ABSTRACT
The Rgg2/3 quorum sensing (QS) system is conserved among all sequenced isolates of group A Streptococcus (GAS; Streptococcus pyogenes). The molecular architecture of the system consists of a transcriptional activator (Rgg2) and a transcriptional repressor (Rgg3) under the control of autoinducing peptide pheromones (SHP2 and SHP3). Activation of the Rgg2/3 pathway leads to increases in biofilm formation and resistance to the bactericidal effects of the host factor lysozyme. In this work, we show that deletion of a small gene, spy49_0414c, abolished both phenotypes in response to pheromone signaling. The gene encodes a small, positively charged, secreted protein, referred to as StcA. Analysis of recombinant StcA showed that it can directly interact with GAS cell wall preparations containing phosphodiester-linked carbohydrate polymers but not with preparations devoid of them. Immunofluorescence microscopy detected antibody against StcA bound to the surface of paraformaldehyde-fixed wild-type cells. Expression of StcA in bacterial culture induced a shift in the electrostatic potential of the bacterial cell surface, which became more positively charged. These results suggest that StcA promotes phenotypes by way of ionic interactions with the GAS cell wall, most likely with negatively charged cell wall-associated polysaccharides.
IMPORTANCE This study focuses on a small protein, StcA, that is expressed and secreted under induction of Rgg2/3 QS, ionically associating with negatively charged domains on the cell surface. These data present a novel mechanism of resistance to the host factor lysozyme by GAS and have implications in the relevance of this circuit in the interaction between the bacterium and the human host that is mediated by the bacterial cell surface.
KEYWORDS: pheromone, surface protein, wall polysaccharides, CHAP domain, S-layers, biofilms, cationic peptides, murein hydrolases
INTRODUCTION
A ubiquitous process in the natural world, interorganismal communication, is a mechanism used to coordinate behaviors among members of a group. In bacteria, the cell-to-cell communication process of quorum sensing (QS) allows microbial populations to synchronize gene expression via the production, secretion, accumulation, and detection of chemical signals in their environment. In some cases, the consequences of quorum sensing can be readily apparent, such as in the production of bioluminescence in many Vibrio species or toxin secretion by Staphylococcus aureus in response to small-molecule signals generated by bacteria (1, 2). However, in other cases the effects of bacterial communication systems remain enigmatic.
Streptococcus pyogenes, also known as group A Streptococcus (GAS), is a Gram-positive pathogen responsible for several diseases that range from uncomplicated superficial infections, such as pharyngitis and impetigo, to severe and life-threatening invasive infections, such as necrotizing fasciitis and toxic shock (3). Additionally, immune-mediated postinfection sequelae can develop in the host, generating pathologies such as rheumatic heart disease and glomerulonephritis, which account for 600,000 deaths globally each year. Antibiotic treatment failure rates, independent of resistance to the antibiotic, are as high as 30% and remain poorly understood mechanistically, although internalization of GAS in epithelial cells and biofilm formation in tonsillar crypts have been shown to contribute to these incidents (4–8).
GAS is also carried asymptomatically in up to 35% of some populations, especially in children (9). In fact, asymptomatic nasopharyngeal carriage of GAS in the general population serves as the reservoir of this pathogen (10–12). Although classically considered an extracellular pathogen, there have been numerous studies identifying the ability of GAS to invade and survive intracellularly in the cytoplasm of immune and epithelial cells (13, 14).
As in other Gram-positive bacteria, QS in S. pyogenes utilizes the activity of posttranslationally processed oligopeptides, termed autoinducing peptides or pheromones. The genomes of GAS isolates contain several QS systems, which control or are implicated with virulence, genetic exchange, and antimicrobial compound production (15, 16). Some of these QS systems are found in only a subset of sequenced serotypes, but the Rgg2/3 QS system is found in all sequenced isolates of GAS. The Rgg2/3 system consists of two transcriptional regulators, an activator (Rgg2) and a repressor (Rgg3), that are modulated by two short hydrophobic peptide (SHP) signals (17, 18). These signals are ribosomally produced and further processed by proteolytic enzymes prior to becoming mature signals capable of diffusion and interaction with nearby cells. At low pheromone concentrations, below threshold levels, pheromones do not strongly interact with the Rggs, and Rgg3 predominates as a negative transcriptional regulator, suppressing transcription from the shp promoters (Pshp2 and Pshp3). When pheromones accumulate to levels above the threshold (∼1 nM in culture) that result in receptor binding, they provoke Rgg3 to dissociate from the DNA and Rgg2 to serve as a transcriptional activator, causing a strong increase in Pshp expression. Thus, the net effect of SHP binding to Rgg2 and Rgg3 is robust transcriptional induction of the operons encoding each SHP, and this induction is sustained by a resulting positive-feedback regulatory loop made possible through further pheromone production. Downstream of each shp are genes that are coexpressed under Pshp activation, namely, genes in regions encompassing spy49_0450 to spy49_0460 and spy49_0412 to spy49_0414c (18). In the latter, the spy49_0412 and spy49_0413 genes are carried on the opposite strand as the Pshp2 promoter and spy49_0414c.
We recently found that the Rgg2/3 system is active under certain environmental conditions, such as in the presence of low metal concentrations or when mannose is a utilized carbon source (19). Conversely, the PepO endopeptidase degrades SHPs and therefore inhibits the Rgg2/3 system (20). Expression of pepO is under the control of the CovRS two-component signal transduction system, in which the CovS kinase senses signals from the environment and, in response, modulates CovR phosphorylation. Under conditions in which CovR is dephosphorylated, several virulence genes are upregulated along with pepO. Many highly virulent isolates contain mutations in the CovRS two-component system, suggesting that elevated levels of pepO expression and, consequently, enhanced degradation of SHP pheromone peptides occur in these strains. We have hypothesized that an inverse relationship exists between expression of CovRS-regulated virulence factors and Rgg2/3 QS, implying a role for this system in asymptomatic colonization of the host (20).
Here, we demonstrate that an Rgg2/3-regulated gene, spy49_0414c, encodes a small, secreted, and positively charged protein that we have named StcA (streptococcal charged A protein). StcA was necessary and sufficient to trigger an increase in biofilm formation and lysozyme resistance. The expression of this protein is dependent on the induction of Rgg2/3 by SHPs and can be observed on the cell surface of paraformaldehyde-fixed cells. The production of StcA induces a shift in the electrostatic charge of the bacterial surface, making the cells more positively charged. The shift in charge is dependent on carbohydrate-based polymers present on the GAS cell surface, indicating that the small, positively charged StcA protein likely interacts with negatively charged polysaccharides to aid the bacteria in evading host factors, such as lysozyme.
RESULTS
A small protein, StcA, mediates Rgg2/3-regulated lysozyme resistance and biofilm formation.
In previously published works, we demonstrated that pheromone signaling via the Rgg2/3 pathway induces a variety of phenotypes in GAS cultures, including increased lysozyme resistance and biofilm formation (17, 19). These phenotypes were observed upon the addition of synthetic peptide pheromone to the medium of growing cultures and were abrogated in strains lacking the transcriptional activator of the system (Δrgg2 strains). However, the downstream mechanism leading to these phenotypes remained uncharacterized. To elucidate the molecular mechanism(s) at work, we generated mutations in target genes controlled by the Rgg2/3 pathway, using biofilm formation as a readout.
