Abstract
The inflammatory properties of the enteric microbiota of Human Immunodeficiency Virus (HIV)-infected individuals are of considerable interest because of strong evidence that bacterial translocation contributes to chronic immune activation and disease progression. Altered enteric microbiota composition occurs with HIV infection but whether altered microbiota composition or increased intestinal permeability alone drives peripheral immune activation is controversial. To comprehensively assess the inflammatory properties of HIV-associated enteric microbiota and relate these to systemic immune activation, we developed methods to purify whole fecal bacterial communities (FBCs) from stool for use in in vitro immune stimulation assays with human cells. We show that the enteric microbiota of untreated HIV-infected subjects induce significantly higher levels of activated monocytes and T cells compared to seronegative subjects. FBCs from anti-retroviral therapy (ART)-treated HIV-infected individuals induced intermediate T cell activation, indicating an only partial correction of adaptive immune cell activation capacity of the microbiome with ART. In vitro activation levels correlated with activation levels and viral load in blood and were particularly high in individuals harboring specific gram-positive opportunistic pathogens. Blockade experiments implicated Tumor Necrosis Factor (TNF)-α and Toll-Like Receptor-2 (TLR2), which recognizes peptidoglycan, as strong mediators of T cell activation; This may contradict a previous focus on lipopolysaccharide as a primary mediator of chronic immune activation. These data support that increased inflammatory properties of the enteric microbiota and not increased permeability alone drives chronic inflammation in HIV.
Keywords: Microbiome, HIV, Chronic inflammation, Immune activation, Fecal bacteria, TLR2
Highlights
-
•
The fecal microbiomes of HIV positive subjects induce higher levels of activated monocytes and T cells in vitro.
-
•
Levels of induced activation in vitro correlate with activation levels and viral load in the blood.
-
•
HIV-associated microbiome immune activation is linked to the pathways involving the inflammatory cytokine TNF-α, and TLR-2.
Chronic immune activation is a hallmark of HIV infection. Although many studies defined gut microbiome dysbiosis with HIV, we have little understanding of whether a pro-inflammatory microbiome underlies systemic immune activation. Here, we show that fecal microbiomes of individuals with HIV induce higher levels of activated monocytes and T cells in vitro, and these levels correlate with activation levels and viral load in blood. We also link this to the inflammatory cytokine TNF-α, TLR2 (which recognizes peptidoglycan), and specific gram-positive opportunistic bacteria. This work heralds toward a deep understanding of mechanisms by which the HIV-associated gut microbiome composition drives disease.
1. Introduction
Individuals with Human Immunodeficiency Virus (HIV)-infection display increased gut and systemic immune activation (French et al., 2009; Liovat et al., 2012). Chronic immune activation influences disease progression and persistence by enhancing viral replication and has also been linked with cardiovascular disease (Nou et al., 2016), chronic obstructive pulmonary disease (Crothers, 2007; Fitzpatrick et al., 2013) and frailty (Desquilbet et al., 2007). Chronic immune activation is thought to be mediated in part by translocation of bacterial products, such as lipopolysaccharide (LPS) and peptidoglycan (PGN) following an HIV-associated breakdown of gut mucosal barrier function (Brenchley and Douek, 2008, Brenchley et al., 2006). Elevated levels of plasma LPS correlate with monocyte and T-cell activation, propagation of HIV infection and T cell depletion (Brenchley and Douek, 2008, Marchetti et al., 2013, Ericsen et al., 2016, Ancuta et al., 2008). It is unclear whether compositional changes of the gut microbiota or other factors, such as damage caused by the virus itself, effect bacterial translocation and chronic immune activation in the periphery.
Culture-independent surveys of stool and mucosal biopsies have established that the enteric microbiome in HIV-infected individuals is compositionally distinct from HIV negative subjects (Lozupone et al., 2013, Lozupone et al., 2014, Mutlu et al., 2014, Vujkovic-Cvijin et al., 2013, Paquin-Proulx et al., 2017, McHardy et al., 2013, Dinh et al., 2015, Perez-Santiago et al., 2013, Noguera-Julian et al., 2016, Dillon et al., 2014, Pinto-Cardoso et al., 2017). However, results of many of these studies are confounded by microbiome differences in men who have sex with men (MSM). Specifically, several reports have linked stool microbiomes relatively high in the genus Prevotella and low in Bacteroides with HIV, but recent reports have demonstrated that a Prevotella rich microbiota is prevalent in MSM regardless of HIV status (Noguera-Julian et al., 2016; Kelley et al., 2017). In cohorts not confounded by MSM, relatively subtle differences in fecal microbiota composition have been found with HIV in the absence of CD4+ T cell counts indicative of AIDS (Noguera-Julian et al., 2016; Monaco et al., 2016), although more pronounced changes may be evident in mucosal biopsy of individuals not on anti-retroviral therapy (ART) (Vujkovic-Cvijin et al., 2013). Several studies demonstrated correlations between specific enteric bacteria and immune activation markers in the gut and blood of HIV-infected subjects (Dillon et al., 2016a; Vujkovic-Cvijin et al., 2013). However, correlations can be driven by indirect factors and do not establish causality. To establish whether correlative bacteria may be direct drivers of inflammation, in vitro stimulations of host peripheral blood mononuclear cells (PBMC) and lamina propria mononuclear cells (LPMC) with heat-killed cultured bacteria that differ with HIV has been utilized (Lozupone et al., 2013; Dillon et al., 2016a). However, these investigations have been limited because they 1) focused on individual bacterial species that represent a very small component of the complex microbiome, 2) obtained these organisms from culture collections rather than patient samples, 3) selected these species as HIV-associated in cohorts confounded by MSM, and 4) never showed that levels of activation induce by bacteria in vitro were related to ex vivo immune activation seen in the study population.
To determine whether gut microbiota with particularly pro-inflammatory components may drive high immune activation in individuals with HIV, we developed a method for purifying the whole set of intact microbial cells from stool. We stimulated PBMC and LPMC with these heat-killed fecal bacterial communities (FBCs) to establish the immune-modulatory properties of their collective components, which could include LPS, PGN, capsular components or other metabolic products. We then compared the results from stimulation with FBCs from MSM, men who have sex with woman (MSW) and women with and without HIV and ART. Immune assays with bacterial components and PBMC are relevant to HIV disease since it is translocation of microbial components to the periphery and not necessarily active growth of translocated bacteria in blood (e.g. bacteremia) that is thought to drive systemic immune activation in individuals with HIV. We then related our in vitro measurements of FBC-induced innate and adaptive immune activation to measurements of immune activation and viral load in the blood of our study population, demonstrating that microbiota with more pro-inflammatory components may be an essential driving factor in chronic immune activation. This system allowed us to further identify microbes and signaling pathways that may be driving factors of chronic inflammation in HIV infected individuals.
2. Methods
2.1. Study Subjects
Stool samples were obtained from seven HIV-infected males who were ART naïve (no prior treatment with antiretrovirals) and eleven HIV-infected males who were ART experienced (antiretroviral treatment with a minimum of three antiretroviral drugs for ≥12 months prior to study entry with plasma HIV-1 RNA below the limit of detection for >6 months). Male HIV-infected individuals enrolled in the study were determined to be MSM using a behavioral questionnaire. Control cohorts included low-risk HIV-seronegative heterosexual males (n = 6), low risk females (n = 5) and high-risk MSM (n = 13) who were recruited from a high-risk cohort assembled for a study of a candidate HIV-1 preventative vaccine (Hammer et al., 2013). Designation of high-risk could have been related to a number of different behaviors including 1) a history of unprotected anal intercourse with one or more male or male-to-female transgender partners 2) anal intercourse with two or more male or male-to-female transgender partners and 3) being in a sexual relationship with a person who has been diagnosed with HIV. All enrolled individuals live in Metropolitan Denver. Individuals were excluded during recruitment if they were pregnant, weighed <110 pounds, or had received antibiotics within the prior 30 days.
2.2. Ethics Statement
Written informed consent was obtained from healthy HIV-seronegative individuals and from HIV-infected individuals and the study protocol was approved by the Colorado Multiple Institution Review Board (COMIRB No: 14-1595). All subjects were adults.
