Abstract
The long non-coding RNA maternally expressed gene 3 (MEG3) is frequently dysregulated in human cancers; however, its roles in colorectal cancer (CRC) development are largely unknown. Here, we reported that MEG3 was down-regulated in CRC tissues and CRC patients with lower MEG3 showed poorer overall survival and disease-free survival than those with higher MEG3 level. MEG3 over-expression represses CRC cells proliferation and migration in vivo and in vitro, while MEG3 knockdown leads to the enhanced proliferation and metastasis of CRC cells. In CRC cells, MEG3 over-expression is related to decreased Clusterin mRNA and the corresponding protein levels, and it also directly binds to Clusterin protein through its 732–1174 region. In further, Clusterin over-expression rescues the compromised abilities of proliferation and metastasis induced by MEG3 over-expression, suggesting that MEG3 inhibits the CRC progression through regulating the Clusterin activities. Additionally, we found that 1α,25-(OH)2D and vitamin D receptor (VDR) stimulate MEG3 expression in CRC cells through directly binding to its promoter. These results suggested that MEG3 functions as a tumor suppressor in CRC via regulating the Clusterin activities and may underlie the anticancer activities of vitamin D on CRC cells. The VDR/MEG3/Clusterin signaling pathway may serve as potential therapeutic targets and prognosis biomarkers for CRC patients in future.
Keywords: lncRNA, CRC, MEG3, Clusterin, Vitamin D
Highlights
-
•
MEG3 serves as a novel CRC prognosis biomarker and a potential therapeutic target.
-
•
MEG3 over-expression represses CRC cells proliferation and metastatic features.
-
•
MEG3 has a role in Clusterin expression and activity down-regulation at transcriptional and post-transcriptional levels.
-
•
VDR activated MEG3 expression via directly binding to MEG3 promoter.
1. Introduction
Colorectal cancer (CRC) is one of the most common malignancies of the digestive system worldwide with the incidence has being increased significantly over the past three decades (Arnold et al., 2017). >700,000 deaths has been estimated to be caused by CRC in 2012, ranking it as the 4th leading cause of cancer-related deaths (Torre et al., 2015). Although much progress has been made for CRC patients for the past decades, such as the advanced chemotherapy methods and targeted therapy including cetuximab or panitumumab, the prognosis of CRC patients is still poor. The 5-year cause specific survival of CRC patients ranges from 90.9% for AJCC 6th TNM stage I to 12.2% for those with stage IV (Lin et al., 2015). Thus, uncovering the underlying mechanisms involved in CRC development and progression may provide potential therapy targets, which may further improve the prognosis of the patients.
The development of colorectal cancer is a multi-stage and multi-step process, involving multiple genomic and epigenetic alterations (Guinney et al., 2015). Clinical studies have reported that mutation in oncogenes or tumor suppressor genes, including APC, KRAS, SMAD4 and TP53 are frequently observed in CRC patients (Yu et al., 2015). Guinney et al. have categorized CRC into four consensus molecular subtypes with distinguishing features and prognoses based on the gene expression pattern and genomic alterations of CRC tissues (Guinney et al., 2015). Recently, increasing evidence has shown that long non-coding RNAs (lncRNAs), the novel regulators in cellular signaling, play vital roles in CRC tumorigenesis and progression (Ohtsuka et al., 2016; Ma et al., 2016). lncRNA is a type of RNA transcripts longer than 200 nucleotides but do not translated into protein in cells. lncRNAs are involved in diverse cel1ular processes including cell proliferation, migration, invasion and transformation etc., and dysregulation of lncRNAs are associated with various types of diseases (Ponting et al., 2009). lncRNAs regulate the gene functions through multiple levels including the chromatin modification, transcription, post-transcription, interaction with RNA-binding proteins, co-activation of transcription factors and repressors (Marchese et al., 2017). Maternally expressed gene 3 (MEG3), located on 14q32.3 of the human genome, encodes a 1.6 kilobase (kb) lncRNA (Zhou et al., 2012). It is expressed in multiple organs, including the brain, stomach, liver, pancreas and ovary. By contrast, MEG3 expression is frequently repressed in tumor tissues (Zhou et al., 2012). In various types of cancer, genomic deletion and abnormal methylation in the promoter of MEG3 were noticed, leading to the down-regulation of MEG3 in tumor tissues (Bando et al., 1999; Yin et al., 2015; Lu et al., 2013). In non-small cell lung cancer (NSCLC) cells, MEG3 inhibits proliferation and induces cell apoptosis through activating p53 and its down-stream signaling pathway (Lu et al., 2013). Interestingly, for tumor cells with p53 deletion, MEG3 over-expression also inhibits tumorigenesis through targeting the microRNAs such as microRNA-421 and microRNA-184 (Zhang et al., 2017; Li et al., 2018). These studies indicated multiple mechanisms underlying the roles of MEG3 in tumor development. Previous studies have reported that a lower MEG3 level was associated with the increased liver metastasis of CRC patients (Kong et al., 2016), and an enhanced CRC cells chemosensitivity to oxaliplatin (Li et al., 2017). However, the underlying mechanisms regarding the tumor suppressor activities of MEG3 are still largely unknown. In the current study, we evaluated the anticancer activities and the underlying mechanisms of MEG3 in CRC development and progression, which may provide potential novel intervention methods for CRC in the future.
2. Materials and Methods
2.1. Tissue Microarray Construction
Tumor specimens used in tissue microarrays (TMAs) were obtained from 371 colorectal cancer patients who underwent curative resection at Changhai Hospital of the Second Military Medical University from January 2001 to December 2010. Patients were selected with the following inclusion and exclusion criteria: (i) pathological confirmed as the primary CRC according to the World Health Organization criteria; (ii) with available formalin-fixed, paraffin-embedded (FFPE) CRC tissue samples; (iii) without any pre-operative anti-cancer treatment and no evidence of distant metastases; (iv) with complete clinicopathologic and follow-up data for the patients. Each participant provided the written informed consent and the study was approved by the Changhai Hospital Ethics Committee. The overall survival (OS) time was defined as the length of time between the surgery date and deaths by any causes. For surviving patients, the data were censored at the last following-up. The disease-free survival (DFS) was defined as the length of time between the date of the surgery and the date of tumor recurrence, metastasis or death. The tissue microarrays (TMAs) were constructed with the FFPE tissues by Shanghai Biochip Co, Ltd., Shanghai, China, following the routine protocols (Cai et al., 2017). For each patient, a 0.75-mm diameter core of the FFPE tumor tissue was punched and arranged in the TMA blocks.
2.2. Immunohistological Chemistry Staining and the in situ Hybridization
Six-micrometer thick TMA sections were used to perform immunohistochemistry staining and in situ hybridization (ISH) following standardized protocols (Pan et al., 2015; Deng et al., 2013). The antibody used for immunohistochemical staining of VDR was purchased from Cell Signaling Technology (Cat# 12550, RRID: AB_2637002). The lncRNA-MEG3 probes were designed and produced by Exiqon (Vedbaek, Denmark). ISH was performed following the manufacturer's guidelines. The immunohistochemical score for each TMA sample was assessed independently by 2 pathologists.
2.3. Cell Culture
The human colorectal cancer cell lines RKO, SW1116, HT29, HCT116, LoVo, SW620, SW480 and 293 T were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences. All cell lines were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 U/mL penicillin, 100 mg/mL streptomycin), in a humidified atmosphere of 5% CO2 at 37 °C. Cell lines were authenticated by short tandem repeat polymerase chain reaction (STR-PCR). Mycoplasma infection status was tested by 4′, 6-diamidino-2-phenylindole (DAPI) staining in the laboratory. All colorectal cancer cell lines were used to investigate MEG3 expression, while RKO, SW1116, and LoVo were used to investigate the biological functions of MEG3. The SW1116 cell line was used to investigate the effects of MEG3 on CLU expression.
2.4. Cellular Proliferation Assay
Cellular proliferation was measured using the Cell Counting Kit-8 (CCK-8, Dojindo, Japan) kit. Cells with modified MEG3 and Clusterin expression or not were seeded at a density of 5 × 103 cells/well in 96-well culture plates and cultured for 24, 48, or 72 h. The cells were then incubated with 10 μL CCK8 for another 4 h at 37 °C. After incubation, the viability of cells was measured at 450 nm using a microplate reader (BioTek, USA), and all experiments were repeated three times. Down-regulation of MEG3 or Clusterin (CLU) was performed by small interfering RNA (siRNA) transfection (MEG3 siRNA, UUAGGUAAGAGGGACAGCUGGCUGG; si-CLU1, CCAGACGGUCUCAGACAAU; si-CLU2, GGUUGACCAGGAAAUACAA; si-CLU3, CCAGGAAGAACCCUAAAUU). Over-expression of MEG3 or Clusterin was performed by infection of lentiviruses expressing MEG3 or Clusterin coding regions, which were obtained from Shanghai Obio technology Company (Shanghai, China).
