crivii S, Allen MD, Haas D, Camara M, Williams P. 2006. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J Mol Biol 355:1026–1036. doi: 10.1016/j.jmb.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 31.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 33.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. 2013. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A 110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valverde C, Lindell M, Wagner EG, Haas D. 2004. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J Biol Chem 279:25066–25074. doi: 10.1074/jbc.M401870200. [DOI] [PubMed] [Google Scholar]
- 36.Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FH. 2014. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 509:588–592. doi: 10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- 37.Bylund GO, Wipemo LC, Lundberg LA, Wikstrom PM. 1998. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol 180:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorger-Domenigg T, Sonnleitner E, Kaberdin VR, Blasi U. 2007. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem Biophys Res Commun 352:769–773. doi: 10.1016/j.bbrc.2006.11.084. [DOI] [PubMed] [Google Scholar]
- 39.McCaw ML, Lykken GL, Singh PK, Yahr TL. 2002. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol 46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 40.Park H, Yakhnin H, Connolly M, Romeo T, Babitzke P. 2015. CsrA participates in a PNPase autoregulatory mechanism by selectively repressing translation of pnp transcripts that have been previously processed by RNase III and PNPase. J Bacteriol 197:3751–3759. doi: 10.1128/JB.00721-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrade MO, Farah CS, Wang N. 2014. The post-transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5′ UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas. PLoS Pathog 10:e1003945. doi: 10.1371/journal.ppat.1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen R, Weng Y, Zhu F, Jin Y, Liu C, Pan X, Xia B, Cheng Z, Jin S, Wu W. 2016. Polynucleotide phosphorylase regulates multiple virulence factors and the stabilities of small RNAs RsmY/Z in Pseudomonas aeruginosa. Front Microbiol 7:247. doi: 10.3389/fmicb.2016.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller CL, Romero M, Karna SL, Chen T, Heeb S, Leung KP. 2016. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions. BMC Microbiol 16:155. doi: 10.1186/s12866-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kay E, Dubuis C, Haas D. 2005. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A 102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamoun S, Tola E, Kamdar H, Kado CI. 1992. Rapid generation of directed and unmarked deletions in Xanthomonas. Mol Microbiol 6:809–816. doi: 10.1111/j.1365-2958.1992.tb01531.x. [DOI] [PubMed] [Google Scholar]
- 47.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 48.Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.