2.3. Sample Collection and Stool Isolate Preparation
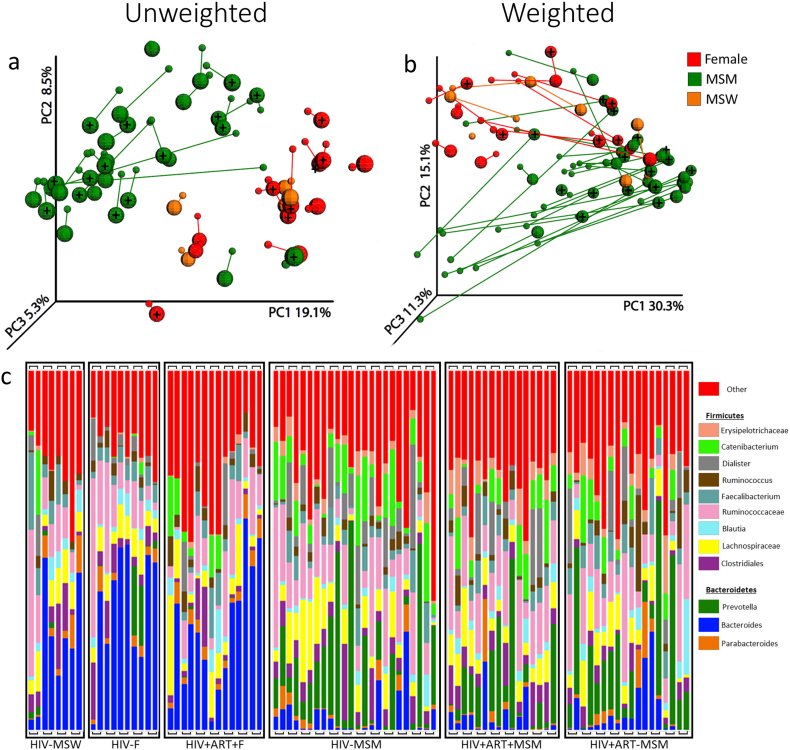
Stool samples were collected by the patient, both on a sterile swab and with a sterile scoop within 48 h of a clinic visit. Patients immediately stored the samples in a cooler with −20 °C freezer packs. After delivery to the clinic, the swab was subsequently stored at −80 °C to await DNA extraction for 16S rRNA targeting sequencing. To separate the microbiota from the rest of the fecal material, stool was subjected to density gradient according to the previously described method (Hevia et al., 2015) with some modifications. Specifically, 2 g of feces was homogenized in 40 ml sterile PBS by aggressive vortex for ~1 min. The homogenized fecal sample was passed through a 100 μm filter. 80% Histodenz (Sigma, St. Louis MO) was prepared in PBS and was sterilized by autoclave at 121 °C for 15 mins. 20 ml of filtered homogenized fecal sample was overlaid on 5 ml of the 80% Histodenz solution in 2 separate tubes and centrifuged at 10,000g for 40 min at 4 °C. The interphase layers corresponding to the microbiota were transferred to a 50 ml tube and were washed, resuspended, overlaid on 5 ml of 80% Histodenz solution and centrifuged again at 10,000g for 20 min at 4 °C. The top layer was discarded and the microbiota layers were extracted to a new tube and washed with 10 ml PBS and centrifuged at 10,000g for 20 min. The visible pellet comprised of white bacteria. The bacteria pellet was resuspended in 25 ml PBS and the presence of bacteria was confirmed by viewing a small aliquot on a glass slide. The number and viability of bacterial cells were determined using the BD Cell Viability and counting Kit (BD Biosciences) and samples that were <5:1 bacteria to debris were excluded. Since we determined that fresh (not previously frozen) sample tended to produce FBCs with less debris, we modified our stool collection procedure; the portion of fecal sample used to produce the FBCs was shipped in an insulated tube so that it remained cold but did not freeze during shipping, and processed the samples immediately upon arrival when possible. Isolated FBCs were resuspended at 500,000 bacteria per μl, subjected to multiple freeze/thaw or autoclave treatment to prevent bacterial growth, and aliquoted and stored at −80 °C. The bacterial composition of the FBCs was evaluated by 16S rRNA sequencing and compared to autologous whole stool (Fig. 2).
Fig. 2.
Principal Coordinates Analysis (PCoA) of 16S rRNA data from whole stool and associated FBCs. The composition of the bacterial communities in stool and fecal bacterial communities (FBC) from females, men who have sex with men (MSM) and men who have sex with women (MSW) with and without HIV were compared. (A) Unweighted and (B) Weighted UniFrac PCoA plots. The large spheres represent FBCs and the smaller spheres whole stool. FBCs are joined to their source stool sample with a line. HIV-infected subjects are indicated with a “+”. (C) Genus level taxa bar charts of 16S rRNA data from whole stool and FBCs. Genera representing <2% of the community are collapsed into the “Other” category (red). Samples are grouped by HIV and treatment status with stool (right) and associated FBCs (left) adjacent to each other and joined with a bridge.
2.4. DNA Sequencing
16S rRNA targeted sequencing was conducted according to earth microbiome project standard protocols (http://www.earthmicrobiome.org). DNA was extracted from the fecal swab and from a 250 μl aliquot of the stool isolate prep using the standard PowerSoil protocol (Qiagen) with one modification; the vortexing step was replaced with bead-beating for 1 min. PCR amplification of the extracted DNA, along with water controls, was conducted with barcoded primers targeted the V4 region of the 16S rDNA gene (515F-806R; FWD:GTGCCAGCMGCCGCGGTAA; REV:GGACTACHVGGGTWTCTAAT). Amplified DNA was quantified using a PicoGreen assay (Invitrogen) and equal amounts of DNA from each sample were pooled and cleaned using the UltraClean PCR Clean-up protocol (Qiagen). The final DNA pool was sequenced using the Illumina MiSeq platform (San Diego, CA) using the V2 2 × 250 kit. All sequences and associated metadata have been deposited at the European Nucleotide Archive (ERP107331).
2.5. Sequence Data Analysis
Raw sequences were assigned to samples based on their barcodes using Qiime 1.9 (Caporaso et al., 2010). The libraries were denoised and grouped by sequence variants using dada2 1.2.2 (Callahan et al., 2016). Samples produced from 12,746 to 122,310 sequences; analyses that contained both fecal samples and FBCs were conducted on the standardized sequence amount of 12,746 sequences per sample. Analyses that only included the FBCs (e.g. correlations with immune assay results) were conducted at 44,866 sequences per sample, as this was the minimum number of sequences that any FBC produced. Principal Coordinates Analysis of unweighted (Lozupone and Knight, 2005) and weighted (Lozupone et al., 2007) UniFrac distance matrices were conducted using QIIME 1.9. Statistical comparisons were conducted to evaluate microbial taxa that differed 1) between stool samples and FBCs, 2) Between HIV negative MSW and MSM, and 3) Between HIV negative and HIV positive MSM with untreated infection and on ART. We also evaluated statistically which taxa in FBCs had relative abundances that correlated with immune activation in stimulations of PBMCs with FBCs. The specific statistical test chosen depended on the data characteristics (e.g. whether dichotomous or continuous variables) and are specified in each data table. Statistical tests were conducted either on tables of bacterial genera or dada2-defined OTUs to achieve maximum taxonomic resolution. The identity of OTUs was determined with the RDP classifier (Wang et al., 2007) trained on the greengenes 3_8 taxonomic database (McDonald et al., 2012) and BLAST to NCBI. Any OTUs/genera that were not detected in at least 20% of the samples were excluded from correlational analysis with immune activation measurements, resulting in the evaluation of 301 OTUs and 85 genera. In all statistical comparisons, the p-values were corrected for multiple comparisons with the False Discovery Rate (FDR) technique (Benjamini et al., 2001).
2.6. Immune Assays
Human PBMCs were isolated by Ficol density gradient centrifugation as previously described from the blood of healthy HIV-seronegative individuals. PBMCs were cultured with 10 μg or 20× isolated fecal bacteria per PBMC for 24-h (for monocyte activation assays) or 4-days (for T cell activation assays) at 37 °C. Six day stimulations for T cell activation assays were also performed and had similar results to those of 4-day stimulations (data not shown). Data presented is the median of 3 PBMC for monocyte activation and 6 PBMC for T cell activation stimulated with the same FBC. Stimulations were performed in the presence of Streptomycin and Penicillin, in aerobic conditions and no bacterial growth was observed. In all cases background levels from unstimulated controls were subtracted out.