2.5. Tran-swell and Invasion Assays
5 × 104 cells in 200 μL serum-free Dulbecco's Modified Eagle Medium (DMEM) medium were placed in upper surface of the Transwell chambers (8 μm, Corning Costar Co., MA, USA) with the pre-coated 1:10 diluted Matrigel (BD Biosciences, CA, USA) for invasion assay or without the Matrigel for migration assay. The lower chamber was filled with 500 μL complete RPMI 1640 medium with 10% FBS. The cells were incubated for 12 to 18 h at 37 °C and then the cells on the top surface of the membrane were removed by wiping with a cotton swab. The cells that had migrated or invaded from the upper surface to the bottom surface of the filter membrane were stained with 0.5% crystal violet solution and photographed in five representative fields per insert. The cell number per field was counted and compared between the groups.
2.6. Xenograft and Metastasis Animal Models
Male BALB/c nude mice aged 4–6 weeks were obtained from Shanghai Laboratory Animal Center of China. For the tumor growth model, 1 × 106 SW1116 cells with or without MEG3 stable over-expression were injected into the axillary subcutaneous tissues of nude mice. Tumor growth was determined by measuring the tumor volume, V = tumor length × tumor width2/2 every 4 days using calipers.
For the tail vein metastasis models, 1 × 106 cells were suspended in 200 μL serum-free RPMI1640 were injected into the tail vein of nude mice. After 7 weeks, all of mice were sacrificed and the lung tissues were dissected and fixed with 10% formalin for at least 72 h. The metastasis nodules in lung tissues were analyzed by hematoxylin and eosin (HE) staining methods. All animal experiments were performed according to the guidelines on the care and use of animals for scientific use and approved by the Institutional Animal Care and Use Committee at Second Military Medical University.
2.7. RNA-pull Down Assay
RNA-pull down assay was performed as previously reported (Yuan et al., 2017). MEG3 was in vitro-transcribed from the vector pSPT19-MEG3 using T7 or Sp6RNA polymerase (Roche) and biotin-labeled with the Biotin RNA Labeling Mix (Roche) and T7 or Sp6 RNA polymerase (Roche). 1 μg whole-cell lysates from SW1116 cells were incubated with 3 μg of purified biotinylated transcripts for 1 h at 25 °C. The complexes were isolated with streptavidin agarose beads (Invitrogen) and send for RT-PCR and western blot analysis.
2.8. Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift analysis (EMSA) was performed as previously reported (Yan et al., 2009). The oligonucleotide 5′-GGGCTTGTCCGTATTTACGTTGAGGCGGGA-3′ and 5′-TCCCGCCTCAACGTAAATACGGACAAGCCC-3′, corresponding to the MEG3 were synthesized. The annealed oligonucleotide was labeled with [γ-32P] dATP using T4 polynucleotide kinase and used for the following binding reaction. The Clusterin protein (5 μg, R&D, 2937-HS-050 USA) and Clusterin antibody (Novus Cat# NBP1–68308-0.05 mg, RRID: AB_11027164) were used.
2.9. Luciferase Reporter Assay
The 293 T cells were plated at a subconfluent density in 24-well plate and co-transfected with 0.05 μg of the reporter plasmid with MEG3 promoter region, 0.5 μg of VDR expression or control vectors, and 0.05 μg of Renilla luciferase pRL-TK as an internal control for transfection efficiency. Cell lysates were prepared 48 h after transfection, and the reporter activity was measured using the Dual-luciferase reporter assay system (Promega).
2.10. Western Blotting
The level of proteins such as secretory Clusterin were investigated using western blotting performed following the general protocols. 20 μg total proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Membranes were blocked in 5% non-fat milk in Tris-buffered saline with 0.05% Tween-20 at room temperature for 2 h and then probed with antibodies against Clusterin (sc-6420; RRID: AB_2083323; Santa Cruz, CA, 1:200 dilution) and beta-actin (Abcam Cat# 1854–1, RRID:AB_764434, 1:1000 dilution), and probed with goat anti-rabbit secondary antibody conjugated with horseradish peroxidase (Abcam Cat# ab7088, RRID:AB_955595). The bands were observed with the enhanced chemiluminescence western blotting detection kit and visualized with a Tanon luminescent imaging workstation (Tanon, Shanghai, China).
2.11. Statistical Analyses
Quantitative variables were analyzed with the Student's t-test or the one-way ANOVA tests. The categorized characteristics between the groups were compared with χ2 test. The univariate and multivariate Cox regression models were applied to assess the association between the clinical characteristics and the overall survival or DFS of the patients. The Kaplan-Meier plots in together with the log-rank tests were used to compare the survival proportion between the groups. All statistical analyses were performed with SPSS 15.0 for Windows (SPSS, Chicago, IL). Two sided P < .05 was considered of statistically significant for all tests.
3. Results
3.1. MEG3 Expression is Decreased in CRC Tissues and Correlated with Poor Prognosis of CRC Patients
To evaluate MEG3 expression in CRC tissues, ISH was performed using MEG3 specific probes (Supplementary Fig. 1). Significantly decreased MEG3 expression (22/27) was observed in CRC tissues compared with that in the corresponding adjacent normal tissues (Wilcoxon test, P = .009; Fig. 1A). For further validation, MEG3 expression in another 61 paired CRC tissues was assessed using real-time PCR (RT-PCR). Approximately 77% (N = 47) of these CRC samples expressed a lower level of MEG3 compared with the corresponding normal tissues, while only 23% (N = 14) of samples showed a higher level of MEG3 (Wilcoxon test, P < .001; Fig. 1B). These results suggested that MEG3 was down-regulated in CRC patients and that it may be involved in CRC tumorigenesis and progression.
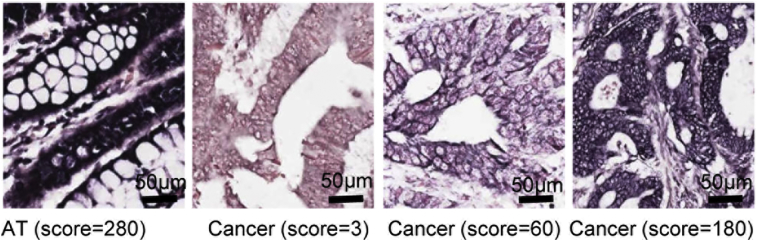
Supplementary Fig. 1.
Representative image of ISH of MEG3 expression in the CRC and the adjacent tissues. Positive staining of MEG3 was found in the cytoplasm and nucleus. Strong expression of MEG3 was noticed in adjacent tissues (AT) and the relatively lower MEG3 expression in CRC tissue (Cancer). The scores were calculated according to staining intensity and area.
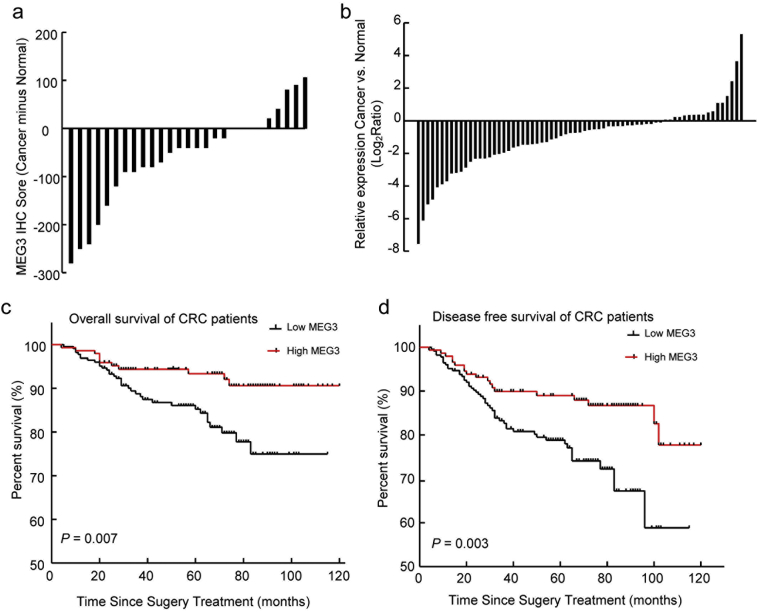
Fig. 1.
Association of MEG3 expression in CRC tissues with the prognosis of CRC patients. (A) Expression level of MEG3 in CRC (Cancer) and adjacent normal tissues (Normal) was determined by ISH (N = 27); (B) Expression level of MEG3 in CRC (Cancer) relative to adjacent normal tissues (Normal) was determined by RT-PCR (N = 61); (C and D) The Kaplan-Meier analysis of the relationship between MEG3 expression level and overall survival (OS) or disease-free survival (DFS) of CRC patients (Log rank-test, P = .007 and P = .003, respectively).
