ABSTRACT
Many organisms possess pathways that regenerate NAD+ from its degradation products, and two pathways are known to salvage NAD+ from nicotinamide (Nm). One is a four-step pathway that proceeds through deamination of Nm to nicotinic acid (Na) by Nm deamidase and phosphoribosylation to nicotinic acid mononucleotide (NaMN), followed by adenylylation and amidation. Another is a two-step pathway that does not involve deamination and directly proceeds with the phosphoribosylation of Nm to nicotinamide mononucleotide (NMN), followed by adenylylation. Judging from genome sequence data, the hyperthermophilic archaeon Thermococcus kodakarensis is supposed to utilize the four-step pathway, but the fact that the adenylyltransferase encoded by TK0067 recognizes both NMN and NaMN also raises the possibility of a two-step salvage mechanism. Here, we examined the substrate specificity of the recombinant TK1676 protein, annotated as nicotinic acid phosphoribosyltransferase. The TK1676 protein displayed significant activity toward Na and phosphoribosyl pyrophosphate (PRPP) and only trace activity with Nm and PRPP. We further performed genetic analyses on TK0218 (quinolinic acid phosphoribosyltransferase) and TK1650 (Nm deamidase), involved in de novo biosynthesis and four-step salvage of NAD+, respectively. The ΔTK0218 mutant cells displayed growth defects in a minimal synthetic medium, but growth was fully restored with the addition of Na or Nm. The ΔTK0218 ΔTK1650 mutant cells did not display growth in the minimal medium, and growth was restored with the addition of Na but not Nm. The enzymatic and genetic analyses strongly suggest that NAD+ salvage in T. kodakarensis requires deamination of Nm and proceeds through the four-step pathway.
IMPORTANCE Hyperthermophiles must constantly deal with increased degradation rates of their biomolecules due to their high growth temperatures. Here, we identified the pathway that regenerates NAD+ from nicotinamide (Nm) in the hyperthermophilic archaeon Thermococcus kodakarensis. The organism utilizes a four-step pathway that initially hydrolyzes the amide bond of Nm to generate nicotinic acid (Na), followed by phosphoribosylation, adenylylation, and amidation. Although the two-step pathway, consisting of only phosphoribosylation of Nm and adenylylation, seems to be more efficient, Nm mononucleotide in the two-step pathway is much more thermolabile than Na mononucleotide, the corresponding intermediate in the four-step pathway. Although NAD+ itself is thermolabile, this may represent an example of a metabolism that has evolved to avoid the use of thermolabile intermediates.
KEYWORDS: archaea, NAD salvage, Thermococcus, hyperthermophiles, metabolism, nicotinamide adenine dinucleotide
INTRODUCTION
NAD+ is a major electron carrier in all three domains of life. Two major pathways are known for the de novo biosynthesis of NAD+ (1–4). The majority of eukaryotes utilize tryptophan as the precursor and synthesize NAD+ via formylkynurenine, kynurenine, 3-hydroxy-kynurenine, 3-hydroxyanthranilic acid, quinolinic acid (Qa), nicotinic acid mononucleotide (NaMN), and nicotinic acid adenine dinucleotide (NaAD) (Fig. 1). Most bacteria utilize another pathway and synthesize NAD+ from aspartic acid via iminoaspartic acid, Qa, NaMN, and NaAD (Fig. 1). Eukaryotes that utilize the pathway from aspartic acid and bacteria that use tryptophan as the starting material are also known (3, 4). Judging from genome sequence data, archaea utilize the pathway initiating from aspartic acid.
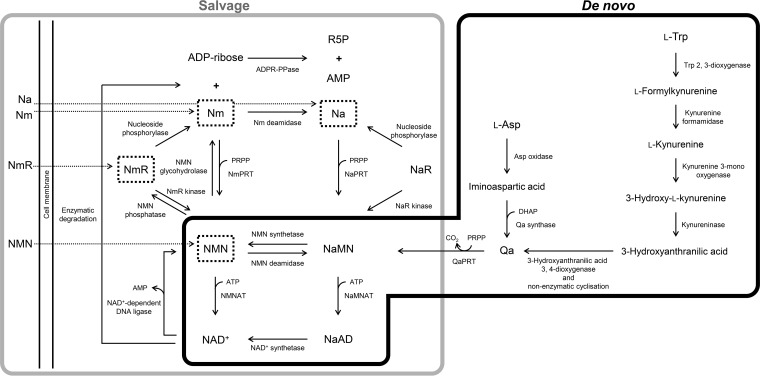
FIG 1.
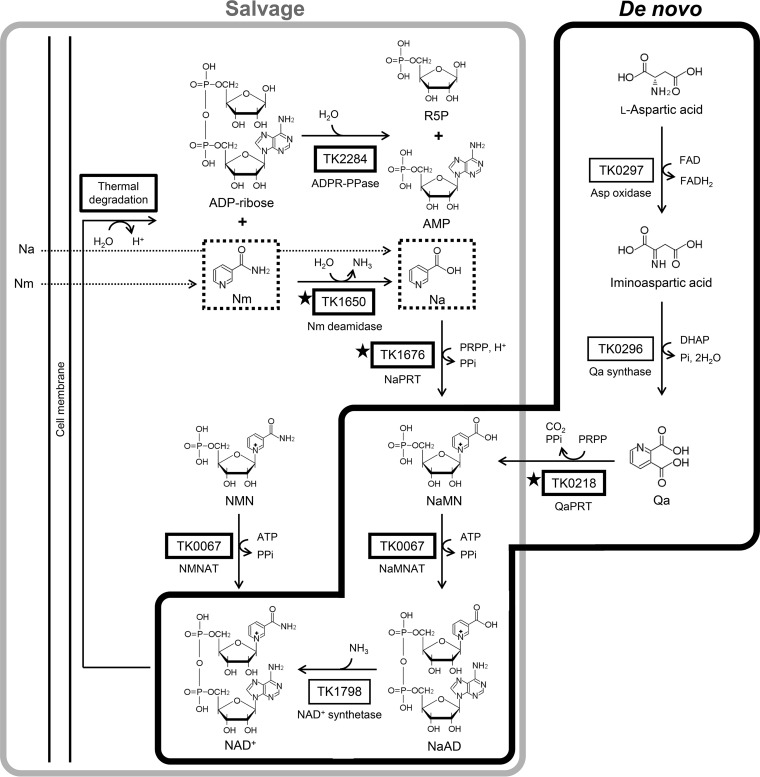
Previously recognized de novo and salvage pathways for NAD+ biosynthesis. Dotted boxes show compounds that can be taken up by the cell. ADPR-PPase, ADP-ribose pyrophosphatase; DHAP, dihydroxyacetone phosphate; Na, nicotinic acid; NaAD, nicotinic acid adenine dinucleotide; NaMN, nicotinic acid mononucleotide; NaMNAT, nicotinic acid mononucleotide adenylyltransferase; NaPRT, nicotinic acid phosphoribosyltransferase; NaR, nicotinic acid riboside; Nm, nicotinamide; NMN, nicotinamide mononucleotide; NMNAT, nicotinamide mononucleotide adenylyltransferase; NmPRT, nicotinamide phosphoribosyltransferase; NmR, nicotinamide riboside; PRPP, phosphoribosyl pyrophosphate; Qa, quinolinic acid; QaPRT, quinolinic acid phosphoribosyltransferase; R5P, ribose 5-phosphate.