To quantify cytokine secretion after 24-h and 4-day stimulations with FBCs, supernatant was collected and subjected to ELISA Ready Set Go! (eBioscience) following manufacturer's instructions. To enumerate monocyte and T cell activation, cells were washed with staining buffer containing PBS, 2% BSA, 2 mM EDTA and 0.09% NaN3 and surface staining was performed. Surface staining for monocyte activation included an APC-Cy7 labelled anti-CD3 (BioLegend Cat# 300318, RRID:AB_314054), anti-CD19 (BioLegend Cat# 302218, RRID:AB_314248), and anti-CD56 (BioLegend Cat# 318332, RRID:AB_10896424) used as a dump gate, Brilliant Violet 421 labelled anti-CD11c (BioLegend Cat# 371511, RRID:AB_2650794), FITC labelled anti-CD14 (Thermo Fisher Scientific Cat# 11–0149-71, RRID:AB_464951), Brilliant Violet 510 labelled anti-CD33 (BioLegend Cat# 366609, RRID:AB_2566402), Pe-Cy7 labelled anti-CD80 (BioLegend Cat# 305217, RRID:AB_1877254) and PerCP-Cy5.5 labelled anti-HLA-DR (Thermo Fisher Scientific Cat# 45-9956-73, RRID:AB_925757). Surface staining for T cell activation was performed with BV785-labelled anti-CD3 antibody (BioLegend Cat# 317329, RRID:AB_11219196), PerCP/Cy5.5-labelled anti-CD4 (BioLegend Cat# 317428, RRID:AB_1186122), APC-labelled anti-CD8 antibody (Thermo Fisher Scientific Cat# 17-0086-71, RRID:AB_49402), APC-Cy7-labelled anti-CD69 (BD Biosciences Cat# 335792, RRID:AB_1937286), Pe-Cy7 labelled anti-HLA-DR (BioLegend Cat# 307616, RRID:AB_493588) and BV605 labelled anti-CD38 (BioLegend Cat# 303531, RRID:AB_2561527).
2.7. Isolation of CD14+ Monocytes and CD4+ T Cells
To purify monocytes, whole PBMC was subjected to the Monocyte Isolation Kit II (Miltenyi Biotec) per manufactures instructions. Efficiency > 95% was confirmed by staining purified monocytes with surface markers APC-Cy7 labelled anti-CD3, anti-CD19, and anti-CD56 (Biolegend), Brilliant Violet 421 labelled anti-CD11c (Biolegend), FITC labelled anti-CD14 (eBioscience), and Brilliant Violet 510 labelled anti-CD33 (Biolegend). CD4+ T cells were purified in a similar manner using the CD4+ T cell Isolation Kit, Human (Miltenyi Biotec). To verify CD4+ T cell purity, isolated cells were stained with BV785-labelled anti-CD3 antibody (BD Biosciences), PerCP/Cy5.5-labelled anti-CD4 (BioLegend), APC-labelled anti-CD8 antibody (Invitrogen), APC-Cy7 labelled anti-CD19 (Biolegend), and FITC labelled anti-CD14 (eBioscience). Stained cells were analyzed on a LSR II flow cytometer.
2.8. Blocking Assays
For TLR and MHC-II blockades, PBMC was pre-treated with 10 μg of either anti-TLR-2 (eBioscience), TLR-4 (eBioscience) or MHC-II (eBioscience) blocking antibodies for 30 min. PBMC were then stimulated with FBCs from 10 HIV + ART-MSM subjects for 4 days and T cell activation was enumerated. For cytokine blockades, PBMC were treated with 10 μg of anti-TNF-α (eBioscience), IFN-γ (eBioscience) or IL-6 (eBioscience) monoclonal antibodies, then stimulated with FBCs and T cell activation was enumerated as above.
2.9. Isolation of Intestinal Lamina Propria Lymphocytes and Flow Cytometry
Resected jejunum gut tissue was obtained from patients undergoing elective abdominal surgery. All patients signed a release to allow the unrestricted use of discarded tissues for research purposes and all protected patient information was de-identified to the laboratory investigators. Tissues were opened longitudinally, and trimmed of excess fat, the mucosal layer and extraneous material, such as stitched portions, damaged tissues or connective tissues. Gut tissue was then washed with PBS supplemented with Streptomycin, Penicillin, and Amphotericin B and incubated at 37 °C with PBS containing 1 mM EDTA for 2 cycles of 45 min to remove epithelial cells. The lamina propria layers were cut into 1 g pieces and incubated while shaking with AIMS V media containing antibiotics, 0.5 mg collagenase D and 10 μg DNase I for 3 cycles of 45 min at 37 °C. The digested tissues were collected and washed with PBS and resuspended in RPMI1650 containing 10% human serum. For analysis of T cell activation, isolated lymphocytes were labelled with the LIVE/DEAD fixable dye (Invitrogen) to exclude dead cells from the analysis. The cells were washed with staining buffer, then surface staining was performed as above with Pe-labelled anti-CD45. Cells were washed 2 times, enumerated with a LSR II (BD Biosciences) and analyzed using FlowJo software (Treestar).
2.10. Statistical Analysis for Difference in Immune Cell Populations/Cytokines
Statistical analysis for difference in immune cell populations/cytokines was performed using GraphPad Prism (GraphPad, San Diego, CA). Mann Whitney, paired Student's t-test, One-way ANOVA, and Pearson correlation analysis were used to determine significance of differences between subject groups. A p value of <0.05 was considered statistically significant.
3. Results
3.1. MSM Behavior Is Associated With Highly Distinctive Fecal Microbiota Compositions; Effects of HIV and Treatment Are Relatively Subtle
Fecal microbiota compositions were characterized for all samples on which we conducted immune assays; these included samples from MSM with treated (n = 11) or non-treated (n = 8) chronic HIV infection, females with treated chronic HIV infection (n = 7), HIV negative MSM (n = 13) and male (n = 6) and female (n = 5) heterosexual HIV negative controls (Table S1). The CD4+ T cell counts of the HIV-infected subjects ranged widely and those on ART had suppressed virus (<40 RNA copies/ml) (Table S1).
Consistent with previous studies (Verstrepen et al., 2016; Kestens et al., 1992), MSM with untreated HIV infection had significantly higher microbial translocation (as assessed by plasma levels of LPS binding protein (LBP)) and higher activation of both peripheral innate (soluble CD14 (sCD14)), and adaptive (CD8+ T cells expressing HLA-DR and CD38) immune cells compared to HIV negative MSW and MSM (Fig. 1). Translocation and peripheral immune activation did not significantly increase in HIV positive MSM on ART compared to HIV negative MSM but trended higher (Fig. 1). HIV positive females on ART had significantly higher levels of sCD14 and LBP compared to HIV negative females (Fig. 1).
Fig. 1.
Levels of sCD14, LPS Binding Protein (LBP) and CD8+ T cell activation in plasma and peripheral blood. (A) Plasma levels of soluble CD14. (B) Plasma levels of LBP. (C) PBMC was stained directly ex vivo for CD8 + HLA-DR + CD38+ activated T cells. Red: FBC from HIV-infected subjects, Black: FBC from HIV negative. ∗ = p < 0.05, ∗∗ = p < 0.005, ∗∗∗ = p < 0.0005.
We characterized fecal microbiota composition in this cohort using gene sequences from the V4 region of 16S ribosomal RNA (rRNA). The most important factor that we measured for explaining differences in fecal microbiome composition across our cohort was sexual behavior; MSM clustered apart from MSW and females regardless of HIV status with both weighted and unweighted UniFrac (Fig. 2a). Many genera and sequence variants (100% Operational Taxonomic Units (OTUs) defined with dada2 (Callahan et al., 2016)) differed significantly between HIV negative MSM and MSW/females (Tables S2 and S3), including taxa previously reported to differ with HIV in cohorts confounded by MSM. Notably, consistent with the findings of others (Noguera-Julian et al., 2016), we observed a predominance of Prevotella rich/Bacteroides poor microbiomes in HIV negative as well as HIV positive MSM (Fig. 2C, Table S2); Women and MSW tended to be Prevotella poor regardless of HIV status (Fig. 2C).