To investigate the association between MEG3 expression level and the prognosis of CRC patients, MEG3 expression was evaluated by ISH in TMAs containing 371 samples of CRC patients. Compared with patients with lower MEG3 levels (score ≤ 150; N = 225), patients with higher MEG3 levels (score > 150; N = 146) have relative better OS as suggested by the Kaplan-Meier plot (log-rank test; P = .007; Fig. 1C). The univariate analyses suggested that the TNM stage, CEA level, CA19-9 level and MEG3 expression levels were associated with the OS of CRC patients, while multivariate analyses indicated that MEG3 expression was an independent prognosis factor of OS for CRC patients (Table 1). In addition, Kaplan-Meier analyses also showed that patients with higher MEG3 expression have better DFS (log-rank test; P = .003; Fig. 1D) than those with lower MEG3 level, and that MEG3 is also an independent prognostic factor for the DFS of CRC patients (Table 1). These results suggest that MEG3 expression is decreased in CRC patients, and lower MEG3 expression level was correlated with poorer prognosis.
Table 1.
Association between the clinical characteristics and MEG3 expression level with the overall survival and disease-free survival of CRC patients (N = 371).
| Overall survival |
Disease-free survival |
|||||||
|---|---|---|---|---|---|---|---|---|
| Variable | Univariate analysis |
Multivariate analysis |
Univariate analysis |
Multivariate analysis |
||||
| HR (95% CI) | P-value | HR (95% CI) | P value | HR (95% CI) | P-value | HR (95% CI) | P value | |
| TNM stage (III vs. I+ II) | 2.34 (1.26–4.32) | 0.007 | 2.48 (1.33–4.60) | 0.004 | 2.26 (1.38–3.69) | 0.001 | 2.32 (1.42–3.80) | <0.001 |
| Depth of invasion [(T3 + T4) vs. (T1 + T2)] | 1.21 (0.60–2.44) | 0.591 | 1.57 (0.92–2.69) | 0.099 | ||||
| Sex (women vs. men) | 1.08 (0.61–1.92) | 0.785 | 0.92 (0.57–1.47) | 0.718 | ||||
| Tumor location (rectum vs.colon) | 0.82 (0.42–1.59) | 0.554 | 1.25 (0.76–2.06) | 0.379 | ||||
| Age (>60 vs. ≤60 years) | 1.32 (0.74–2.35) | 0.352 | 1.01 (0.63–1.62) | 0.967 | ||||
| Differentiation | ||||||||
| Moderately vs. well | 1.74 (0.42–7.25) | 0.444 | 1.25 (0.45–3.48) | 0.659 | ||||
| Poorly vs. well | 1.65 (0.27–9.91) | 0.585 | 1.28 (0.34–4.78) | 0.713 | ||||
| Treatment (yes vs. no) | 1.85 (0.73–4.68) | 0.195 | 2.13 (0.97–4.67) | 0.058 | ||||
| CEA (>20 ng/mL vs. <20 ng/mL) | 2.91 (1.48–5.72) | 0.002 | 2.83 (1.28–6.25) | 0.010 | 2.08 (1.12–3.88) | 0.021 | 2.11 (1.03–4.32) | 0.041 |
| CA199 (>22.32 U/mL vs. <22.32 U/mL) | 1.79 (1.01–3.18) | 0.047 | 1.41 (0.75–2.67) | 0.289 | 1.30 (0.80–2.12) | 0.284 | ||
| MEG3 (high vs. low) | 0.37 (0.19–0.74) | 0.005 | 0.34 (0.16–0.71) | 0.004 | 0.45 (0.27–0.78) | 0.004 | 0.43 (0.25–0.76) | 0.004 |
3.2. MEG3 Inhibits Cellular Proliferation and Tumor Growth of CRC Cells in vitro and in vivo
We next intended to investigate the biological activities of MEG3 in CRC cells. The mRNA level of MEG3 in 7 CRC cell lines was determined, among which RKO expressed the highest and SW1116 and LoVo showed the lowest MEG3 levels (Fig. 2A). In RKO cells transfected with siRNA of MEG3 significantly reduced its mRNA levels (Supplementary Fig. 2A). MEG3 knockdown led to a significant increased cellular proliferation compared with the blank or negative control group (Fig. 2B). In contrast, MEG3 expressing lentiviral transfection for SW1116 and LoVo cells significantly increased the MEG3 expression levels (Supplementary Fig. 2B), and led to the reduced cellular proliferation compared to control lentivirus as suggested by the CCK-8 assay (Fig. 2C).
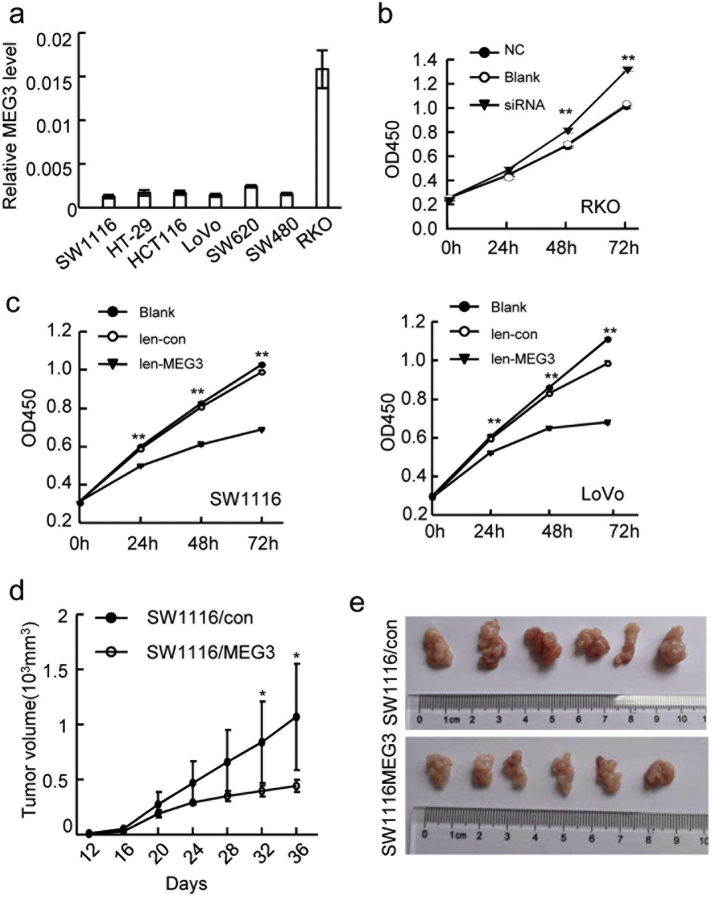
Fig. 2.
MEG3 inhibits the cellular proliferation and tumor growth in vitro and in vivo. (A) Relative expression of MEG3 in 7 CRC cell lines was determined by RT-PCR. (B) The proliferation of RKO cells with siRNA transfection or not was assessed by CCK-8; (C) The proliferation of SW1116 (left) and LoVo (right) cells with MEG3 over-expression or not was assessed by CCK-8 assays. (D) The growth curves of the xenograft tumors among the different groups (SW1116/MEG3 and SW1116/con) (*P < .05, **P < .01); (E) The image of tumors derived from SW1116/MEG3 or SW1116/con 36 days after injection. The data are representative of at least three independent experiments.
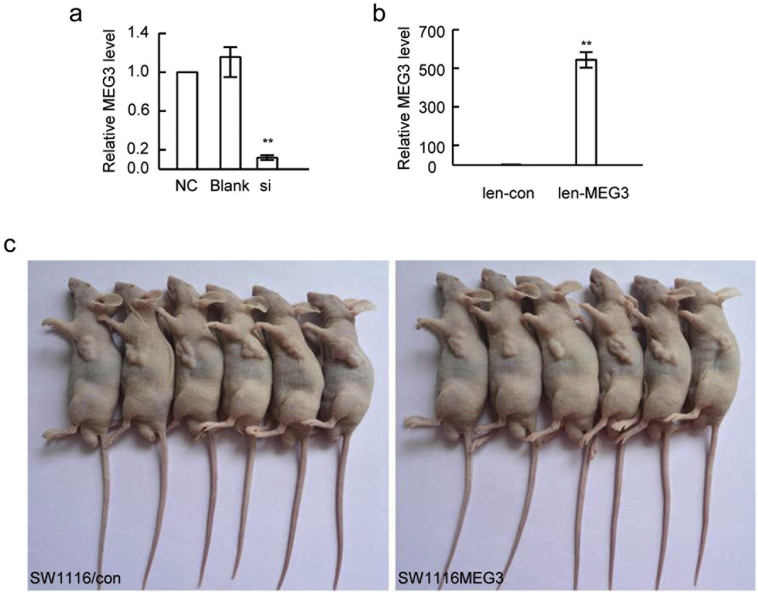
Supplementary Fig. 2.
