ABSTRACT
The oral cavity is home to a wide variety of bacterial species, both commensal, such as various streptococcal species, and pathogenic, such as Porphyromonas gingivalis, one of the main etiological agents of periodontal disease. Our understanding of how these bacteria ultimately cause disease is highly dependent upon understanding how they coexist and interact with one another in biofilm communities and the mechanisms by which biofilms are formed. Our research has demonstrated that the DNABII family of DNA-binding proteins are important components of the extracellular DNA (eDNA)-dependent matrix of bacterial biofilms and that sequestering these proteins via protein-specific antibodies results in the collapse of the biofilm structure and release of the resident bacteria. While the high degree of similarity among the DNABII family of proteins has allowed antibodies derived against specific DNABII proteins to disrupt biofilms formed by a wide range of bacterial pathogens, the DNABII proteins of P. gingivalis have proven to be antigenically distinct, allowing us to determine if we can use anti-P. gingivalis HUβ antibodies to specifically target this species for removal from a mixed-species biofilm. Importantly, despite forming homotypic biofilms in vitro, P. gingivalis must enter preexisting biofilms in vivo in order to persist within the oral cavity. The data presented here indicate that antibodies derived against the P. gingivalis DNABII protein, HUβ, reduce by half the amount of P. gingivalis organisms entering into preexisting biofilm formed by four oral streptococcal species. These results support our efforts to develop methods for preventing and treating periodontal disease.
IMPORTANCE Periodontitis is one of the most prevalent chronic infections, affecting 40 to 50% of the population of the United States. The root cause of periodontitis is the presence of bacterial biofilms within the gingival space, with Porphyromonas gingivalis being strongly associated with the development of the disease. Periodontitis also increases the risk of secondary conditions and infections such as atherosclerosis and infective endocarditis caused by oral streptococci. To induce periodontitis, P. gingivalis needs to incorporate into preformed biofilms, with oral streptococci being important binding partners. Our research demonstrates that targeting DNABII proteins with an antibody disperses oral streptococcus biofilm and prevents P. gingivalis entry into oral streptococcus biofilm. These results suggest potential therapeutic treatments for endocarditis caused by streptococci as well as periodontitis.
KEYWORDS: DNABII, Porphyromonas gingivalis, antibody, biofilm, streptococci
INTRODUCTION
The oral cavity contains one of the most diverse microbial communities found in the human body, with an estimated 500 to 700 different species that comprise the oral microbiome (1). While diverse, the bacteria present within the oral cavity colonize and form biofilm communities in a predictable and organized manner. Initial colonizers are comprised of many streptococcal species capable of attaching to the salivary pellicle of the tooth surface and forming a base that subsequent colonizers can attach to and build upon. Late-colonizing bacteria are primarily Gram-negative anaerobic species comprising various periodontal pathogens (2). Primary among those oral pathogens is Porphyromonas gingivalis, considered a predominant etiological agent of periodontitis. Periodontitis is a chronic inflammatory disease characterized by inflammation of the soft tissue surrounding the tooth and subsequent underlying bone loss resulting in an increase in the gingival pocket and, if left untreated, loss of the tooth (3). While tooth loss is problematic, severe periodontitis can also increase an individual's risk for additional chronic diseases, such as rheumatoid arthritis, atherosclerosis, and infective endocarditis (4–8). Infective endocarditis can be a particular risk, as the chronic inflammation and bleeding characteristic of periodontal disease can provide a readily accessible access point to the bloodstream for the oral streptococcal species, such as Streptococcus gordonii, Streptococcus mitis, and Streptococcus oralis, commonly associated with this disease.
While the individual species present within a biofilm vary widely, one component is common to all biofilms: the presence of an extracellular polymeric substance (EPS). EPS is a self-forming matrix of bacterial and environmental components that functions in part to protect the bacteria within the biofilm from environmental hazards, including antimicrobials and host defenses. While protective, the EPS still allows for nutrient exchange, intracellular signaling, and communication, as well as both the entrance into and exit of homologous and heterologous bacteria. One of the most universal components of EPS is extracellular DNA (eDNA) (9). eDNA can be either primarily prokaryotic or derived from other sources, such as neutrophil extracellular traps produced by polymorphonuclear neutrophils at infection sites (10). Our laboratory has established that in order to maintain the highly complex and organized structure of eDNA, bacteria rely on the DNABII family of proteins to bind to and stabilize the mesh-like structure (11–18).
The DNABII family of proteins are ubiquitously expressed by all eubacteria and consist of histone-like protein (HU) and integration host factor (IHF), which has been found only in proteobacteria (19). HU and IHF bind to and bend double-stranded DNA, displaying particular affinity for structured DNA such as Holliday junctions, replication forks, or sites of DNA damage (20–22). With regard to the EPS of a biofilm, DNABII proteins have been located at intersections of eDNA, where they stabilize the structure of the eDNA component of the EPS (15).
Early work from our lab demonstrated that antiserum derived against a single DNABII protein, the IHF protein from Escherichia coli (EcIHF), was capable of disrupting monospecies biofilms formed by multiple pathogenic bacteria and polymicrobial samples such as cystic fibrosis sputum (11–13, 15–17) and, more recently, that antibodies delivered in poly(lactic-co-glycolic acid) microspheres could induce resolution of an experimental form of peri-implantitis (14). It is important to note that the use of this polyclonal antiserum results not in the killing or death of the bacteria but rather a shift in the partition from the biofilm state to a planktonic state (13). Our current data indicate that anti-DNABII antibodies are capable of sequestering unbound DNABII proteins, preventing them from entering into the biofilms and stabilizing the structure of the eDNA. Removal of a sufficient fraction of DNABII protein causes a shift in the equilibrium from an eDNA-bound state to an unbound state, resulting in a collapse of the eDNA structure and release of the biofilm resident bacteria (12).