We observed few significantly differing taxa in HIV positive MSM with untreated (Tables S4 and S5) or treated (Tables S6 and S7) infection compared to HIV negative MSM after correcting for multiple comparisons. Unclassified taxa within the family Erysipelotrichaceae increased with untreated HIV infection (Table S4).
3.2. Microbiota Composition in Stool Samples versus Stool Isolate Preps
To gain insight into whether a gut microbiota with particularly pro-inflammatory components may be a driver of high immune activation in individuals with HIV, we developed a method for purifying the whole set of intact microbial cells from stool. We used 16S rRNA sequencing to compare the composition of FBCs to that of whole stool. The FBCs maintained very similar microbiota community membership to whole stool (Fig. 2A) but had systematic divergence in relative abundances (Fig. 2B); Specifically, Coprococcus and the gram-negative genera Prevotella and Sutterella were significantly reduced in FBC while several gram-positive genera in the Firmicutes and Actinobacteria lineages were increased (Table S8). However, the overall clustering of the microbiome composition by MSM sexual behavior was maintained in FBC (Fig. 2).
3.3. FBCs From MSM With and Without HIV Infection Activate Peripheral Monocyte Populations
To model the effects of bacterial translocation on peripheral monocyte activation, we stimulated PBMCs from healthy controls with FBC for 24 h and evaluated activated monocytes as CD33 positive cells that have shedding of CD14 and increased expression of CD80 (Gomes et al., 2010; Fleischer et al., 1996) (Fig. 3A). We found a significant increase in activated monocytes (Fig. 3A,B) and sCD14 (Fig. 3B) levels in the supernatant with FBCs isolated from both HIV negative and positive MSM compared to HIV negative MSW. sCD14 levels strongly correlated with the frequency of membrane associated CD14+ cells (r = 0.65, ci = 0.23–0.86, p = 0.006).
Fig. 3.
FBCs from HIV-infected MSM induce monocyte activation and inflammatory cytokines. Healthy PBMC was stimulated for 24 h with FBC isolated from 6 HIV negative MSW, 13 HIV negative MSM, 8 HIV positive MSM on ART and 11 HIV positive untreated MSM. (A) Representative dot plots and accumulative data showing percent CD14 shedding of CD33+ cells after stimulation with FBCs. Percent shedding was calculated by subtracting the percent of CD14+ of CD33+ cells in the unstimulated control by the percent of CD14+ of CD33+ cells in the FBC stimulated cells. (B) Percent CD80+ of CD33+ cells after stimulation with FBC. (C) Levels of soluble CD14 (sCD14) and (D) cytokines in the supernatant. Red: FBC from HIV-infected subjects, Black: FBC from HIV negative. ∗ = p < 0.05, ∗∗ = p < 0.005, ∗∗∗ = p < 0.0005.
However, FBCs from treated and untreated HIV positive MSM induced significantly higher levels of pro-inflammatory TNF-α and IL-6 compared to FBC from HIV negative MSW and MSM. Unlike phenotypic monocyte activation, there were no differences in these cytokines between HIV negative MSW and MSM (Fig. 3D). MSM and HIV infection potentially had an additive effect on IFN-γ levels, with significant differences only being observed between HIV negative MSW and HIV positive untreated MSM, although the levels of IFN-γ were much lower than TNF-α or IL-6 (Fig. 3D). There was no difference in anti-inflammatory IL-10 between any of the groups. Overall, our results indicate that although there was no significant increase in phenotypic monocyte activation between MSM with and without HIV, there was increased production of pro-inflammatory cytokines with FBCs from both treated and untreated HIV positive MSM.
To verify that the monocytes were directly responding to the FBC, CD14+ monocytes were purified prior to culture. As with whole PBMC, purified monocytes stimulated for 24 h with FBCs from untreated HIV positive MSM exhibited increased CD80 expression and inflammatory cytokines compared with FBCs from HIV negative MSW (Fig. S1). These findings support that in these in vitro assays monocytes are most likely the primary cells within PBMC producing inflammatory cytokines after FBC stimulation.
3.4. FBCs From HIV Positive MSM but Not From HIV Negative MSM Activate Peripheral T Cells
To assess the effect of FBC on adaptive immunity, PBMC were cultured with FBCs for 4 days and HLA-DR and CD38 expression was measured on CD4 and CD8+ T cells. FBCs from HIV positive individuals, whether untreated or on ART, stimulated significantly increased percentages of activated (HLA-DR + CD38+) CD4+ T cells compared to HIV negative MSW. Compared to HIV negative MSM, FBCs from individuals with untreated but not treated HIV infection had significantly increased CD4+ T cell activation (Fig. 4A) and levels of IFN-γ and IL-17 (Fig. 4B) in the supernatant. FBC from HIV negative MSM did not induce significantly more CD4+ T cell activation or cytokine production compared to HIV negative MSW. A similar result was seen with CD8+ T cell activation (Fig. S2B). To determine whether FBCs were directly activating CD4+ T cells, we stimulated purified CD4+ T cells with FBCs from HIV positive untreated MSM subjects for 4 days. In the absence of other immune cell subsets the FBCs did not induce CD4+ T cells activation (Fig. 4C) nor elicit cytokine production (data not shown). Since heat killing FBCs may denature protein, potentially impacting our stimulation assays, we also wanted to evaluated whether non-heat killed bacterial communities induce different activation patterns between cohorts. Consistent with our results from autoclaved FBCs, freeze-thawed FBCs from HIV positive MSM had a significant increase in the frequency of activated CD4+ T cells compared to stimulations with FBCs from HIV negative MSW (Fig. S3).
Fig. 4.
FBCs from HIV-infected MSM induce high levels of T cell activation in PBMC. Healthy PBMC was stimulated with FBC from 6 SN MSW, 13 SN MSM, 8 HIV+ART+MSM or 11 HIV+ART−MSM subjects for 4 days. (A) Representative dot plots and accumulative data showing HLA-DR+ CD38+ CD4+ activated T cells. (B) Levels of cytokines in the supernatant. Each dot represents the median of 6 different PBMC stimulated with the same FBC. (C) Whole PBMC or isolated CD4+ T cells were stimulated with FBC isolated from 10 HIV+ART−MSM for 4 days. Red: FBC from HIV-infected subjects, Black: FBC from HIV negative. ∗ = p < 0.05, ∗∗ = p < 0.005, ∗∗∗ = p < 0.0005.
Because immune cell subsets in different compartments behave differently, we cultured LPMC isolated from healthy sections of resected gut tissue with FBCs for 4 days. Representative dot plots of activated CD4+ and CD8+ T cells in LPMC are shown (Fig. S4A). As with PBMC, the culture of LPMC with FBCs from HIV infected MSM resulted in higher frequencies of activated CD4+ T cells compared with FBCs from HIV negative MSW and MSM (Fig. S4B). Overall, our results indicate that the microbiota of HIV-infected individuals increased T cell activation and cytokine production compared to HIV negative controls via a mechanism that requires mediation by other immune cell subsets.
3.5. T Cell Activation Is TLR and TNF-α Mediated
To investigate the mechanism of HIV-associated FBC-induced T cell activation we treated whole PBMC induced by FBCs from HIV positive untreated MSM with either TLR2, TLR4 or MHC-II (HLA-DR) or with IL-6, IFN-γ or TNF-α blocking monoclonal antibodies. For TLR2, TLR4 and cytokine blockades, PBMC was cultured with FBCs for 4 days and HLA-DR and CD38 expression on T cells was examined. For MHC-II blockade, only CD38 expression was recorded due to interference with HLA-DR staining. CD4+ T cell activation was most significantly reduced with TLR2 blockade followed by TLR4 blockade (Fig. 5A). A similar trend was seen for CD8+ T cells (Fig. S2C). MHC-II blockade also reduced the frequency of CD4 + CD38+ T cells (Fig. 5B). As expected MHC-II blockade did not influence the frequency of CD38+ cells of MHC class I-restricted CD8+ T cells (Fig. S2D). Blockade of all three inflammatory cytokines reduced the frequency of activated CD4+ T cells but only blockade of TNF-α reached statistical significance (Fig. 5C). Importantly, the frequency of CD33+ cells which had shed CD14+ at 24 h was strongly associated with the frequency of activated CD4+ T cells at 4 days (Fig. 5D). A similar correlation was seen with CD80 + CD33+ cells at 24 h compared with the frequency of activated CD4+ T cells at 4 days (Fig. S5). These findings suggest that the FBC isolated from HIV-infected MSM induce peripheral monocytes through TLRs, and to a lesser extent MHC-II, to release inflammatory cytokines, especially TNF-α, resulting in T cell activation.