MEG3 expression in CRC cells with MEG3 over-expression or down-regulationand image of mice model. (A) down-regulation of MEG3 in RKO cells by siRNA transfection (si) (**P < .01); (B) MEG3 over-expression in SW1116 cells infected by lentivirus expressing MEG3 (len-MEG3) (**P < .01); (C) Smaller tumors were observed in the nude mice injected withSE1116 cells with MEG3 over-expression (SW1116/MEG3 cells, right) compared with control group (SW1116/con, left) after 36 days.
To further evaluate the roles of MEG3 in tumor growth in vivo, SW1116 cells with or without stable MEG3 over-expression (SW1116/MEG3 or SW1116/con) were subcutaneously injected into nude mice. For every four days, the tumor size was measured and the tumor growth curve suggested that SW1116 cells with MEG3 over-expression was significantly inhibited compared to control group (N = 6 for each group, Fig. 2D). At the end of the experiment, the tumor size was also significantly reduced in the mice injected with SW1116/MEG3 compared with those injected with control cells in the xenograft animal models (Fig. 2E, Supplementary Fig. 2C). These results indicated that MEG3 inhibited the cellular proliferation and tumor growth of CRC cells in vitro and in vivo.
3.3. MEG3 Suppresses CRC Cell Migration and Metastasis in vitro and in vivo
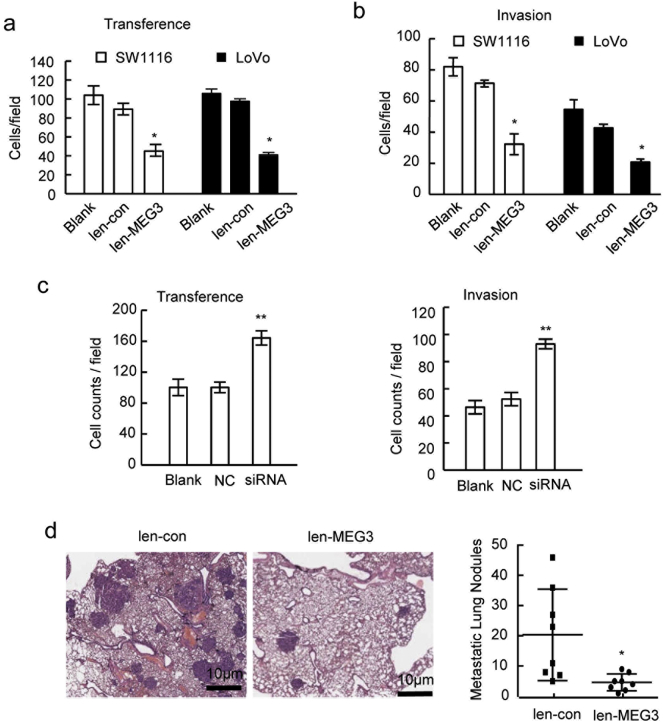
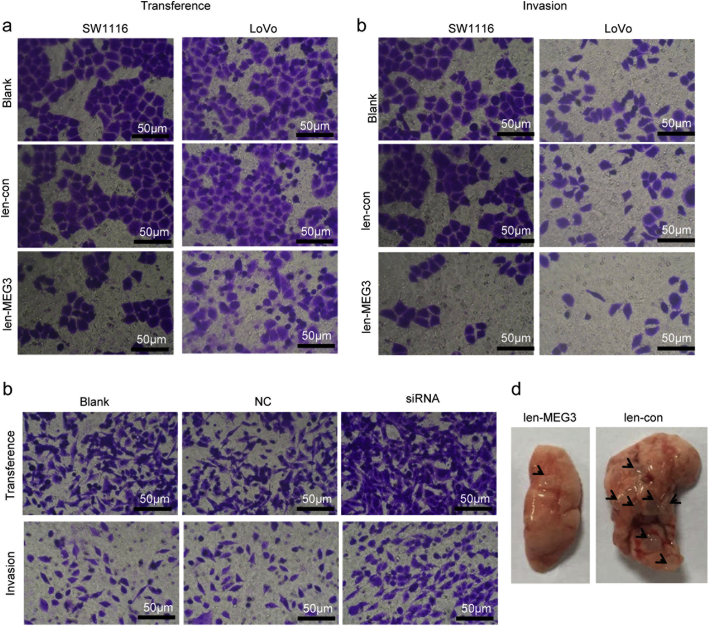
We also evaluated the influences of MEG3 on the migration activities of the CRC cells in vitro and in vivo. The trans-well migration assays showed that MEG3 over-expression in SW1116 and LoVo cells significantly inhibited the migration capacities of tumor cells (Fig. 3A, Supplementary Fig. 3A). The invasion assays suggested that MEG3 over-expression significantly inhibits the invasiveness abilities of the two cell lines (Fig. 3B, Supplementary Fig. 3B). By contrast, when MEG3 was knocked-down in RKO cells, the migration and invasion abilities of tumor cells were significantly enhanced (Fig. 3C, Supplementary Fig. 3C).
Fig. 3.
MEG3 suppresses the CRC cell migration and metastasis in vitro and in vivo. The cell migration (A) and invasion (B) ability of SE1116 and LoVo with MEG3 over-expression was significantly decreased compared with the blank or vector group (*P < .05); (C) Cell migration and invasion ability of RKO cells with MEG3 down-regulation was significantly higher than that of negative control or blank control group (**P < .01); (D) the represent image of lung tissues with tumor node (left, HE staining) and the number of lung metastases nodes in groups of SW1116/MEG3 and SW1116/con (right, *P < .05). The data are representative of at least three independent experiments.
Supplementary Fig. 3.
MEG3 inhibits the cellular migration and invasion in vitro and in vivo. Representative image of the trans-well assay to investigate migration ability (A) and invasion ability (B) of SW1116 and LoVo cells with MEG3 over-expression or not. (C) Representative image of the trans-well assay to investigate migration ability (upper) and invasion ability (lower) of RKO cells with MEG3 down-regulaton or not. (D) Representative image of lung tissues from tumor metastasis mice model. Tumor modals indicated by arrows.
For further verification, the in vivo tumor metastasis mice model was established through injecting SW1116/MEG3 cells into nude mice via the tail vein. Seven weeks later, the mice were sacrificed and the metastasis loci in the lungs were monitored by the HE staining. Fewer metastasis loci were observed in lung tissues from mice injected with SW1116/MEG3 cells than in those from the control group (Fig. 3D, Supplementary Fig. 3D). These results suggested that MEG3 inhibits the metastatic features of CRC cells both in vitro and in vivo.