A number of compounds are produced from the intracellular degradation of NAD+. In bacteria, nicotinamide mononucleotide (NMN) is generated as a by-product of the NAD+-dependent DNA ligase reaction (5). In eukaryotes and some bacteria, nicotinamide (Nm) is generated as a by-product when NAD+ is used for ADP-ribosylation of proteins (6, 7). Nm is also generated by NAD+ glycohydrolase (NADase), which acts in regulating and maintaining intracellular concentrations of NAD+ (8, 9). Additionally, it is well known that NAD+ is a thermolabile molecule (10, 11) and that its degradation results in the generation of ADP-ribose and Nm (12, 13). This is particularly relevant in thermophilic organisms, and several studies have examined the mechanisms that are involved in regenerating NAD+ from its degradation products (13, 14).
Organisms in many cases harbor salvage pathways that can regenerate NAD+ from these degradation products. In terms of Nm salvage, there are two major pathways that convert Nm to NAD+ (2, 3). In one pathway (the two-step pathway), Nm is directly converted to NMN by an Nm phosphoribosyltransferase (NmPRT). NMN is then converted to NAD+ via an adenylyltransferase. In the second pathway (the four-step pathway), which is also called the Preiss-Handler pathway (15, 16), the amide group of Nm is initially hydrolyzed through the function of Nm deamidase, producing nicotinic acid (Na) and ammonia. Na is then converted to NaMN by Na phosphoribosyltransferase (NaPRT), which is then converted to NAD+ via NaAD. Most bacteria utilize the four-step pathway, whereas the two pathways are well distributed among eukaryotes. In terms of NMN, many bacteria can hydrolyze NMN to ribose 5-phosphate and Nm, and Nm enters the four-step pathway. Many bacteria can also directly convert NMN to NaMN (1, 17). In addition, organisms can utilize exogenous pyridinium compounds for NAD+ salvage. The uptake of Na and Nm in Escherichia coli was reported over 40 years ago, and de novo synthesis is suppressed in their presence (18). Uptake of NMN has been reported in Salmonella enterica serovar Typhimurium (17). The uptake of exogenous nicotinamide riboside (NmR) has also been demonstrated in Haemophilus species (19, 20).
NMN adenylyltransferase (NMNAT) is present in all three domains of life. NMNAT found in eukaryotes differs in structure from those found in bacteria and archaea (21, 22). The bacterial and archaeal NMNAT proteins, along with most of the enzymes from eukaryotes, display significant adenylyltransferase activities toward both NaMN and NMN. On the other hand, NaMN adenylyltransferase (NaMNAT) is only found in bacteria and exhibits relevant activity only toward NaMN (23, 24). The presence of NMNAT in bacteria is not widespread, and bacteria that harbor an NMNAT also possess an NmPRT homolog (Gloeobacter violaceus is an exception). This suggests that, although a minority, some bacteria utilize the two-step pathway in NAD+ salvage (25).
Thermococcus kodakarensis is a hyperthermophilic archaeon with an optimum growth temperature of 85°C (26). The genome sequence of this archaeon has been determined (27), and versatile genetic systems have been developed (28–32). The organism possesses a number of enzymes that utilize NAD+ (33–35), and, judging from the genome sequence data, harbors a complete set of homologs involved in the de novo synthesis of NAD+ from aspartate. The homolog presumed to be responsible for the conversion of NaMN to NaAD is encoded by TK0067, which is a homolog of NMNAT that displays activity toward both NMN and NaMN. We have previously shown experimentally that the TK0067 protein actually displays activity toward both NMN and NaMN (13). An NaMNAT homolog that functions only on NaMN cannot be found on the T. kodakarensis genome. Although the presence of a homolog of NAD+ synthetase (TK1798) that catalyzes the amide formation of NaAD suggests that T. kodakarensis (and other archaea) utilizes the four-step pathway, the presence or absence of the two-step pathway is still unclear, particularly as this organism utilizes an NMNAT that displays activity toward NMN in addition to NaMN. In this study, we addressed this problem with biochemical and genetic analyses on enzymes/genes related to this metabolism in T. kodakarensis.
RESULTS
Preparation of the recombinant TK1676 protein.
As described in the introduction, the TK0067 protein from T. kodakarensis displays adenylyltransferase activity toward both NaMN and NMN (13). We thus set out to examine the substrate specificity of the TK1676 protein, an NaPRT homolog predicted to display phosphoribosyltransferase activity toward Na. Among archaeal NaPRT proteins, the crystal structure of a NaPRT homolog from Thermoplasma acidophilum has already been determined (36). However, it has not been examined whether NaPRT homologs from archaea display activity toward Nm. The TK1676 protein is 49% identical to the NaPRT homolog from T. acidophilum (Ta1145). The TK1676 gene was cloned and expressed in E. coli, and the recombinant protein was purified to apparent homogeneity, as judged from the result of SDS-PAGE electrophoresis (see Fig. S1 in the supplemental material).
Enzyme assay of the TK1676 protein.