Fig. 5.

TLR, MHC-II and TNF-α blockade ameliorate T cell activation induced by FBCs from HIV positive untreated MSM and CD4+ T cell activation is correlated to monocyte activation. Healthy PBMC was pretreated with (A) TLR2 or TLR4 or (B) HLA-DR (MHC-II) blockade for 30 min prior to stimulation with FBC isolated from 10 HIV positive untreated MSM for 4 days. Frequency of CD4 + HLA-DR + CD38+ T cells is shown for TLR blockades while frequency of CD38+ CD4+ T cells is shown for HLA-DR blockade. (C) FBC stimulated PBMC was treated with IL-6, IFN-γ or TNF-α blockade for 4 days. (D) Correlation between CD14 shedding of CD33+ cells and CD4 + HLA-DR + CD38+ T cells after stimulation with FBCs. Red: FBC from HIV-infected subjects, Black: FBC from HIV negative. ∗ = p < 0.05, ∗∗ = p < 0.005, ∗∗∗ = p < 0.0005.
3.6. FBCs from HIV-infected Females Induce Immune Cell Activation
When stimulating PBMC for 24 h with FBC from HIV positive females on ART, there was a significant increase in CD14+ shedding of CD33+ cells (Fig. 6A) and a trend toward an increase in CD80 (p = 0.10) compared to HIV negative females. Additionally, FBCs of HIV positive females on ART also induced a significantly increased frequency of activated CD4+ T cells in PBMC (Fig. 6B) and LPMC (Fig. S4B) and higher levels of IFN-γ and IL-17 (Fig. 6C) but not IL-10 (data not shown). These data demonstrate that FBCs from HIV positive females on ART are more inflammatory than those found in HIV negative.
Fig. 6.
FBCs isolated from HIV-infected females induce monocyte and T cell activation. Healthy PBMC was stimulated with FBCs isolated from 5 HIV-negative, and 7 HIV-infected treated females. (A) Percent change of CD14+ and CD80+ CD33+ cells after 24 h stimulation. (B) Frequency of CD4 + HLA-DR + CD38+ T cells after 4 days of culture. (C) Levels of IFN-γ and IL-17 in PBMC after 4 days of culture in the supernatant. Red: FBC from HIV-infected subjects, Black: FBC from HIV negative. ∗ = p < 0.05, ∗∗ = p < 0.005, ∗∗∗ = p < 0.0005.
3.7. Associations Between Measures of In Vitro and Ex Vivo Activation and Plasma Viral Load
To assess potential relevance of our in vitro measurements, we compared in vitro levels of sCD14 and T cell activation with ex vivo levels measured in the peripheral blood of the same individuals. Interestingly, there was a strong correlation between in vitro and ex vivo levels of both sCD14 (Fig. 7A) and percent of activated CD8+ T cells (Fig. 7B). In contrast, we did not find a significant association with in vitro and ex vivo levels of CD4+ T cell activation (Fig. S6). One reason for this may be that HIV preferentially infects and depletes activated CD4+ T cells, confounding the effect in HIV-infected subjects. The plasma viral load of HIV positive untreated MSM also strongly correlated with in vitro FBC induced CD4+ and CD8+ HLA-DR + CD38+ T cell percentages (Fig. 7C). Taken together, these data indicate that inflammatory properties of the fecal microbiota are associated with levels of peripheral innate and adaptive immune activation and viral load in the blood, which are markers of clinical outcomes in HIV infection (Sandler et al., 2011), further supporting a potential role of pro-inflammatory gut bacteria in HIV pathogenesis and chronic immune activation.
Fig. 7.
Immune cell activation induced by FBCs in vitro is correlated to ex vivo peripheral sCD14 and T cell activation levels and HIV viral load. (A) Correlation of sCD14 induced in vitro by FBC to ex vivo sCD14 levels in the plasma of the fecal donor. (B) The frequency of CD8 + HLA-DR + CD38+ activated T cells induced by FBCs in vitro was correlated to ex vivo levels from the same subjects who donated sample. (C) Correlation of peripheral viral load in HIV+ART− subjects with CD4 and CD8 activated T cells induced by FBCs from the same donor in vitro. Red: FBC from HIV-infected subjects, Black: FBC from HIV negative.
3.8. Microbes That Correlate With Immune Activation
We determined whether any OTUs in the FBCs correlated with T cell activation and cytokine supernatant measurements from our in vitro stimulations of PBMCs (Table S9). Unlike correlations conducted between microbes and immune phenotypes measured in vivo, these correlations are less likely to be driven by indirect host factors. Overall, the most significant correlations found were with the frequency of activated CD8+ and CD4+ T cells; more limited correlations were found with innate immune cells and their cytokines.
Of particular interest were OTUs that both positively correlated with T cell activation and were increased in MSM with untreated HIV infection, as these may be drivers of increased T cell activation by FBC of untreated HIV positive versus negative MSM. The most highly significant OTU with this property (sequence variant 277) correlated strongly with both CD4+ and CD8+ T cell activation (Table S9), and was also positively correlated with levels of IL-17 and IFN-γ in the supernatants after 4 days of stimulation (Tables S5, S9). BLAST against 16S rRNA sequences of type species in the Ribosomal Database Project (RDP II) indicated that this OTU has 99.6% sequence identity to Terrisporobacter glycolicus ATCC 14880 (Table S9). The only other OTUs that both positively correlated with T cell activation and were increased with untreated HIV infection was an OTU with a 99.6% identical 16S rRNA sequence to Turicibacter sanguinis and an OTU that had an identical 16S rRNA sequence to Bifidobacterium gallicum DSM 20093.
Several OTUs that positively correlated with activated T cells were increased in HIV negative MSM compared to MSW/females. These included two OTUs highly related to Eubacterium biforme and OTUs related to strains of Colinsella aerofaciens (98.4% ID), Slackia isoflavoniconvertans (100% ID), Mitsuokella jaludinii (100% ID), and Hungatella hathawayi (94.6% ID). An OTU with an identical 16S rRNA sequence to the Senegalimassilia anaerobia type strain was increased in the FBCs of HIV negative MSM versus MSW and positively correlated with %CD14 shedding. OTUs highly related to the type species of Parabacteroides distasonis and Clostridium methylpentosum (95.1% ID) negatively correlated with percent CD14 shedding.
4. Discussion
We demonstrate higher adaptive and innate immune activating components in the fecal microbiota of individuals with HIV infection compared to HIV negative MSM. A strong correlation between the in vitro immune-stimulatory ability of these FBCs and levels of ex vivo peripheral immune activation and viral load from the same subjects supports the relevance of these findings and suggests that the composition of the gut microbiome is an important contributing factor to the level of peripheral immune activation and HIV disease progression.
Increased levels of translocation with HIV infection has been clearly established (Marchetti et al., 2013; Brenchley et al., 2006) leading to the possibility that chronic inflammation may be driven in the absence of gut microbiome dysbiosis due to increased translocation levels or the preferential translocation of particular gut microbes in the context of infection (Klase et al., 2015). Our results, however, suggest that the presence of more pro-inflammatory bacteria in the gut microbiome does contribute to increased peripheral immune activation.
