Abstract
Depending on the length of their carbon backbone and their saturation status, natural fatty acids have rather distinct biological effects. Thus, longevity of model organisms is increased by extra supply of the most abundant natural cis-unsaturated fatty acid, oleic acid, but not by that of the most abundant saturated fatty acid, palmitic acid. Here, we systematically compared the capacity of different saturated, cis-unsaturated and alien (industrial or ruminant) trans-unsaturated fatty acids to provoke cellular stress in vitro, on cultured human cells expressing a battery of distinct biosensors that detect signs of autophagy, Golgi stress and the unfolded protein response. In contrast to cis-unsaturated fatty acids, trans-unsaturated fatty acids failed to stimulate signs of autophagy including the formation of GFP-LC3B-positive puncta, production of phosphatidylinositol-3-phosphate, and activation of the transcription factor TFEB. When combined effects were assessed, several trans-unsaturated fatty acids including elaidic acid (the trans-isomer of oleate), linoelaidic acid, trans-vaccenic acid and palmitelaidic acid, were highly efficient in suppressing autophagy and endoplasmic reticulum stress induced by palmitic, but not by oleic acid. Elaidic acid also inhibited autophagy induction by palmitic acid in vivo, in mouse livers and hearts. We conclude that the well-established, though mechanistically enigmatic toxicity of trans-unsaturated fatty acids may reside in their capacity to abolish cytoprotective stress responses induced by saturated fatty acids.
Keywords: Fasting, Ketogenic diet, Immune response, Immunosurveillance, Obesity, Aging, Cytoprotection, Systems biology
Graphical Abstract

Highlights
-
•
The molecular geometry of fatty acids dictates their biological function
-
•
Saturated fats induce autophagy through a BECN1-dependent pathway
-
•
Cis fats trigger cytoprotection via the induction of non-canonical autophagy, trans fats are toxic as they inhibit the stress response induced by saturated fats
Cellular responses to fatty acids are distinct according to their unsaturation status and carbon backbone; saturated & cis fats can induce various cellular stresses, including autophagy (through a BECN1-dependent pathway for the first category only), whereas trans fats are toxic by specifically inhibiting saturated fats-elicited stress responses.
1. Introduction
Autophagy is a phylogenetically conserved cellular stress response in which portions of the cytoplasm are sequestered in two-membraned autophagosomes that fuse with lysosomes for bulk degradation of their luminal content (Morel et al., 2017; Bento et al., 2016). Autophagy is induced by various stimuli including nutrient shortage (that triggers the digestion of macromolecules to generate energy), adaptation to changing environmental conditions (that may induce the destruction of portions of the cell to rebuild new organelles), as well as sublethal damage (that requires the recycling of dysfunctional organelles and repair responses to increase cellular fitness) (Kaur and Debnath, 2015). As a result, periodic or chronic stimulation of autophagy acts as a cytoplasmic rejuvenation mechanism that increases the longevity of model organisms including yeast, nematodes, flies and mice (Melendez et al., 2003; Tavernarakis et al., 2008; Rubinsztein et al., 2011; Pyo et al., 2013; Green and Levine, 2014; Eisenberg et al., 2016; Ho et al., 2017). Several clinically relevant syndromes leading to premature neurodegeneration and general aging are linked to autophagy defects, (Menzies et al., 2015; Lopez-Otin et al., 2016) suggesting that autophagy may play a general health-improving role in humans as well.
Recently, the impact of fatty acids (FAs) on autophagy regulation has attracted some attention. The most abundant natural FAs present in the human body and food are the saturated FA palmitic acid (PA) and the mono-cis-unsaturated oleic acid (OL). Strikingly, these two FAs have rather distinct cellular effects in the sense that PA causes the activation of the diabetogenic stress kinase Jun N-terminal kinase-1 (JNK1), while OL rather inhibits PA-induced JNK1 activation (Holzer et al., 2011). Moreover, PA stimulates a canonical pathway of autophagy that, downstream of JNK1, depends on the enzymatic activity of the Beclin 1 (BCLN1)/phosphatidylinositol 3-kinase, catalytic subunit type 3 (PIK3C3) complex generating phosphatidylinositol (3)-phosphate (PI3P), while OL elicits the JNK1 and BCLN1/PIK3C3-independent association of the autophagosome-linked microtubule-associated protein 1A/1B light chain 3 (MAP1LC3, best known as LC3) with the Golgi apparatus (Niso-Santano et al., 2015b; Bankaitis, 2015; Niso-Santano et al., 2015a). This difference in pro-autophagic signals elicited by PA and OL is evolutionarily conserved in Saccharomyces cerevisiae, Caenorhabditis elegans, mice and cultured human cells (Niso-Santano et al., 2015b; Enot et al., 2015). Moreover, it extends to other saturated FAs (that behave like PA) and mono- or poly-cis-unsaturated FAs (that behave like OL) with respect to JNK1 and PIK3C3 activation (Niso-Santano et al., 2015b).
Unsaturated FAs that are contained in vegetables and human tissues are cis-isomers, meaning that the unsaturation causes a kink in the FA chain (Duplus et al., 2000). Trans-unsaturated FAs, which lack such a kink, meaning that their physicochemical properties are rather different, are generated during industrial food processing, for example during partial hydrogenation of fats resulting in the isomerization of cis-unsaturated FAs into their trans-unsaturated isomers (Tzeng and Hu, 2014; Kadhum and Shamma, 2017). Thus, elaidic acid (EL), the most abundant trans-monoene contained in hydrogenated vegetable oil, results from the isomerization of OL; palmitelaidic acid, another trans-monoene, from the isomerization of its natural precursor palmitoleic acid; and linoelaidic acid, the most abundant trans-polyunsaturated FA, from the isomerization of linoleic acid (Tzeng and Hu, 2014, Kadhum and Shamma, 2017). Another source of trans-unsaturated FAs are the milk and body fat of ruminants (such as cattle and sheep) that contains up to 8% of trans fats (Craig-Schmidt, 2006). Trans-vaccenic acid (VA) is the predominant trans-monoene formed during rumination, as a result of bacterial fermentation processes (Lock and Bauman, 2004).
Regardless of their origin, trans-unsaturated FAs are considered to have negative effects on human health, especially at the cardiovascular level (Brostow et al., 2012). Thus, the nutritional uptake or the plasma concentration of trans-unsaturated FAs are positively correlated with the severity of coronary arteriosclerosis (Hadj Ahmed et al., 2018), inflammatory biomarkers (Mazidi et al., 2017), as well as shortening of telomers in circulating leukocytes, which is a proxy of accelerated aging (Mazidi et al., 2018). Accordingly, trans-unsaturated FAs precipitate cardiovascular disease in animal models (Monguchi et al., 2017). Moreover, FAs may favor signs of inflammation in tissues (Oteng et al., 2017). Based on major epidemiological studies, (Oomen et al., 2001; Mozaffarian et al., 2006) the US Federal Drug Administration has recommended banning all sources of trans FAs from human consumption (Brownell and Pomeranz, 2014).
Free FAs including saturated FAs (and to a lesser degree cis-unsaturated FAs) are well known for their potential toxicity, a phenomenon that may contribute to the pathogenesis of lipotoxic diseases (Unger, 2002) as well as to the induction of chronic inflammation contributing to metabolic syndrome (Ralston et al., 2017). Free FAs including PA can induce endoplasmic reticulum (ER) stress or even trigger mitochondrial or lysosomal permeabilization, leading to subsequent caspase activation and apoptosis (Li et al., 2008; Malhi and Kaufman, 2011; Szeto et al., 2016). However, less information is available on the specific toxicity of trans-unsaturated FAs.