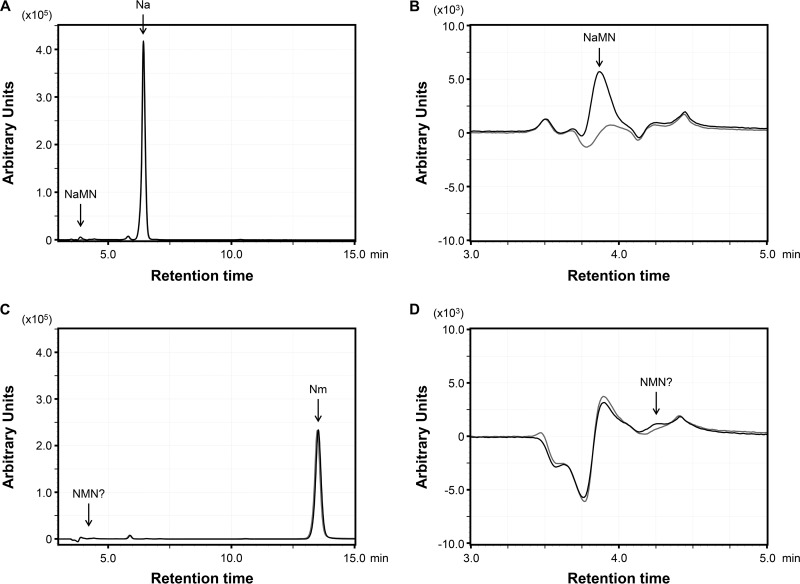
In order to examine the substrate specificity of the TK1676 protein, we monitored the production of NaMN or NMN from Na or Nm, respectively, using high-performance liquid chromatography (HPLC). Under the applied conditions, we were able to separate the four compounds (Fig. S2). With 2 mM Na and 2 mM PRPP as the substrates in the presence of 1 μg of TK1676 protein, we clearly observed the formation of a peak that corresponds to the retention time of NaMN after a 30-min incubation at 85°C (Fig. 2A and B). This peak was not observed in the absence of the TK1676 protein. The concentration of NaMN calculated from the difference of the two peak areas was 34 ± 4 μM. By carrying out these reactions for different periods of time, we were able to determine the initial velocity of the NaPRT activity of the TK1676 protein, which was 0.44 ± 0.04 μmol · mg−1 · min−1 in the presence of 2 mM Na and 2 mM PRPP. This value can be considered comparable to the activity levels observed for the human NaPRT (0.012 ± 0.001 μmol · mg−1 · min−1) in the presence of 1 mM substrates at 37°C (37). In order to examine whether the protein could also recognize Nm, we performed the same experiments but with 2 mM Nm instead of 2 mM Na. We could not detect a formation of a peak corresponding to NMN (data not shown). We thus increased the amount of protein added to the reaction by 20-fold to 20 μg. In this case also, we could not detect a clear formation of a peak corresponding to NMN, which should display a retention time between 4.0 min and 4.5 min (Fig. 2C and D).
FIG 2.
HPLC analysis of the products obtained after reaction with the TK1676 protein. (A) The reaction was carried out using Na and PRPP as the substrates. (B) Enlargement of the area including the NaMN peak in panel A. (C) The reaction was performed using Nm and PRPP as the substrates. (D) Enlargement of the area in which the standard NMN peak can be detected in panel C. Black and gray lines represent the products of reactions with and without TK1676 protein, respectively.
Growth properties of gene disruptants.
Our activity measurements suggested that the TK1676 protein displayed significant activity toward Na but not toward Nm. In order to clarify whether the TK1676 protein displayed activity toward Nm and to evaluate the function of this protein in vivo, we designed and constructed gene disruption strains that would enable us to distinguish the contributions of the two-step and four-step salvage pathways in T. kodakarensis.
First of all, in order to remove the effects of de novo synthesis, we disrupted the TK0218 gene, which encodes Qa phosphoribosyltransferase (QaPRT) (Fig. 1). The genotype of a selected transformant was examined by PCR analysis using one primer set annealing outside the flanking regions for homologous recombination and another set within the target gene (Fig. S3A and B). As expected, we observed a shorter amplified DNA fragment in the ΔTK0218 transformant using the primer set annealing outside the homologous region (Fig. S3A) and no product using the primer set that anneals within the coding region (Fig. S3B). We also confirmed the absence of unintended mutations in the homologous regions by DNA sequencing analysis.
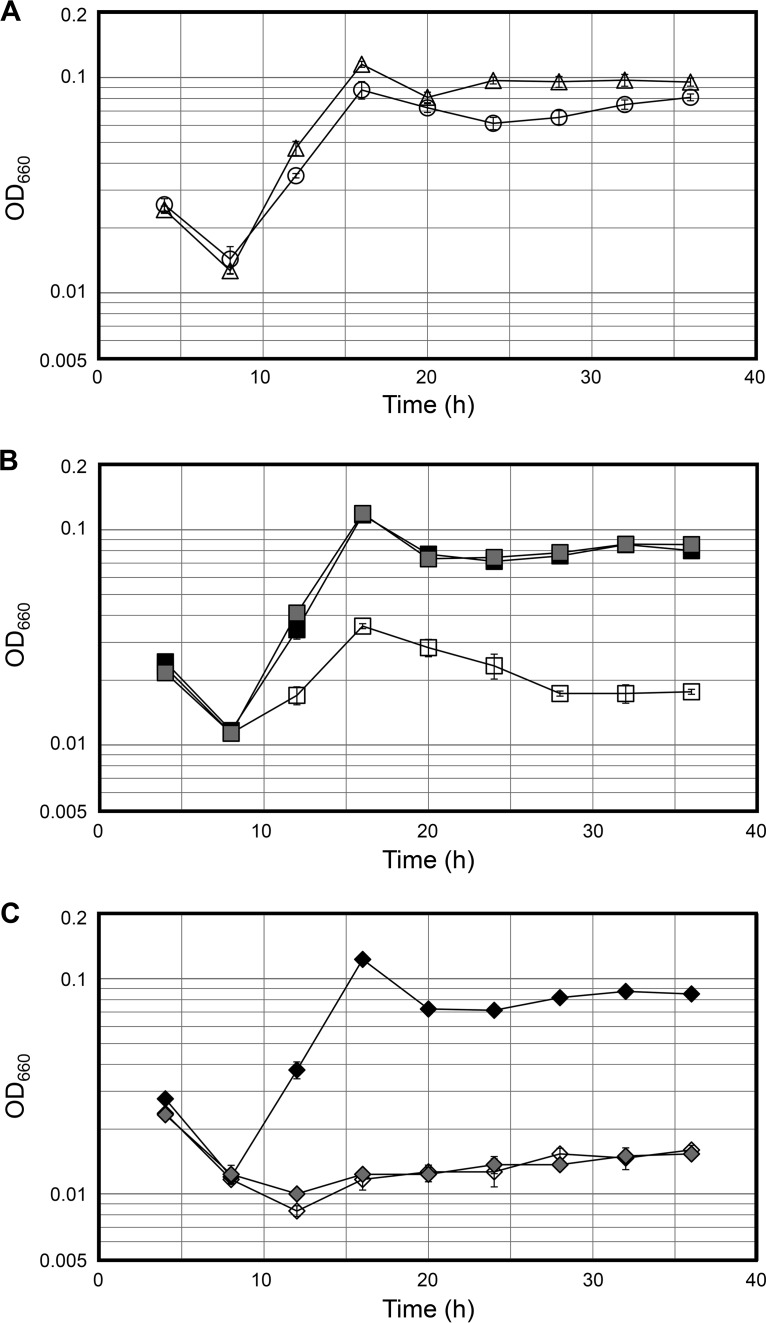
The growth characteristics of the ΔTK0218 mutant strain were examined in a synthetic medium based on 0.8× artificial seawater, a mixture of 20 amino acids, modified Wolfe's trace minerals, a vitamin mixture and 2.0 g · liter−1 elemental sulfur supplemented with 2.5 g · liter−1 sodium pyruvate, 10 μg · ml−1 uracil, and 10 μM tungsten (ASW-AA-S0-Pyr-Ura-W) and compared with those of the host strain. The ΔTK0218 mutant cells displayed growth that was indistinguishable from that of the host strain when Na or Nm was supplemented to the medium. The concentrations of these compounds could be decreased to 2 μM, which still supported almost the same growth of ΔTK0218 mutant cells as that of the host strain (Fig. 3A and B). However, in the absence of these compounds, the growth of ΔTK0218 mutant cells was slower and ceased after a slight increase in cell density (Fig. 3B). This is the expected result for a disruption of a gene required for de novo synthesis of NAD+, which is further supported by the fact that the addition of Na or Nm to the medium can complement the growth defect. We also disrupted the TK1676 gene and confirmed the genotype of a selected transformant with procedures applied for the TK0218 gene (Fig. S3C and D). The ΔTK1676 mutant cells did not show any growth defects in ASW-AA-S0-Pyr-Ura-W medium without Na and Nm (Fig. 3A). This agrees well with the presumption that this gene functions in a salvage pathway for NAD+ that is not essential when de novo synthesis is intact. The involvement of TK1676 in a (lone) salvage pathway was further supported by the fact that we could not obtain a strain disrupted of both TK1676 and TK0218, which would abolish both the de novo and salvage pathways for NAD+ biosynthesis, although the same disruption plasmids used for individual gene disruption strains were used for transformation.