Our previous work focused on the identification of variations within DNABII proteins and the specificities of antisera derived against DNABII proteins to allow us to further exploit this technology. To that end, we determined that the DNABII proteins of P. gingivalis were antigenically distinct from all other DNABII proteins previously examined. Antisera derived against the HUα and HUβ proteins (anti-PgHUα and anti-PgHUβ, respectively) were able to recognize their cognate proteins only by Western blotting, although the HUβ protein (PgHUβ) was the only protein of the two involved in biofilm formation (18). However, the antiserum derived against the HU protein of S. gordonii (anti-SgHU) demonstrated a limited specificity in that it recognized any HU protein tested but did not recognize any IHF proteins. While SgHU and PgHUβ proteins were antigenically distinct from one another, they were still capable of functionally complementing one another within the biofilm EPS (18); i.e., the DNABII proteins could be removed from a biofilm of one species and replaced with the DNABII protein from a second species. For example, the SgHU protein could be removed from an S. gordonii biofilm with anti-SgHU and replaced with PgHUβ and the biofilm would maintain its structure.
Following up on these data, two approaches were taken to further develop potential treatments for periodontitis. First, using the anti-SgHU antiserum, which has been shown to have broad reactivity toward HU proteins, we determined if other potentially pathogenic oral streptococci could be dispersed in a manner similar to that for S. gordonii. Second, taking advantage of the specificity that anti-PgHUβ displayed for PgHUβ, we explored the potential of this antiserum to prevent the spread of P. gingivalis within preformed biofilms, mimicking the route of colonization this important periodontal pathogen takes within the oral cavity.
In this report, we present data which indicate that other potentially pathogenic oral streptococci, similar to S. gordonii, utilized DNABII proteins to support the structural integrity of the eDNA component of their EPS and, further, that anti-SgHU had an ability to disrupt these biofilms similar to its ability to disrupt S. gordonii biofilm. We also present data that suggest that pretreatment of P. gingivalis with anti-PgHUβ has the ability to prevent P. gingivalis from entering and expanding within preexisting biofilms formed by multiple oral streptococci. This presents a potential method for the prevention and removal of pathogenic biofilms from the oral cavity.
RESULTS
Antiserum derived against S. gordonii HU recognized DNABII proteins expressed by other oral streptococci.
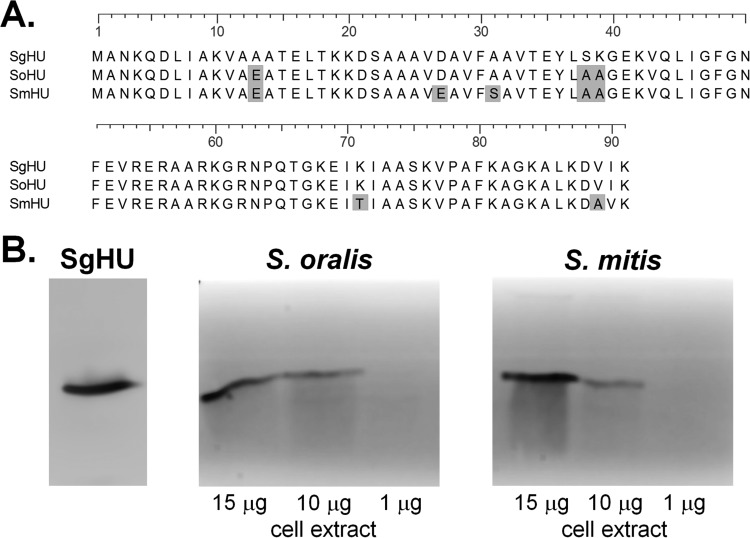
We have established the ability of anti-E. coli IHF (anti-EcIHF) antiserum to disrupt a wide variety of bacterial biofilms (11–13, 15–17), although to date we have not fully explored the effectiveness of this approach on biofilms formed by bacteria of the oral cavity. We have previously established that antisera directed against the S. gordonii HU protein (SgHU), the only DNABII protein present in S. gordonii, and the HUβ protein of P. gingivalis (PgHUβ), the P. gingivalis DNABII protein involved in biofilm formation, have the ability to disrupt established biofilms in vitro (18). However, we have not examined the efficacy of anti-DNABII antiserum against any other potentially pathogenic oral bacterial species. As the antiserum derived against the SgHU protein (anti-SgHU) has been previously shown to have the ability to recognize HU proteins from other bacteria (18), we tested the ability of this antiserum to recognize DNABII proteins from S. oralis and S. mitis, both early colonizers of the oral cavity similar to S. gordonii. As can be seen in Fig. 1A, an amino acid alignment of the DNABII proteins expressed in both of these bacteria indicates that these proteins are highly similar to the SgHU protein, with 97% sequence identity for S. oralis and 91% sequence identity for S. mitis and each species containing only a single HU allele. Therefore, we hypothesized that these proteins would likely be recognized by antiserum directed against SgHU. Western blot analysis of whole-cell extracts identified proteins of the size corresponding to DNABII proteins in both S. mitis and S. oralis (Fig. 1B). These results suggest that anti-SgHU antiserum was capable of recognizing DNABII proteins of both S. mitis and S. oralis.
FIG 1.
Antisera to the SgHU protein recognize DNABII proteins from additional oral streptococci. (A) Alignment of S. gordonii HU (SgHU), S. oralis HU (SoHU), and S. mitis HU (SmHU) sequences. Amino acid differences from the S. gordonii sequence are shaded. Sequences were aligned using the ClustalW program. (B) Western blot analysis of 1, 10, and 15 μg of soluble cell extracts of S. oralis, S. mitis, and SgHU protein probed with anti-SgHU, demonstrating the cross-reactivity of anti-SgHU antiserum with the DNABII proteins from the two bacteria.
HU proteins are present within biofilms formed by oral streptococci.