Our methods allow for establishing immune modulatory properties of the components of entire microbiomes using in vitro stimulations. Using the full FBC in immune stimulations rather than individual isolates allows for the complicated interactions between immune-modulatory factors in the microbiome to be accounted for. This is of interest given that many taxa are present in different contexts; for example, the Prevotella genus has been associated with inflammatory diseases such as rheumatoid arthritis in humans (Scher et al., 2013) and osteomyelitis in mice (Lukens et al., 2014), yet are found in health in those who consume more plant and carbohydrate based meals (Wu et al., 2011) including those living in agrarian cultures (Yatsunenko et al., 2012). These conflicting associations emphasize how complex the interactions between host and microbiota are, and how impact of specific taxa may be dependent on the overall community structure as well as host factors. The methods for isolating and evaluating immune-modulatory properties of FBCs developed here may be valuable for studying other diseases where translocation of bacterial components may induce immune activation, including disease of the kidney (Ramezani and Raj, 2014), liver (Seki and Schnabl, 2012), joints (Bjarnason et al., 1984), and lung (Dickson et al., 2016).
We found that the FBC preparations resembled whole stool bacteria in community membership, however, relative abundance of several genera of gram positive bacteria were systemically enriched. Despite these differences, our FBCs appeared to adequately capture relevant components of the gut microbiome, because we observed a strong correlation between in vitro immune phenotypes elicited by these FBCs and ex vivo immune phenotypes in our study cohort. However, further development of FBC preparation methods is needed, as these changes in relative abundance of microbiome constituents have the potential to be important.
Consistent with the findings of others (Noguera-Julian et al., 2016; Kelley et al., 2017), we support that microbiomes characterized by increased Prevotella and decreased Bacteroides, which has been used as a hallmark of HIV-mediated dysbiosis, was found in MSM independent of HIV status. Furthermore, we found that HIV negative MSM FBCs induced significantly higher activation of CD14+ monocytes compared to HIV negative MSW, indicating that the MSM-associated gut microbiome may be pro-inflammatory. Interestingly, LPS levels have been shown to be elevated in the plasma of HIV negative MSM compared to MSW and correlate with markers of innate immune activation including TNF-α, IP-10 and MCP-1 (Palmer et al., 2014).
Previous studies have also reported differences in adaptive immune phenotypes in MSM such as a decrease in the CD4/CD8 T cell ratio (Palmer et al., 2014) and increased frequencies of activated CD4+ and CD8+ T cells (Gianella et al., 2012). Although we saw no significant differences between HIV negative MSM and MSW in adaptive immune activation with autoclaved (heat-killed) FBCs, we did see a trend in elevated T cell activation using non-autoclaved FBCs (Fig. S3). We use autoclaved bacteria in our assays to ensure no bacterial growth. However, we acknowledge that this approach potentially denatures immune modulatory factors. Whether the highly divergent microbiome observed in HIV negative MSM versus MSW is a driving factor of reported MSM immune phenotypes is an important area to study further, since immune activation levels in MSM will influences both acquisition of HIV and viral replication.
When controlling for MSM, we found few significant differences in gut microbiome composition with HIV infection in this small cohort. In one study that used a considerably larger cohort in Europe and a high-risk MSM control population, HIV infection was associated with a decrease in alpha diversity of the fecal microbiome, but this result was largely driven by individuals on unsuccessful ART; only subtle changes in the relative abundances of particular microbial taxa were observed with HIV infection (Noguera-Julian et al., 2016). Another study conducted in Sub-Saharan Africa, where populations are dominated by heterosexual transmission, had similar results, with HIV infection associating with decreased alpha diversity of the fecal microbiome, particularly with viral mediated decline of CD4+ T cells, but not with strong compositional differences (Monaco et al., 2016). Our results are thus consistent with the results of these previously published studies.
Despite only subtle compositional differences in the gut microbiome with HIV status in our cohort, we found that the FBC from HIV positive untreated MSM dramatically increased the frequency of activated CD4+ and CD8+ T cells in PBMC and that in vitro and ex vivo levels of activated CD8+ T cells correlated. Furthermore, plasma HIV RNA concentrations in individuals with untreated HIV infection was also correlated with in vitro FBC-induce T cell activation suggesting that the magnitude of HIV replication in vivo is also linked with the microbiota composition. Together these findings suggest an important role for microbiome composition in disease progression.
Our findings also support an important role for gut-microbiome mediated induction of inflammatory cytokines by myeloid cells in inducing T cell activation. Notably, when T cells isolated from PBMC were stimulated with bacteria in the absence of other immune cell populations, activation was not observed. FBC from HIV-infected subjects induced increased production of TNF-α and IL-6 by monocytes and blocking TNF-α ameliorated T cell activation. TNF-α itself, and the signaling pathway, has long been established as having a role in HIV infection (Herbein and Khan, 2008; Pasquereau et al., 2017) and TNF-α blockade is used clinically to treat other inflammatory conditions of the gastrointestinal tract (Sandborn, 2005; Danese et al., 2013). We also found that high levels of sCD14 in the plasma, a marker of poor clinical outcomes (Verstrepen et al., 2016), were correlated with high levels of sCD14 in the supernatant of PBMC cultured with isolated fecal bacterial cells.
We evaluated immune activation of FBCs from both women and MSM who were on ART and found evidence of a partial correction of certain pro-inflammatory components of the HIV-associated gut microbiome with ART. Specifically, CD4+ and CD8+ T cell activation, and IFN-γ and IL-17 levels were significantly reduced in ART treated MSM compared to untreated HIV positive MSM. However, HIV positive MSM on ART still had higher CD4+ T cell activation compared to HIV negative MSM and TNF-α and IL-6 production by monocytes in 24 h stimulations of PBMC was just as high in MSM with treated and untreated HIV infection.
FBCs from HIV positive females on ART also stimulated significantly increased levels of innate and adaptive immune activation compared to HIV negative females despite effective virus suppression. That only a partial correction of immune cell activation capacity of the microbiome occurs with ART suggests the possibility for microbiome involvement in the peripheral immune activation that has been shown to persist over time with ART and contribute to various co-morbidities in this population (Klatt et al., 2013).
Despite our failure to observe many significantly differing taxa between HIV positive and negative MSM, we did find several OTUs to be highly correlated with levels of immune activation in in vitro assays with FBCs, and some of these were significantly enriched in HIV without the FDR correction that we applied for our more general search for HIV-associated taxa differences. Almost all of these correlations were positive (pro-inflammatory), with the exception of 2 OTUs that negatively correlated with shedding of CD14; one highly related to Parabacteroides distasonis (99.2% ID) and one more distantly related (95.1% ID) to Clostridium methylpentosum (99.2% ID). Antigens of P. distasonis have previously been shown to protect against intestinal inflammation in mouse models (Kverka et al., 2011).
Receptor blockade in our assays indicated that TLR2, which recognizes peptidoglycan and thus gram-positive bacteria, was a stronger modulator of T cell activation in our HIV positive untreated MSM FBCs than TLR4, which recognizes LPS of gram-negative bacteria. Interestingly, all of the OTUs that positively correlated with T cell activation were in gram positive lineages, including diverse Firmicutes and Actinobacteria. Although, gram negative Prevotella has been positively correlated with T cell activation in human mucosal biopsy in HIV-infected individuals (Dillon et al., 2016b), we found no such correlation. We note that although Prevotella was preferentially lost in the FBCs versus fecal matter, this genus still composed an average of 4% of bacteria in the FBCs. Two OTUs that positively correlated with T cell activation were highly related to E. biforme; These OTUs both had increased relative abundance in HIV negative MSM compared to MSW but were not significantly enriched with HIV infection. E. biforme was previously targeted by our group because of a positive association with HIV in a cohort not controlled for MSM behavior. Consistent with pro-inflammatory correlations detected here, E. biforme had a significantly higher TNF-α to IL-10 ratio after a 24 h stimulation of PBMC than LPS, P. copri or two different species in the Bacteroides genus (Lozupone et al., 2013). C. aerofaciens positively correlated with T cell activation, and has been previously shown to increase gut permeability and disease severity in experimental arthritis (Chen et al., 2016).