In spite of the recognized chronic toxicity of trans fats, the mechanisms explaining why they provoke arteriosclerosis are largely elusive. Autophagy has recently been recognized as a major mechanism to counteract cardiovascular aging and to diminish arteriosclerosis (Torisu et al., 2016; Ho et al., 2017). Based on this consideration, we decided to systematically investigate the capacity of saturated, cis-unsaturated and trans-unsaturated FAs to modulate autophagy and other cellular stress responses in vitro, in cultured human cells. Our results reveal the unexpected finding that trans-unsaturated FAs can inhibit autophagy induction by saturated FAs.
2. Materials and Methods
2.1. Cell Culture and Chemicals
Culture media and supplements for cell culture were purchased from Gibco-Life Technologies (Carlsbad, CA, USA) and plasticware from Greiner Bio-One (Monroe, CA, USA). All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin G sodium and 100 μg/ml streptomycin sulfate at 37 °C under 5% CO2. All FAs were purchased from Larodan (Malmö, Sweden); golgicide A, rapamycin, thapsgargin and tunicamycin were obtained from Sigma-Aldrich (St. Louis, MO, USA). Each fatty acid was dissolved at a concentration of 100 mM in an appropriate volume of 100% ethanol pre-warmed at 37 °C. The obtained solution was then used to treat the cells, with final concentrations ranging from 125 to 1000 μM. The SCREEN-WELL® Autophagy library (BML-2837) was purchased from Enzo Life Sciences (Farmingdale, NY, USA).
2.2. Yeast Clonogenic Survival Assay
All yeast experiments were carried out in the BY4741 (MATa his3∆1 leu2∆0 met15∆0 ura3∆0) wild type strain background. Yeast cells were grown in SC medium containing 0.17% yeast nitrogen base (Difco), 0.5% (NH4)2SO4 and 30 mg/l of all amino acids (except 80 mg/l histidine and 200 mg/l leucine), 30 mg/l adenine, and 320 mg/l uracil and 2% glucose. All yeast cultures were inoculated from a stationary overnight culture to an OD600 of 0.1 and then grown at 28 °C and 145 rpm shaking for the indicated time. Chronological lifespan was assessed as described before (Buttner et al., 2007). In brief: cultures were inoculated from fresh overnight cultures to an OD600 of 0.1 with the culture volume being 10% of flask volume. Aliquots were taken to detect clonogenic survival at the indicated time points (starting from day 2, where aging starts) with 500 cells being plated on YPD agar. FAs (purchased as sodium salts from Sigma-Aldrich) were added to growth media from 1% stock solutions in ddH2O with 10% tergitol shortly before inoculation to a final concentration of 1.8 mM.
2.3. Fly Kaplan-Meier survival assay
Unless otherwise specified standard laboratory breeding was carried out at 25 °C, 65–70% humidity and a 12:12 h light/dark cycle as described previously (Sigrist et al., 2003). Standard fly food was prepared according to the “Sigrist” recipe as a semi-defined cornmeal-molasses medium with slight modifications (0.9 l H2O, 4.2 g agar-agar, 12.5 ml molasses, 85 g malt extract, 8.3 g soy, 66.7 g cornmeal, 1.33 g p-hydroxy-benzoic acid methyl ester dissolved in ethanol, 5.25 ml propionic acid). All experiments were performed in D. melanogaster isow1118 background (Hazelrigg et al., 1984). Parental flies were transferred to new vials every third day and only 1–3 days old progenitor flies from the F1 generation were used for experiments. The flies were anesthetized on a porous pad by CO2 application for 8 min as a maximum, separated into males and females and 20–40 flies each were subsequently transferred to a fresh vial. For assessment of fly-lifespans the flies were sex-separated and transferred to small vials (20 flies per vial) containing either standard corn food or fly food supplemented with 0.05% FA. At least 120 flies per sex and per genotype were analyzed to determine lifespans and fresh food was supplied every other day. Concomitantly dead individuals were counted until no more flies were alive. Kaplan-Meier survival was analyzed by GraphPad Prism 5 software applying a Log-rank test.
2.4. High-Content Screening Microscopy
U2OS or MEF cells stably expressing GFP-LC3, GFP-TFEB, GALT1-GFP, GFP-ATF4, GFP-XBP1, and FYVE-RFP were seeded in 384-well black microplates for 24 h. After treatment, cells were fixed with 4% paraformaldehyde (PFA, w/v in PBS) for 20 min at room temperature and stained with 10 μg/ml Hoechst 33342 in PBS. Image acquisition was performed using an ImageXpress Micro XL automated microscope (Molecular Devices, Sunnyvale, CA, USA). A minimum of 4 viewfields were captured per well. Upon acquisition, images were analyzed using the Custom Module Editor functionality of the MetaXpress software (Molecular Devices). Briefly, cells were segmented and divided into nuclear and cytoplasmic regions based on the Hoechst staining and GFP or RFP cytoplasmic signals. GFP-LC3 and FYVE-RFP dots were detected using an automated threshold, and their number and surface were measured in the cytoplasmic compartment. GFP-TFEB, ATF4-GFP, XBP1-Venus, and GALT1-GFP intensities were systematically measured in both compartments. Data processing and statistical analyses were performed using the R software (http://www.r-project.org/).
2.5. Immunofluorescence
GFP-LC3 stable expressing U2OS cells were seeded in 384-well microplates for 24 h. After experimental treatments, cells were fixed with 4% paraformaldehyde for 20 min at room temperature and permeabilized with 0.1% Triton X-100 (v:v in PBS) for 10 min on ice. Thereafter, cells were maintained in 5% bovine serum albumin (BSA, w/v in PBS) for 1 h to block non-specific binding, followed by overnight incubation at 4 °C with phosphoneoepitope-specific eIF2α antibody (ab32157, Abcam, Cambridge, UK). After several washing steps with PBS, cells were incubated in AlexaFluor™ conjugates (Life Technologies) against the primary antibody for 2 h at room temperature. Nuclear staining was achieved by incubation with 10 μg/ml Hoechst 33342 in PBS. Images were acquired and analyzed as described before.
2.6. Immunoblotting
After treatment, cells were collected and lysed in RIPA lysis and extraction buffer (ThermoFisher, Carlsbad, CA, USA) supplemented with Pierce protease and phosphatase inhibitor mini tablet (ThermoFisher) on ice for 40 min. After centrifugation at 12,000 g for 15 min, supernatants were heated in sample buffer (ThermoFisher) at 100 °C for 10 min. Protein samples were separated on pre-cast 4–12% polyacrylamide NuPAGE Bis-Tris gels (Life Technologies) and electro-transferred to PVDF membranes (Millipore Corporation, Billerica, MA, USA). Membranes were probed overnight at 4 °C with primary antibodies specific for LC3 (#2775, Cell Signaling Technology), p62 (ab56416, Abcam), Atg5 (A2859, Sigma-Aldrich), GAPDH (ab8254, Abcam), XBP1s (BLE619502, Biolegend, San Diego, CA, USA), P-p38 (#9211, Cell Signaling Technology), p38 (#9212, Cell Signaling Technology), followed by incubation with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (Southern Biotech, Birmingham, AL, USA). Immunoreactive bands were visualized with ECL prime western blotting detection reagent (Sigma-Aldrich) by means of an ImageQuant LAS4000 (GE Healthcare, Little Chalfont, UK).
2.7. Molecular Descriptors Calculation
For each FA, 319 descriptors were calculated using the Chemistry Development Kit implemented in the R rcdk package (available on CRAN). The obtained data set was refined by removing irrelevant descriptors (discarding redundant parameters and those with a median absolute deviation lower than 10−4), resulting in 25 discriminatory descriptors.