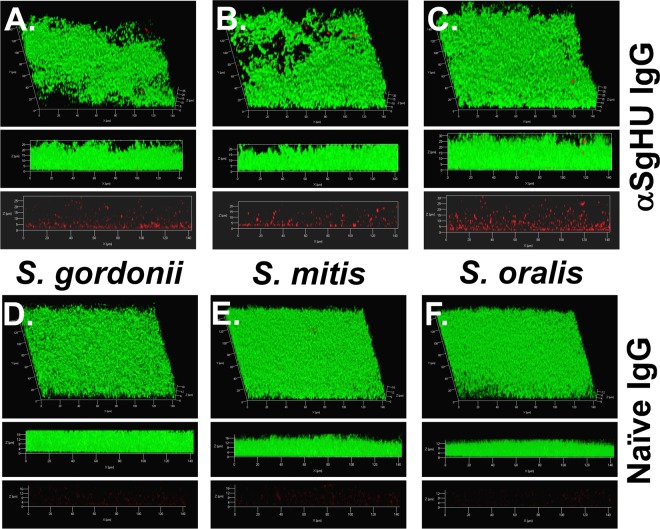
Having shown that anti-SgHU antiserum is capable of recognizing the DNABII proteins of other oral streptococcal species, we aimed to determine if DNABII proteins were present within the extracellular matrix of biofilms formed by S. mitis and S. oralis. Preformed 24-h biofilms were labeled with 1.5 μg of purified rabbit anti-SgHU IgG and imaged using confocal scanning laser microscopy. In Fig. 2, biofilm biomass (labeled with 5- or 6-carboxyfluorescein diacetate succinimidyl ester [CFDA]) can be seen in green, while HU protein labeling can be seen in red. Both the S. mitis (Fig. 2B) and the S. oralis (Fig. 2C) biofilms have architecture similar to that of S. gordonii (Fig. 2A), and HU proteins can be detected throughout each biofilm. The marked lack of signal when biofilms are probed with IgG purified from a preimmune serum (Fig. 2D to F) provides additional evidence that HU proteins are indeed abundant within the biofilm EPS architecture of all three bacteria.
FIG 2.
Localization of HU proteins within streptococcal biofilms. Biofilm growth was maintained for 24 h, and unfixed S. gordonii (A), S. mitis (B), and S. oralis (C) biofilms were incubated with anti-SgHU and then treated with goat anti-rabbit IgG conjugated to Alexa Fluor 594 (HU; red). Biofilms treated with preimmune sera are in panels D (S. gordonii), E (S. mitis), and F (S. oralis). The presence of HU proteins can be readily observed in the biofilms probed with anti-SgHU, while minimal signal is detectable with preimmune serum. Cells were stained with CFDA (green). The biofilms were then imaged using a 63× objective on a Zeiss 510 Meta laser scanning confocal microscope. The top image is a 3-dimensional reconstruction, the middle image is a side view of the reconstruction, and the bottom image shows the distribution of DNABII in the absence of cell signal.
Antibodies derived against S. gordonii HU disrupt biofilms of oral streptococci.
Having established that anti-SgHU can recognize DNABII proteins from both S. mitis and S. oralis, we tested the ability of the antibody as well as IgG isolated from anti-EcIHF antiserum to disrupt monospecies biofilms of these bacteria. Biofilms were exposed to antibodies both at the time of initial seeding of the bacteria (seeding) and after 16 h of growth (treatment) and to two different concentrations of antibodies (30 μg/ml and 100 μg/ml). After 24 h, the biofilms were stained and imaged.
Analysis of the confocal microscopy of biofilms with the COMSTAT program revealed that both anti-SgHU and anti-EcIHF possess the ability to disrupt biofilms of S. mitis and S. oralis (Table 1). The lower concentration of antibodies (30 μg/ml) had only a minimal effect on the biofilms parameters measured. A statistically significant decrease in average thickness was observed when anti-EcIHF was used at seeding for the S. mitis biofilms and for anti-SgHU treatment of S. oralis biofilms. However, when 100 μg/ml of antibody was added, large and significant decreases were seen in average thickness of the biofilms as well as total biofilm biomass under both seeding and treatment conditions for S. mitis and S. oralis (Table 1). Additionally, the dose response demonstrates that the observed disruption was likely dependent on the titration of DNABII proteins away from the biofilms and that collapse occurs only when a sufficient amount of protein has been sequestered from the biofilm EPS.
TABLE 1.
Effect of antibodies on S. mitis and S. oralis biofilmsa
| Antibody use and amt | % reduction after seeding or treatment |
|||||||
|---|---|---|---|---|---|---|---|---|
|
S. mitis |
S. oralis |
|||||||
| Anti-SgHU |
Anti-EcIHF |
Anti-SgHU |
Anti-EcIHF |
|||||
| Avg thickness | Biomass | Avg thickness | Biomass | Avg thickness | Biomass | Avg thickness | Biomass | |
| Seeding | ||||||||
| 30 μg/ml | <15 | 20 | 25* | 25 | 20 | 25 | <15 | <15 |
| 100 μg/ml | 45** | 50** | 50**** | 55**** | 60**** | 60*** | 55*** | 60*** |
| Treatment | ||||||||
| 30 μg/ml | 25 | 25 | <15 | 20 | 15* | 20 | <15 | <15 |
| 100 μg/ml | 45**** | 50**** | 40** | 40*** | 50** | 55** | 50**** | 50** |
Biofilms were either treated at the time of seeding with antibody (seeding) or grown for 16 h before addition of antibody (treatment). Antibodies (anti-SgHU or anti-EcIHF) were added at the indicated amounts. Significant reductions in average thickness and biofilm biomass can be observed with the addition of increasing amounts of antibody. Biofilms were imaged and analyzed with COMSTAT to obtain average thickness and total biomass parameters. Statistically significant values compared to those for preimmune sera are in bold. *, P ≤ 0.05; **, P ≤ 0.01; ***; P ≤ 0.001; ****, P ≤ 0.0001.
Preincubation of P. gingivalis with antibodies directed against the PgHUβ protein prevents P. gingivalis from entering biofilms formed by oral streptococci.