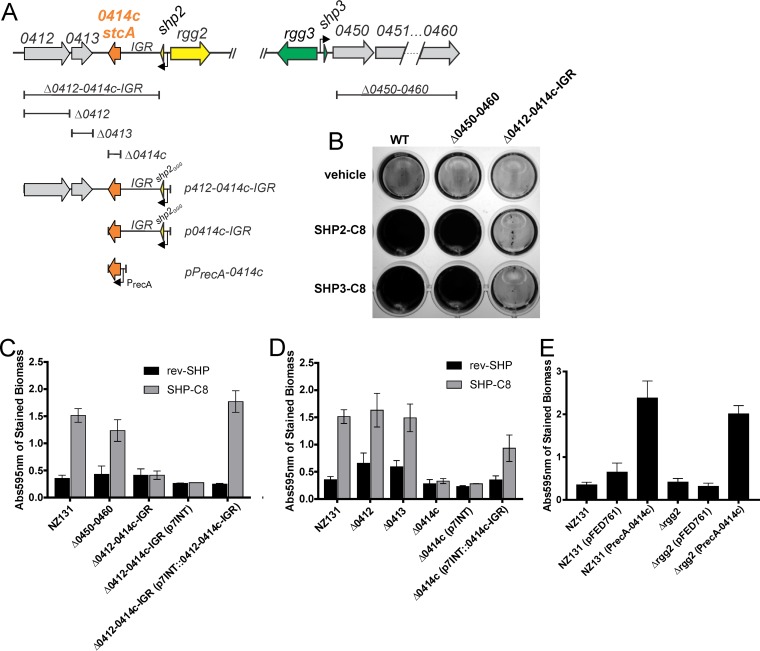
The Rgg2/3 system regulates expression of at least two mRNA transcripts for the pheromone genes shp2 and shp3, as well as additional downstream genes of unknown function (Fig. 1A) (10). The polycistronic mRNA for shp3 includes additional genes (spy49_0450 to spy49_0460) (21). The region downstream of shp2 comprises a region of several hypothetical genes. The genes spy49_0414c and shp2 are carried on the same DNA strand and are predicted to be controlled by the Pshp2 promoter but are separated by a 913-bp intergenic region (referred to in this work as IGR). Systematic mutation of loci located downstream of the shp2 and shp3 genes was carried out to test their involvement as factors contributing to biofilm formation. Mutant strains in which the operon from spy49_0450 to spy49_0460 or the region encompassing IGR-spy49_0412 was deleted were generated. Neither of the deletions interfered with pheromone signaling, as determined by use of a previously developed luciferase reporter that responds to SHP pheromones (see Fig. S1 in the supplemental material) (10). When the biofilm production of the mutants was measured, only deletion of the IGR-spy49_0412 region disrupted the phenotype generated by addition of peptide pheromones (Fig. 1B and C). To identify the precise gene(s) required for this disruption, we subsequently generated strains with the individual deletion of spy49_0414c, spy49_0413, or spy49_0412. Only the mutant with a deletion of the spy49_0414c gene was incapable of developing biofilms when SHP pheromones were supplemented into the cultures. Complementation of the ΔIGR-spy49_0412 and Δspy49_0414c mutants either with the entire Pshp3-IGR-spy49_0412 locus or with Pshp3-IGR-spy49_0414c, provided in a single gene copy from the chromosomally integrated p7INT plasmid, restored the pheromone-dependent biofilm increase to these mutants (Fig. 1C and D). Additionally, a multicopy plasmid carrying spy49_0414c expressed from a heterologous recA promoter but not including the IGR was generated and transferred into the wild type (WT) and a Δrgg2 mutant. spy49_0414c provided in trans but not the vector alone was able to stimulate biofilm formation in the wild type in the absence of SHP stimulation and in the Δrgg2 mutant, which is nonresponsive to SHP pheromones (Fig. 1E). Thus, spy49_0414c is necessary and sufficient to regulate biofilm development and works downstream of the Rgg2/3 signaling system.
FIG 1.
Biofilm formation experiments carried out following systematic mutation of genes regulated by the Rgg2/3 QS system. (A) Targets of Rgg2/3 regulation revealed by previous transcriptional analysis. Rgg2 and Rgg3 antagonistically compete to regulate two target promoters, Pshp3 and Pshp2, which promote transcription of their corresponding pheromone and downstream genes (17, 18). (B) Representative image showing biofilm formation with either the SHP2-C8 or SHP3-C8 pheromone. (C) Biofilm formation was examined with and without pheromone in strains with mutations in the operons controlled by the QS system, and the spy49_0412 to spy49_0414c region was complemented on a p7INT plasmid under the control of its native, pheromone-responsive promoter. (D) Genes in the spy49_0412 to spy49_0414c region were systematically knocked out, and spy49_0414c was reintroduced on a p7INT plasmid under the control of its native, pheromone-responsive promoter. (E) Expression of the spy49_0414c gene from a constitutive PrecA promoter generates an increase in biofilm formation in the absence of QS signaling. Quantification of biofilm formation was performed in both a wild-type background and a Δrgg2 mutant background; the latter was unable to activate the QS system. The error bars represent the standard deviations from three independent experiments, each carried out in duplicate.
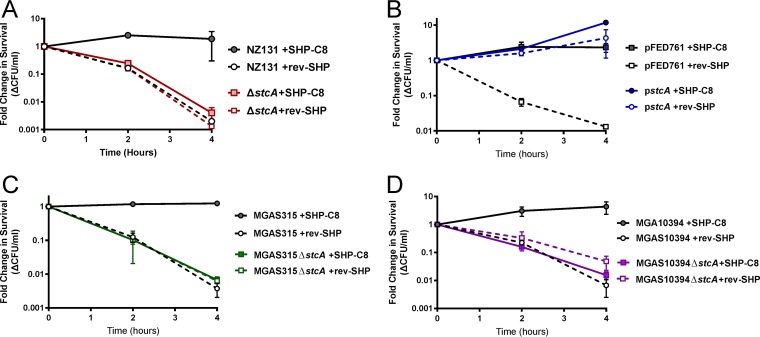
Previous work also showed that activation of the Rgg2/3 pathway promotes the survival of GAS in response to the bactericidal effects of lysozyme (19). Prestimulation of wild-type GAS with SHP for 30 min led to enhanced lysozyme resistance, whereas untreated cultures had >99% of cells killed in 4 h (Fig. 2A). To investigate whether spy49_0414c was also associated with lysozyme resistance, we tested the Δspy49_0414c mutant in the killing assay. In contrast to the wild type, the Δspy49_0414c mutant was unable to resist killing by lysozyme, even when prestimulated with SHP pheromones (Fig. 2A). The rev-SHP pheromone, which is made up of the same amino acids as SHP3-C8 but in the reversed N-C orientation, was used as a control. Conversely, providing the Δspy49_0414c mutant with the plasmid expressing spy49_0414c from the constitutive PrecA promoter led to survival in the presence of lysozyme challenge, even without pretreatment of SHP pheromones (Fig. 2B). When mutations in spy49_0414c orthologs were generated in other GAS isolates, MGAS315 (serotype M3) and MGAS10394 (serotype M6), these strains also lost their ability to resist the lysozyme challenge after SHP-C8 induction, supporting the conserved function of this protein within the species (Fig. 2C and D).
FIG 2.
Pheromone-dependent lysozyme resistance is attributable to StcA across GAS serotypes. (A) Pheromone-dependent challenge in 2 mg ml−1 lysozyme after pretreatment with 100 nM SHP-C8 or rev-SHP, which is the control peptide, in WT and ΔstcA strains. (B) Pheromone-dependent challenge in 2 mg ml−1 lysozyme in a strain containing the stcA gene on plasmid pFED761 under the control of the constitutive promoter PrecA and a strain containing the empty pFED761 vector. Error bars indicate standard deviations. (C) Lysozyme challenge of isolate MGAS315 of the M3 serotype after pretreatment with 100 nM SHP-C8 or rev-SHP. The concentration of lysozyme used for MGAS315 challenge was 50 mg ml−1. The mean and standard deviation represent biological duplicates done in technical duplicate. (D) Lysozyme challenge of isolate MGAS10394 of the M6 serotype after pretreatment with 100 nM SHP-C8 or Rev-SHP peptide. The concentration of lysozyme used for MGAS10394 challenge was 20 mg ml−1. The bars represent the standard deviations of biological duplicates done in technical duplicate.
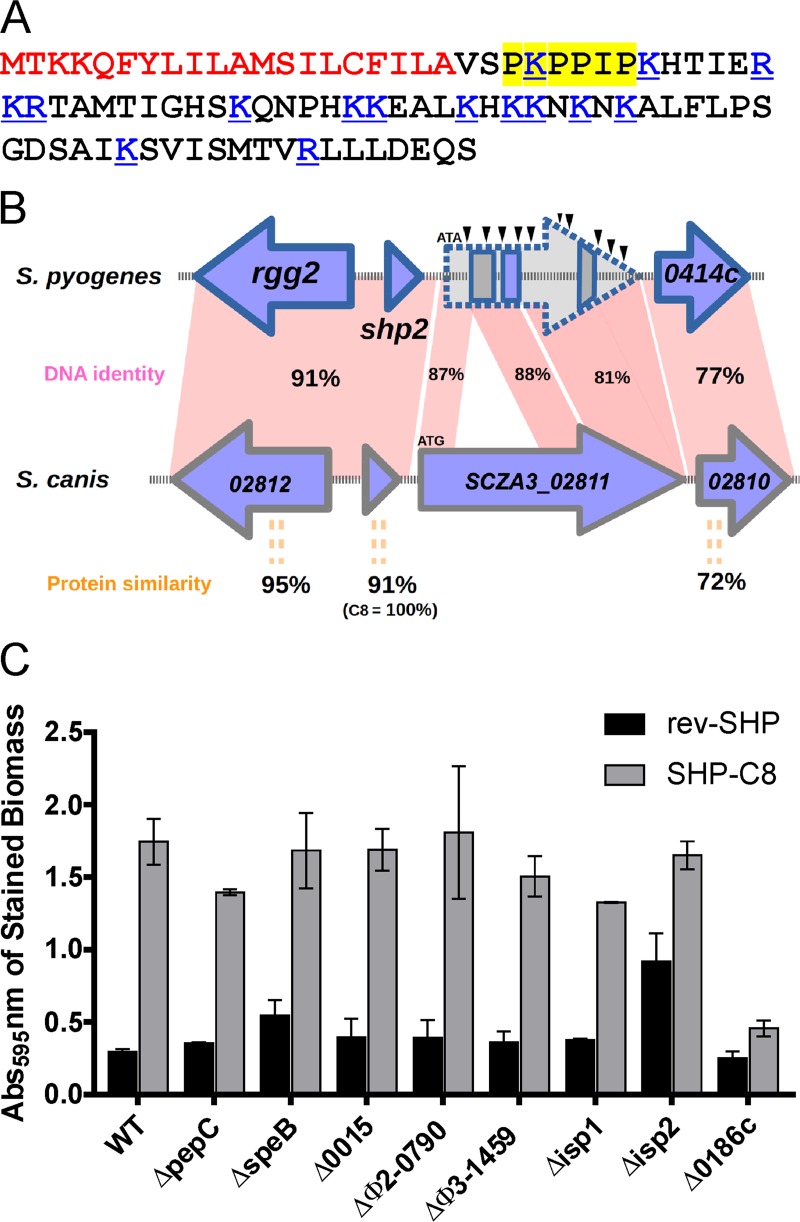
The predicted amino acid sequence of the spy49_0414c gene product indicates an N-terminal secretion signal sequence, and the remainder contains a striking number of lysine and arginine residues. Fifteen of the 69 amino acids that follow the signal sequence are positively charged (Fig. 3A). Therefore, we henceforth refer to the spy49_0414c product as StcA, for the streptococcal charged A protein.