We are particularly interested in identifying OTUs that both positively correlated with levels of adaptive immune activation and increased in relative abundance in HIV positive untreated MSM compared to HIV negative MSM as these may be driving an overall higher level of immune activation. Only 3 OTUs fit these criteria and all 3 were highly related to type species, specifically T. glycolicus, T. sanguinis and B. gallicum. T. glycolicus (Cheng et al., 2016; Cai et al., 2012) and T. sanguinis (Bosshard et al., 2002; Justesen et al., 2010) are both opportunistic pathogen in humans, which is intriguing since opportunistic infections are very important and prevalent in later stages of HIV infection (Lifson et al., 1994). However, these OTUs only make up a very minor component of the full microbiome, for instance with the T. glycolicus OTU only making up at most 0.2% of the reads in any one sample. Further study will be needed to determine whether minor gut microbiome components with potent immune-modulatory phenotypes may be a driver of high immune activation of FBCs of untreated HIV positive MSM. Other possible explanations are strain level variation or differential expression of pro-inflammatory bacterial components in the gut of HIV positive versus HIV negative individuals. Intestinal microbes can alter the expression of surface/capsular components in different contexts (Backhed et al., 2005; Comstock and Kasper, 2006). Further interrogation of microbiome composition with shotgun metagenomes and metatranscriptomes will be needed to differentiate between these alternate hypotheses.
There is substantial interest in elucidating how enteric microbiota influence immune homeostasis in the periphery, as this process has important implications for the treatment of many inflammatory disorders and diseases. Our findings provide insight into how translocating bacteria from the gut can differentially activate peripheral monocyte through TLR which activate T cells though the release of inflammatory cytokines such as TNF-α. Our findings also suggest that novel therapeutic targets, such as the modulation of enteric bacteria to create less inflammatory fecal microbiome or blockade of TNF-α, should be evaluated as a strategy to reduce the systemic inflammation that persists with HIV infection and antiretroviral treatment.
Funding Sources
This work was funded from NIH R01 DK104047 (BEP and CL) and NIH RO1 DK108366 (BEP and CL). Dr. Neff was supported by NIH T32 AI007405, Dr. Li Was supported by NIH T32 AI007447-25 and Abigail Armstrong was supported by NIH T32 AI05266.
Conflicts of Interest
The authors have no conflicts of interest to report.
Author Contributions
C.P.N., C.A.L and B.E.P. designed research; C.P.N., O.K., K.X., S.A., N.N., J.M.S., A.C., A.A., and S.L. performed research; C.P.N., C.A.L. and B.E.P. analyzed data; M.D.M. provided tissues and T.B·C recruited subjects. C.P.N., C.A.L., and B.E.P. wrote the manuscript.
Acknowledgments
Authors acknowledge study participants for providing blood, gut and stool samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.03.024.
Contributor Information
Catherine A. Lozupone, Email: Catherine.Lozupone@ucdenver.edu.
Brent E. Palmer, Email: brent.palmer@ucdenver.edu.
Appendix A. Supplementary data
Supplementary material
Supplementary tables
References
- Ancuta P., Kamat A., Kunstman K.J., Kim E.Y., Autissier P., Wurcel A., Zaman T., Stone D., Mefford M., Morgello S., Singer E.J., Wolinsky S.M., Gabuzda D. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Bjarnason I., Williams P., So A., Zanelli G.D., Levi A.J., Gumpel J.M., Peters T.J., Ansell B. Intestinal permeability and inflammation in rheumatoid arthritis: effects of non-steroidal anti-inflammatory drugs. Lancet. 1984;2:1171–1174. doi: 10.1016/s0140-6736(84)92739-9. [DOI] [PubMed] [Google Scholar]
- Bosshard P.P., Zbinden R., Altwegg M. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 2002;52:1263–1266. doi: 10.1099/00207713-52-4-1263. [DOI] [PubMed] [Google Scholar]
- BRENCHLEY J.M., DOUEK D.C. The mucosal barrier and immune activation in HIV pathogenesis. Curr. Opin. HIV AIDS. 2008;3:356–361. doi: 10.1097/COH.0b013e3282f9ae9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Price D.A., Schacker T.W., Asher T.E., Silvestri G., Rao S., Kazzaz Z., Bornstein E., Lambotte O., Altmann D., Blazar B.R., Rodriguez B., Teixeira-Johnson L., Landay A., Martin J.N., Hecht F.M., Picker L.J., Lederman M.M., Deeks S.G., Douek D.C. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Cai D., Sorokin V., Lutwick L., Liu W., Dalal S., Sandhu K., Ouyang J., Pincus M. C. glycolicum as the sole cause of bacteremia in a patient with acute cholecystitis. Ann. Clin. Lab. Sci. 2012;42:162–164. [PubMed] [Google Scholar]
- Callahan B.J., Mcmurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., Mcdonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wright K., Davis J.M., Jeraldo P., Marietta E.V., Murray J., Nelson H., Matteson E.L., Taneja V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.P., Domingo M.C., Levesque S., Yansouni C.P. A case report of a deep surgical site infection with Terrisporobacter glycolicus/T. Mayombei and review of the literature. BMC Infect. Dis. 2016;16:529. doi: 10.1186/s12879-016-1865-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock L.E., Kasper D.L. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–850. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Crothers K. Chronic obstructive pulmonary disease in patients who have HIV infection. Clin. Chest Med. 2007;28:575–587. doi: 10.1016/j.ccm.2007.06.004. (vi) [DOI] [PubMed] [Google Scholar]
- Danese S., Colombel J.F., Peyrin-Biroulet L., Rutgeerts P., Reinisch W. Review article: the role of anti-TNF in the management of ulcerative colitis — past, present and future. Aliment. Pharmacol. Ther. 2013;37:855–866. doi: 10.1111/apt.12284. [DOI] [PubMed] [Google Scholar]
- Desquilbet L., Jacobson L.P., Fried L.P., Phair J.P., Jamieson B.D., Holloway M., Margolick J.B., Multicenter A.C.S. HIV-1 infection is associated with an earlier occurrence of a phenotype related to frailty. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:1279–1286. doi: 10.1093/gerona/62.11.1279. [DOI] [PubMed] [Google Scholar]
- Dickson R.P., Singer B.H., Newstead M.W., Falkowski N.R., Erb-Downward J.R., Standiford T.J., Huffnagle G.B. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S.M., Lee E.J., Kotter C.V., Austin G.L., Dong Z., Hecht D.K., Gianella S., Siewe B., Smith D.M., Landay A.L., Robertson C.E., Frank D.N., Wilson C.C. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S.M., Lee E.J., Donovan A.M., Guo K., Harper M.S., Frank D.N., Mccarter M.D., Santiago M.L., Wilson C.C. Enhancement of HIV-1 infection and intestinal CD4+ T cell depletion ex vivo by gut microbes altered during chronic HIV-1 infection. Retrovirology. 2016;13:5. doi: 10.1186/s12977-016-0237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S.M., Lee E.J., Kotter C.V., Austin G.L., Gianella S., Siewe B., Smith D.M., Landay A.L., Mcmanus M.C., Robertson C.E., Frank D.N., Mccarter M.D., Wilson C.C. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 2016;9:24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh D.M., Volpe G.E., Duffalo C., Bhalchandra S., Tai A.K., Kane A.V., Wanke C.A., Ward H.D. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J. Infect. Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsen A.J., Lauck M., Mohns M.S., Dinapoli S.R., Mutschler J.P., Greene J.M., Weinfurter J.T., Lehrer-Brey G., Prall T.M., Gieger S.M., Buechler C.R., Crosno K.A., Peterson E.J., Reynolds M.R., Wiseman R.W., Burwitz B.J., Estes J.D., Sacha J.B., Friedrich T.C., Brenchley J.M., O'connor D.H. Microbial translocation and inflammation occur in hyperacute immunodeficiency virus infection and compromise host control of virus replication. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick M., Crothers K., Morris A. Future directions: lung aging, inflammation, and human immunodeficiency virus. Clin. Chest Med. 2013;34:325–331. doi: 10.1016/j.ccm.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer J., Soeth E., Reiling N., Grage-Griebenow E., Flad H.D., Ernst M. Differential expression and function of CD80 (B7-1) and CD86 (B7-2) on human peripheral blood monocytes. Immunology. 1996;89:592–598. doi: 10.1046/j.1365-2567.1996.d01-785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French M.A., King M.S., Tschampa J.M., Da Silva B.A., Landay A.L. Serum immune activation markers are persistently increased in patients with HIV infection after 6 years of antiretroviral therapy despite suppression of viral replication and reconstitution of CD4+ T cells. J. Infect. Dis. 2009;200:1212–1215. doi: 10.1086/605890. [DOI] [PubMed] [Google Scholar]