FIG 3.
Growth properties of T. kodakarensis host strain KU216 and three gene disruption strains. (A) Growth of the host KU216 strain (circles) and ΔTK1676 mutant (triangles) were examined in a synthetic amino acid medium without Na and Nm (minimal medium, ASW-AA-S0-Pyr-Ura-W). (B) Growth of ΔTK0218 mutant in the minimal medium and medium supplemented with Na or Nm are shown. (C) Growth of the ΔTK0218 ΔTK1650 mutant strain in the minimal medium and medium supplemented with Na or Nm. Open, filled, and gray symbols indicate ASW-AA-S0-Pyr-Ura-W, ASW-AA-S0-Pyr-Ura-W-Na(+), and ASW-AA-S0-Pyr-Ura-W-Nm(+) media, respectively. Each value is an average of those from three independent growth experiments. The vertical axis is represented on a logarithmic scale.
We next disrupted the TK1650 gene, which is predicted to encode an Nm deamidase that converts Nm to Na. As expected, the strain did not show noticeable growth defects compared to the host strain (data not shown). In the case of the TK1650 gene, we were able to obtain double-gene-disruption strains lacking both TK1650 and TK0218 (ΔTK0218 ΔTK1650 mutant) in a medium supplemented with NAD+, Nm, and Na (Fig. S3E to H). The ΔTK0218 ΔTK1650 mutant strain did not grow at all in ASW-AA-S0-Pyr-Ura-W medium without Na and Nm (Fig. 3C). The growth defect was more severe than that observed for the ΔTK0218 mutant cells in the same medium. We next grew ΔTK0218 ΔTK1650 mutant cells in the presence of 2 μM Na or Nm (Fig. 3C). The growth of the strain was completely complemented in the presence of 2 μM Na. Most importantly, however, the addition of 2 μM Nm did not lead to a recovery of growth. Compared to the growth characteristics of the ΔTK0218 mutant strain grown in the same medium, the result indicates that the lack of growth of the ΔTK0218 ΔTK1650 mutant strain in a medium with 2 μM Nm is due to the lack of Nm deamidase activity, and the Nm is utilized for NAD+ salvage via Na, i.e., the four-step pathway.
Detection of Nm deamidase activity in cell extracts of T. kodakarensis.
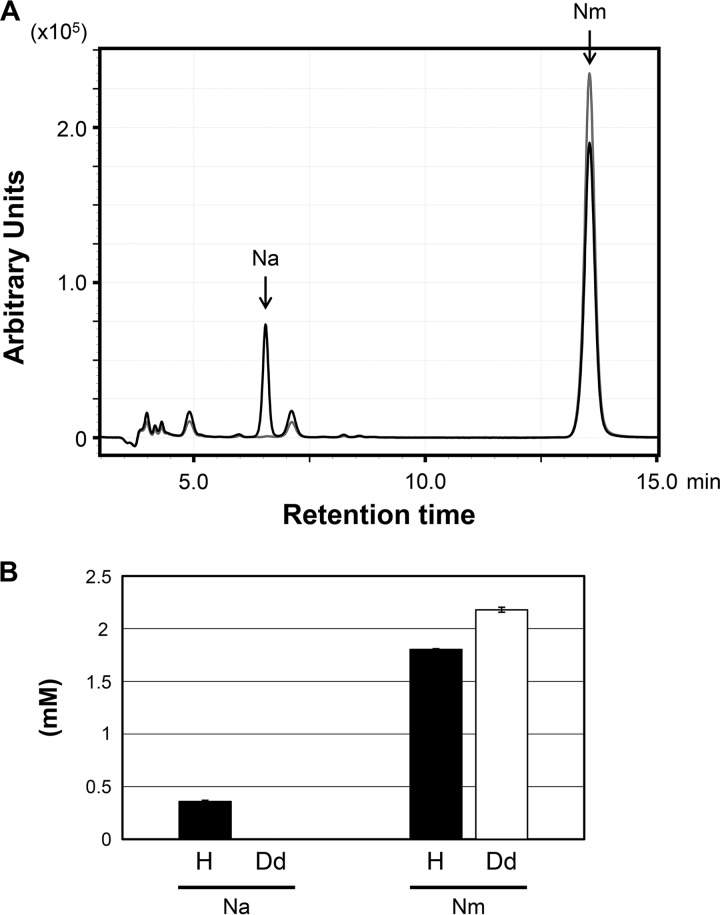
In order to confirm that TK1650 actually encodes an Nm deamidase, we examined Nm deamidase activity in cell extracts of T. kodakarensis. Cell extracts from the host strain KU216 and the ΔTK0218 ΔTK1650 mutant strain were prepared, and 2 mM Nm was added to the cell extracts (100 μg). The mixtures were incubated at 85°C for 30 min, and the reaction products were analyzed by HPLC (Fig. 4A). When the area of the peaks corresponding to Na and Nm were quantified (Fig. 4B), we clearly observed an accumulation of Na in the host strain. In contrast, no such accumulation was observed in the ΔTK0218 ΔTK1650 mutant strain, indicating that the strain lost its capability to generate Na from Nm, most likely due to the lack of TK1650.
FIG 4.
Deaminase activity converting Nm to Na in cell extracts. (A) Black and gray lines show reaction products in cell extracts from the KU216 host strain and the ΔTK0218 ΔTK1650 mutant strain, respectively. The experiment was performed three times with cell extracts obtained from three independent cultures for each strain, and a representative result is shown here. (B) The concentrations of Na and Nm were quantified based on peak area in the HPLC analysis. Error bars indicate standard deviations. Filled bars (H) indicate results with the cell extracts of the KU216 host strain, and open bars (Dd) indicate those of the ΔTK0218 ΔTK1650 mutant strain.