FIG 3.
(A) The 89-amino-acid sequence of StcA, showing the predicted 20-amino-acid secretion signal in red, the 15 positively charged amino acids in blue, and the highlighted polyproline region, with the flanking basic lysines underlined. (B) Alignment of the Rgg2 regulons of GAS and S. canis. In S. canis FSL-Z3, a putative transglutaminase is encoded downstream of the SHP2 pheromone. Alignment of the GAS NZ131 IGR shows high homology to the transglutaminase. (C) Biofilm formation of the wild type and nine cysteine proteinase deletion mutants. Quantification of biofilm formation in three experiments in the presence or absence of the SHP3-C8 pheromone, with the bars displaying standard deviations.
StcA-dependent lysozyme resistance is independent of known mechanisms.
As several mechanisms that enhance resistance to lysozyme through the modification of peptidoglycan have been described, we hypothesized that StcA may work through one of these known pathways. These include the enzymes PgdA, a deacetylase that modifies GlcNAc at the C-2 position, and OatA, an O-acetyltransferase that modifies MurNAc at the C-6 position (22). Additionally, the incorporation of d-alanine into lipoteichoic acids of the cell wall via the dlt operon alters lysozyme resistance in a variety of Gram-positive bacteria, including GAS (23). A BLASTp search was performed in the GAS NZ131 genome using PgdA or OatA amino acid sequences from Streptococcus pneumoniae or S. aureus, respectively, and putative GAS homologues were identified (pgdA = spy49_1092c; oatA = spy49_0035). To determine whether these genes or dlt was regulated in response to Rgg-SHP signaling, luciferase reporters were constructed for the relevant promoters and introduced into wild-type NZ131, and transcription in response to exogenous SHP pheromone was measured. The luciferase activity of the reporter strains did not increase in response to peptide; in fact, the reporters exhibited a slight decrease upon the addition of the SHP3-C8 peptide relative to the controls treated with the reverse peptide (Fig. S2A to C). Deletion strains in which each of the genes of interest was replaced by an antibiotic resistance marker were also constructed, and the resulting mutants were tested for changes in sensitivity to lysozyme following induction of the Rgg-SHP system by the addition of synthetic peptide. Both the ΔpgdA and ΔoatA mutants responded to pheromone like the wild type, with uninduced cells exhibiting a >1,000-fold decrease in the number of CFU after 4 h of exposure to lysozyme (Fig. S2D and E). As reported previously, the deletion of the dlt and pgdA genes led to an increase in lysozyme sensitivity relative to that of the wild type (14) (Fig. S2D and F). However, when dlt cells were challenged with a lower concentration of lysozyme, an SHP-dependent increase in overall survival relative to that of cells treated with the reverse peptide was observed (Fig. S2F). Taken together, these data indicate that the increased lysozyme resistance observed in response to Rgg-SHP signaling occurs through mechanisms unrelated to PgdA, OatA, or Dlt activities. We also tested the ability of StcA to induce resistance to another antimicrobial that acts upon the cell surface, the cationic antimicrobial peptide LL-37, and found no increased resistance (Fig. S3).
Phylogenetic comparisons infer StcA function.
With a high number of positively charged residues, the predicted mature form of StcA stands among 1% of all bacterial proteins having an isoelectric point of greater than 10 (24), and this characteristic is implicated as a key factor for the protein's function. Additionally, a proline-rich motif (PKPPIPK) is present at the N terminus of the 69-amino-acid mature protein. In eukaryotes, the PXPPXP domain is well documented in its binding to eukaryotic SH3 domain-containing proteins (25–27).
Sequence homology-based searches revealed that stcA homologs exist in three species, S. pyogenes, Streptococcus dysgalactiae, and Streptococcus canis, all of which are in the pyogenic group of streptococci. In S. dysgalactiae and S. canis, the SCZA3_02811 open reading frame (ORF) residing between shp2 and SCZA3_02810 (stcA) encodes a predicted cysteine protease of the transglutaminase-like family. An ORF homologous to SCZA3_02811 is not present in S. pyogenes; instead, a start codon mutation (ATG to ATA), several nonsense mutations, and a 740-bp deletion of the coding sequence render this remnant gene nonfunctional in GAS (Fig. 3B). These mutations are well conserved in other sequenced GAS isolates, as the IGR has 98% sequence identity across 53 assembled GAS genomes. The stcA gene is also highly conserved in GAS, with other strains showing only 2 to 6% variation in the 89 amino acids of the full-length protein.
We considered the possibility that since StcA is a small protein with no apparent enzymatic domain, it is a modulator of a partnering enzyme, in a fashion analogous to that for small proteins that inhibit the activity of cysteine proteases in Streptomyces, Staphylococcus, and Enterococcus (28). Based on the premise that stcA is located next to a silenced cysteine protease pseudogene in S. pyogenes, we hypothesized that the stcA product could modulate the activity of other cysteine proteases encoded by the GAS NZ131 genome. We reasoned that if this were the case, deleting the target protease of StcA would recapitulate the effects seen when stcA is expressed, providing a constitutively high level of biofilm formation.
The GAS NZ131 strain has 10 genes encoding members of the cysteine proteinase superfamily (SSF54001) (Table S1). These include the well-studied papain-like protease SpeB, the Ig protease Mac, and six genes encoding cysteine- and histidine-dependent aminohydrolase/peptidase (CHAP) domains, which are predicted to be involved in the processing of peptide cross-linkages in peptidoglycan. We generated mutant strains with mutations in all members of the cysteine protease superfamily. Most of these strains exhibited a normal pattern of growth, except the mutant with a mutation in spy49_0026c, which encodes a protease involved in cell division processes. The spy49_0026c mutant was strongly impaired for growth in chemically defined medium (CDM) and was excluded from further analysis. When all protease mutants were tested for their biofilm-forming activity (Fig. 3C), all required SHP3-C8 to increase biofilm production, except the mutant with a mutation in immunogenic secreted protein 2 (spy49_1407c, isp2), which partially recapitulated the pheromone-dependent increase in biofilm production. Unexpectedly, a mutation in spy49_0186c, which encodes a putative S-layer transglutaminase, exhibited a diminished ability to increase biofilm formation in response to pheromone signaling. We confirmed that spy49_0186c expression does not affect transcription of the Rgg2/3 regulon (Fig. S4A) and the diminished ability of the mutant to form biofilms in the presence of pheromone was successfully complemented (Fig. S4B). Deletion of spy49_0186c also diminished the ability of the bacteria to form biofilms in the presence of constitutively transcribed stcA (Fig. S4C). The mutant with the deletion of spy49_0186c also exhibited a decreased resistance to lysozyme in the presence of both pheromone and constitutively transcribed stcA (Fig. S4D).
Deletion of isp2 partially phenocopies pheromone-induced biofilm formation and lysozyme resistance.