2.8. In vivo Experiments
The protocols described below have been approved by the Institutional Animal Care and Use Committee of the Rutgers New Jersey Medical School. All mice were maintained in a temperature-controlled and pathogen free environment with 12 h light/dark cycles, with free access to food and water. C57BL/6 mice were obtained from RIKEN BioResource Center (Ibaraki, Japan). Six-week-old female wild type C57BL/6 mice were intraperitoneally injected with 100 mg/kg palmitic acid (PA)(n = 2), 100 mg/kg elaidic acid (EL)(n = 2), or the combination of the two FAs (n = 2) in 10% bovine serum albumin (BSA, w:v in PBS) respectively. An equivalent volume of vehicle was injected as a control (n = 2). Two hours later, the mice were sacrificed and organs were collected and processed for immunoblotting. Alternatively, 8-week-old male cardiac heterozygous GFP-LC3 transgenic (Tg-GFP-LC3) mice, containing a rat LC3-EGFP fusion under the control of the chicken β-actin promoter (Maejima et al., 2013), were treated as described above for immunoblotting, with 3 mice per group. Hearts were thereafter collected and processed for the detection of GFP-LC3 (Maejima et al., 2013).
2.9. Histological Analysis
Tissue samples from GFP-LC3 transgenic mice were embedded in Tissue-Tek OCT compound (Sakura Finetechnical, Tokyo, Japan) and stored at −80 °C. The samples were sectioned at 10 μm thickness with a cryostat (CM 3050 S, Leica, Mannheim, Germany), air-dried for 30 min and fixed with 4% paraformaldehyde. The fluorescence of GFP-LC3 was observed under a fluorescence microscope as previously described (Maejima et al., 2013).
2.10. Widely-targeted Analysis of Intracellular Metabolites gas Chromatography (GC) Coupled to a Triple Quadrupole (QQQ) Mass Spectrometer
Wild-type U2OS cells were plated in 6-well plates and left to adapt at 37 °C for 24 h. After treatment, cells were washed 5 times with DMEM supplemented with 10% FBS, and 3 times with Phosphate-Buffered Saline (PBS). After supernatant removal, 500 μl from a water/methanol (70% water; 30% methanol; v/v) mix was added to the cells for lysis and FAs extraction. Three-hundred and ten microlitres were transferred for injection. Ten microlitres were collected from each biological sample and pooled to create a quality control (QC) sample. Biological and QC samples were evaporated and dried extracts were resuspended in 80 μl of N-Trimethylsilyl-N-methyl trifluoroacetamide (MSTFA) at 40 °C for 30 min. Following samples were injected into a 7890B gas chromatograph (Agilent Technologies, Waldbronn, Germany) coupled to a triple quadrupole 7000C (Agilent Technologies) equipped with a high sensitivity electronic impact source (EI) operating in positive mode. The collision gas used was nitrogen and the scan mode was the MRM for biological samples. Peak detection, integration and quantification of the analytes were performed using the Agilent Mass Hunter quantitative software (B.07.01).
The concentration from each of the 19 FAs was then calculated with the calibration curve using the area under the curve of each metabolite. These concentrations were thereafter corrected by subtracting baseline concentrations determined in blanks. FA fractions were calculated based on the sum of determined concentrations for all 19 FAs in each sample.
3. Results
3.1. Differential Longevity Effects of FAs on Yeast and Flies
When administered to mice, PA and OL elicit differential changes in the circulating metabolome as well as in the metabolomic profile of the liver, heart, and skeletal muscle that we interpreted as an OL-selective increase of anti-aging metabolites (such as nicotinamide adenine dinucleotide and polyamines) (Enot et al., 2015). We therefore tested the effects of long-term feeding of yeast and flies with palmitate and oleate. While OL was able to increase the chronological lifespan of Saccharomyces cerevisiae, PA used at equimolar doses failed to do so (Fig. S1A). Similarly, OL, but not PA, extended the longevity of Drosophila melanogaster (Fig. S1B). Hence, distinct FAs have differential effects on the aging process. Driven by this result, we decided to test 19 distinct FAs that differ in their number of carbon atoms (C14 to C22), their number of saturations (0 to 4) and their configuration (cis or trans) to determine their effects on cultured human cells (Table S1).
3.2. Differential Effects of FAs on Autophagy-linked Pathways
The internalization of different FAs (all used at 500 μM) by U2OS osteosarcoma cells was determined by means of gas chromatography coupled to mass spectrometry (Fig. 1A, B), while the formation of autophagosomes was determined by means of a biosensor cell line expressing a GFP-LC3 fusion protein and quantitated as cytoplasmic GFP-LC3-positive puncta (Fig. 1C, D). The two parameters, lipid uptake and formation of GFP-LC3 puncta did not correlate (Fig. 5B). Importantly, however, the three trans-monounsaturated FAs included in the collection (elaidic acid, linoelaidic acid, trans-vaccenic acid) were unable to induce GFP-LC3B puncta, although entering the cells (Fig. 1A–D). The cis-isomer of EL, OL, as well as the cis-isomer of trans-vaccenic acid, (cis-)vaccenic acid, however, did induce GFP-LC3 dots (Fig. 1C,D). Accordingly, PA and OL, but not EL, stimulated LC3 lipidation (Fig. 1E), in an ATG5-dependent fashion (Fig. 1F).
Fig. 1.
Cellular uptake and autophagy induction by selected FAs. (A, B) Human osteosarcoma U2OS cells were treated with the indicated FAs (saturated FAs in black, trans-unsaturated FAs in blue, cis-unsaturated FAs in red, and unsaturated FAs with both trans- and cis- bonds in green) or were left untreated (A, Control in B) for 6 h. Intracellular FA content was measured by GC/MS after cell lysis, and the fraction of each FA was calculated. Fractions of basal FA levels in untreated conditions are reported as a pie chart in A, as relative concentrations summing up to 100%. Changes in the relative concentrations upon addition of each FA are shown as a bar chart in B, the black part of the columns indicating the increase. (C, D) U2OS cells stably expressing GFP-LC3 were treated with FAs as in A. After fixation the cells were imaged and the area of GFP-LC3+ dots was assessed as an indicator for autophagy. Data are means ± SEM of at least three independent experiments (* = p < 0.05; **=p < 0.01;***=p < 0.001). Representative images are shown in C. Scale bar equals 10 μm. (E) U2OS cells were treated with 500 μM OL, EL, or PA as indicated or were left untreated (Control) in the presence or absence of chloroquine for 6 h. Then, cells were processed to measure LC3 lipidation and p62 degradation by SDS-PAGE and immunoblot. G3PD was measured as a loading control. (F) U2OS WT and ATG5 knockout cells (ATG5KO) were treated with 500 μM OL, EL, or PA for or were left untreated (Control) for 6 h and were then collected for protein separation by SDS-PAGE and immunoblot. LC3 lipidation and p62 degradation was assessed by specific antibodies and GAPDH was measured as a loading control.
Fig. 5.
Systemic analysis of the biological effects of FAs. (A) FAs were hierarchically clustered after Z-score calculation of each measured parameter, then reported in heatmap (GOH, percent of cells with normal Golgi morphology; VIAB, healthy cell count; ACE acetylated protein intensity; GOS, percent of cells showing fragmented Golgi apparatus; GFP-TFEBnuc/cyto intensity ratio; XBP1/ATF4, percent of cells with high venus/GFP intensity; GOV, percent of cells with degraded Golgi apparatus; INT, fractional increase of cellular intake). (B) Correlation matrix was built from Z-scores; the diagonal shows the distribution of Z-scores for each parameter; upper panel shows the Pearson correlation coefficient (*=p < 0.05;**=p < 0.01;***=p < 0.001); lower panel depicts bi-parametric plots together with linear regression (red line). (C) A set of chemical descriptors (MW, molecular weight; RBC, rotable bound count; BSC, bond stereocenter count; NCU, number of cis-unsaturations; other abbreviations are reported in the web page http://sysbiolab.bio.ed.ac.uk/wiki/index.php/CDK_Small_Molecule_Descriptors) was calculated for each FA, and then correlated with the Z-scores of measured biological parameters (doi:10.17632/3d2zvjbh7z.1); Pearson correlation coefficients are depicted on the correlation matrix as dots, with size representing the absolute value, and colour the real value (ranging from −1, red to +1), blue. (D) Coordinates from the two main dimensions of a principal component analysis are plotted with the axes reporting the percentage of inertia. Polygons represent groups in which FAs were classified after K-Means analysis. Numbers depicted above each dot represents the carbon chain length as well as NCU for each corresponding FA.