DISCUSSION
Here, we provide biochemical and genetic evidence that clearly indicates that T. kodakarensis utilizes only the four-step pathway for NAD+ salvage, as shown in Fig. 5. Although this organism harbors an NMNAT homolog (TK0067) that exhibits activities toward both NaMN and NMN, the TK1676 protein exhibits little activity toward Nm. Moreover, when TK1650 was disrupted, exogenous Nm could no longer be utilized for NAD+ salvage, providing in vivo evidence that Nm is utilized via an Nm deamidase reaction to produce Na, which is subsequently converted to NaMN by the NaPRT activity of the TK1676 protein.
FIG 5.
Predicted de novo and salvage pathways for NAD+ biosynthesis in T. kodakarensis. Thick-lined boxes represent proteins or reactions that have been characterized in T. kodakarensis. Those with stars indicate proteins examined in this study. Dotted boxes show compounds that can be taken up by the cell, based on the results of this study. Other reactions in this figure are based on studies on homologous proteins from other archaea, such as TK0297 homologs from Pyrococcus horikoshii (40), from Sulfolobus tokodaii (41), and from Thermococcus litoralis (42), TK0296 homologs from P. horikoshii (43) and from Pyrococcus furiosus (44), and a TK1798 homolog from Methanocaldococcus jannaschii (38). ADPR-PPase, ADP-ribose pyrophosphatase; DHAP, dihydroxyacetone phosphate; FAD, flavin adenine dinucleotide; NaAD, nicotinic acid adenine dinucleotide; Na, nicotinic acid; NaPRT, nicotinic acid phosphoribosyltransferase; NaMNAT, nicotinic acid mononucleotide adenylyltransferase; Nm, nicotinamide; NmPRT, nicotinamide phosphoribosyltransferase; NMNAT, nicotinamide mononucleotide adenylyltransferase; PRPP, phosphoribosyl pyrophosphate; Qa, quinolinic acid; QaPRT, quinolinic acid phosphoribosyltransferase; R5P, ribose 5-phosphate.
During our analyses, we were also able to actually observe the contribution of NAD+ salvage toward the growth of T. kodakarensis. The growth curves of the ΔTK0218 and ΔTK0218 ΔTK1650 mutant strains in the absence of Na or Nm are shown in Fig. 3B and C, respectively. The curves illustrate the differences in growth of cells with and without a salvage system, respectively, when de novo synthesis is inactivated. Although slower than that of the host strain, the growth of ΔTK0218 mutant cells was observed for 16 h, which can be presumed to be due to the use of Nm, a major thermodegradation product of NAD+ (12, 13). No such growth was observed with the ΔTK0218 ΔTK1650 mutant cells.
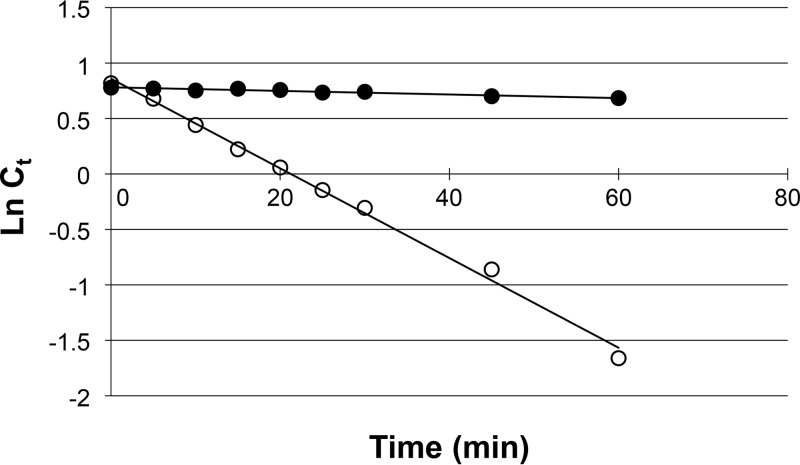
Although our evidence demonstrates that T. kodakarensis utilizes the four-step pathway for NAD+ salvage, we were interested in why the organism had not come to utilize the two-step pathway. We previously reported that NAD+ displays thermal degradation, with a half-life (t1/2) of 24.2 min at 85°C, resulting in the formation of ADP-ribose and Nm (13). ADP-ribose, Nm, and Na display extreme thermostability (Fig. 4A) (13). Direct conversion of Nm through the two-step pathway would proceed via NMN, whereas conversion through the four-step pathway would generate NaMN. We thus compared the stability of NMN and NaMN at 85°C (Fig. 6). We observed a stark difference in thermostability, with NaMN displaying a t1/2 of 431 min, whereas NMN was relatively susceptible to degradation, with a t1/2 of 17.1 min. Moreover, the degradation of NMN resulted in the generation of Nm, indicating that the NmPRT reaction and thermal degradation of NMN would result in a futile cycle that would potentially waste PRPP. As NAD+ is also susceptible to thermal degradation, the advantages may be limited, but this raises the possibility that pathways in organisms living under high temperature, such as T. kodakarensis, evolved to synthesize NAD+ through pathways that utilize relatively thermostable intermediates or that avoid thermolabile compounds as much as possible.
FIG 6.
Thermal degradation rates of NaMN and NMN at 85°C. Ct indicates the concentration of NaMN or NMN after heat treatment for t min. Ln Ct denotes natural logarithm of Ct. Filled and open circles indicate NaMN and NMN, respectively.
The presence of adenylyltransferase activity of the TK0067 protein toward NMN raises the possibility that the TK1798 protein generates the amide group on NaMN and generates NMN, or in other words, the amide-forming reaction precedes the adenylyltransferase reaction. We presume that this is unlikely, as there is no reason to convert the thermostable NaMN to the labile NMN prior to the synthesis of NAD+. Most importantly, this possibility has been addressed in the enzymes from Methanocaldococcus jannaschii, which grows at temperatures similar to those of T. kodakarensis and whose NAD+ synthetase protein is 49% identical to the TK1798 protein. The M. jannaschii protein only recognizes NaAD and does not utilize NaMN as a substrate (38). Taking these findings together, we presume that the activity toward NMN in archaeal NMNAT proteins is not physiologically relevant.
We examined the distribution of the four-step pathway in a number of selected archaeal genomes (see Table S1 in the supplemental material). Homologs of the enzymes related to the de novo and salvage pathways were identified on all of the Thermococcales genomes. Although the degrees of similarity were lower, homologs were also found in Archaeoglobus fulgidus, Halobacterium salinarum, and Sulfolobus species. Interestingly, homologs of enzymes for de novo synthesis are not found in Thermoplasma species and Desulfurococcus amylolyticus. These organisms harbor homologs for NAD+ salvage and may utilize structurally distinct enzymes for de novo synthesis or may require Na or Nm for growth. On the other hand, the methanogens harbor the de novo pathway but do not seem to possess salvage pathways. The situation for Thermoproteus tenax is interesting. The organism harbors a putative NaPRT homolog, but an Nm deamidase homolog is absent. This might be explained by the use of the two-step pathway. Bacterial NmPRT, which displays activity toward Nm, is also called NadV. In a previous study, the distribution of NadV homologs was examined throughout the three domains of life (3). NadV homologs are only present in a limited number of bacteria and eukaryotes. These organisms also harbor NMNAT, suggesting that these organisms utilize the two-step pathway. However, NadV is not present in T. tenax, so the function of NaPRT in this organism may be to utilize exogenous Na, or a structurally novel Nm deamidase may be present.