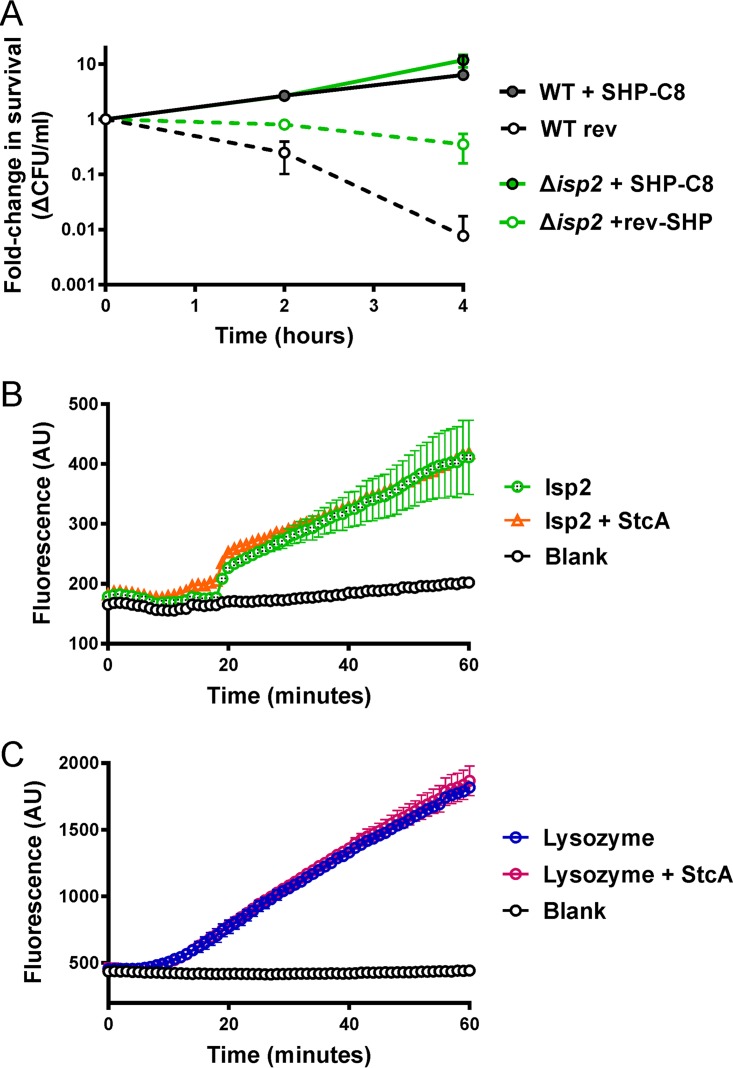
The partial phenocopying by the isp2 deletion in the biofilm assay (Fig. 3C) led us to test the resistance of the strain with this deletion to lysozyme. When treated with lysozyme, the Δisp2 strain exhibited only a partial increase in resistance in comparison to the level observed for the stcA strain (Fig. 4A). Despite the partial phenotype and the possibility that the Isp2 phenocopying is simply a serendipitous coincidence, we pursued testing whether Isp2 and StcA directly interact, as we postulated that Isp2 might be only one of multiple protein partners of StcA.
FIG 4.
Investigation of Isp2-StcA interaction. (A) Pheromone-dependent challenge in 2 mg ml−1 lysozyme after pretreatment with 100 nM SHP-C8 or rev-SHP in WT and Δisp2 strains. Bars display standard deviations for biological duplicates done in technical duplicate. (B) FITC-labeled cell wall was incubated with recombinant Isp2, and cleavage of the cell wall was observed over the course of 1 h. Addition of 10 nM recombinant StcA resulted in no significant change to the hydrolytic capability of Isp2. (C) Cell wall was incubated with lysozyme in the same manner, with or without the addition of 10 nM StcA. AU, arbitrary units.
To test the in vitro activities of Isp2 and StcA, recombinant versions of both proteins that lack signal sequences but contain N-terminal 6× His tags were expressed in Escherichia coli and purified by Ni-nitrilotriacetic acid affinity chromatography. We anticipated that if Isp2, a predicted murein hydrolase, were susceptible to inhibition by StcA, Isp2-dependent hydrolysis of S. pyogenes cell wall preparations would be blocked in the presence of StcA. To observe the hydrolysis of peptidoglycan by recombinant Isp2 (rIsp2), GAS NZ131 cell wall preparations were covalently labeled with fluorescein isothiocyanate (FITC) by a method previously used to demonstrate lysozyme hydrolysis of Micrococcus luteus peptidoglycan; degradation of the labeled substrate generates an activity-dependent increase in fluorescence intensity (29). With this reagent, we observed hydrolysis of cell wall preparations when treated with lysozyme as a positive control (Fig. 4C). Although Isp2 was not as efficient as lysozyme, an increase in fluorescence intensity was also observed during Isp2 incubation with the FITC-labeled cell wall (Fig. 4B). However, addition of recombinant StcA (rStcA) protein at molar excess concentrations failed to interfere with either reaction (Fig. 4B and C). These results indicate that StcA is unable to directly interact with Isp2, at least under these conditions, nor does it appear that StcA directly inhibits lysozyme to confer resistance. We were initially concerned that the purified StcA preparation may not be active, but later experiments suggested that this is not the case (see Fig. 5). Taken together, these results suggest that to enhance lysozyme resistance, StcA works with at least one additional partner or by a mechanism that is independent of Isp2.
FIG 5.
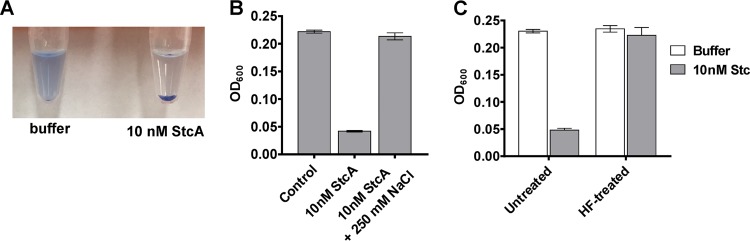
In vitro aggregation of purified GAS cell wall upon addition of recombinant StcA. Remazol brilliant blue-labeled GAS cell wall with the addition of buffer or 10 nM recombinant StcA after shaking at room temperature for 30 min. (B) Aggregation of Remazol brilliant blue-labeled GAS cell wall with StcA was disrupted with 250 mM NaCl. (C) Cell wall treated with hydrofluoric acid (HF) to remove lipoteichoic acid was incubated with 10 nM StcA before and after treatment. Each experiment was performed in triplicate, with bars representing standard deviations.
StcA induces robust aggregation of S. pyogenes cell wall preparations.
Wall preparations of S. pyogenes comprise large peptidoglycan polymers with integrated lipoteichoic acids (LTA) and carbohydrates. These substances form a suspension in aqueous solution that, when left undisturbed, eventually settles over the course of hours or days. During incubation of cell wall preparations with recombinant StcA, we noticed that rates of aggregation increased. To quantify this phenomenon, Remazol brilliant blue-labeled cell wall preparations in phosphate-buffered saline (PBS) were vigorously stirred in the presence or absence of StcA for 30 min. Samples containing StcA aggregated, whereas samples treated only with buffer remained in suspension (Fig. 5A). Addition of 250 mM NaCl to the PBS eliminated the aggregation, suggesting an ionic interaction between StcA and the cell wall (Fig. 5B). Furthermore, cell wall preparations treated with 40% hydrofluoric acid, to hydrolyze and remove the phosphodiester bonds of peptidoglycan-linked polysaccharides or intercalating lipoteichoic acid (LTA) (30–32), no longer displayed enhanced aggregation in the presence of StcA. This implicated carbohydrate-containing polymers as the component of the cell wall to which StcA bound and caused aggregation (Fig. 5C). Hydrolysis of the cell wall preparations by phage lysin PlyC was unaffected by hydrofluoric acid, confirming that the wall preparations remained a biologically accessible substrate after acid treatment (Fig. S5).
StcA is found on the surface of SHP-induced bacterial cells.
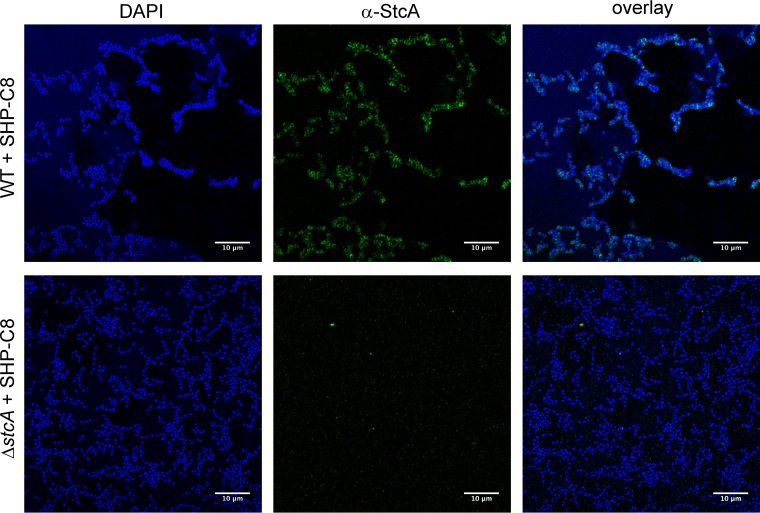
To confirm the localization of the StcA protein on the surface of the bacterium, a polyclonal antibody against StcA was used to stain paraformaldehyde-fixed WT and ΔstcA cells for microscopy (Fig. 6A). WT but not ΔstcA cells were often found in dense aggregates with thick layers of StcA-positive space between each cell. As imaging was conducted on intact cells and immunostaining was found primarily around their periphery, we interpret the primary location of StcA to be extracellular.