3.4. MEG3 Inhibits Proliferation and Metastasis through Regulating Clusterin
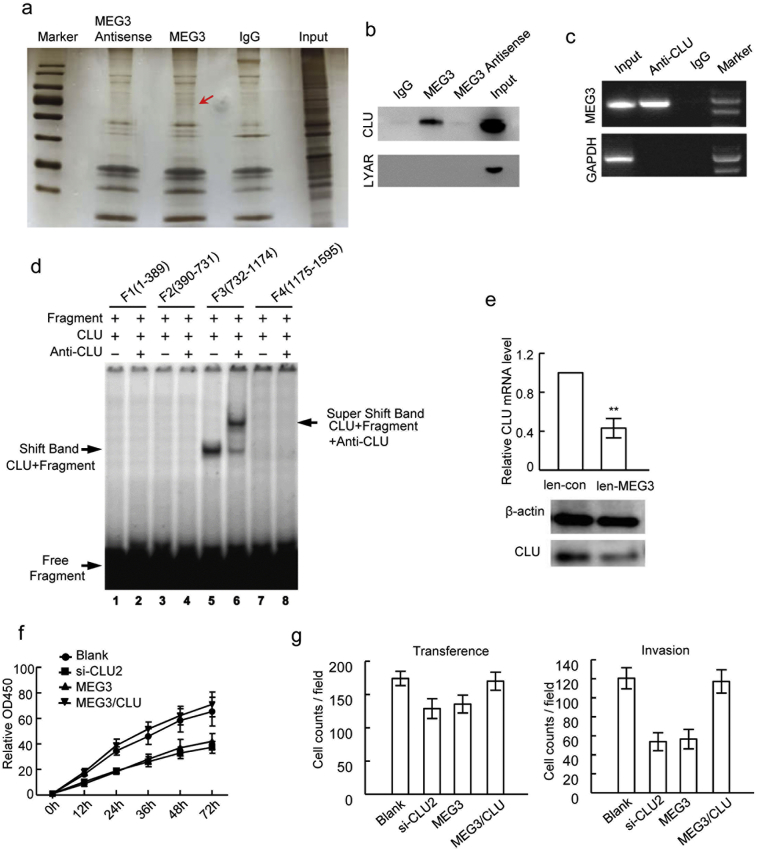
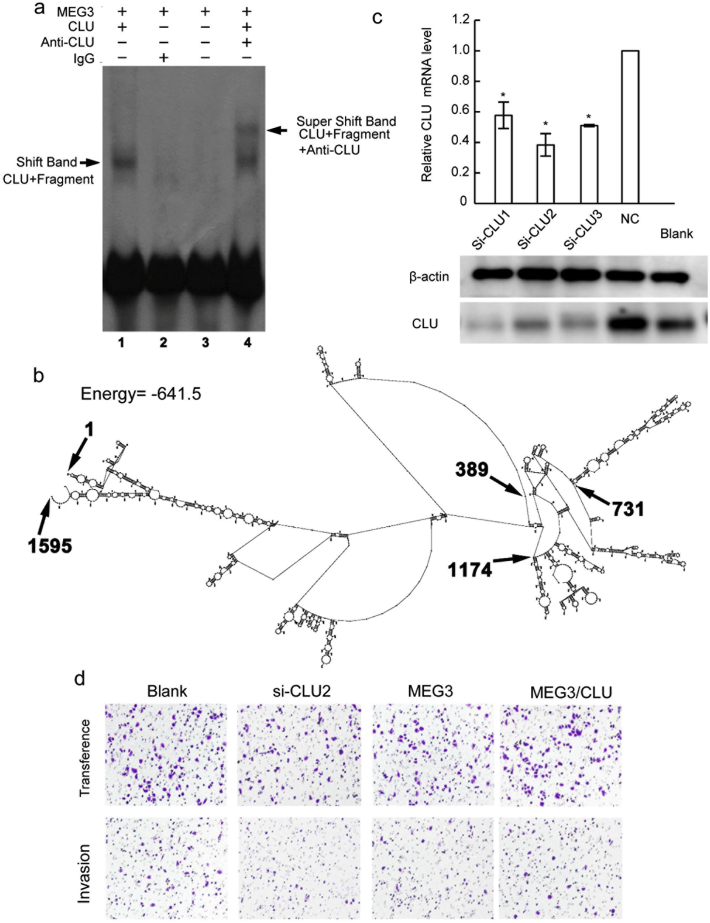
lncRNAs could bind to the target proteins to modulate gene expression and biological processes (Zhu et al., 2013). To identify target proteins interacting with MEG3 in CRC cells, the RNA-pull down assays were performed using biotin labeled MEG3, and the precipitated proteins were applied for SDS-PAGE electrophoresis. Those protein bands specifically evident in MEG3 precipitates, but not in the IgG and MEG3 antisense controls, were then analyzed by mass spectrometry (Fig. 4A). 68 proteins were identified by mass spectrometry, which may potentially bind to lncRNA MEG3 (Supplementary Table S1). Among them, secretory Clusterin (CLU) and cell growth-regulating nucleolar protein (LYAR) were selected for further verification due to their tight association with tumors (Wu et al., 2015; Chen et al., 2003). The precipitate from the MEG3 RNA-pull down was analyzed by western blotting using CLU or LYAR antibody. The secretory CLU was found specifically in the MEG3 precipitate but not in that from the IgG and MEG3 antisense control, while LYAR was not found in all three groups (Fig. 4B), suggesting there could be direct physical binding between MEG3 and CLU. The following RIP using CLU antibody and EMSA also suggested MEG3 was directly bind with CLU (Fig. 4C, Supplementary Fig. 4A). To identify the binding sites of MEG3 with CLU, four MEG3 mRNA fragments (F1:1–389; F2:390–731; F3:732–1174, and F4:1175–1595), truncated based on the predicted RNA structure of MEG3, were cloned and used for EMSA analyses (Supplementary Fig. 4B). In the EMSA assays, a significant band shift was noticed for the F3 fragment of MEG3 mRNA, suggesting that this segment (732–1174 nucleotide acids) contained the binding sites for CLU (Fig. 4D). We next determined whether MEG3 affects the expression level of secretory CLU in CRC cells. Decreased CLU mRNA level was observed when MEG3 was over-expressed in SW1116 cells (Fig. 4E) and the corresponding secretory CLU protein level was also significantly decreased in the SW1116 cells as suggested by western blotting (Fig. 4E).
Fig. 4.
MEG3 interacts with Clusterin and inhibites proliferation and metastasis. (A) The represent image of SDS-PAGE with precipitated proteins from MEG3 RNA pull down. Gel indicated by arrow was obtained for mass spectrometry analysis; (B) The represent image of western blotting for detection of LYAR and CLU in precipitated proteins from MEG3 RNA pull down; (C) The represent image of agarose gel for MEG3 detection in the precipitation from RIP by CLU antibody; (D) The represent image of EMSA to investigate the binding site of CLU protein in MEG3.MEG3 was truncated into four fragments: F1–F4; (E) Up-regulation of CLU by MEG3 over-expression (**P < .01); (F and G) The cellular proliferation, migration and invasion abilities of SW1116 cells transfected with CLU siRNAs (SW1116/si-CLU), SW1116 cells with MEG3 over-expression (SW1116/MEG3), SW1116 cells with MEG3 and CLU over-expression (SW1116/MEG3/CLU) and blank control cells detected by CCK8 (F) and trans well assays (G). The data are representative of at least three independent experiments.
Supplementary Fig. 4.
MEG3 inhibits the CLU through directly binding to CLU protein. (A) MEG3 directly binds to CLU protein as the CLU antibody formed RNA-protein-antibody complexes suggested by EMSA assay; (B) The MEG3 spatial structure predicted by RNA structure (version 3.7). MEG3 was divided into four segments according to prediction, F1:1–389, f2: 390–731, F3: 732–1174, F4: 1175–1598; (C) C expression of LU mRNA and protein in SW1116 transfected with CLU siRNAs or not etermined by RT-PCR and western-blot (*P < .05); (D) Representative image of the trans-well assay to investigate migration ability (upper) and invasion ability (lower) of CRC cells with CLU down-regulation(si-CLU2), MEG3 over-expression (MEG3), MEG3 over-expression combinated with CLU over-expression (MEG3/CLU) or not (Blank).
Moreover, whether secretory CLU interferes the anticancer activities of MEG3 in CRC was determined through knockdown of CLU by siRNAs (Supplementary Fig. 4C). The proliferation and metastasis properties of SW1116 cells with CLU down-regulation (si-CLU2), MEG3 overexpression (MEG3) and MEG3 and CLU co-overexpression (MEG3/CLU) were determined. The CCK-8 assays suggested that SW1116 cells with CLU down-regulation was correlated with reduced proliferation capacities than control cells and that MEG3 over-expression significantly inhibited SW1116 cells proliferation (Fig. 4F). When MEG3 and CLU were simultaneously over-expressed, the SW1116 cells show the similar proliferation capacities with the control cells (Fig. 4F). The migration and invasion assays showed that both CLU down-regulation and MEG3 over-expression could reduce the migration abilities of SW1116 cells and that CLU over-expression weakened the anti-metastasis activities of MEG3 in SW1116 cells (Fig. 4G; Supplementary Fig. 4D). Collectively, these data suggested that CLU inhibits the anticancer activities of the MEG3 in CRC cells.