MATERIALS AND METHODS
Strains, media, and culture conditions.
T. kodakarensis KOD1 (wild type), KU216 (ΔpyrF mutant) (29), and its derivative strains were cultured under anaerobic conditions at 85°C in a nutrient-rich medium (ASW-YT) or a synthetic medium (ASW-AA). ASW-YT-S0 and ASW-YT-Pyr media were composed of 0.8× artificial seawater (ASW), 5.0 g · liter−1 yeast extract, 5.0 g · liter−1 tryptone, and 0.8 mg · liter−1 resazurin with elemental sulfur (2.0 g · liter−1) and sodium pyruvate (5.0 g · liter−1), respectively. The ASW-AA-S0 medium consisted of 0.8× ASW, a mixture of 20 amino acids, modified Wolfe's trace minerals, a vitamin mixture and 2.0 g · liter−1 elemental sulfur (28, 39). Na2S·9H2O was added to the medium until it became colorless. In the growth experiments, the amount of Na2S·9H2O added was constant at 12.5 mg · liter−1. For solid medium used to isolate transformants, elemental sulfur and Na2S·9H2O were replaced with 2 ml · liter−1 of a polysulfide solution (10 g of Na2S·9H2O and 3 g of sulfur flowers in 15 ml of H2O), and 10 g · liter−1 of Gelrite was added to solidify the medium. The ASW-AA-S0 medium was slightly modified when used for the enrichment of transformants and growth experiments, as stated below. E. coli strains DH5α used for plasmid construction and BL21 for gene expression were cultivated at 37°C in lysogeny broth (LB) medium containing ampicillin (100 mg · liter−1). All chemical reagents were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan), unless stated otherwise.
Overexpression of T. kodakarensis NaPRT gene and purification of the product.
To construct the TK1676 (T. kodakarensis NaPRT) gene expression vector (pETK1676), the TK1676 gene was amplified by PCR from genomic DNA of T. kodakarensis KOD1 with the primer set eTK1676-F/eTK1676-R (see Table S2 in the supplemental material). The primers contained 14 bp- and 15 bp sequences flanking the NdeI and BamHI restriction sites of pET21a(+) (EMD Millipore, Billerica, MA), respectively. The amplified fragment and pET21a(+) digested with NdeI/BamHI were fused by fusion PCR using the In-Fusion HD cloning kit (TaKaRa Bio, Shiga, Japan). After confirming the absence of unintended mutations by DNA sequencing analysis, pETK1676 was introduced into E. coli strain BL21-CodonPlus(DE3)-RIL (Agilent Technologies, Santa Clara, CA). The transformant was cultivated until the optical density at 660 nm (OD660) reached 0.3 to ∼0.6, and isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added at a final concentration of 0.1 mM to induce gene expression. After a further 4 h of culture, cells were harvested, resuspended in 50 mM Tris-HCl buffer (pH 7.5), and disrupted by sonication. After centrifugation (4°C, 5,000 × g, 10 min), the soluble cell extract was incubated at 85°C for 20 min to remove thermolabile proteins. After centrifugation (4°C, 5,000 × g, 20 min), the supernatant was applied to an anion-exchange column, ResourceQ (GE Healthcare, Chicago, IL), and proteins were eluted with a linear gradient of NaCl (0 to 1.0 M) in 50 mM Tris-HCl (pH 7.5) at a flow rate of 2.0 ml · min−1. After concentrating relevant fractions using an Amicon Ultra centrifugal filter unit (molecular weight cutoff [MWCO], 30,000; EMD Millipore), the proteins were separated with a Superdex 200 Increase 10/300 gel filtration column (GE Healthcare), with a mobile phase of 50 mM Tris-HCl (pH 7.5) with 150 mM NaCl at a flow rate of 0.4 ml · min−1. Protein concentration was determined with the Protein assay system (Bio-Rad, Hercules, CA) using bovine serum albumin (Thermo Fisher Scientific, Waltham, MA) as a standard.
Enzyme assay of the recombinant T. kodakarensis NaPRT.
NaPRT and NmPRT activities were measured in 100-μl reaction mixtures containing 50 mM HEPES-NaOH (pH 7.0 at 85°C), 10 mM MgCl2, 2 mM Na or Nm, 2 mM PRPP, and 1 to 20 μg of the purified TK1676 protein. For determining the presence of NaPRT and NmPRT activities, reaction mixtures were incubated at 85°C for 30 min. For examining the initial velocity of the TK1676 reaction toward Na, the reaction mixture without Na was preincubated at 85°C for 1 min, and the reaction was initiated by adding Na. The reactions were carried out at 85°C for 1, 3, and 5 min. In both cases, reactions were terminated by rapid cooling on ice for 10 min, followed by removal of protein with an Amicon Ultra centrifugal filter unit (MWCO, 10,000). NMN or NaMN present in the filtered reaction mixtures was quantified by HPLC using a Cosmosil 5-μm C18 (5C18)-PAQ column (Nacalai Tesque, Kyoto, Japan) with 50 mM sodium phosphate (pH 6.8) as the mobile phase. The flow rate was set to 0.8 ml · min−1, and the column temperature was set at 40°C. Absorbance at 254 nm (A254) was monitored for the detection of compounds, including NMN and NaMN.
Construction of gene disruption strains of T. kodakarensis.
To construct TK0218 (QaPRT), TK1650 (Nm deamidase), and TK1676 (NaPRT) gene disruption vectors, the respective genes with their 5′- and 3′-flanking regions were amplified by PCR with the primer sets dTK0218-F/dTK0218-R, dTK1650-F/dTK1650-R, and dTK1676-F/dTK1676-R (Table S2), respectively, and were inserted into the HincII site of plasmid pUD3, which harbors a pyrF gene. The coding regions of each gene were removed by inverse PCR with the primer sets invdTK0218-F/invdTK0218-R, invdTK1650-F/invdTK1650-R, and invdTK1676-F/invdTK1676-R (Table S2), and the amplified DNA fragments were self-ligated. The three disruption vectors were named pDTK0218, pDTK1650, and pDTK1676.