FIG 6.
Immunofluorescence microscopy. WT and ΔstcA cells were induced with 100 nM SHP-C8 pheromone for 1 h before being fixed with paraformaldehyde. The cells were then stained with a rabbit polyclonal anti-StcA antibody and an Αlexa Fluor 488-coupled anti-rabbit goat immunoglobulin antibody. WT and ΔstcA cell imaging showing anti-StcA binding in the green fluorescent protein channel and DAPI (cytoplasm) in the blue channel is shown. Magnifications, ×126.
In vivo expression of StcA induces a change in the overall surface charge of S. pyogenes.
To confirm that the in vitro findings were relevant in the physiology of quorum sensing activation, we performed an assay to monitor electrostatic changes to the surface of the cells treated with active pheromone (SHP-C8) or a control peptide (rev-SHP). A cytochrome c binding assay (23) showed that when cells were treated with pheromone, diminished amounts of positively charged cytochrome c bound to the cells, indicating a net increase in positive charges on the cell surface. In the ΔstcA mutant, the difference in the amount of bound cytochrome c between cells treated with pheromone or the control peptide was not significant (Fig. 7). Cytochrome c was used previously to show that deletion of the gene that leads to d-alanylation of LTA produces a more negatively charged cell surface and, consequently, increases the amount of cytochrome c able to bind (23). Treatment of the Δdlt mutant with pheromone subsequently led to a decreased amount of bound cytochrome c, but this did not occur in a ΔdltD ΔstcA strain. The difference in the percentage of bound cytochrome c in response to pheromone was similar between the WT and Δdlt strains, suggesting that the potential interaction of StcA and surface structures is unaffected by LTA d-alanylation.
FIG 7.
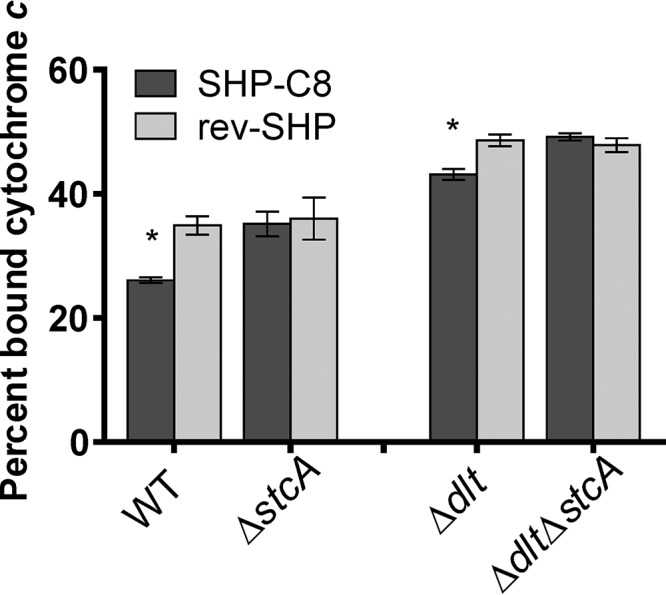
Cytochrome c binding assay. NZ131 and mutant cells were grown to an OD600 of 0.3 with 50 nM SHP-C8 pheromone or 50 nM rev-SHP pheromone before incubation with 1 mg ml−1 cytochrome c. The percentage of the total bound cytochrome c was quantified spectrophotometrically at 535 nm. Asterisks indicate conditions that were significant at a P value of <0.01. Experiments were performed in biological triplicate, and bars represent standard deviations.
DISCUSSION
In this work, we identified the genetic basis of Rgg-SHP-induced lysozyme resistance and biofilm formation (Fig. 1). These phenotypes are produced by the expression of a small, positively charged secreted protein, StcA, which associates with the surface of the bacterium. This is a novel mechanism of lysozyme resistance that has not previously been described.
The results presented here suggest that the positively charged StcA protein associates with the cell surface of GAS. The overall charge of the cell surface becomes more positive when StcA is expressed upon stimulation by pheromone (Fig. 7), indicating that negative charges of the envelope are obscured as the positively charged protein associates with the cell surface. The cell wall component of GAS that contributes most to the negative charge of the surface is provided by phosphates of covalently bound carbohydrate polysaccharides and especially the large number of phosphates present in LTA. S. pyogenes does not encode the TagO enzyme, which functions to attach teichoic acid covalently to peptidoglycan, producing wall teichoic acid (WTA). Therefore, it is thought that S. pyogenes possesses LTA but not WTA. Hydrolysis of phosphodiester bonds by hydrofluoric acid abrogated the interaction of StcA with the cell wall preparations (Fig. 5). We had expected that a strain deficient in d-alanylation of lipoteichoic acids (a Δdlt strain), which has fewer alanine residues to counteract the negatively charged phosphate groups in the polymer, would bind larger amounts of StcA, leading to a larger difference in the percentage of bound cytochrome c when treated with SHP and rev-SHP (Fig. 6). However, we found that the differences in the percentages of bound cytochrome c were similar in the WT and Δdlt strains, which suggests that alanylation does not affect the interaction between StcA and the wall and could indicate that StcA associates with a different surface polymer or an LTA moiety that is discrete from alanylation sites.
We initially hypothesized that StcA may be acting as an inhibitor of the Isp2 peptidoglycan hydrolase (Fig. 3). The genomes of Gram-positive bacteria encode many accessory peptidoglycan hydrolases whose roles remain perplexing. The increase in biofilm formation and resistance to lysozyme in the Δisp2 mutant was consistent with this hypothesis; however, the phenotypes were only partially restored by the Δisp2 mutant, and recombinant StcA did not affect the hydrolytic activity of recombinant Isp2 (Fig. 4). Furthermore, we have no evidence that Isp2 is differentially regulated under conditions where Rgg2/3 QS is activated. It is plausible that inactivation of Isp2 independently induces a surface change that partially phenocopies the effect of Stc expression for biofilm formation and lysozyme resistance characteristics. Serendipitously, the experiments with recombinant Isp2 led us to discover a direct interaction between StcA and cell wall preparations (Fig. 5). Although our analysis has not ruled out the possibility that Isp2 activity is affected by StcA, we no longer favor this hypothesis, but we felt that it was important to report the data pertaining to Isp2 activity to further understanding of these generally uncharacterized hydrolases. Our in vitro findings support a notion that StcA associates with components of the cell wall via electrostatic interactions, but the precise mechanisms underlying the enhanced biofilm production and lysozyme resistance that we observed remain under investigation.
There could be a variety of potential benefits that result from StcA associating with the cell surface. The most obvious is that modulation of surface molecules could contribute to immune evasion or resistance to host-produced antimicrobial peptides or enzymes. We have already found that expression of StcA increases the resistance to lysozyme, a host enzyme found in mucosal secretions and in neutrophilic granules. LTA plays an important antigenic role, activating macrophages and T cells via the Toll-like 2 (TLR2) receptor. In fact, exposure to exogenous LTA has been shown to eliminate colonization of nasopharyngeal GAS cells in a mouse model (33). Thus, if StcA can obstruct the LTA antigen from being detected, it might increase successful colonization of the bacterium.
An additional peculiarity of the StcA protein is the N-terminal PXPPXP motif, which is common in eukaryotic SH3-binding sequences and important for many protein-protein interactions. SH3-binding, proline-rich motifs are often flanked by a basic amino acid that allows interaction with the RT loop of the SH3 domain; a lysine flanks the PXPPXP domain in StcA (34). The motifs are also most commonly seen near the terminal ends of proteins, and for StcA, it is within the first few amino acids of the putative signal peptidase cleavage site. Eukaryotic SH3 domains are almost exclusively found in the cytoplasm of host cells, which raises the question of whether StcA enters host cells and plays a role during invasion and/or intracellular survival of GAS in eukaryotic cells. Supporting this hypothesis, the confined space of bacterium-containing vacuoles presents an opportunity for pheromone concentrations to accumulate to levels that would trigger bacterial responses. This concept was recently demonstrated to be employed by a quorum sensing system in Listeria monocytogenes (35). We wonder if the Rgg2/3 quorum sensing pathway is induced during GAS cellular invasion, leading to StcA expression in vacuolar or cytoplasmic compartments of the cell to interact with host proteins.