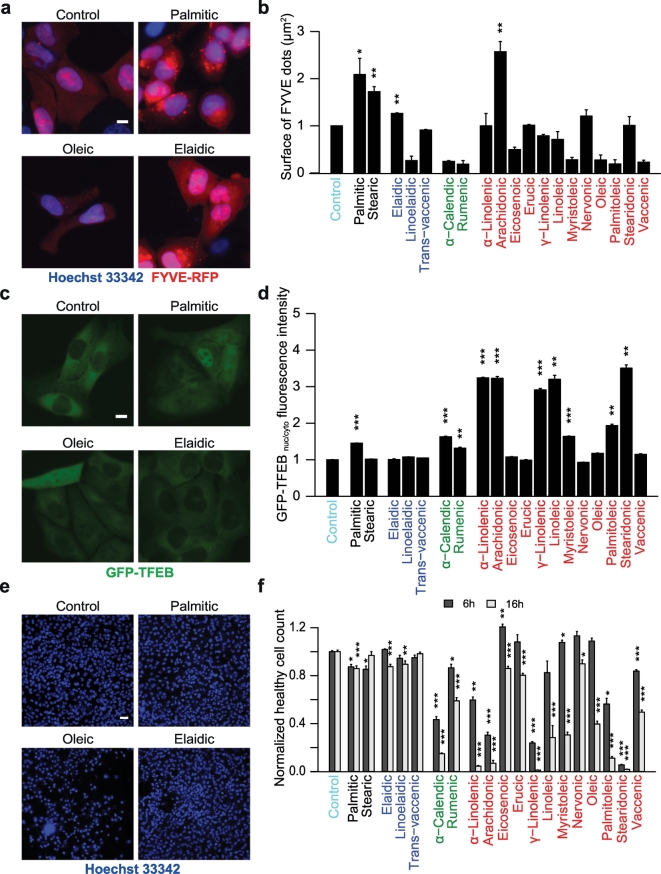
Most of the cis-unsaturated FAs (with the exception of arachidonic acid) failed to induce FYVE dots (which constitute a measurement of phosphatidyl inositol-3-phosphate production) (Yakhine-Diop et al., 2017), and this applies to mixed cis-trans-unsaturated FAs such as α-calendic and rumenic acids as well (Fig. 2A, B), even though many of them induced GFP-LC3 puncta (Fig. 1C, D). These data recapitulate our observation that PA and stearic acid induce GFP-LC3 puncta in a Beclin-1/phosphatidylinositol 3-kinase catalytic subunit type 3-dependent fashion, while unsaturated FAs can do so without the formation of phosphatidyl inositol-3-phosphate (Niso-Santano et al., 2015b). In contrast, most of the FAs able to induce GFP-LC3 puncta also stimulated the translocation of a GFP-TFEB reporter construct from the cytosol to the nucleus (Fig. 2C,D), yielding a positive correlation between the two parameters (r = 0.45, p < 0.05, Fig. 5B). We found a negative correlation (r = −0.65, p < 0.01, Fig. 5B) between autophagy induction (measured at 6 h) and later loss in cell viability (measured at 16 h) (Fig. 2E, F), which could be explained, in line with the idea that autophagy is a major cytoprotective mechanism (Moreau et al., 2010), as an attempt from cells to attenuate stress signals induced by high doses (500 μM) of FAs.
Fig. 2.
Autophagy-related signaling pathways elicited by FAs. (A-F) FYVE-RFP stable expressing U2OS cells (A, B), GFP-TFEB stably expressing U2OS cells (C, D), and wild-type U2OS cells (E, F) were treated with 500 μM FAs as indicated for 6 h (A-F) and 16 h (E,F) . Images from fixed cells were acquired and the area of FYVE-RFP+ dots was measured as an indicator for autophagy induction (B). Nuclear and cytoplasmic GFP-TFEB fluorescence intensities were measured and the ratio between nuclear and cytoplasmic values (GFPnuc/cyto ratio) was calculated to indicate TFEB nuclear translocation (D). Nuclei were stained with Hoechst 33342 and the number of cells harboring normal nuclei (i.e. non pyknotic, “healthy cells”) was determined (F). Data are means ± SEM of at least three independent experiments (*=p < 0.05;**=p < 0.01;*** = p < 0.001). Representative images of FYVE-RFP, GFP-TFEB, and viability are shown in A, C, E respectively. Scale bar equals 10 μm in A, C, and 50 μm in E.
Altogether, it appears that FAs vary in their pro-autophagic and cytotoxic potential as they elicit GFP-LC3 puncta through rather divergent mechanisms.
3.3. Differential Effects of FAs on the Golgi and the Endoplasmic Reticulum
Using a panel of biosensor cell lines, we evaluated the effects of FAs on the morphology of the Golgi apparatus and the endoplasmic reticulum (ER) stress response. For this, we used the biosensor GALT1-GFP (which co-localizes with the Golgi, allowing to monitor morphological alterations) (Niso-Santano et al., 2015b), a GFP protein that is expressed under the control of the ATF4 promoter (which is only expressed if ATF4 is activated) (van Galen et al., 2014) and a XBP1-Venus fusion protein (which is usually out-of-frame, yet is spliced in-frame and hence expressed to yield the expression of Venus, a GFP derivative, upon activation of the XBP1-activatory factor IRE1) (Iwawaki et al., 2004). Multiple FAs could induce signs of ER stress. However, the three trans-monounsaturated FAs (elaidic, linoelaidic and trans-vaccenic acid) were rather inert in this parameter, contrasting with the effects of multiple saturated, cis-monounsaturated or cis-trans-polyunsaturated acids many of which elicited the activation of the two ER stress-linked transcription factors ATF4 and XBP1 (Fig. 3A-D). The failure of EL to induce LC3 lipidation and XBP1 maturation was confirmed by immunoblot analyses of two distinct cell lines (Fig. 3E,F). Several cis and cis-trans-unsaturated FAs were highly efficient in disrupting the Golgi apparatus, causing its spread from its orthodox perinuclear location to the cytoplasm or its dissolution (Fig. 4A, B). While saturated and trans-monounsaturated FAs did not relevantly affect Golgi morphology, cis-monounsaturated, cis-polyunsaturated or cis-trans-polyunsaturated perturbed this organelle (Fig. 4A, B). To explore the possibility that some FAs might induce autophagy by reducing protein acetylation, as this has been shown for so-called caloric restriction mimetics (Marino et al., 2014; Madeo et al., 2014), we also measured protein acetylation; however, none of the tested FAs induced major protein deacetylation (Fig. 4C, D), suggesting a different mode of autophagy induction.
Fig. 3.
Effects of FAs on the endoplasmic reticulum stress response. pSMALB-ATF4.5rep (A, B) and XBP1-ΔDBD-venus (C, D) stable expressing U2OS cells were treated with 500 μM FAs as indicated or 3 μM thapsgargin or tunicamycin as positive controls for 16 h. After fixation, ATF4-GFP and XBP1-venus cytoplasmic intensities were measured and then the percentage of cells with cytoplasmic GFP intensity higher than threshold was calculated (B, D). Data are means ± SEM of at least three independent experiments (*=p < 0.05; **=p < 0.01; ***=p < 0.001). Representative images of ATF4-GFP or XBP1-venus are respectively shown in (A,C), scale bar equals 10 μm. (E, F) U2OS (E) and HepG2 (F) cells were treated with 250 μM or 500 μM FAs as indicated or 3 μM thapsigargin as a positive control for 16 h. Then, proteins were separated by SDS PAGE and immunoblots were performed to detect spliced XBP1 (XBP1s), phosphorylated p38 (also known as mitogen-activated protein kinases (MAPK)) and lipidated LC3.