3.5. Vitamin D Signaling Pathway Regulates MEG3 Expression in CRC Cells
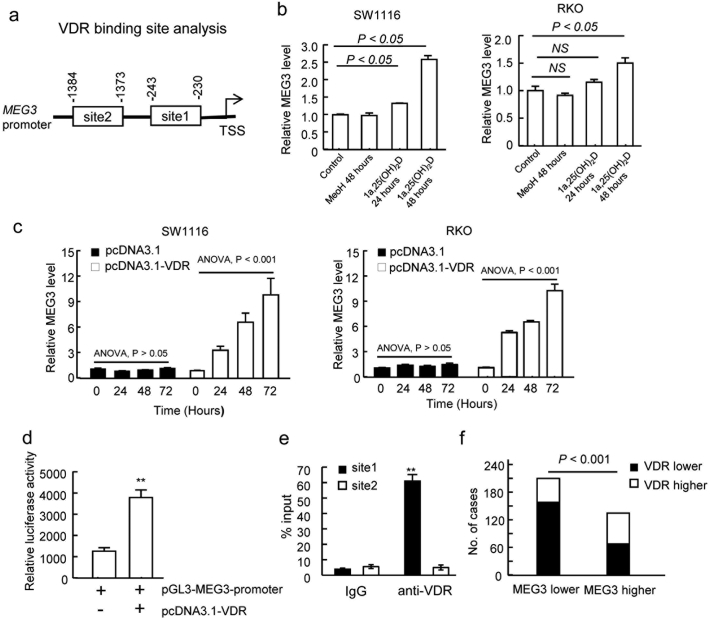
Evidence shows that the vitamin D levels are inversely associated with CRC risk and positively associated with improved overall and CRC-specific survival (Morales-oyarvide et al., 2016). Vitamin D and its nuclear transcriptional receptor VDR have chemo-prevention activities for CRC (Aggarwal and Kallay, 2016). Vitamin D supplementation could decrease the inflammation-associated colorectal tumors incidence in mice, suggesting the pivotal roles of vitamin D in CRC development (Elimrani et al., 2017). To determine whether vitamin D regulates MEG3 transcription in CRC cells, we analyzed the transcriptional factor binding sites in the promoter region of MEG3 (−1500 bp to 0 bp) and found two potential VDR binding motifs (site 1: −230~−243; site2: −1373~−1384; Fig. 5A) according to the genome-wide ChIP-seq study performed by Ramagopalan et al. (2010). To elucidate whether vitamin D could activate MEG3 transcription, SW1116 cells were treated with 1α,25-(OH)2D, the VDR ligand in cells. Significantly increased MEG3 levels were observed 24 and 48 h after the treatment compared with the baseline or ethanol vehicle treatments (Fig. 5B). Similar results were observed in the RKO cells that received 1α, 25-(OH)2D treatment (Fig. 5B). In addition, over-expression of VDR increased MEG3 expression both in SW1116 and RKO cells compared to the vector control with time-dependent manner (Fig. 5C). These results suggested that VDR might stimulate the transcription of MEG3 in CRC cells. In further, we applied the dual luciferase reporter assay and ChIP assay to verify whether VDR directly binds to the MEG3 promoter. The promoter of MEG3 (−1500–1) was cloned into the pGL3 vector (pGL3-MEG3-promoter) and then co-transfected with VDR expression vector (pcDNA3.1-VDR) or control vector into 293 T cells. The pGL3-MEG3-promoter showed significantly higher relative luciferase activities when co-transfected with pcDNA3.1-VDR (Fig. 5D), suggesting the potential bind of VDR on the MEG3 promoter. The following ChIP assays using the VDR antibody found that the site 1 segment, but not the site 2 segment, was enriched in the ChIP precipitate (Fig. 5E), suggesting that VDR may binds to the site 1 (−230 ~ − 243) region of the MEG3 promoter. These results suggested that VDR may stimulate MEG3 transcription through direct binding to its promoter at the −230 ~ − 243 region.
Fig. 5.
VDR regulates MEG3 through directly binding to the promoter region of MEG3 in CRC cells. (A) Schematic of VDR binding site (site1 and site2) in the promoter of MEG3 by bioinformatics analysis; (B) up-regulated MEG3 expression in SW1116 and RKO cells by 1α,25-(OH)2D treatment for 24 and 48 h determined by RT-PCR; (C) MEG3 expression in SW1116 and RKO cells with VDR over-expression (pcDNA3.1-VDR) or not (pcDNA3.1 vector); (D) Enhancement of MEG3 promoter activity by VDR assessed by the luciferase reporter assay; (E) Binding of VDR with MEG3 promoter verified by the ChIP assay using VDR antibody (**P < .01); (F) Correlation between expression level of VDR and MEG3 in colorectal cancer tissues assessed by IHC and ISH staining. The data are representative of at least three independent experiments.
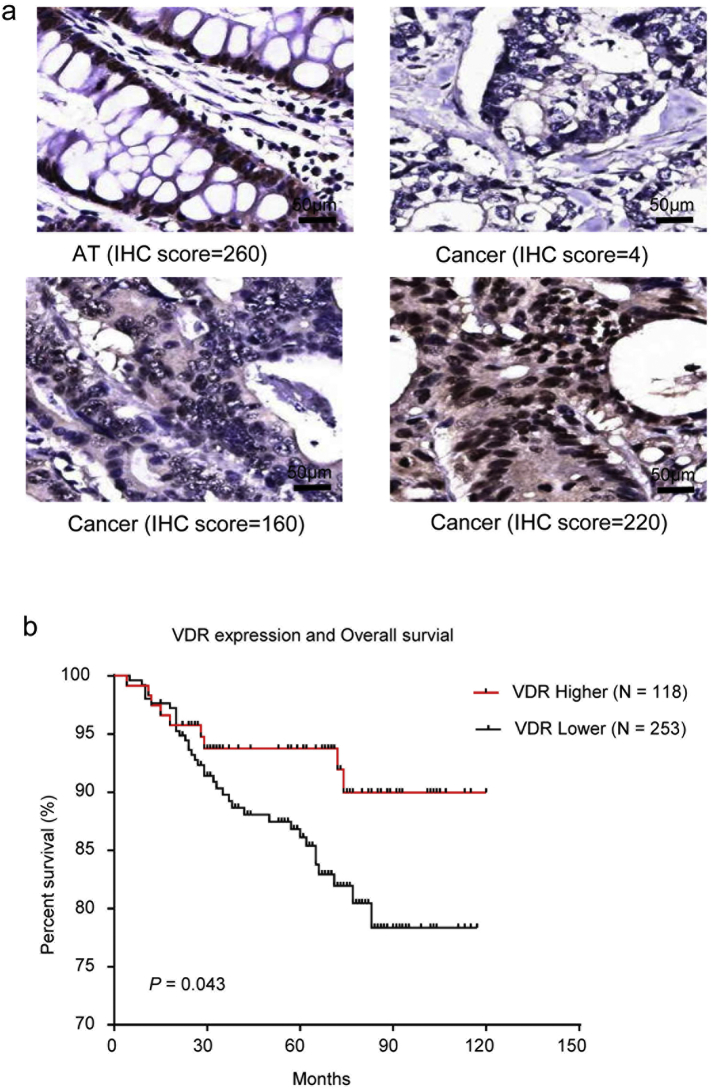
Furthermore, we determined the association between the MEG3 and VDR expression levels in CRC tissues. A significantly positive correlation between the VDR protein level and MEG3 mRNA levels was noticed (P < .001, N = 371, Fig. 5F, Supplementary Fig. 5A). Additionally, Kaplan-Meier analysis showed that lower VDR expression was associated with poorer overall survival of CRC patients (log-rank test, P = .043; Supplementary Fig. 5B), suggesting that loss of function for VDR may lead to poorer prognosis of CRC patients through down-regulating the MEG3 levels.
Supplementary Fig. 5.
VDR expression level was associated with overall survival of CRC patients. (A) Representativeative images of VDR expression in the CRC tissues (Cancer) and the adjacent tissues (AT) by IHC. The scores were calculated according to staining intensity and area. (B) Higher VDR expression level was associated with better overall survival for CRC patient (Log rank-test, P = .043).
4. Discussion
Although the mortality of CRC has decreased steadily in recent years, it remains the fourth most common cancer deaths globally (Arnold et al., 2017). Uncovering the genetic and epigenetic alterations that are involved in the pathogenesis of CRC may guide the prevention and clinical treatments for patients (Walsh and Terdiman, 2003). Recently, dysregulation of lncRNAs were involved in the tumorigenesis and progression of various types of cancers including CRC, which might serve as potential biomarkers or targets for disease prevention or treatment (Luo et al., 2017). MEG3 is a novel lncRNA that functions as a tumor suppressor gene in glioma, gastric cancer, lung cancer and liver cancer (Zhang et al., 2003). Bando T et al. firstly reported heterogeneous deletions of the 14q32 region encoding MEG3 in CRC tissues, suggesting the potential involvement of MEG3 in CRC development (Bando et al., 1999). Previous studies have reported that MEG3 served as a prognostic factor for CRC and modulated the chemotherapy sensitivity of CRC cells; however, the underlying mechanisms remain unclear (Kong et al., 2016; Li et al., 2017). In the current study, we confirmed that MEG3 expression was frequently down-regulated in CRC tissues and MEG3 knockdown stimulated the migration and proliferation of CRC cell lines. Moreover, we found that MEG3 was also an independent prognostic factor for the overall survival and disease-free survival of CRC patients as suggested by the multivariate Cox regression models. These results suggested that MEG3 could serve as a novel prognosis biomarker for CRC and might be a potential therapeutic target for CRC patients.