Aspects of the hypothesis that StcA acts as a cell wall binding protein also support the potential for its classification as an S-layer protein. The term S-layer protein refers to a protein that self-assembles into a two-dimensional crystalline lattice over the surface of a cell (36). Characterized S-layer proteins bear very little homology to one another in terms of amino acid sequence and thus have been difficult to predict. They include a cell wall binding domain and often have attached functional domains, such as proteinases. Although no S-layer proteins have been described yet in streptococci, there is one predicted S-layer lipoprotein in the GAS genome, which is encoded by spy49_0186c. This protein shares 31% identity with Bacillus anthracis and Bacillus cereus S-layer proteins, and, interestingly, a mutation in spy49_0186c disrupted quorum sensing-dependent biofilm formation and lysozyme resistance (see Fig. S4 in the supplemental material). S-layers are often made up of multiple proteins, so it is conceivable that the protein encoded by spy49_0186c and StcA associate together to form an S-layer on the surface of the cell. The possibility that StcA serves as an S-layer protein is attractive, considering that S-layer activities in other Gram-positive bacteria have been attributed to biofilm formation, cellular aggregation, peptidoglycan remodeling, and permeability barriers that impart resistance to host factors (36), each of which is consistent with the characteristics that we have described for StcA. Furthermore, two documented examples of S-layer proteins being regulated by quorum sensing have been characterized in Serratia liquefaciens and Bacillus anthracis, where the luxS-autoinducer 2 system mediates expression (37, 38).
In Gram-positive bacteria, S-layer proteins noncovalently bind to components of the cell wall (39). For example, the S-layer protein of Lactobacillus acidophilus, SlpA, contains a C-terminal domain (SAC) that is required for the protein to associate with lipoteichoic acid for surface association (40). Like StcA, the SAC domain has many positively charged residues, and binding of the protein is severely reduced in the presence of salt concentrations greater than 250 mM NaCl (30). The S-layer proteins of Lactobacillus generally have isoelectric points (pI) of greater than 9 and many lysines in the cell wall binding C-terminal domain (41).
Until recently, the smallest S-layer proteins known were upwards of 100 kDa (42). However, the positively charged S-layer proteins in Lactobacillus were more recently discovered and have molecular masses of between 25 and 70 kDa and isoelectric points of between 9 and 10.4 (41). Still, StcA, at 8 kDa, is small in comparison to these proteins, but when amino acid sequences are compared, StcA is most similar to the domains of Lactobacillus S-layer proteins that bind the cell wall (30). These domains, also referred to as S-layer homology domains (SLHDs), are typically between 50 and 70 amino acids, a size comparable to StcA's 69 amino acids when its secretion signal is excluded. Both StcA and these Lactobacillus SLHDs have a large number of lysines, leading to their high isoelectric points. It is feasible that sequences upstream of stcA, containing the IGR (Fig. 3B), once encoded a functional portion of the protein, but now the cell wall binding domain has evolved to act on its own or interact with another protein partner. We plan to explore the idea of StcA as a lattice-forming S-layer protein in future studies.
The physiological consequences of the expression of StcA are unknown, as the only screen that has identified this protein found it to be expressed in group C streptococcal strains isolated from bovine mastitis (43). As we continue to dissect the mechanistic nuances of the StcA protein, we will also begin to examine the role of the protein in the host-bacterium interaction. In conclusion, the selective modulation of a cell wall binding protein by a quorum sensing system is an intriguing finding, and this study has identified a novel mechanism of lysozyme resistance attributed to a unique, conserved protein of pyogenic streptococci.
MATERIALS AND METHODS
Bacterial strains and media.
All strains used in this work are listed in Table S2 in the supplemental material, and construction of the various mutant and recombinant strains is described in detail below. GAS strains were routinely grown in Todd-Hewitt medium supplemented with 2% (wt/vol) yeast extract (THY). For all pheromone signaling-related studies, GAS was grown in a chemically defined medium (CDM) (17, 49) containing 1% (wt/vol) glucose. When needed, antibiotics were added at the following concentrations: chloramphenicol (Cm), 3 μg ml−1; erythromycin (Erm), 0.5 μg ml−1; spectinomycin (Sp), 100 μg ml−1; kanamycin (Kan), 100 μg ml−1. When required, starter cultures were prepared as follows to minimize differences in lag phase. GAS strains of interest were inoculated into THY containing appropriate antibiotics and grown overnight at 30°C. The next morning, cultures were diluted 1:100 into CDM and grown at 37°C until mid-exponential phase (optical density at 600 nm [OD600] = 0.4 to 0.7). At this point, glycerol was added to a final concentration of 20% and aliquots were stored at −80°C. E. coli strains were grown in Luria broth (LB). When required, antibiotics were added at the following concentrations: chloramphenicol (Cm), 10 μg ml−1; erythromycin (Erm), 500 μg ml−1; spectinomycin (Sp), 100 μg ml−1, kanamycin (Kan), 100 μg ml−1.
Construction of plasmids and mutant strains.
All plasmids and primers used in this study are listed in Tables S3 and S4, respectively. The majority of GAS strains in this study were derived from the serotype M49 strain NZ131 (44). Plasmids for GAS allelic replacement were constructed as previously reported (17). Briefly, fragments encompassing the upstream and downstream regions flanking the gene of interest were amplified from NZ131 genomic DNA by PCR and cloned into the pFED760 temperature-sensitive plasmid (45) by the use of restriction enzymes. When necessary, antibiotic resistance cassettes for kanamycin (aph3A), amplified from pOskar (46), or chloramphenicol (cat), amplified from pEVP3 (47), were cloned between the upstream and downstream fragments, to generate plasmids for selective allelic replacement. All deletion vectors were electroporated into competent GAS cells, and a two-step temperature-dependent selection process was used to isolate the desired mutants (48). This process was repeated to generate double mutants. Complementation plasmids were constructed by cloning the gene of interest and its putative promoter region into the respective backbone plasmids.
Synthetic pheromone peptides.
Synthetic peptides of ∼95% purity were purchased from Neo-Peptide (Cambridge, MA), reconstituted as 1 mM stocks in dimethyl sulfoxide (DMSO), and stored at −80°C. Dilutions for working stocks were made in DMSO and stored at −20°C. The SHP3-C8 peptide has the sequence DIIIIVGG, whereas the reverse peptide (rev-SHP) has the sequence GGVIIIID.
Biofilm formation assays.
GAS strains were grown overnight in THY at 30°C, as previously reported (17). In the morning, the strains were diluted 1:25 into fresh CDM and 500 μl was dispensed in duplicate into cell culture-treated 24-well polystyrene plates. Synthetic pheromones (25 nM) were added to each well, and plates were incubated statically at 37°C with 5% CO2 for 20 h. The medium was aspirated, the wells were washed once with 300 μl of 0.9% NaCl, and the biofilms were dry fixed at 37°C for at least 6 h. Biofilms were stained with a 0.2% crystal violet solution, washed three times with 0.9% NaCl in 10% ethanol, and quantified by measurement of the absorbance at 595 nm by area scan of the wells in a Synergy 2 BioTek plate reader. A minimum of three experimental replicates was performed for each strain and condition.
Luciferase reporter assays.
Luciferase reporter assays were conducted as previously reported (17). Briefly, starter cultures were diluted into fresh CDM and grown at 37°C. Every 30 min, 80-μl samples were taken from each culture, transferred to a white-bottom 96-well plate (Greiner), and exposed to decyl aldehyde (Sigma) fumes, and luminescence counts per second were measured using a Turner BioSystems microplate luminometer. The OD600 of the culture was measured at each time point using a Spectronic 20D spectrophotometer (Thermo), and relative light units (RLU) were calculated by normalizing the counts per second to the OD600. For the analysis of peptide pheromone signaling, 25 nM synthetic peptides were added at the beginning of growth.
Lysozyme challenge assays.
Lysozyme challenge assays were carried out as previously reported (19). Briefly, cultures were grown to an OD of 0.1 in CDM, after which SHP3-C8 pheromone or the reverse control peptide (rev-SHP) was added to the culture and the cells were incubated for 1 h. Lysozyme from chicken egg white (Sigma) was added to the cells, which were incubated at 37°C, and every hour aliquots were taken, serially diluted, and plated on THY plates. Plates were evaluated for the number of CFU.
Bioinformatic analysis.
For the comparison of the DNA sequences of S. pyogenes NZ131 and S. canis FSL-Z3, alignments were generated to assess DNA identity (by use of the BLASTn program [https://blast.ncbi.nlm.nih.gov/Blast]) and protein similarity (by use of EMBOSS matcher [https://www.ebi.ac.uk/Tools/psa/emboss_matcher/]). Stop codon analysis and putative ORFs from the IGR of NZ131 were predicted using the Clustal Omega program and the ORF prediction tools from CLC Sequence Viewer software.
Recombinant protein purification.