The TK0218 disruption strain (ΔTK0218 mutant), TK1676 disruption strain (ΔTK1676 mutant), and TK0218-TK1650 double-disruption strain (ΔTK0218 ΔTK1650 mutant) were prepared as follows. For the ΔTK0218 and ΔTK1676 mutants, T. kodakarensis KU216 cells were grown in ASW-YT-S0 for 12 h, harvested, resuspended in 200 μl of 0.8× ASW, and kept on ice for 30 min. pDTK0218 or pDTK1676 (3 to 5 μg) was added to the cells, and the mixtures were kept on ice for over 1 h. After heat shock at 85°C for 45 s, the mixtures were kept on ice for 10 min. Cells were inoculated into ASW-AA-S0 liquid medium supplemented with 10 μM tungsten (ASW-AA-S0-W) and incubated at 85°C for over 1 day twice in order to enrich transformants harboring the pyrF gene due to single-crossover recombination. Cells were then grown at 85°C for 3 to 5 days on ASW-AA solid medium with 0.75% 5-fluoroorotic acid (FOA), 10 μg · ml−1 uracil, 0.1 mM NAD+, 0.1 mM Na, and 0.1 mM Nm to select transformants in which target genes were removed along with the pyrF gene due to a second recombination. Genotypes of the transformants were examined by PCR with primer sets that anneal within the target genes (idTK0218-F/idTK0218-R or idTK1676-F/idTK1676-R) and outside the homologous regions for homologous recombination (odTK0218-F/odTK0218-R or odTK1676-F/odTK1676-R) (Table S2). DNA sequencing analysis was also carried out to confirm the absence of unintended mutations within the homologous regions. To construct the ΔTK0218 ΔTK1650 double mutant, the ΔTK0218 mutant was transformed with pDTK1650 with the same method described above. The genotypes of the transformant were examined by PCR with the primer sets idTK0218-F/idTK0218-R, idTK1650-F/idTK1650-R, odTK0218-F/odTK0218-R, and odTK1650-F/odTK1650-R (Table S2), and the absence of mutations in the homologous regions was confirmed by DNA sequencing.
Growth measurements of T. kodakarensis.
The growth properties of the host strain KU216 and the constructed gene disruption strains were mainly examined in ASW-AA-S0 supplemented with 2.5 g · liter−1 sodium pyruvate, 10 μg · ml−1 uracil, and 10 μM tungsten, but without Na, which is usually present in the vitamin mixture (39). This minimal medium (ASW-AA-S0-Pyr-Ura-W) contains neither Na nor Nm. When necessary, this medium was supplemented either with 2 μM Na or 2 μM Nm [ASW-AA-S0-Pyr-Ura-W-Na(+) or ASW-AA-S0-Pyr-Ura-W-Nm(+), respectively]. Cells were grown in ASW-YT-S0 medium for 12 to 16 h until the stationary phase, harvested, resuspended in 200 μl of 0.8× ASW, and inoculated into each synthetic medium. Growth experiments were performed at 85°C and in triplicate, and the OD660 was monitored.
Assay of Nm deamidase activity in cell extracts of T. kodakarensis.
KU216 and ΔTK0218 ΔTK1650 mutant cells after cultivation in ASW-YT-Pyr for 16 h were harvested by centrifugation (4°C, 5,800 × g, 15 min) and washed with 0.8× ASW. After centrifugation (4°C, 5,800 × g, 15 min), cells were lysed in approximately a 1/20 volume of 50 mM HEPES-NaOH (pH 7.0) containing 0.1% Triton X-100. After pipetting and mixing with a vortex, the supernatant after centrifugation (4°C, 20,400 × g, 15 min) was used as the cell extract. To examine Nm deamidase activity, a reaction mixture containing 50 mM HEPES-NaOH (pH 7.0), 100 μg of cell extract, 10 mM MgCl2, and 2 mM Nm was incubated at 85°C for 30 min. After proteins were removed with an Amicon Ultra centrifugal filter unit (MWCO, 3,000), Na and Nm accumulation levels were quantified by HPLC using the same methods described above.
Determination of the degradation rates of NaMN and NMN at 85°C.
To examine the degradation rates of NaMN and NMN, 2 mM NaMN and 2 mM NMN in 50 mM HEPES-NaOH (pH 7.0) were incubated at 85°C for 5, 10, 15, 20, 25, 30, 45, and 60 min. The residual concentrations of NaMN and NMN were quantified by HPLC using the same methods described above.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially funded by the Core Research for Evolutional Science and Technology program of the Japan Science and Technology Agency to H.A. within the research area Creation of Basic Technology for Improved Bioenergy Production through Functional Analysis and Regulation of Algae and Other Aquatic Microorganisms.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00785-17.
REFERENCES
- 1.Foster JW, Moat AG. 1980. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev 44:83–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katoh A, Hashimoto T. 2004. Molecular biology of pyridine nucleotide and nicotine biosynthesis. Front Biosci 9:1577–1586. doi: 10.2741/1350. [DOI] [PubMed] [Google Scholar]
- 3.Gazzaniga F, Stebbins R, Chang SZ, McPeek MA, Brenner C. 2009. Microbial NAD metabolism: lessons from comparative genomics. Microbiol Mol Biol Rev 73:529–541. doi: 10.1128/MMBR.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi J, Wang H, Xie J. 2011. Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets. J Cell Physiol 226:331–340. doi: 10.1002/jcp.22419. [DOI] [PubMed] [Google Scholar]
- 5.Lehman IR. 1974. DNA ligase: structure, mechanism, and function. Science 186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 6.Hassa PO, Haenni SS, Elser M, Hottiger MO. 2006. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassa PO, Hottiger MO. 2008. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 8.Aksoy P, White TA, Thompson M, Chini EN. 2006. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun 345:1386–1392. doi: 10.1016/j.bbrc.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 9.Chini EN. 2009. CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr Pharm Des 15:57–63. doi: 10.2174/138161209787185788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan NO, Colowick SP, Barnes CC. 1951. Effect of alkali on diphosphopyridine nucleotide. J Biol Chem 191:461–472. [PubMed] [Google Scholar]
- 11.Anderson BM, Anderson CD. 1963. The effect of buffers on nicotinamide adenine dinucleotide hydrolysis. J Biol Chem 238:1475–1478. [PubMed] [Google Scholar]
- 12.Honda K, Hara N, Cheng M, Nakamura A, Mandai K, Okano K, Ohtake H. 2016. In vitro metabolic engineering for the salvage synthesis of NAD+. Metab Eng 35:114–120. doi: 10.1016/j.ymben.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Hachisuka SI, Sato T, Atomi H. 2017. Metabolism dealing with thermal degradation of NAD+ in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol 199:e00162-17. doi: 10.1128/JB.00162-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi H, Sungwallek S, Chotchuang P, Okano K, Honda K. 2017. A key enzyme of the NAD+ salvage pathway in Thermus thermophilus: characterization of nicotinamidase and the impact of its gene deletion at high temperatures. J Bacteriol 199:e00359-17. doi: 10.1128/JB.00359-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preiss J, Handler P. 1958. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem 233:488–492. [PubMed] [Google Scholar]