In our earlier work, we demonstrated that while antisera directed against their respective cognate DNABII proteins could disrupt S. gordonii biofilms as well as those of the periodontal pathogen P. gingivalis, the DNABII proteins of P. gingivalis were antigenically distinct, allowing P. gingivalis biofilms to be disrupted only by anti-PgHUβ (18). Additional previous work identified a role for the EcIHF proteins in both the invasion of and maintenance of uropathogenic E. coli (UPEC) community architecture within epithelial cells (23). Indeed, anti-EcIHF inhibited attachment of UPEC to epithelial cells (23). We performed experiments to determine if these results could translate into attachment to bacterial biofilms by testing whether anti-PgHUβ could specifically prevent the pathogenic bacteria from entering into preformed biofilms of various oral streptococci.
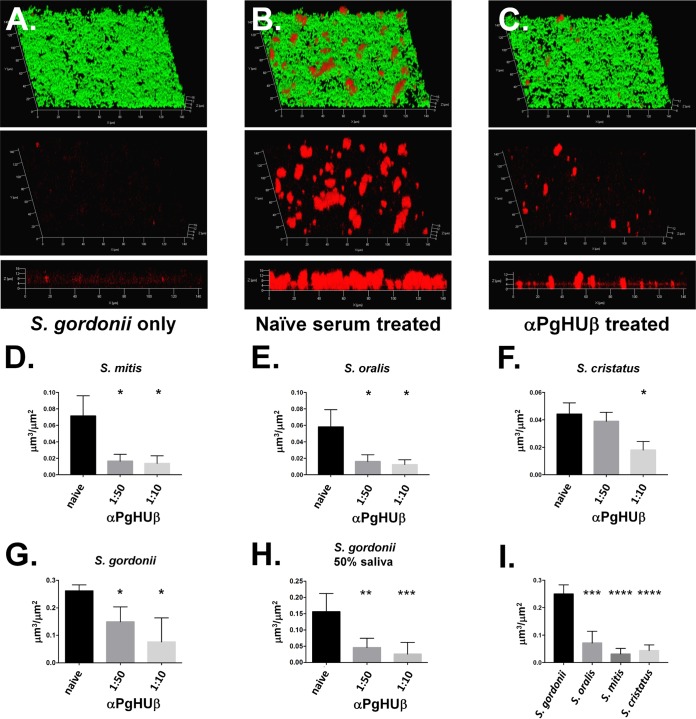
P. gingivalis (∼108 CFU) was treated with either a preimmune serum or anti-PgHUβ for 1 h. Treated cells were then added to preformed biofilms of S. mitis, S. oralis, S. gordonii, or Streptococcus cristatus, an additional oral streptococcus that appears to have inhibitory effects on P. gingivalis biofilm formation. The biofilms were then grown for an additional 16 h before imaging. P. gingivalis cells were then labeled with a polyclonal antiserum directed against the FimA fimbrial protein, and the biofilm was stained with CFDA. As can be seen in the representative images in Fig. 3A to C for S. gordonii, pretreatment of P. gingivalis cells before addition to preformed streptococcal biofilms resulted in significant decreases in the amount of P. gingivalis organisms entering into the biofilm. COMSTAT analysis of the biomass of P. gingivalis present within the biofilm revealed a significant decrease in the ability of this bacterium to enter into a biofilm formed by either S. mitis (Fig. 3D), S. oralis (Fig. 3E), S. cristatus (Fig. 3F), or S. gordonii (Fig. 3G). Additionally, dose-dependent responses can be easily observed with both S. gordonii and S. cristatus.
FIG 3.
Preincubation of P. gingivalis with anti-PgHUβ prevents P. gingivalis from entering into streptococcal biofilm. Biofilms were grown for 24 h before addition of P. gingivalis. P. gingivalis was incubated with a 1:50 or 1:10 dilution of anti-PgHUβ antisera for 1 h before addition. Representative biofilm images show results for S. gordonii alone (A) as well as naive serum (B) and a 1:50 dilution of anti-PgHUβ-treated P. gingivalis (C). All cells (green) and P. gingivalis (red) are shown in the top image, while P. gingivalis-only labeling is shown in in the middle image. The bottom image is a side view of the P. gingivalis-only-labeled biofilm. A no-P. gingivalis control biofilm (A) shows little to no red signal, while that treated with naive serum shows significant amounts of P. gingivalis (B). Treatment with a 1:50 dilution of anti-PgHUβ results in a significant decrease in P. gingivalis detected within the biofilm (C). Graphical representations of decrease in P. gingivalis entering into S. mitis (D), S. oralis (E), S. cristatus (F), or S. gordonii (G) indicate dose-dependent decreases in detection of P. gingivalis. Experiments with S. gordonii were repeated with 50% pooled human saliva (H). Total naive serum-treated P. gingivalis entering streptococcal biofilms is shown in panel I, which shows that significantly more P. gingivalis cells enter an S. gordonii biofilm than the other oral streptococci tested. Dual-species biofilms were grown for 16 h in THBHK. Cells were stained with carboxyfluorescein succinimidyl ester (CFSE). P. gingivalis was immunofluorescently labeled with a polyclonal antifimbrial primary antisera and a secondary goat anti-rabbit Alexa Fluor 647-conjugated antibody and visualized using confocal scanning laser microscopy. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To better characterize the ability of antiserum to specifically prevent P. gingivalis entry into extant S. gordonii biofilms, we eliminated the antiserum-P. gingivalis preincubation step or washed away free antiserum after the preincubation. The former shows the kinetics of antibody finding its target, while the latter prevents free antibody from acting on the extant oral streptococcus biofilms. Elimination of the 1-h incubation of P. gingivalis and antiserum resulted in a decrease in the ability of the antiserum to prevent P. gingivalis entry into streptococcal biofilm at a 1:50 dilution but had no effect at a 1:10 dilution (see Fig. S1 in the supplemental material). Additionally, when P. gingivalis cells were incubated for 1 h with antiserum and subsequently washed to remove unbound antibody before addition to oral strep biofilms, there was again a decrease in the ability to prevent entry into biofilms (Fig. S2). However, the decrease observed was not as severe as that seen with the elimination of the 1-h incubation, suggesting that the preincubation is important for prevention of P. gingivalis entry into preformed biofilms of oral streptococci.