Fig. 4.
Effects of FAs on the Golgi apparatus and deacetylases expression. (A, B) Wild-type and GALT1-GFP stably expressing U2OS cells were treated with 500 μM FAs as indicated or 10 μM golgicide A as positive controls for 16 h. Golgi apparatus from fixed cells was clustered into “fragmented” or “degraded” according to the area of the fluorescent signal, cytoplasmic intensity and distribution within the cytoplasm; the percentage of cells harboring one or the other phenotype was thereafter calculated (B). Representative images of control, palmitate, oleate, and elaidic acid are shown in A. Scale bar equals 10 μm. (C, D) U2OS cells were treated with 500 μM FAs or left untreated as indicated for 6 h. After fixation, cells were treated with specific antibodies to block acetylated tubulin, followed by immunofluorescence employing antibodies specific for proteins with acetylated lysines. Acetylated lysine cytoplasmic fluorescence intensities were measured. Data are means ± SEM of at least three independent experiments (*=p < 0.05; **=p < 0.01) (D). Representative images of control, palmitate, oleate, and elaidic acid are shown in C. Scale bar equals 10 μm.
The three trans-monounsaturated FAs included in this study (elaidic, linoelaidic and trans-vaccenic acid) resembled three relatively large (≥20 carbon atoms) cis-monounsaturated FAs (nervonic, erucic, eicosenoic acid) in their relative failure to induce major biological effects on cultured cells, as revealed by non-supervised hierarchical clustering of the results (Fig. 5A). Moreover, careful analysis of all data and their correlations among measured parameters (Fig. 5B), revealed that transcriptional responses leading to activation of ATF4, XBP1 and TFEB correlated among each other. Moreover, both XBP1 and TFEB activation were associated with the fragmented Golgi phenotype. Lipid uptake by the cells, however, failed to correlate with the parameters measured here (Fig. 5B). Altogether, these results underscore the complexity of lipid effects on cultured cells.
3.4. Physicochemical Characteristics Dictate Biological Effects of FAs
Principal component analyses of the biological lipid effects led to the classification of FAs into three clusters (Fig. 5D, S2). One cluster is composed by unsaturated FAs (PA and stearic acid) trans-monounsaturated FAs (elaidic, linoelaidic and trans-vaccenic acid), and large (≥20 carbon atoms) cis-monounsaturated FAs (nervonic, erucic, eicosenoic acid). Another cluster comprises four highly unsaturated (3–4 unsaturations) FAs (arachidonic, α-linoleic, γ-linoleic and stearidonic acid). The third cluster contains the two cis-trans polyunsaturead FAs (α-calendic and rumenic acid) as well as 5 FAs with up to 18 carbon molecules bearing one single cis-unsaturated bond (myristoleic, oleic, palmitoleic, vaccenic acid) or two cis-unsaturations (linoleic acid). Driven by these findings, which suggest correlations between biological function and chemical structures, we attempted a systematic correlation between multiple physicochemical parameters retrieved for each lipid (Steinbeck et al., 2003) and its biological effects. Indeed, multiple significant correlations emerged (Fig. 5C). One of the strongest correlations concerned the number of cis-unsaturations (NCU), which describes the bending of the carbon backbone of each lipid (Table S1). This observation paired with the K-means analysis performed on biological parameters, which resulted in three clusters obviously related to the NCU (Fig. 5D), in line with the interpretation that cis-unsaturations indeed dictate the biological effects of different lipid species. In addition, the length of carbon chains can discriminate the properties of cis-monounsaturated acids that fall into the two distinct clusters (Fig. 5D). Altogether, these results underscore the importance of the overall lipid structure in determining the biological effects of FAs.
3.5. Functional Interactions Between Elaidic Acid (EL) and Other FAs
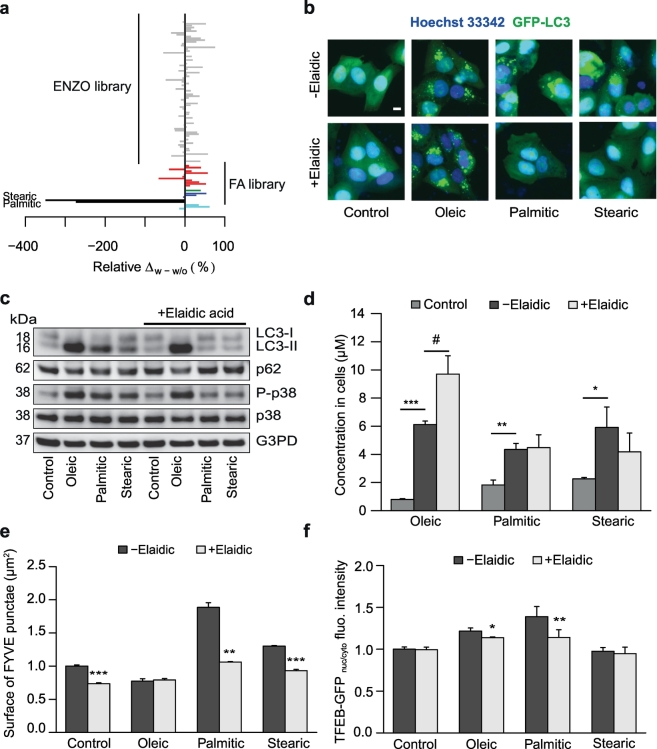
In contrast to its cis-isomer oleic acid, the trans-monounsaturated FA elaidic acid (EL) appeared unable to stimulate major stress responses including autophagy, ER stress and Golgi perturbations (Fig. 1, Fig. 2, Fig. 3, Fig. 4). Based on the consideration that trans-unsaturated FAs have been associated with negative health effects including reduced lifespan in humans (Oomen et al., 2001; Mozaffarian et al., 2006) and that autophagic flux is one of the major determinants of health span and longevity (Melendez et al., 2003; Tavernarakis et al., 2008; Rubinsztein et al., 2011; Pyo et al., 2013; Green and Levine, 2014; Eisenberg et al., 2016; Ho et al., 2017), we attempted to define conditions in which EL, which is particularly abundant in industrial food (Chajes et al., 2015), might inhibit autophagy. For this, we tested multiple autophagy inducers from the ENZO SCREEN-WELL library and all FAs included in this study for their capacity to stimulate GFP-LC3 puncta in U2OS cells in the presence or absence of EL (Fig. 6A). Strikingly, EL strongly inhibited autophagy stimulated by PA and stearic acid but not by any other agent including rapalogs and OL (Fig. 6A-C, Fig. S5). Although EL did not decrease the uptake of lipids (Fig. 6D), it inhibited all stress signals elicited by palmitate and/or stearate including p38 MAPK phosphorylation (Fig. 6C), formation of phosphatidyl inositol-3-phosphate (Fig. 6E) and translocation of TFEB to the nucleus (Fig. 6F). Careful dose response and kinetic analyses confirmed the capacity of even comparatively low doses of EL (125 μM) to inhibit induction of autophagy, ER stress (measured as eIF2a phosphorylation) and cell death induction by PA (Fig. 7A). However, EL failed to affect any of the effects elicited by OL (Fig. 7B). The capacity of EL to inhibit palmitate effects on cultured cells was shared by several additional trans-FAs including linoelaidic, trans-vaccenic, and palmitelaidic acid (Fig. 7C). Moreover, EL prevented the induction of signs of autophagy such as LC3B lipidation and STQM1/p62 depletion in the liver and in the heart of mice after intraperitioneal injection of PA (Fig. 7D, Fig. S3). In contrast, EL failed to prevent autophagy induction by OL in vivo (Fig. S4). The capacity of EL to inhibit autophagy induction by PA was confirmed using mice expressing transgenic GFP-LC3, in which PA induced formation of autophagic puncta in the myocardium was inhibited by EL (Fig. 7E). Importantly, PA (but not OL) was able to induce autophagy in mouse embryonic fibroblasts (MEF) bearing a knockin mutation in eIF2α (S51A) that renders the protein unphosphorylable (Scheuner et al., 2001) and the autophagy-inhibitory effect of EL persisted in these cells (Fig. 7F).