Recombinant StcA and Isp2 were purified in the same manner as previously described in our paper on the PepO peptidase (20). In brief, stcA lacking the secretion signal (stcA Δsec) was cloned into pET15b (pJJ286). isp2 Δsec was cloned into pET21 (pJJ285). Constructs were verified by sequencing and transformed into BL21(DE3) cells for protein expression. E. coli cells containing the indicated plasmids were grown with shaking to an OD of ∼0.5, treated with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and grown at 28°C for an additional 5 h. Cells were pelleted by centrifugation and lysed in buffer A (0.02 M phosphate, 0.0054 M potassium chloride, 0.274 M sodium chloride, 20 mM imidazole, 0.07% β-mercaptoethanol) with the addition of 1× EDTA-free protease inhibitor cocktail (Pierce) via sonication. The resulting lysate was applied to a nickel resin column and washed with buffer A to remove unbound proteins. Following the washes, rIsp2 was eluted in a stepwise fashion using increasing concentrations of buffer B (buffer A, 500 mM imidazole), primarily eluting between 116 mM and 308 mM imidazole. Fraction purity was assessed via SDS-PAGE, and samples were pooled. As rStcA lacks any aromatic amino acids, elution detection via UV-visible light was not possible. Therefore, rStcA was subjected to a linear gradient of buffer B ranging from 20 mM imidazole to 500 mM imidazole. All fractions were assessed via SDS-PAGE, with the majority of StcA eluting at 500 mM. After the samples were concentrated and dialyzed, the concentration of rIsp2 was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher), and the concentration of rStcA was determined by measuring the absorbance of the protein at 205 nm using a previously described protocol (42). The extinction coefficient for StcA at 205 nm (327,849 M−1 cm−1) was calculated using an online web tool developed in the described protocol.
Purification of cell wall preparations from S. pyogenes.
Overnight cultures of wild-type GAS strain NZ131 were inoculated 1:100 into 400 ml THY and grown overnight at 37°C. Cells were pelleted (12,000 × g, 10 min) and resuspended in a solution of 0.25% SDS in 100 ml 0.1 M Tris-HCl, pH 6.8, boiled for 45 min, and repelleted (12,000 × g, 10 min). The pellet was washed four times with 100 mM Tris-HCl, pH 6.8, and resuspended in 30 ml suspension buffer (0.1 M Tris, pH 7.5, 0.02 M MgSO4, 200 μg/ml RNase A, 100 μg/ml DNase I). This suspension was mechanically disrupted at high pressure (15,000 lb/in2) using an EmulsiFlex-C5 homogenizer (Avestin) and then incubated for 2 h with shaking (37°C, 250 rpm). Finally, trypsin (final concentration, 1 mg ml−1) and CaCl2 (final concentration, 10 mM) were added to the suspension and the suspension was incubated overnight with shaking (37°C, 250 rpm). The suspension was pelleted (12,000 × g, 10 min) and dried on a heat block before being stored at −20°C. Using methods previously described for removal of lipoteichoic acid (30), 100 mg of the sample was subjected to incubation with 40% hydrofluoric acid (96 h, 4°C) to facilitate removal of the lipoteichoic acid component. After completion of the incubation period, saturated NaHCO3 was added to neutralize the hydrofluoric acid and the resulting pellet was washed two times in deionized H2O (dH2O).
FITC labeling and digestion of GAS cell wall preparations.
An NZ131 cell wall preparation (6.5 mg) was suspended in 500 μl water, and in another tube, 3 mg FITC isomer I was added to 500 μl 1 M carbonate buffer, pH 9.3. The two solutions were mixed, covered in foil, and shaken at 37°C for 4 h. After incubation, the reaction mixture was washed two times with each of the following solutions with pelleting between each wash (12,000 × g, 5 min): carbonate buffer, pH 9.3, deionized H2O, acetone, and ethanol. After the final wash, the pellet was dried and stored in the dark at 4°C. To observe digestion of the FITC-labeled peptidoglycan, the sample was resuspended at 100 μg ml−1 in PBS, added to a 96-well half-area assay plate (Costar), and read on a plate reader (BioTek Synergy 2) at 37°C at an excitation wavelength of 485 nm and an emission wavelength of 528 nm, reading kinetically every 1 min for an hour, with 10 s of medium shaking before each read. The indicated amounts of lysozyme, rIsp2, and recombinant Spy49_0414c (rSpy49_0414c) were added at the start of each experiment.
Remazol brilliant blue labeling and sedimentation of cell wall preparations.
The cell wall preparations were incubated with 20 mM Remazol brilliant blue (Sigma) in 0.25 M NaOH overnight at 37°C. After neutralization of the reaction with HCl, the preparations were washed with dH2O and pelleted until the supernatant was clear. The pellet was stored in PBS at 4°C. Sedimentation assays were performed by adding 100 μl of 5 mg ml−1 purified cell wall to a 500-μl microcentrifuge tube and placing the tube on a plate shaker at 25 rpm. Recombinant StcA protein was added at a concentration of 10 nM to the solution, and after 20 min, the OD600 of the top 50 μl of supernatant was read on a spectrophotometer (SmartSpec 3000; Bio-Rad) in low-volume cuvettes (trUView; Bio-Rad). Assays under each condition were repeated in triplicate.
Immunofluorescence microscopy.
Wild-type and ΔstcA cells were grown in CDM to an OD of 0.1 before being induced with 100 nM SHP-C8 for 1 h. Cells were then spun down and resuspended in PBS to an OD of 2.2. A 100-μl drop of this suspension was placed on a pretreated poly-l-lysine coverslip and allowed to sit for 10 min before washing with dH2O and fixing with 4% paraformaldehyde. A polyclonal anti-StcA antibody raised in a rabbit (Abclonal) was incubated with paraformaldehyde-fixed ΔstcA bacteria in a poly-l-lysine-treated 24-well plate prior to being added to the coverslips in order to adsorb nonspecific polyclonal antibodies. The primary anti-StcA antibody was diluted to 1:100 in phosphate buffered saline plus 1% BSA protein (PBSP) and incubated for 30 min at room temperature on the coverslips. The slips were then washed five times in dH2O before addition of the Alexa Fluor 488-coupled anti-rabbit goat immunoglobulin secondary antibody (Abcam) at a dilution of 1:500 in 5% goat serum in PBS. After incubation for 30 min in the dark, the coverslips were washed and mounted on glass slides using Vectashield with DAPI (4′,6-diamidino-2-phenylindole) hard mounting medium (Vector Laboratories). Imaging utilized a Zeiss LSM 710 confocal laser scanning microscope with a 63× objective, and images were amplified 2 times for an ultimate ×126 zoom. The green wavelength used was 488 nm, and the DAPI wavelength was 405 nm.
Cytochrome c binding assays.
Using the protocol outlined by Kristian et al. (23), we performed the cytochrome c binding assay as follows: S. pyogenes strains were inoculated at a 1:20 dilution from 1-ml THY overnight cultures into 10 ml CDM and grown to an OD600 of 0.1, and then 50 nM synthetic SHP3-C8 or SHP3-C8rev was added to the medium and the cells were allowed to continue growing to an OD600 of 0.3. The culture was spun down and resuspended to a final OD600 of 7 in MOPS (morpholinepropanesulfonic acid) buffer containing 1 mg ml−1 cytochrome c. This mixture was incubated at room temperature for 10 min and pelleted by centrifugation (12,000 × g, 5 min). The cytochrome c remaining in the supernatants was quantified by measuring the A535. The assay was performed in biological duplicate and technical triplicate for each strain and analyzed for significance by the two-tailed t test.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UIC Research Resources Center for use of the Zeiss microscope and EmulsiFlex-C5 homogenizer. We also thank Bobbi Xayarath for guidance with preparation of planktonic bacterial cells for immunofluorescence microscopy.
This work was supported by grants from the NIH (AI091779) and the Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Disease.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00701-17.