The ability of anti-PgHUβ to prevent P. gingivalis from entering into biofilms formed by these oral streptococci suggests either a role for DNABII proteins in establishment of P. gingivalis within preexisting biofilms or that there is sufficient surface-associated PgHUβ that addition of the antibody prevents sufficient interaction between the two species.
To ensure the efficacy of the response under more physiologic conditions, P. gingivalis cells were preincubated with antiserum in pooled human saliva before addition to the streptococcal biofilms. As can be seen in Fig. 3H, there was no decrease in the ability of anti-PgHUβ to prevent P. gingivalis from entering into S. gordonii biofilms, suggesting that the antibodies still function in the presence of saliva.
The data in Fig. 3 also demonstrate the differing abilities of P. gingivalis to readily form communities with different oral streptococci. Figure 3I compares the amounts of P. gingivalis biomass detected within biofilms of the different streptococcal species when P. gingivalis has been treated with preimmune antiserum. The biomass of P. gingivalis measured in the S. gordonii biofilms was significantly greater than that observed in either the S. mitis, S. oralis, or S. cristatus biofilms, suggesting that P. gingivalis is better able to form biofilms with S. gordonii than other oral streptococci. This result is consistent with S. gordonii and P. gingivalis being natural coaggregation partners (24, 25).
DISCUSSION
Some of the earliest colonizers of the oral cavity consist of various species of Gram-positive streptococci which attach to the pellicle on the tooth surface and provide an additional substrate to which subsequent bacteria can attach (26, 27). Predominant among the early-colonizing bacteria are species such as S. gordonii, S. oralis, S. mitis, and S. cristatus (27). Late-colonizing Gram-negative bacteria such as the red-complex bacteria P. gingivalis, Treponema denticola, and Tannerella forsythia are established periodontal pathogens which significantly contribute to the chronic inflammation that is characteristic of periodontal disease (27–30). This chronic inflammation is proving to be associated with increased risk of more systemic diseases such as rheumatoid arthritis, osteoporosis, atherosclerosis, and cardiovascular disease, resulting from the increased circulation of proinflammatory compounds (4–8, 31, 32). While late-colonizing bacteria are more well-established pathogens, the chronic inflammation and tissue damage caused by periodontal disease can provide early-colonizing streptococcal species, considered mostly commensal in the oral cavity, access to the bloodstream and areas of the body they are not normally exposed to, increasing the likelihood of an infection (33). In fact, periodontal disease is considered a risk factor for the possibility of infective endocarditis (34).
We have demonstrated that like S. gordonii, other oral streptococci (such as S. mitis and S. oralis) utilize the DNABII protein HU within the EPS of their biofilms (Fig. 2). We have also shown that the HU proteins of S. mitis and S. oralis are sufficiently similar in sequence and structure to the SgHU protein that antibodies derived against the SgHU protein are capable of recognizing HU proteins from these bacteria both by Western blotting (Fig. 1) and within a biofilm (Fig. 2). With the ability to recognize these oral streptococcus HU proteins, anti-SgHU antibodies also demonstrate an ability to disrupt biofilms of both S. mitis and S. oralis (Table 1). The ability to disrupt biofilms of viridans group streptococci like S. mitis and S. oralis after they have left the oral cavity and colonized different locations, such as heart valves, is of great importance, as this group is one of the primary causes of infective endocarditis (35). Because of the biofilm nature of the disease, treatment of infective endocarditis generally requires high-dose intravenous antibiotics to be effective. Targeting the DNABII proteins of a biofilm and releasing the bacteria has the potential to allow for lower doses of antibiotic to be used, as releasing nontypeable Haemophilus influenzae from a biofilm results in a concurrent decrease in the MIC of antibiotic needed to effectively eliminate the bacteria (12).
Additionally, as oral streptococci are the predominant species that function as initial colonizers of the oral cavity, the ability to remove them from a biofilm, or even prevent their initial attachment, should allow for the reduction of total biofilm within the oral cavity or alteration of the biofilm composition. Recent work was able to determine that the prevalence of the periodontal pathogen P. gingivalis was reduced when initial colonizers were omitted from a multispecies biofilm model (36). Controlling the levels of initial colonizers within an oral biofilm may be a method to indirectly control the presence of pathogenic species as well. Methods to prevent or reduce the levels of initial colonizing bacteria should result in a subsequent decrease in the levels of later-colonizing bacteria, as they would have less preformed biofilm surface to attach to.
A second method for reducing the presence of P. gingivalis within a biofilm is demonstrated in Fig. 3, which shows that pretreating P. gingivalis with anti-PgHUβ antiserum before addition to preformed biofilms reduced the total amount of P. gingivalis detected within the biofilm. These results suggest that DNABII proteins could play a role in attachment of P. gingivalis to preexisting biofilms. Or, if the bacteria are still able to attach, sequestering PgHUβ could prevent P. gingivalis from expanding and growing the biofilm. It is also possible that antibodies binding to PgHUβ physically block the ability of P. gingivalis from interacting with the preformed biofilm. Previous work has shown that pretreatment with anti-EcIHF was capable of preventing UPEC from attaching to epithelial cells efficiently, possibly through steric hindrance of the antibody binding to surface-associated DNABII proteins (23), so it should not be unexpected that anti-PgHUβ antibodies are capable of preventing P. gingivalis from binding to a streptococcal biofilm. However, it is unclear if by binding to DNABII proteins, the antibody is physically blocking the P. gingivalis from interacting with the streptococcal cells, i.e., antibody binding the DNABII proteins is able to prevent the fimbriae from interacting with the SspA/B proteins of S. gordonii (24). The noted decrease in the ability of the antibodies to prevent P. gingivalis entry into preformed biofilms seen when the preincubation step was removed (see Fig. S1 in the supplemental material) suggests a kinetic effect of antibodies binding to P. gingivalis-associated PgHUβ, since increasing the amount of antibody overcomes this reduction. Additionally, washing the P. gingivalis cells after incubation with antisera also affects entry into streptococcal biofilms (Fig. S2), although it appears to have less of an effect than elimination of the incubation step. This result again is evidence of the importance of the antibody binding to P. gingivalis-associated PgHUβ before exposure to the streptococcal biofilms for efficient prevention of P. gingivalis entry into the biofilm.