Fig. 6.
Inhibitory effects of elaidic acid on autophagy induction by saturated FAs. (A, B) U2OS cells stably expressing GFP-LC3 were treated with 500 μM FA, or 10 μM of the autophagy-inducer subset from the ENZO SCREEN-WELL autophagy library, in the presence or absence of 500 μM EL for 6 h. Autophagy was assessed after fixation by measuring the area of LC3 dots within each cell. The relative difference with and without EL co-treatment was calculated and is reported as barchart in A. Representative images are shown in B. Scale bar equals 10 μm. (C) U2OS cells were treated with either vector (ethanol 0.5%), 500 μM OL, PA, or stearate in the presence or absence of 500 μM elaidate for 6 h. Thereafter, LC3 lipidation and p62 degradation and p38 activation were measured by immunoblot as indicators for autophagy. (D) U2OS cells treated as in C with OL, PA, and stearate in the presence or absence of elaidate, were harvested and analyzed with GC/MS to measure the intracellular FA concentration and to test the effect of EL on FA uptake into cells. Data are means ± SEM of at least three independent experiments (*=p < 0.05, **=p < 0.01, ***=p < 0.001, FA concentration in cells treated with a single FA compared to cells treated with vector control; #=p < 0.05, FA concentration in cells receiving EL co-treatment versus cells treated with single FA). (E, F) FYVE-RFP (E) and GFP-TFEB (F) stable expressing U2OS cells were treated as in C. After fixation, images were acquired and the area of FYVE+ dots and TFEB nuclear translocation were analyzed. Data are means ± SEM of at least three independent experiments (* = p <0.05, **=p < 0.01, ***=p < 0.001, FYVE dots surface or GFP-TFEBnuc/cyto ratio in cells with EL as compared to cells that were not co-treated with EL).
Fig. 7.
Dose response and kinetic analysis of the effects of elaidic acid on autophagy induction by various FAs. (A, B, C) U2OS cells stably expressing GFP-LC3 were treated with increasing concentrations of EL (A,B,C), linoelaidic acid, transvaccenic acid, or palmitelaidic acid (C), and co-treated or not with the indicated increasing concentrations of PA (A,C) or OL (B) for 2, 4, 6, 16 (A,B) or 24 h (A, B, C). After fixation, phosphorylated eIF2α was visualized by immunofluorescence. For each treatment, the degree of autophagy was measured by the quantifying the area of LC3 dots. ER-Stress was monitored by measuring the phosphorylation eIF2α by immunofluorescence, and the loss of viability was quantitated by enumerating the number of cells depicting a healthy morphotype. Data is depicted as a heatmap with maximum effects depicted in red and minimal effects in blue. (D) C57BL/6 mice were injected intraperitoneally with vehicle only, 100 mg/kg PA, 100 mg/kg EL, or the combination of the two FAs. 2 h later, mice were euthanized and heart and liver were collected for immunoblotting. LC3 lipidation and p62 degradation were detected to indicate autophagy activity. GAPDH levels were monitored to ensure equal loading. (E) Transgenic mice harboring GFP-LC3 reporter were treated with either PA, EL, or their combination. Frozen sections were made from hearts and thereafter imaged on a fluorescent microscope. Mean numbers of GFP-LC3 dots per cell were determined on the obtained images. Results indicated on the bar chart represent the means from three mice. *p < 0.05. Representative micrographs of GFP-LC3 dots are shown together with barchart; dots local contrast was enhanced for better visibility. Scale bar represents 10 μm. (F) MEFs wildtype (WT) or carrying a non phoshorylable knock in mutation in eIF2α (KI) both stably expressing GFP-LC3 were treated with increasing concentrations of OL or PAL, in the presence or the absence of 500 μM EL for 2, 4 and 6 h. Cells were imaged after fixation and autophagy levels were measured by measuring the total area of LC3 dots per cell. Quantifications are depicted as a heatmap with maximum effects in red and minimal effects in blue.
Altogether, these results support the idea that trans-FAs can selectively interfere with the cell stress-inducing effects of saturated FAs.
4. Discussion
The present study constitutes a systematic attempt to compare the cellular effects of multiple FAs that differ in their carbon chain length, and the presence of cis or trans unsaturations. When determining multiple cellular parameters linked to autophagy (GFL-LC3 puncta, FYVE dots reflecting the formation of PI3P, nuclear translocation of TFEB), Golgi morphology (GALT1-GFP), the unfolded protein stress response (activation of the eIF2α phosphorylation/ATF4 and the IRE1/XBP1 pathways) and viability for some 19 distinct FAs, a pattern of differences emerged. Such a pattern could be best resolved by means of principal component analysis, leading to the conclusion that one of the most important physicochemical differences dictating the biological activity of FAs is indeed the presence or absence of a trans-unsaturated bond. Hence, the cis-unsaturated FAs oleic acid and linoleic acid are rather distinct in their biological activities from their trans-isomers, which are elaidic acid and linoelaidic acid, respectively. Generally, it appeared that trans-unsaturated FAs were less active in eliciting cellular stress than cis-unsaturated FAs, as well as saturated FAs. Most strikingly, all trans-unsaturated FAs tested here were unable to stimulate an autophagic response.
Intrigued by the incapacity of trans-unsaturated FA to induce autophagy, we investigated the possibility that they might inhibit autophagy. EL, the most abundant FA generated by industrial food processing, failed to inhibit autophagy induced by starvation as well as a panel of all major autophagy inducers that comprises rapamycin, rapalogs, as well as caloric restriction mimetics such as spermidine and resveratrol (Eisenberg et al., 2009; Marino et al., 2014). However, EL and all other trans-unsaturated FAs used in this study were highly efficient in inhibiting autophagy induction by saturated FAs. This effect was specific, meaning that EL only prevents autophagy and endoplasmic reticulum stress induced by the saturated FA PA but not by the unsaturated FA OL. Moreover, it could be confirmed in vivo, in a mouse model in which PA stimulated autophagy in the heart and in the liver, and this effect was abolished by co-injection of EL. The exact molecular mechanisms through which trans-FAs inhibit PA-induced autophagy remain elusive. Of note, this inhibitory effect was independent of the sequence of administration (trans-FA before or after PA), and the potency of the inhibitory effect was distinct for different trans-FAs, arguing against direct interactions among the FAs. Moreover, the inhibitory effect of EL on PA-induced autophagy could be uncoupled from the inhibition of eIF2α phosphorylation, because both PA-induced autophagy and its inhibition by EL were still observed in cells bearing a non-phosphorylable eIF2α mutant. At this stage, it is tempting to speculate that yet-to-be-characterized changes in cellular membrane properties (Payet et al., 2013) might account for the autophagy-inhibitory effects of trans-FAs.
Autophagy constitutes a major cytoprotective (and in particular cardioprotective) mechanism that comes into action to protect cells against acute stress, dampens inflammatory responses and counteracts aging (Linton et al., 2015; Nishida and Otsu, 2016; Shirakabe et al., 2016; Eisenberg et al., 2016; Tong and Hill, 2017). As a result, the capacity of trans-FAs to reduce autophagy in specific circumstances warrants further scrutiny to unveil the mechanisms through which they increase the likelihood of cardiovascular morbidity and mortality.