Clusterin is frequently overexpressed in multiple types of cancer, including prostate, breast, lung and colorectal cancer (Lee et al., 2016). It has been reported that Clusterin inhibits TNF-α induced apoptosis in breast cancer cells through activating the NF-κB and Bcl-2 signaling pathways (Wang et al., 2012). It also stimulates the migration and metastasis of the cells through activating the EIF3I/Akt/MMP13 signaling pathways in hepatocellular carcinoma (Wang et al., 2015). In CRC cells, Clusterin knockdown mimics the anticancer activities of MEG3, and Clusterin over-expression weakens the inhibition activities of MEG3 in cellular proliferation and metastasis, suggesting that Clusterin partially underlies the anti-cancer activities of MEG3 in CRC cells. In the present study, we reported that Clusterin was directly bound to the lncRNA MEG3. EMSA found that MEG3 directly binds to Clusterin at its 732–1174 nucleotide acid region. As the binding between Clusterin and MMP-9 or VEGF is critical in the metastasis in nasopharyngeal carcinoma (Li et al., 2016) and colon cancer (Radziwon-Balicka et al., 2014), MEG3 binding may impede the interactions between Clusterin and its target proteins. However, whether the directly binding with MEG3 could influence the Clusterin function need to be resolved with more investigation. Interestingly, MEG3 over-expression was also found to correlate with Clusterin transcription inhibition, suggesting that MEG3 might inhibit Clusterin activities through multiple mechanisms. As cAMP was reported to stimulate MEG3 expression and down-regulate clusterin expression (Zwain and Amato, 2001; Zhao et al., 2006), MEG3 may possibly rule clusterin regulation through cAMP signaling pathways. It has also been reported that MEG3 regulates gene expression via binding to distal regulatory elements in DNA or recruiting miRNAs or proteins to destabilize target gene mRNA (Mondal et al., 2015; Su et al., 2016; Modali et al., 2015), suggesting a possible regulation of clusterin by MEG3 at transcript level. In addition, other proteins, except CLU and LYAR, were also detected in the MEG3 pull-down assay. Whether these proteins are involved in the anti-cancer activities of MEG3 in CRC remain unclear. Elucidating their roles may provide further insights into the mechanisms of MEG3 in CRC development and progression.
We also reported that VDR may directly regulate the MEG3 level in colorectal cells through binding to its promoter. Vitamin D is an essential vitamin that was found to be associated with CRC development and a poor prognosis (Li et al., 2014). Multiple genes, including lncRNAs encoding genes such as H19 and HOTAIR have been found to be regulated by Vitamin D through VDR in multiple cancers (Jiang and Bikle, 2014a; Jiang and Bikle, 2014b). Here, we found that vitamin D treatment or VDR over-expression up-regulated MEG3 expression in CRC cells. VDR may activate MEG3 expression via direct binding to its promoter at the −230 ~ − 243 promoter region. Accordingly, Menigatti et al. reported that MEG3 was epigenetically silenced in precancerous colorectal lesions, possibly leading to the reduced transcriptional level of MEG3 by VDR (Menigatti et al., 2013). In clinic, VDR level was positively correlated with the MEG3 level in CRC tissues, and patients with higher VDR were associated with better OS. These results suggested that the regulation of MEG3 expression by vitamin D/VDR can partially explain why CRC patients with vitamin D deficiency showed poorer prognosis (Duffy et al., 2017). The down-regulation of MEG3 might also confer the increased risk of CRC development under the vitamin D deficiency status, which needs to be addressed with more investigations.
In conclusion, we found that MEG3 expression was frequently down-regulated in CRC tissues and CRC patients with lower MEG3 level showed poorer prognosis. MEG3 inhibited metastasis and proliferation of CRC tumor cells both in vivo and in vitro, which may be caused through down-regulating clusterin expression level and its directly binding to CLU protein. In addition, vitamin D activated MEG3 transcription in CRC tumor cells via direct binding to the MEG3 promoter by VDR. These results suggest the potential roles of MEG3 in CRC development and progression and provide further insights into the mechanisms of the anti-cancer activities of vitamin D in CRC development and progression.
Funding Sources
This work was supported by the grants of China National Funds for National Natural Science Fund (No. 81672899), Science and Technology Commission of Shanghai Municipality (16140900703 and 16411966800), Shanghai Municipal Commission of Health and Family Planning (No. 2015ZB0202 and 20164Y0250). The funders had no role in study design, data collection, data analysis, interpretation, and writing of the report.
Conflicts of Interest
None.
Author Contributions
The conception and design of the study: Jianming Zheng, Shupeng Liu, Peizhan Chen and Yan Zhu; Acquisition of data: Yan Zhu, Peizhan Chen, Yisha Gao, Na Ta, Yunshuo Zhang, Jialin Cai and Yong Zhao; Analysis and interpretation of data: Yan Zhu, Peizhan Chen and Shupeng Liu; Drafting the article: Yan Zhu, Peizhan Chen and Shupeng Liu; Revising it critically for important intellectual content: Yan Zhu, Peizhan Chen, Shupeng Liu and Jianming Zheng; Final approval of the version to be submitted: Shupeng Liu and Jianming Zheng.
Yan Zhu and Peizhan Chen contributed equally to this article.
Research in context
The mechanisms underlying colorectal cancer (CRC) development and progression are largely unknown. Here, we found that MEG3 was down-regulated in CRC cells and its expression level was an independent prognostic factor for CRC patients in clinic. MEG3 over-expression suppressed CRC cell proliferation and metastasis in vivo and in vitro. MEG3 reduced the transcription of Clusterin and the corresponding protein level in CRC cells. MEG3 also directly bind to Clusterin protein via its 732–1174 nucleotide acid region. In further, Clusterin overexpression could impede the anti-proliferation and anti-metastasis activities of MEG3 in CRC cells. Moreover, 1α,25-(OH)2D could activate MEG3 expression in CRC cells via its nuclear receptor VDR (vitamin D receptor), providing further insights into how Vitamin D inhibits CRC development and progression in clinic. These results indicate that VDR/MEG3/Clusterin signaling pathway may serve as potential therapeutic targets and prognosis biomarkers for CRC.
The following are the supplementary data related to this article.
Potential interacting proteins of MEG3 identified by the mass spectrometry.
Contributor Information
Shupeng Liu, Email: lshu_p@aliyun.com.
Jianming Zheng, Email: jmzheng1962@163.com.
References
- Aggarwal A., Kallay E. Cross talk between the calcium-sensing receptor and the vitamin D system in prevention of cancer. Front. Physiol. 2016;7:451. doi: 10.3389/fphys.2016.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- Bando T., Kato Y., Ihara Y., Yamagishi F., Tsukada K., Isobe M. Loss of heterozygosity of 14q32 in colorectal carcinoma. Cancer Genet. Cytogenet. 1999;111:161–165. doi: 10.1016/s0165-4608(98)00242-8. [DOI] [PubMed] [Google Scholar]
- Cai J., Li B., Zhu Y., Fang X., Zhu M., Wang M., Liu S., Jiang X., Zheng J., Zhang X., Chen P. Prognostic biomarker identification through integrating the gene signatures of hepatocellular carcinoma properties. EBioMedicine. 2017;19:18–30. doi: 10.1016/j.ebiom.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Halberg R.B., Ehrhardt W.M., Torrealba J., Dove W.F. Clusterin as a biomarker in murine and human intestinal neoplasia. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9530–9535. doi: 10.1073/pnas.1233633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., He M., Chen L., Chen C., Zheng J., Cai Z. The loss of miR-26a-mediated post-transcriptional regulation of cyclin E2 in pancreatic cancer cell proliferation and decreased patient survival. PLoS One. 2013;e76450:8. doi: 10.1371/journal.pone.0076450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M.J., Murray A., Synnott N.C., O'donovan N., Crown J. Vitamin D analogues: potential use in cancer treatment. Crit. Rev. Oncol. Hematol. 2017;112:190–197. doi: 10.1016/j.critrevonc.2017.02.015. [DOI] [PubMed] [Google Scholar]
- Elimrani I., Koenekoop J., Dionne S., Marcil V., Delvin E., Levy E., Seidman E.G. Vitamin D reduces colitis- and inflammation-associated colorectal Cancer in mice independent of NOD2. Nutr. Cancer. 2017;69:276–288. doi: 10.1080/01635581.2017.1263346. [DOI] [PubMed] [Google Scholar]