If binding of antibodies to DNABII proteins does not prevent interaction with S. gordonii cells within a biofilm, then it may be that DNABII proteins are necessary for attachment and or entry into the biofilm or that once in a biofilm, P. gingivalis needs DNABII proteins to allow the organism to proliferate within the biofilm. Future experiments will address whether anti-PgHUβ can prevent not only initial cell attachment but also aggregation and expansion of P. gingivalis within the heterogeneous biofilm. Nonetheless, this antigen-specific blocking of P. gingivalis creates conditions where the oral commensal streptococci in their “healthy” state can be protected from invasion by P. gingivalis and any subsequent dysbiosis and creation of a pathogenic state.
The work presented here expands our understanding of how DNABII proteins are utilized within a biofilm, as well as identifying additional Gram-positive bacterial biofilms that are capable of being disrupted by antibodies derived against DNABII proteins. The effectiveness of anti-DNABII antibodies against these opportunistic pathogens presents a potential method to treat these biofilm-based infections. Additionally, our experiments demonstrating our ability to prevent P. gingivalis from entering into preexisting biofilms provide an avenue to develop new therapeutics capable of either preventing P. gingivalis colonization of the oral cavity or, if biofilms causing periodontal disease have already formed, halting growth of these biofilms, thereby reducing the severity and progression of the disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis strain 381 was maintained on Todd-Hewitt agar (1.5%) supplemented with hemin (5 μg/ml) and menadione (1 μg/ml) (THBHK) and grown under anaerobic conditions (5% hydrogen, 10% carbon dioxide, and 85% nitrogen) at 37°C. Broth cultures of P. gingivalis were grown in THBHK under anaerobic conditions at 37°C. S. gordonii strain Chalis CH1 (DL1), S. mitis ATCC 33399, S. oralis ATCC 10557, and S. cristatus ATCC 49999 were maintained on THB 1.5% agar plates at 37°C in an atmosphere of 5% CO2. Broth cultures of streptococci were grown in THB at 37°C in an atmosphere of 5% CO2. Biofilms of S. mitis and S. oralis were grown in THB at 37°C in an atmosphere of 5% CO2. Biofilms of S. gordonii were grown in a chemically defined medium (CDM) at 37°C in an atmosphere of 5% CO2 (37).
Western blot analysis.
Soluble cell extracts were prepared as follows. Ten milliliters of early stationary-phase cell culture was harvested by centrifugation at 10,000 × g for 5 min. Cell pellets were then resuspended in 50 mM Tris HCl and 100 mM NaCl (pH 7.5) and broken via French press (Constant Systems cell disruptor). Cell debris was removed by centrifugation at 30,000 × g for 30 min. Soluble cell protein was quantified and protein was resolved using 13% SDS-PAGE and transferred to a nitrocellulose membrane for 45 min at 40 V. After transfer, the membranes were blocked using a solution of TBS-T (20 mM Tris HCl, 1 mM NaCl, and 0.1% Tween) containing 5% Blotto nonfat dry milk overnight at 4°C. The membranes underwent three 5-min washes with TBS-T and were probed with the primary antibody at a dilution of 1:1,000 for 1 h at room temperature. The membranes then underwent three 5-min washes with TBS-T and were probed with a horseradish peroxidase-linked goat anti-rabbit IgG antibody (Cell Signaling) at a 1:10,000 dilution for 1 h at room temperature. The membranes then underwent three 5-min washes with TBS-T and were developed with the Pierce ECL-2 Western blotting substrate (Thermo Fisher) and were imaged using a Typhoon FLA 7000 laser scanner (GE Healthcare Life Sciences).
Purification of IgG from serum.
IgG from rabbit antiserum was purified with HiTrap protein G HP columns (GE Healthcare) using an AKTA Pure fast-pressure liquid chromatograph (GE Healthcare). Serum derived against individual DNABII proteins was dialyzed against 20 mM potassium phosphate buffer (pH 7.0) before loading onto the column. The column was then washed with 15 column volumes of buffer before a linear gradient of 2 column volumes and elution of the protein with 6 column volumes of glycine (pH 2.7). Fractions (1 ml) were collected into 200 μl of Tris (pH 9.0) and screened for the presence of purified IgG by 16% SDS-PAGE. The IgG enriched fractions were dialyzed against 150 mM Tris (pH 7.4)–150 mM KCl and concentrated using an Amicon Ultra-4 centrifugal filter device (GE Healthcare). IgG concentration was determined with a Pierce bicinchoninic acid (BCA) protein assay kit (Fisher Scientific) and confirmed by SDS-PAGE. IgG was stored at −80°C in 150 mM Tris (pH 7.4)–150 mM KCl–10% glycerol.
In vitro biofilm analysis.
Streptococci were grown in THB overnight at 37°C in an atmosphere of 5% CO2. Cultures were diluted to an optical density at 490 nm (OD490) of 0.65, diluted 1:4 in THB, and grown statically at 37°C until an OD490 of 0.65 was reached. Cultures were then diluted 1:2,500 in THB, and 200 μl of this culture was used to inoculate each well of an eight-well chambered glass coverslip (Thermo Scientific). The cultures were grown at 37°C with 5% CO2 for 24 h to allow for biofilm formation.
Purified IgG preparations derived from antisera against specific DNABII proteins were added either at seeding of the biofilms or after 16 h of growth. Biofilms were then grown for an additional 8 h after IgG addition.