Acknowledgments
Acknowledgments
This study was supported by a research grant from Institut Mérieux. GK is supported by the Ligue contre le Cancer (équipe labelisée); Agence Nationale de la Recherche (ANR) – Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Institut National du Cancer (INCa); Institut Universitaire de France; Fondation pour la Recherche Médicale (FRM); the European Commission (ArtForce); the European Research Council (ERC); a donation by Elior; the LeDucq Foundation; the LabEx Immuno-Oncology; the Seerave Foundation; the RHU Torino Lumière, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); and the Paris Alliance of Cancer Research Institutes (PACRI). P.R. is an Erwin Schrödinger research fellow of the Austrian Science Fund (FWF). FM is grateful to BioTechMed-Graz for the flagship project “EPIAge”, to the Austrian Science Fund FWF (Austria) for grants P29262, P29203, P27893, and “SFB Lipotox” (F3012), as well as to BMWFW and the Karl-Franzens University for grants ‘Unkonventionelle Forschung’ and flysleep (80.109/0001 -WF/V/3b/2015)
Conflict of Interest
There are no conflict of interest to be declared.
Author Contributions
Conceptualization, G.K. and O.K.; Methodology, G.K., G.C., P.R., F.M. and J.S.; Investigation, G.C., G.Ce., K.M., J.F., F.A., S.D., M.L. and M.T.; Formal Analysis, A.S and M.L.; Software, A.S.; Writing – Original Draft, G.K. and A.S.; Writing –Review & Editing, O.K. and P.R.; Funding Acquisition, G.K., J.S. and F.M.; Resources, L.B.; Supervision, G.K.
Contributor Information
Oliver Kepp, Email: captain.olsen@gmail.com.
Guido Kroemer, Email: kroemer@orange.fr.
Appendix A. Supplementary data
Supplementary material
References
- Bankaitis V.A. Unsaturated fatty acid-induced non-canonical autophagy: unusual? Or unappreciated? EMBO J. 2015;34:978–980. doi: 10.15252/embj.201591392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento C.F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M., Menzies F.M., Rubinsztein D.C. Mammalian autophagy: how does it work? Annu. Rev. Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- Brostow D.P., Hirsch A.T., Collins T.C., Kurzer M.S. The role of nutrition and body composition in peripheral arterial disease. Nat. Rev. Cardiol. 2012;9:634–643. doi: 10.1038/nrcardio.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell K.D., Pomeranz J.L. The trans-fat ban—food regulation and long-term health. N. Engl. J. Med. 2014;370:1773–1775. doi: 10.1056/NEJMp1314072. [DOI] [PubMed] [Google Scholar]
- Buttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., Ruckenstuhl C., Sigrist C., Wissing S., Kollroser M., Frohlich K.U., Sigrist S., Madeo F. Endonuclease G regulates budding yeast life and death. Mol. Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Chajes V., Biessy C., Ferrari P., Romieu I., Freisling H., Huybrechts I., Scalbert A., Bueno De Mesquita B., Romaguera D., Gunter M.J., Vineis P., Hansen C.P., Jakobsen M.U., Clavel-Chapelon F., Fagherazzi G., Boutron-Ruault M.C., Katzke V., Neamat-Allah J., Boeing H., Bachlechner U., Trichopoulou A., Naska A., Orfanos P., Pala V., Masala G., Mattiello A., Skeie G., Weiderpass E., Agudo A., Huerta J.M., Ardanaz E., Sanchez M.J., Dorronsoro M., Quiros J.R., Johansson I., Winkvist A., Sonested E., Key T., Khaw K.T., Wareham N.J., Peeters P.H., Slimani N. Plasma elaidic acid level as biomarker of industrial trans fatty acids and risk of weight change: report from the EPIC study. PLoS One. 2015;e0118206:10. doi: 10.1371/journal.pone.0118206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig-Schmidt M.C. World-wide consumption of trans fatty acids. Atheroscler. Suppl. 2006;7:1–4. doi: 10.1016/j.atherosclerosissup.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Duplus E., Glorian M., Forest C. Fatty acid regulation of gene transcription. J. Biol. Chem. 2000;275:30749–30752. doi: 10.1074/jbc.R000015200. [DOI] [PubMed] [Google Scholar]
- Eisenberg T., Knauer H., Schauer A., Buttner S., Ruckenstuhl C., Carmona-Gutierrez D., Ring J., Schroeder S., Magnes C., Antonacci L., Fussi H., Deszcz L., Hartl R., Schraml E., Criollo A., Megalou E., Weiskopf D., Laun P., Heeren G., Breitenbach M., Grubeck-Loebenstein B., Herker E., Fahrenkrog B., Frohlich K.U., Sinner F., Tavernarakis N., Minois N., Kroemer G., Madeo F. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Eisenberg T., Abdellatif M., Schroeder S., Primessnig U., Stekovic S., Pendl T., Harger A., Schipke J., Zimmermann A., Schmidt A., Tong M., Ruckenstuhl C., Dammbrueck C., Gross A.S., Herbst V., Magnes C., Trausinger G., Narath S., Meinitzer A., Hu Z., Kirsch A., Eller K., Carmona-Gutierrez D., Buttner S., Pietrocola F., Knittelfelder O., Schrepfer E., Rockenfeller P., Simonini C., Rahn A., Horsch M., Moreth K., Beckers J., Fuchs H., Gailus-Durner V., Neff F., Janik D., Rathkolb B., Rozman J., de Angelis M.H., Moustafa T., Haemmerle G., Mayr M., Willeit P., von Frieling-Salewsky M., Pieske B., Scorrano L., Pieber T., Pechlaner R., Willeit J., Sigrist S.J., Linke W.A., Muhlfeld C., Sadoshima J., Dengjel J., Kiechl S., Kroemer G., Sedej S., Madeo F. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016;22:1428–1438. doi: 10.1038/nm.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enot D.P., Niso-Santano M., Durand S., Chery A., Pietrocola F., Vacchelli E., Madeo F., Galluzzi L., Kroemer G. Metabolomic analyses reveal that anti-aging metabolites are depleted by palmitate but increased by oleate in vivo. Cell Cycle. 2015;14:2399–2407. doi: 10.1080/15384101.2015.1064206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R., Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadj Ahmed S., Kharroubi W., Kaoubaa N., Zarrouk A., Batbout F., Gamra H., Najjar M.F., Lizard G., Hininger-Favier I., Hammami M. Correlation of trans fatty acids with the severity of coronary artery disease lesions. Lipids Health Dis. 2018;17:52. doi: 10.1186/s12944-018-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T., Levis R., Rubin G.M. Transformation of white locus DNA in drosophila: dosage compensation, zeste interaction, and position effects. Cell. 1984;36:469–481. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- Ho T.T., Warr M.R., Adelman E.R., Lansinger O.M., Flach J., Verovskaya E.V., Figueroa M.E., Passegue E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer R.G., Park E.J., Li N., Tran H., Chen M., Choi C., Solinas G., Karin M. Saturated fatty acids induce c-Src clustering within membrane subdomains, leading to JNK activation. Cell. 2011;147:173–184. doi: 10.1016/j.cell.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T., Akai R., Kohno K., Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Kadhum A.A., Shamma M.N. Edible lipids modification processes: a review. Crit. Rev. Food Sci. Nutr. 2017;57:48–58. doi: 10.1080/10408398.2013.848834. [DOI] [PubMed] [Google Scholar]