- Guinney J., Dienstmann R., Wang X., de Reynies A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., Bot B.M., Morris J.S., Simon I.M., Gerster S., Fessler E., de Sousa E.M.F., Missiaglia E., Ramay H., Barras D., Homicsko K., Maru D., Manyam G.C., Broom B., Boige V., Perez-Villamil B., Laderas T., Salazar R., Gray J.W., Hanahan D., Tabernero J., Bernards R., Friend S.H., Laurent-Puig P., Medema J.P., Sadanandam A., Wessels L., Delorenzi M., Kopetz S., Vermeulen L., Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.J., Bikle D.D. LncRNA profiling reveals new mechanism for VDR protection against skin cancer formation. J. Steroid Biochem. Mol. Biol. 2014;144(Pt A):87–90. doi: 10.1016/j.jsbmb.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Jiang Y.J., Bikle D.D. LncRNA: a new player in 1alpha, 25(OH)(2) vitamin D(3)/VDR protection against skin cancer formation. Exp. Dermatol. 2014;23:147–150. doi: 10.1111/exd.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H., Wu Y., Zhu M., Zhai C., Qian J., Gao X., Wang S., Hou Y., Lu S., Zhu H. Long non-coding RNAs: novel prognostic biomarkers for liver metastases in patients with early stage colorectal cancer. Oncotarget. 2016;7:50428–50436. doi: 10.18632/oncotarget.10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Kim H.J., Rho S.B., Lee S.H. eIF3f reduces tumor growth by directly interrupting clusterin with anti-apoptotic property in cancer cells. Oncotarget. 2016;7:18541–18557. doi: 10.18632/oncotarget.8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chen P., Li J., Chu R., Xie D., Wang H. Review: the impacts of circulating 25-hydroxyvitamin D levels on cancer patient outcomes: a systematic review and meta-analysis. J. Clin. Endocrinol. Metab. 2014;99:2327–2336. doi: 10.1210/jc.2013-4320. [DOI] [PubMed] [Google Scholar]
- Li Y., Lu J., Zhou S., Wang W., Tan G., Zhang Z., Dong Z., Kang T., Tang F. Clusterin induced by N,N′-Dinitrosopiperazine is involved in nasopharyngeal carcinoma metastasis. Oncotarget. 2016;7:5548–5563. doi: 10.18632/oncotarget.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shang J., Zhang Y., Liu S., Peng Y., Zhou Z., Pan H., Wang X., Chen L., Zhao Q. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol. Rep. 2017;38:1383–1392. doi: 10.3892/or.2017.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zi Y., Wang W., Li Y. LncRNA MEG3 inhibits cell proliferation and metastasis in chronic myeloid leukemia via targeting MiR-184. Oncol. Res. 2018;26:297–305. doi: 10.3727/096504017X14980882803151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Qiu M., Xu R., Dobs A.S. Comparison of survival and clinicopathologic features in colorectal cancer among African American, Caucasian, and Chinese patients treated in the United States: results from the surveillance epidemiology and end results (SEER) database. Oncotarget. 2015;6:33935–33943. doi: 10.18632/oncotarget.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.H., Li W., Liu X.H., Sun M., Zhang M.L., Wu W.Q., Xie W.P., Hou Y.Y. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Qu J., Wu D.K., Lu Z.L., Sun Y.S., Qu Q. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget. 2017;8:22187–22202. doi: 10.18632/oncotarget.14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Yang Y., Wang F., Moyer M.P., Wei Q., Zhang P., Yang Z., Liu W., Zhang H., Chen N., Wang H., Wang H., Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menigatti M., Staiano T., Manser C.N., Bauerfeind P., Komljenovic A., Robinson M., Jiricny J., Buffoli F., Marra G. Epigenetic silencing of monoallelically methylated miRNA loci in precancerous colorectal lesions. Oncogene. 2013;2 doi: 10.1038/oncsis.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modali S.D., Parekh V.I., Kebebew E., Agarwal S.K. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol. Endocrinol. 2015;29:224–237. doi: 10.1210/me.2014-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E., Moustakas A., Gyllensten U., Jones S.J., Gustafsson C.M., Sims A.H., Westerlund F., Gorab E., Kanduri C. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-oyarvide V., Meyerhardt J.A., Ng K. Vitamin D and physical activity in patients with colorectal Cancer: epidemiological evidence and therapeutic implications. Cancer J. 2016;22:223–231. doi: 10.1097/PPO.0000000000000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka M., Ling H., Ivan C., Pichler M., Matsushita D., Goblirsch M., Stiegelbauer V., Shigeyasu K., Zhang X., Chen M., Vidhu F., Bartholomeusz G.A., Toiyama Y., Kusunoki M., Doki Y., Mori M., Song S., Gunther J.R., Krishnan S., Slaby O., Goel A., Ajani J.A., Radovich M., Calin G.A. H19 noncoding RNA, an independent prognostic factor, regulates essential Rb-E2F and CDK8-beta-catenin signaling in colorectal Cancer. EBioMedicine. 2016;13:113–124. doi: 10.1016/j.ebiom.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Cao F., Guo A., Chang W., Chen X., Ma W., Gao X., Guo S., Fu C., Zhu J. Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis. Br. J. Cancer. 2015;113:763–772. doi: 10.1038/bjc.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Radziwon-Balicka A., Santos-Martinez M.J., Corbalan J.J., O'sullivan S., Treumann A., Gilmer J.F., Radomski M.W., Medina C. Mechanisms of platelet-stimulated colon cancer invasion: role of clusterin and thrombospondin 1 in regulation of the P38MAPK-MMP-9 pathway. Carcinogenesis. 2014;35:324–332. doi: 10.1093/carcin/bgt332. [DOI] [PubMed] [Google Scholar]
- Ramagopalan S.V., Heger A., Berlanga A.J., Maugeri N.J., Lincoln M.R., Burrell A., Handunnetthi L., Handel A.E., Disanto G., Orton S.M., Watson C.T., Morahan J.M., Giovannoni G., Ponting C.P., Ebers G.C., Knight J.C. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Han D., Wu J., Huo X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumour Biol. 2016;37:3925–3931. doi: 10.1007/s13277-015-4151-2. [DOI] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Walsh J.M., Terdiman J.P. Colorectal cancer screening: scientific review. JAMA. 2003;289:1288–1296. doi: 10.1001/jama.289.10.1288. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang X., Zhao H., Liang B., Du Q. Clusterin confers resistance to TNF-alpha-induced apoptosis in breast cancer cells through NF-kappaB activation and Bcl-2 overexpression. J. Chemother. 2012;24:348–357. doi: 10.1179/1973947812Y.0000000049. [DOI] [PubMed] [Google Scholar]
- Wang C., Jin G., Jin H., Wang N., Luo Q., Zhang Y., Gao D., Jiang K., Gu D., Shen Q., Huo X., Hu F., Ge T., Zhao F., Chu W., Shu H., Yao M., Cong W., Qin W. Clusterin facilitates metastasis by EIF3I/Akt/MMP13 signaling in hepatocellular carcinoma. Oncotarget. 2015;6:2903–2916. doi: 10.18632/oncotarget.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Liu M., Li Z., Wu X.B., Wang Y., Wang Y., Nie M., Huang F., Ju J., Ma C., Tan R., Zen K., Zhang C.Y., Fu K., Chen Y.G., Wang M.R., Zhao Q. LYAR promotes colorectal cancer cell mobility by activating galectin-1 expression. Oncotarget. 2015;6:32890–32901. doi: 10.18632/oncotarget.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.L., Xue G., Mei Q., Wang Y.Z., Ding F.X., Liu M.F., Lu M.H., Tang Y., Yu H.Y., Sun S.H. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009;28:2719–2732. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D.D., Liu Z.J., Zhang E., Kong R., Zhang Z.H., Guo R.H. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36:4851–4859. doi: 10.1007/s13277-015-3139-2. [DOI] [PubMed] [Google Scholar]
- Yu J., Wu W.K., Li X., He J., Li X.X., Ng S.S., Yu C., Gao Z., Yang J., Li M., Wang Q., Liang Q., Pan Y., Tong J.H., To K.F., Wong N., Zhang N., Chen J., Lu Y., Lai P.B., Chan F.K., Li Y., Kung H.F., Yang H., Wang J., Sung J.J. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64:636–645. doi: 10.1136/gutjnl-2013-306620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.H., Liu X.N., Wang T.T., Pan W., Tao Q.F., Zhou W.P., Wang F., Sun S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 2017;19:820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhou Y., Mehta K.R., Danila D.C., Scolavino S., Johnson S.R., Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- Zhang W., Shi S., Jiang J., Li X., Lu H., Ren F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed Pharmacother. 2017;91:312–319. doi: 10.1016/j.biopha.2017.04.085. [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhang X., Zhou Y., Ansell P.J., Klibanski A. Cyclic AMP stimulates MEG3 gene expression in cells through a cAMP-response element (CRE) in the MEG3 proximal promoter region. Int. J. Biochem. Cell Biol. 2006;38:1808–1820. doi: 10.1016/j.biocel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhang X., Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Fu H., Wu Y., Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013;56:876–885. doi: 10.1007/s11427-013-4553-6. [DOI] [PubMed] [Google Scholar]
- Zwain I.H., Amato P. cAMP-induced apoptosis in granulosa cells is associated with up-regulation of P53 and bax and down-regulation of clusterin. Endocr. Res. 2001;27:233–249. doi: 10.1081/erc-100107184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Potential interacting proteins of MEG3 identified by the mass spectrometry.