Biofilms were stained with LIVE/DEAD stain (Molecular Probes, Eugene, OR) according to the manufacturer's protocols, washed once with 200 μl of sterile phosphate-buffered saline (PBS), and fixed with fixative solution (1.6% paraformaldehyde, 0.025% glutaraldehyde, and 4% acetic acid in 0.1 M sodium phosphate buffer [pH 7.4]). Biofilms were imaged on a Zeiss 510 Meta laser scanning confocal microscope (Carl Zeiss) using a 63× water objective. Three-dimensional z-stack images were reconstructed using AxioVision Rel. 4.8 (Carl Zeiss), and the average biofilm thickness and total biofilm biomass parameters were determined using the COMSTAT analysis program running on MatLab software (38). All biofilm conditions were tested in a minimum of 3 independent experiments conducted on different days, with each experiment performed in duplicate for each condition and 3 images captured and averaged from each well.
Immunofluorescence of DNABII proteins within in vitro biofilms.
Streptococcal biofilms were formed as described above. Unfixed 24-h biofilms were labeled with 1.5 μg of purified IgG isolated from polyclonal rabbit antiserum directed against the S. gordonii HU protein in sterile PBS for 1 h, washed with sterile PBS, and then incubated with goat anti-rabbit IgG conjugated to Alexa Fluor 594 (Molecular Probes) at a 1:200 dilution in sterile PBS for 1 h. Bacterial cells were labeled with 5- or 6-carboxyfluorescein diacetate succinimidyl ester (CFDA) (2.6 μg/ml) for 1 h. Biofilms were washed with 200 μl of PBS again before imaging. The biofilms were imaged using a 63× objective on a Zeiss 510 Meta laser scanning confocal microscope (Carl Zeiss, Thornwood, NY). Three-dimensional images were reconstructed with AxioVision Rel. 4.8 (Carl Zeiss).
Inhibition of P. gingivalis from entering preestablished streptococcal biofilms.
Streptococcal biofilms were grown for 24 h as described above before addition of P. gingivalis. P. gingivalis was grown overnight anaerobically in THBHK, diluted 1:2 in THBHK, and grown for an additional 6 h anaerobically. Culture medium was removed from streptococcal biofilms, and 200 μl of a P. gingivalis culture diluted to an OD490 of 0.1 (5 × 108 CFU/ml) was added to the biofilms and allowed to grow for an additional 16 h. When treated with antiserum, P. gingivalis cultures were diluted to an OD490 of 0.1 and incubated with antiserum for 1 h before addition to streptococcal biofilms. For the 0-min preincubation, P. gingivalis cultures were mixed with antiserum and immediately added to biofilms. For experiments with washed P. gingivalis cells, cultures were diluted to an OD490 of 0.1 and incubated with antiserum for 1 h. Cultures were then centrifuged at 5,000 × g for 5 min. Cell pellets were then resuspended in 200 μl of sterile PBS and centrifuged a second time at 500 × g for 5 min. Cell pellets were then resupended in the appropriate volume of THBHK, added to streptococcal biofilms, and allowed to grow anaerobically for 16 h before imaging.
Cells were stained with the cytoplasmic stain CFDA (2.6 μg/ml). P. gingivalis was immunofluorescently labeled with a polyclonal anti-FimA fimbrial primary antiserum at a 1:200 dilution in sterile PBS for 1 h, followed by labeling with a 1:200 dilution of a secondary goat anti-rabbit IgG Alexa Fluor 647-conjugated antibody in sterile PBS. Biofilms were washed with 200 μl of sterile PBS between each step and visualized using confocal scanning laser microscopy. Three-dimensional z-stack images were reconstructed using AxioVision Rel. 4.8 (Carl Zeiss), and P. gingivalis biomass was determined using the COMSTAT analysis program running on MatLab software.
For experiments performed in saliva, P. gingivalis culture was diluted into 50% filtered, pooled human saliva (Lee Biosolutions) and 50% THBHK and treated with the desired amount of antisera for 1 h before addition to preformed streptococcal biofilms.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by R01 DC011818 to S.D.G. and L.O.B.
Antisera derived against the P. gingivalis fimbrial proteins were a gift from Mary Ellen Davey at the University of Florida. We thank Lauren Mashburn-Warren for her critical reading of the manuscript.
L. O. Bakaletz and S. D. Goodman own equity in a company, ProclaRx, outside the reported work. In addition, L. O. Bakaletz and S. D. Goodman have a patent, “Compositions and methods for the removal of biofilms,” issued to ProclaRx.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00790-17.
REFERENCES
- 1.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teles RP, Gursky LC, Faveri M, Rosa EA, Teles FR, Feres M, Socransky SS, Haffajee AD. 2010. Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. J Clin Periodontol 37:313–323. doi: 10.1111/j.1600-051X.2010.01534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen PE, Ogawa H. 2005. Strengthening the prevention of periodontal disease: the WHO approach. J Periodontol 76:2187–2193. doi: 10.1902/jop.2005.76.12.2187. [DOI] [PubMed] [Google Scholar]
- 4.Janket SJ, Baird AE, Chuang SK, Jones JA. 2003. Meta-analysis of periodontal disease and risk of coronary heart disease and stroke. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 95:559–569. doi: 10.1067/moe.2003.107. [DOI] [PubMed] [Google Scholar]
- 5.Cairo F, Gaeta C, Dorigo W, Oggioni MR, Pratesi C, Pini Prato GP, Pozzi G. 2004. Periodontal pathogens in atheromatous plaques. A controlled clinical and laboratory trial. J Periodontal Res 39:442–446. [DOI] [PubMed] [Google Scholar]
- 6.Couper DJ, Beck JD, Falkner KL, Graham SP, Grossi SG, Gunsolley JC, Madden T, Maupome G, Offenbacher S, Stewart DD, Trevisan M, Van Dyke TE, Genco RJ. 2008. The Periodontitis and Vascular Events (PAVE) pilot study: recruitment, retention, and community care controls. J Periodontol 79:80–89. doi: 10.1902/jop.2008.070216. [DOI] [PubMed] [Google Scholar]
- 7.Chun YH, Chun KR, Olguin D, Wang HL. 2005. Biological foundation for periodontitis as a potential risk factor for atherosclerosis. J Periodontal Res 40:87–95. doi: 10.1111/j.1600-0765.2004.00771.x. [DOI] [PubMed] [Google Scholar]