- Kaur J., Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- Li Z., Berk M., Mcintyre T.M., Gores G.J., Feldstein A.E. The lysosomal-mitochondrial axis in free fatty acid-induced hepatic lipotoxicity. Hepatology. 2008;47:1495–1503. doi: 10.1002/hep.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton P.J., Gurney M., Sengstock D., Mentzer R.M., Jr., Gottlieb R.A. This old heart: cardiac aging and autophagy. J. Mol. Cell. Cardiol. 2015;83:44–54. doi: 10.1016/j.yjmcc.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock A.L., Bauman D.E. Modifying milk fat composition of dairy cows to enhance fatty acids beneficial to human health. Lipids. 2004;39:1197–1206. doi: 10.1007/s11745-004-1348-6. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C., Galluzzi L., Freije J.M., Madeo F., Kroemer G. Metabolic control of longevity. Cell. 2016;166:802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- Madeo F., Pietrocola F., Eisenberg T., Kroemer G. Caloric restriction mimetics: towards a molecular definition. Nat. Rev. Drug Discov. 2014;13:727–740. doi: 10.1038/nrd4391. [DOI] [PubMed] [Google Scholar]
- Maejima, et al. Nat Med. 2013 Nov;19(11):1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H., Kaufman R.J. Endoplasmic reticulum stress in liver disease. J. Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G., Pietrocola F., Madeo F., Kroemer G. Caloric restriction mimetics: natural/physiological pharmacological autophagy inducers. Autophagy. 2014;10:1879–1882. doi: 10.4161/auto.36413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazidi M., Gao H.K., Kengne A.P. Inflammatory markers are positively associated with serum trans-fatty acids in an adult American population. J. Nutr. Metab. 2017;2017:3848201. doi: 10.1155/2017/3848201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazidi M., Banach M., Kengne A.P. Association between plasma trans fatty acids concentrations and leucocyte telomere length in US adults. Eur. J. Clin. Nutr. 2018 doi: 10.1038/s41430-017-0065-y. (epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Menzies F.M., Fleming A., Rubinsztein D.C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- Monguchi T., Hara T., Hasokawa M., Nakajima H., Mori K., Toh R., Irino Y., Ishida T., Hirata K.I., Shinohara M. Excessive intake of trans fatty acid accelerates atherosclerosis through promoting inflammation and oxidative stress in a mouse model of hyperlipidemia. J. Cardiol. 2017;70(2):121–127. doi: 10.1016/j.jjcc.2016.12.012. [DOI] [PubMed] [Google Scholar]
- Moreau K., Luo S., Rubinsztein D.C. Cytoprotective roles for autophagy. Curr. Opin. Cell Biol. 2010;22:206–211. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel E., Mehrpour M., Botti J., Dupont N., Hamai A., Nascimbeni A.C., Codogno P. Autophagy: a Druggable process. Annu. Rev. Pharmacol. Toxicol. 2017;57:375–398. doi: 10.1146/annurev-pharmtox-010716-104936. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D., Katan M.B., Ascherio A., Stampfer M.J., Willett W.C. Trans fatty acids and cardiovascular disease. N. Engl. J. Med. 2006;354:1601–1613. doi: 10.1056/NEJMra054035. [DOI] [PubMed] [Google Scholar]
- Nishida K., Otsu K. Autophagy during cardiac remodeling. J. Mol. Cell. Cardiol. 2016;95:11–18. doi: 10.1016/j.yjmcc.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Niso-Santano M., Bravo-San Pedro J.M., Maiuri M.C., Tavernarakis N., Cecconi F., Madeo F., Codogno P., Galluzzi L., Kroemer G. Novel inducers of BECN1-independent autophagy: cis-unsaturated fatty acids. Autophagy. 2015;11:575–577. doi: 10.1080/15548627.2015.1017222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niso-Santano M., Malik S.A., Pietrocola F., Bravo-San Pedro J.M., Marino G., Cianfanelli V., Ben-Younes A., Troncoso R., Markaki M., Sica V., Izzo V., Chaba K., Bauvy C., Dupont N., Kepp O., Rockenfeller P., Wolinski H., Madeo F., Lavandero S., Codogno P., Harper F., Pierron G., Tavernarakis N., Cecconi F., Maiuri M.C., Galluzzi L., Kroemer G. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025–1041. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen C.M., Ocke M.C., Feskens E.J., van Erp-Baart M.A., Kok F.J., Kromhout D. Association between trans fatty acid intake and 10-year risk of coronary heart disease in the Zutphen Elderly Study: a prospective population-based study. Lancet. 2001;357:746–751. doi: 10.1016/s0140-6736(00)04166-0. [DOI] [PubMed] [Google Scholar]
- Oteng A.B., Bhattacharya A., Brodesser S., Qi L., Tan N.S., Kersten S. Feeding Angptl4(−/−) mice trans fat promotes foam cell formation in mesenteric lymph nodes without leading to ascites. J. Lipid Res. 2017;58:1100–1113. doi: 10.1194/jlr.M074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet L.A., Pineau L., Snyder E.C., Colas J., Moussa A., Vannier B., Bigay J., Clarhaut J., Becq F., Berjeaud J.M., Vandebrouck C., Ferreira T. Saturated fatty acids alter the late secretory pathway by modulating membrane properties. Traffic. 2013;14:1228–1241. doi: 10.1111/tra.12117. [DOI] [PubMed] [Google Scholar]
- Pyo J.O., Yoo S.M., Ahn H.H., Nah J., Hong S.H., Kam T.I., Jung S., Jung Y.K. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston J.C., Lyons C.L., Kennedy E.B., Kirwan A.M., Roche H.M. Fatty acids and NLRP3 Inflammasome-mediated inflammation in metabolic tissues. Annu. Rev. Nutr. 2017;37:77–102. doi: 10.1146/annurev-nutr-071816-064836. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Marino G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B., Mcewen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R.J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Shirakabe A., Ikeda Y., Sciarretta S., Zablocki D.K., Sadoshima J. Aging and autophagy in the heart. Circ. Res. 2016;118:1563–1576. doi: 10.1161/CIRCRESAHA.116.307474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S.J., Reiff D.F., Thiel P.R., Steinert J.R., Schuster C.M. Experience-dependent strengthening of Drosophila neuromuscular junctions. J. Neurosci. 2003;23:6546–6556. doi: 10.1523/JNEUROSCI.23-16-06546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck C., Han Y., Kuhn S., Horlacher O., Luttmann E., Willighagen E. The chemistry development kit (CDK): an open-source java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto H.H., Liu S., Soong Y., Alam N., Prusky G.T., Seshan S.V. Protection of mitochondria prevents high-fat diet-induced glomerulopathy and proximal tubular injury. Kidney Int. 2016;90:997–1011. doi: 10.1016/j.kint.2016.06.013. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N., Pasparaki A., Tasdemir E., Maiuri M.C., Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- Tong D., Hill J.A. Spermidine promotes Cardioprotective autophagy. Circ. Res. 2017;120:1229–1231. doi: 10.1161/CIRCRESAHA.117.310603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisu K., Singh K.K., Torisu T., Lovren F., Liu J., Pan Y., Quan A., Ramadan A., Al-Omran M., Pankova N., Boyd S.R., Verma S., Finkel T. Intact endothelial autophagy is required to maintain vascular lipid homeostasis. Aging Cell. 2016;15:187–191. doi: 10.1111/acel.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng Y.Z., Hu C.H. Radical-induced Cis-trans isomerization of fatty acids: a theoretical study. J. Phys. Chem. A. 2014;118:4554–4564. doi: 10.1021/jp502434t. [DOI] [PubMed] [Google Scholar]
- Unger R.H. Lipotoxic diseases. Annu. Rev. Med. 2002;53:319–336. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- van Galen P., Kreso A., Mbong N., Kent D.G., Fitzmaurice T., Chambers J.E., Xie S., Laurenti E., Hermans K., Eppert K., Marciniak S.J., Goodall J.C., Green A.R., Wouters B.G., Wienholds E., Dick J.E. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- Yakhine-Diop S.M., Martinez-Chacon G., Gonzalez-Polo R.A., Fuentes J.M., Niso-Santano M. Fluorescent FYVE chimeras to quantify PtdIns3P synthesis during autophagy. Methods Enzymol. 2017;587:257–269. doi: 10.1016/bs.mie.2016.09.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material