- 16.Preiss J, Handler P. 1958. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J Biol Chem 233:493–500. [PubMed] [Google Scholar]
- 17.Foster JW, Kinney DM, Moat AG. 1979. Pyridine nucleotide cycle of Salmonella Typhimurium: isolation and characterization of pncA, pncB, and pncC mutants and utilization of exogenous nicotinamide adenine dinucleotide. J Bacteriol 137:1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaren J, Ngo DT, Olivera BM. 1973. Pyridine nucleotide metabolism in Escherichia coli. III. Biosynthesis from alternative precursors in vivo. J Biol Chem 248:5144–5149. [PubMed] [Google Scholar]
- 19.Gingrich W, Schlenk F. 1944. Codehydrogenase I and other pyridinium compounds as V-factor for Hemophilus [sic] influenzae and H. parainfluenzae. J Bacteriol 47:535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godek CP, Cynamon MH. 1990. In vitro evaluation of nicotinamide riboside analogs against Haemophilus influenzae. Antimicrob Agents Chemother 34:1473–1479. doi: 10.1128/AAC.34.8.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau C, Niere M, Ziegler M. 2009. The NMN/NaMN adenylyltransferase (NMNAT) protein family. Front Biosci 14:410–431. doi: 10.2741/3252. [DOI] [PubMed] [Google Scholar]
- 22.Pfoh R, Pai EF, Saridakis V. 2015. Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides. Acta Crystallogr D Biol Crystallogr 71:2032–2039. doi: 10.1107/S1399004715015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olland AM, Underwood KW, Czerwinski RM, Lo MC, Aulabaugh A, Bard J, Stahl ML, Somers WS, Sullivan FX, Chopra R. 2002. Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase. J Biol Chem 277:3698–3707. doi: 10.1074/jbc.M109670200. [DOI] [PubMed] [Google Scholar]
- 24.Gerdes SY, Kurnasov OV, Shatalin K, Polanuyer B, Sloutsky R, Vonstein V, Overbeek R, Osterman AL. 2006. Comparative genomics of NAD biosynthesis in cyanobacteria. J Bacteriol 188:3012–3023. doi: 10.1128/JB.188.8.3012-3023.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang N, Sorci L, Zhang X, Brautigam CA, Li X, Raffaelli N, Magni G, Grishin NV, Osterman AL, Zhang H. 2008. Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: structure and function in bacterial NAD metabolism. Structure 16:196–209. doi: 10.1016/j.str.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267. doi: 10.1155/2004/204953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res 15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol 185:210–220. doi: 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol 71:3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumi R, Manabe K, Fukui T, Atomi H, Imanaka T. 2007. Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J Bacteriol 189:2683–2691. doi: 10.1128/JB.01692-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santangelo TJ, Cubonova L, Reeve JN. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl Environ Microbiol 74:3099–3104. doi: 10.1128/AEM.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santangelo TJ, Cubonova L, Reeve JN. 2010. Thermococcus kodakarensis genetics: TK1827-encoded β-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl Environ Microbiol 76:1044–1052. doi: 10.1128/AEM.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia B, Linh le T, Lee S, Pham BP, Liu J, Pan H, Zhang S, Cheong GW. 2011. Biochemical characterization of glyceraldehyde-3-phosphate dehydrogenase from Thermococcus kodakarensis KOD1. Extremophiles 15:337–346. doi: 10.1007/s00792-011-0365-4. [DOI] [PubMed] [Google Scholar]
- 34.Tomita H, Imanaka T, Atomi H. 2013. Identification and characterization of an archaeal ketopantoate reductase and its involvement in regulation of coenzyme A biosynthesis. Mol Microbiol 90:307–321. doi: 10.1111/mmi.12363. [DOI] [PubMed] [Google Scholar]
- 35.Wu X, Zhang C, Orita I, Imanaka T, Fukui T, Xing XH. 2013. Thermostable alcohol dehydrogenase from Thermococcus kodakarensis KOD1 for enantioselective bioconversion of aromatic secondary alcohols. Appl Environ Microbiol 79:2209–2217. doi: 10.1128/AEM.03873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin DH, Oganesyan N, Jancarik J, Yokota H, Kim R, Kim SH. 2005. Crystal structure of a nicotinate phosphoribosyltransferase from Thermoplasma acidophilum. J Biol Chem 280:18326–18335. doi: 10.1074/jbc.M501622200. [DOI] [PubMed] [Google Scholar]
- 37.Galassi L, Di Stefano M, Brunetti L, Orsomando G, Amici A, Ruggieri S, Magni G. 2012. Characterization of human nicotinate phosphoribosyltransferase: kinetic studies, structure prediction and functional analysis by site-directed mutagenesis. Biochimie 94:300–309. doi: 10.1016/j.biochi.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 38.De Ingeniis J, Kazanov MD, Shatalin K, Gelfand MS, Osterman AL, Sorci L. 2012. Glutamine versus ammonia utilization in the NAD synthetase family. PLoS One 7:e39115. doi: 10.1371/journal.pone.0039115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robb F, Place A. 1995. Media for thermophiles, p 167–168. In Robb F, Place A (ed), Archaea: a laboratory manual–thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Sakuraba H, Satomura T, Kawakami R, Yamamoto S, Kawarabayasi Y, Kikuchi H, Ohshima T. 2002. l-Aspartate oxidase is present in the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii OT-3: characteristics and role in the de novo biosynthesis of nicotinamide adenine dinucleotide proposed by genome sequencing. Extremophiles 6:275–281. doi: 10.1007/s00792-001-0254-3. [DOI] [PubMed] [Google Scholar]
- 41.Sakuraba H, Yoneda K, Asai I, Tsuge H, Katunuma N, Ohshima T. 2008. Structure of l-aspartate oxidase from the hyperthermophilic archaeon Sulfolobus tokodaii. Biochim Biophys Acta 1784:563–571. doi: 10.1016/j.bbapap.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Washio T, Oikawa T. 2018. Thermostable and highly specific l-aspartate oxidase from Thermococcus litoralis DSM 5473: cloning, overexpression, and enzymological properties. Extremophiles 22:59–71. doi: 10.1007/s00792-017-0977-4. [DOI] [PubMed] [Google Scholar]
- 43.Sakuraba H, Tsuge H, Yoneda K, Katunuma N, Ohshima T. 2005. Crystal structure of the NAD biosynthetic enzyme quinolinate synthase. J Biol Chem 280:26645–26648. doi: 10.1074/jbc.C500192200. [DOI] [PubMed] [Google Scholar]
- 44.Soriano EV, Zhang Y, Colabroy KL, Sanders JM, Settembre EC, Dorrestein PC, Begley TP, Ealick SE. 2013. Active-site models for complexes of quinolinate synthase with substrates and intermediates. Acta Crystallogr D Biol Crystallogr 69:1685–1696. doi: 10.1107/S090744491301247X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.