- 8.Kaur S, White S, Bartold PM. 2013. Periodontal disease and rheumatoid arthritis: a systematic review. J Dent Res 92:399–408. doi: 10.1177/0022034513483142. [DOI] [PubMed] [Google Scholar]
- 9.Lappann M, Claus H, T van Alen Harmsen M, Elias J, Molin S, Vogel U. 2010. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol Microbiol 75:1355–1371. doi: 10.1111/j.1365-2958.2010.07054.x. [DOI] [PubMed] [Google Scholar]
- 10.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 11.Brandstetter KA, Jurcisek JA, Goodman SD, Bakaletz LO, Das S. 2013. Antibodies directed against integration host factor mediate biofilm clearance from Nasopore. Laryngoscope 123:2626–2632. doi: 10.1002/lary.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockson ME, Novotny LA, Mokrzan EM, Malhotra S, Jurcisek JA, Akbar R, Devaraj A, Goodman SD, Bakaletz LO. 2014. Evaluation of the kinetics and mechanism of action of anti-integration host factor-mediated disruption of bacterial biofilms. Mol Microbiol 93:1246–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devaraj A, Justice SS, Bakaletz LO, Goodman SD. 2015. DNABII proteins play a central role in UPEC biofilm structure. Mol Microbiol doi: 10.1111/mmi.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freire MO, Devaraj A, Young A, Navarro JB, Downey JS, Chen C, Bakaletz LO, Zadeh HH, Goodman SD. 2017. A bacterial-biofilm-induced oral osteolytic infection can be successfully treated by immuno-targeting an extracellular nucleoid-associated protein. Mol Oral Microbiol 32:74–88. doi: 10.1111/omi.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. 2011. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol 4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 16.Gustave JE, Jurcisek JA, McCoy KS, Goodman SD, Bakaletz LO. 2013. Targeting bacterial integration host factor to disrupt biofilms associated with cystic fibrosis. J Cyst Fibros 12:384–389. doi: 10.1016/j.jcf.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novotny LA, Amer AO, Brockson ME, Goodman SD, Bakaletz LO. 2013. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One 8:e67629. doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocco CJ, Davey ME, Bakaletz LO, Goodman SD. 2017. Natural antigenic differences in the functionally equivalent extracellular DNABII proteins of bacterial biofilms provide a means for targeted biofilm therapeutics. Mol Oral Microbiol 32:118–130. doi: 10.1111/omi.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swinger KK, Rice PA. 2004. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol 14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Kamashev D, Rouviere-Yaniv J. 2000. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J 19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnefoy E, Takahashi M, Yaniv JR. 1994. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J Mol Biol 242:116–129. [DOI] [PubMed] [Google Scholar]
- 22.Pontiggia A, Negri A, Beltrame M, Bianchi ME. 1993. Protein HU binds specifically to kinked DNA. Mol Microbiol 7:343–350. [DOI] [PubMed] [Google Scholar]
- 23.Justice SS, Li B, Downey JS, Dabdoub SM, Brockson ME, Probst GD, Ray WC, Goodman SD. 2012. Aberrant community architecture and attenuated persistence of uropathogenic Escherichia coli in the absence of individual IHF subunits. PLoS One 7:e48349. doi: 10.1371/journal.pone.0048349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, Hackett M, Yoshimura F, Demuth DR, Lamont RJ. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun 73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda K, Nagata H, Nonaka A, Kataoka K, Tanaka M, Shizukuishi S. 2004. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect 6:1163–1170. doi: 10.1016/j.micinf.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Nyvad B, Kilian M. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res 95:369–380. [DOI] [PubMed] [Google Scholar]
- 27.Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ Jr, Kolenbrander PE. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol 72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62:1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossi SG, Zambon JJ, Ho AW, Koch G, Dunford RG, Machtei EE, Norderyd OM, Genco RJ. 1994. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol 65:260–267. [DOI] [PubMed] [Google Scholar]
- 30.Dzink JL, Socransky SS, Haffajee AD. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol 15:316–323. [DOI] [PubMed] [Google Scholar]
- 31.Williams RC, Barnett AH, Claffey N, Davis M, Gadsby R, Kellett M, Lip GY, Thackray S. 2008. The potential impact of periodontal disease on general health: a consensus view. Curr Med Res Opin 24:1635–1643. doi: 10.1185/03007990802131215. [DOI] [PubMed] [Google Scholar]
- 32.Kornman KS. 2008. Mapping the pathogenesis of periodontitis: a new look. J Periodontol 79:1560–1568. doi: 10.1902/jop.2008.080213. [DOI] [PubMed] [Google Scholar]
- 33.van der Meer JTM, van Vianen W, Hu E, van Leeuwen WB, Valkenburg HA, Thompson J, Michel MF. 1991. Distribution, antibiotic susceptibility and tolerance of bacterial isolates in culture-positive cases of endocarditis in The Netherlands. Eur J Clin Microbiol Infect Dis 10:728–734. [DOI] [PubMed] [Google Scholar]
- 34.Beck JD, Offenbacher S. 1998. Oral health and systemic disease: periodontitis and cardiovascular disease. J Dent Educ 62:859–870. [PubMed] [Google Scholar]
- 35.Murdoch DR, Corey GR, Hoen B, Miro JM, Fowler VG Jr, Bayer AS, Karchmer AW, Olaison L, Pappas PA, Moreillon P, Chambers ST, Chu VH, Falco V, Holland DJ, Jones P, Klein JL, Raymond NJ, Read KM, Tripodi MF, Utili R, Wang A, Woods CW, Cabell CH, International Collaboration on Endocarditis-Prospective Cohort Study. 2009. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Arch Intern Med 169:463–473. doi: 10.1001/archinternmed.2008.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ammann TW, Belibasakis GN, Thurnheer T. 2013. Impact of early colonizers on in vitro subgingival biofilm formation. PLoS One 8:e83090. doi: 10.1371/journal.pone.0083090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Part 10):2395–2407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.