REFERENCES
- 1.Mok KC, Wingreen NS, Bassler BL. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J 22:870–881. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarwood JM, Schlievert PM. 2003. Quorum sensing in Staphylococcus infections. J Clin Invest 112:1620–1625. doi: 10.1172/JCI200320442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 13:470–511. doi: 10.1128/CMR.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterlund A, Popa R, Nikkila T, Scheynius A, Engstrand L. 1997. Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope 107:640–647. doi: 10.1097/00005537-199705000-00016. [DOI] [PubMed] [Google Scholar]
- 5.Neeman R, Keller N, Barzilai A, Korenman Z, Sela S. 1998. Prevalence of internalisation-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet 352:1974–1977. doi: 10.1016/S0140-6736(97)12452-7. [DOI] [PubMed] [Google Scholar]
- 6.Conley J, Olson ME, Cook LS, Ceri H, Phan V, Davies HD. 2003. Biofilm formation by group A streptococci: is there a relationship with treatment failure? J Clin Microbiol 41:4043–4048. doi: 10.1128/JCM.41.9.4043-4048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez CJ, Mende K, Beckius ML, Akers KS, Romano DR, Wenke JC, Murray CK. 2013. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis 13:47. doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts AL, Connolly KL, Kirse DJ, Evans AK, Poehling KA, Peters TR, Reid SD. 2012. Detection of group A Streptococcus in tonsils from pediatric patients reveals high rate of asymptomatic streptococcal carriage. BMC Pediatr 12:3. doi: 10.1186/1471-2431-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan EL, Top FH, Dudding BA, Wannamaker LW. 1971. Differentiation of active pharyngitis: diagnosis of streptococcal child infection from the carrier state in the symptomatic. J Infect Dis 123:490–501. doi: 10.1093/infdis/123.5.490. [DOI] [PubMed] [Google Scholar]
- 10.Wollein Waldetoft K, Råberg L. 2014. To harm or not to harm? On the evolution and expression of virulence in group A streptococci. Trends Microbiol 22:7–13. doi: 10.1016/j.tim.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Passàli D, Lauriello M, Passàli GC, Passàli FM, Bellussi L. 2007. Group A streptococcus and its antibiotic resistance. Acta Otorhinolaryngol Ital 27:27–32. [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen A, Valdórsson O, Frimodt-Møller N, Hollingshead S, Kilian M. 2015. Commensal streptococci serve as a reservoir for β-lactam resistance genes in Streptococcus pneumoniae. Antimicrob Agents Chemother 59:3529–3540. doi: 10.1128/AAC.00429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischetti VA, Dale JB. 2016. One more disguise in the stealth behavior of Streptococcus pyogenes. mBio 7:e00661-16. doi: 10.1128/mBio.00661-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynskey NN, Lawrenson RA, Sriskandan S. 2011. New understandings in Streptococcus pyogenes. Curr Opin Infect Dis 24:196–202. doi: 10.1097/QCO.0b013e3283458f7e. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez JC, Federle MJ. 2014. Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol 4:127. doi: 10.3389/fcimb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Do H, Makthal N, VanderWal AR, Rettel M, Savitski MM, Peschek N, Papenfort K, Olsen RJ, Musser JM, Kumaraswami M. 2017. Leaderless secreted peptide signaling molecule alters global gene expression and increases virulence of a human bacterial pathogen. Proc Natl Acad Sci U S A 114:e8498–e8507. doi: 10.1073/pnas.1705972114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog 7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasarre B, Aggarwal C, Federle MJ. 2013. Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes. mBio 3:e00333-12. doi: 10.1128/mBio.00333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang JC, Jimenez JC, Federle MJ. 2015. Induction of a quorum sensing pathway by environmental signals enhances group A streptococcal resistance to lysozyme. Mol Microbiol 97:1097–1113. doi: 10.1111/mmi.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkening RV, Chang JC, Federle MJ. 2016. PepO, a CovRS-controlled endopeptidase, disrupts Streptococcus pyogenes quorum sensing. Mol Microbiol 99:71–87. doi: 10.1111/mmi.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaSarre B, Chang JC, Federle MJ. 2013. Redundant group A streptococcus signaling peptides exhibit unique activation potentials. J Bacteriol 195:4310–4318. doi: 10.1128/JB.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis KM, Weiser JN. 2011. Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect Immun 79:562–570. doi: 10.1128/IAI.00651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. 2005. d-Alanylation of teichoic acids promotes group A streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol 187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozlowski LP. 2017. Proteome-pI: proteome isoelectric point database. Nucleic Acids Res 45:D1112–D1116. doi: 10.1093/nar/gkw978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faucher S, Dimock K, Wright KE. 2002. Characterization of the Cyno-EBV LMP1 homologue and comparison with LMP1s of EBV and other EBV-like viruses. Virus Res 90:63–75. doi: 10.1016/S0168-1702(02)00144-2. [DOI] [PubMed] [Google Scholar]
- 26.Meloni A, Incani F, Corda D, Cao A, Rosatelli MC. 2008. Role of PHD fingers and COOH-terminal 30 amino acids in AIRE transactivation activity. Mol Immunol 45:805–809. doi: 10.1016/j.molimm.2007.06.156. [DOI] [PubMed] [Google Scholar]
- 27.Qiu Y, Kung H-J. 2000. Signaling network of the Btk family kinases. Oncogene 19:5651–5661. doi: 10.1038/sj.onc.1203958. [DOI] [PubMed] [Google Scholar]
- 28.Rzychon M, Sabat A, Kosowska K, Potempa J, Dubin A. 2003. Staphostatins: an expanding new group of proteinase inhibitors with a unique specificity for the regulation of staphopains, Staphylococcus spp. cysteine proteinases. Mol Microbiol 49:1051–1066. doi: 10.1046/j.1365-2958.2003.03613.x. [DOI] [PubMed] [Google Scholar]
- 29.Maeda H. 1980. A new lysozyme assay based on fluorescence polarization or fluorescence intensity utilizing a fluorescent peptidoglycan substrate. J Biochem 88:1185–1191. doi: 10.1093/oxfordjournals.jbchem.a133073. [DOI] [PubMed] [Google Scholar]
- 30.Smit E, Pouwels PH. 2002. One repeat of the cell wall binding domain is sufficient for anchoring the Lactobacillus acidophilus surface layer protein. J Bacteriol 184:4617–4619. doi: 10.1128/JB.184.16.4617-4619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greene NG, Narciso AR, Filipe SR, Camilli A. 2015. Peptidoglycan branched stem peptides contribute to Streptococcus pneumoniae virulence by inhibiting pneumolysin release. PLoS Pathog 11:e1004996. doi: 10.1371/journal.ppat.1004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pancholi V, Boël G, Jin H. 2010. Streptococcus pyogenes Ser/Thr kinase-regulated cell wall hydrolase is a cell division plane-recognizing and chain-forming virulence factor. J Biol Chem 285:30861–30874. doi: 10.1074/jbc.M110.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bessen D, Fischetti VA. 1988. Passive acquired mucosal immunity to group A streptococci by secretory immunoglobulin A. J Exp Med 167:1945–1950. doi: 10.1084/jem.167.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kay BK, Williamson MP, Sudol M. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14:231–241. doi: 10.1096/fasebj.14.2.231. [DOI] [PubMed] [Google Scholar]
- 35.Xayarath B, Alonzo F, Freitag NE. 2015. Identification of a peptide-pheromone that enhances Listeria monocytogenes escape from host cell vacuoles. PLoS Pathog 11:e1004707. doi: 10.1371/journal.ppat.1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fagan RP, Fairweather NF. 2014. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol 12:211–222. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- 37.Riedel K, Ohnesorg T, Krogfelt KA, Hansen TS, Omori K, Givskov M, Eberl L. 2001. N-Acyl-l-homoserine lactone-mediated regulation of the lip secretion system in Serratia liquefaciens MG1. J Bacteriol 183:1805–1809. doi: 10.1128/JB.183.5.1805-1809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones MB, Peterson SN, Benn R, Braisted JC, Jarrahi B, Shatzkes K, Ren D, Wood TK, Blaser MJ. 2010. Role of luxS in Bacillus anthracis growth and virulence factor expression. Virulence 1:72–83. doi: 10.4161/viru.1.2.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleytr UB, Schuster B, Egelseer E-MM, Pum D. 2014. S-layers: principles and applications. FEMS Microbiol Rev 38:823–864. doi: 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selle K, Goh YJ, Johnson BR, O'Flaherty S, Andersen JM, Barrangou R, Klaenhammer TR. 2017. Deletion of lipoteichoic acid synthase impacts expression of genes encoding cell surface proteins in Lactobacillus acidophilus. Front Microbiol 8:553. doi: 10.3389/fmicb.2017.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hynönen U, Palva A. 2013. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biotechnol 97:5225–5243. doi: 10.1007/s00253-013-4962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anthis NJ, Clore GM. 2013. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci 22:851–858. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rato MG, Nerlich A, Bergmann R, Bexiga R, Nunes SF, Vilela CL, Santos-Sanches I, Chhatwal GS. 2011. Virulence gene pool detected in bovine group C Streptococcus dysgalactiae subsp. dysgalactiae isolates by use of a group A S. pyogenes virulence microarray. J Clin Microbiol 49:2470–2479. doi: 10.1128/JCM.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, Jia H, Kenton S, Najar F, Wu H, Scott J, Roe BA, Savic DJ. 2008. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J Bacteriol 190:7773–7785. doi: 10.1128/JB.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Breton Y, Mistry P, Valdes KM, Quigley J, Kumar N, Tettelin H, McIver KS. 2013. Genome-wide identification of genes required for fitness of group A streptococcus in human blood. Infect Immun 81:862–875. doi: 10.1128/IAI.00837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claverys JP, Dintilhac A, Pestova EV, Martin B, Morrison DA. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123–128. doi: 10.1016/0378-1119(95)00485-O. [DOI] [PubMed] [Google Scholar]
- 48.Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect Immun 68:2441–2448. doi: 10.1128/IAI.68.5.2441-2448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van De Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.