- Gianella S., Strain M.C., Rought S.E., Vargas M.V., Little S.J., Richman D.D., SPINA C.A., Smith D.M. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J. Virol. 2012;86:1307–1315. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes N.E., Brunialti M.K., Mendes M.E., Freudenberg M., Galanos C., Salomao R. Lipopolysaccharide-induced expression of cell surface receptors and cell activation of neutrophils and monocytes in whole human blood. Braz. J. Med. Biol. Res. 2010;43:853–858. doi: 10.1590/s0100-879x2010007500078. [DOI] [PubMed] [Google Scholar]
- Hammer S.M., Sobieszczyk M.E., Janes H., Karuna S.T., Mulligan M.J., Grove D., Koblin B.A., Buchbinder S.P., Keefer M.C., Tomaras G.D., Frahm N., Hural J., ANUDE C., Graham B.S., Enama M.E., Adams E., Dejesus E., Novak R.M., Frank I., Bentley C., Ramirez S., Fu R., Koup R.A., Mascola J.R., Nabel G.J., Montefiori D.C., Kublin J., Mcelrath M.J., Corey L., Gilbert P.B., Team H.S. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N. Engl. J. Med. 2013;369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G., Khan K.A. Is HIV infection a TNF receptor signalling-driven disease? Trends Immunol. 2008;29:61–67. doi: 10.1016/j.it.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Hevia A., Delgado S., Margolles A., Sanchez B. Application of density gradient for the isolation of the fecal microbial stool component and the potential use thereof. Sci. Rep. 2015;5:16807. doi: 10.1038/srep16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justesen U.S., Skov M.N., Knudsen E., Holt H.M., Sogaard P., Justesen T. 16S rRNA gene sequencing in routine identification of anaerobic bacteria isolated from blood cultures. J. Clin. Microbiol. 2010;48:946–948. doi: 10.1128/JCM.02075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley C.F., Kraft C.S., De Man T.J., Duphare C., Lee H.W., Yang J., Easley K.A., Tharp G.K., Mulligan M.J., Sullivan P.S., Bosinger S.E., Amara R.R. The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: implications for HIV transmission and prevention. Mucosal Immunol. 2017;10:996–1007. doi: 10.1038/mi.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestens L., Vanham G., Gigase P., Young G., Hannet I., Vanlangendonck F., Hulstaert F., Bach B.A. Expression of activation antigens, HLA-DR and CD38, on CD8 lymphocytes during HIV-1 infection. AIDS. 1992;6:793–797. doi: 10.1097/00002030-199208000-00004. [DOI] [PubMed] [Google Scholar]
- Klase Z., Ortiz A., Deleage C., Mudd J.C., Quinones M., Schwartzman E., Klatt N.R., Canary L., Estes J.D., Brenchley J.M. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 2015;8:1009–1020. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt N.R., Chomont N., Douek D.C., Deeks S.G. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol. Rev. 2013;254:326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kverka M., Zakostelska Z., Klimesova K., Sokol D., Hudcovic T., Hrncir T., Rossmann P., Mrazek J., Kopecny J., Verdu E.F., Tlaskalova-Hogenova H. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin. Exp. Immunol. 2011;163:250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifson A.R., Olson R., Roberts S.G., Poscher M.E., Drew W.L., Conant M.A. Severe opportunistic infections in AIDS patients with late-stage disease. J. Am. Board Fam. Pract. 1994;7:288–291. [PubMed] [Google Scholar]
- Liovat A.S., Rey-Cuille M.A., Lecuroux C., Jacquelin B., Girault I., Petitjean G., Zitoun Y., Venet A., Barre-Sinoussi F., Lebon P., Meyer L., Sinet M., Muller-Trutwin M. Acute plasma biomarkers of T cell activation set-point levels and of disease progression in HIV-1 infection. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Hamady M., Kelley S.T., Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Li M., Campbell T.B., Flores S.C., Linderman D., Gebert M.J., Knight R., Fontenot A.P., Palmer B.E. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Rhodes M.E., Neff C.P., Fontenot A.P., Campbell T.B., Palmer B.E. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5:562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- Lukens J.R., Gurung P., Vogel P., Johnson G.R., Carter R.A., Mcgoldrick D.J., Bandi S.R., Calabrese C.R., Vande Walle L., Lamkanfi M., Kanneganti T.D. Dietary modulation of the microbiome affects autoinflammatory disease. Nature. 2014;516:246–249. doi: 10.1038/nature13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti G., Tincati C., Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin. Microbiol. Rev. 2013;26:2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., Desantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy I.H., Li X., Tong M., Ruegger P., Jacobs J., Borneman J., Anton P., Braun J. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco C.L., Gootenberg D.B., Zhao G., Handley S.A., Ghebremichael M.S., Lim E.S., Lankowski A., Baldridge M.T., Wilen C.B., Flagg M., Norman J.M., Keller B.C., Luevano J.M., Wang D., Boum Y., Martin J.N., Hunt P.W., Bangsberg D.R., Siedner M.J., Kwon D.S., Virgin H.W. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu E.A., Keshavarzian A., Losurdo J., Swanson G., Siewe B., Forsyth C., French A., Demarais P., Sun Y., Koenig L., Cox S., Engen P., Chakradeo P., Abbasi R., Gorenz A., Burns C., Landay A. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera-Julian M., Rocafort M., Guillen Y., Rivera J., Casadella M., Nowak P., Hildebrand F., Zeller G., Parera M., Bellido R., Rodriguez C., Carrillo J., Mothe B., Coll J., Bravo I., Estany C., Herrero C., Saz J., Sirera G., Torrela A., Navarro J., Crespo M., Brander C., Negredo E., Blanco J., Guarner F., Calle M.L., Bork P., Sonnerborg A., Clotet B., Paredes R. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou E., Lo J., Grinspoon S.K. Inflammation, immune activation, and cardiovascular disease in HIV. AIDS. 2016;30:1495–1509. doi: 10.1097/QAD.0000000000001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.D., Tomassilli J., Sirignano M., Romero-Tejeda M., Arnold K.B., Che D., Lauffenburger D.A., Jost S., Allen T., Mayer K.H., Altfeld M. Enhanced immune activation linked to endotoxemia in HIV-1 seronegative MSM. AIDS. 2014;28:2162–2166. doi: 10.1097/QAD.0000000000000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin-Proulx D., Ching C., Vujkovic-Cvijin I., Fadrosh D., Loh L., Huang Y., Somsouk M., Lynch S.V., Hunt P.W., Nixon D.F., Sengupta D. Bacteroides are associated with GALT iNKT cell function and reduction of microbial translocation in HIV-1 infection. Mucosal Immunol. 2017;10:69–78. doi: 10.1038/mi.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquereau S., Kumar A., Herbein G. Targeting TNF and TNF receptor pathway in HIV-1 infection: from immune activation to viral reservoirs. Viruses. 2017;9 doi: 10.3390/v9040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Santiago J., Gianella S., Massanella M., Spina C.A., Karris M.Y., Var S.R., Patel D., Jordan P.S., Young J.A., Little S.J., Richman D.D., Smith D.M. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. 2013;27:1921–1931. doi: 10.1097/qad.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Cardoso S., Lozupone C., Briceno O., Alva-Hernandez S., Tellez N., Adriana A., Murakami-Ogasawara A., Reyes-Teran G. Fecal bacterial communities in treated HIV infected individuals on two antiretroviral regimens. Sci. Rep. 2017;7:43741. doi: 10.1038/srep43741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A., Raj D.S. The gut microbiome, kidney disease, and targeted interventions. J. Am. Soc. Nephrol. 2014;25:657–670. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandborn W.J. New concepts in anti-tumor necrosis factor therapy for inflammatory bowel disease. Rev. Gastroenterol. Disord. 2005;5:10–18. [PubMed] [Google Scholar]
- Sandler N.G., Wand H., Roque A., Law M., Nason M.C., Nixon D.E., Pedersen C., Ruxrungtham K., Lewin S.R., Emery S., Neaton J.D., Brenchley J.M., Deeks S.G., Sereti I., Douek D.C., Group I.S.S. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J. Infect. Dis. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher J.U., Sczesnak A., Longman R.S., Segata N., Ubeda C., Bielski C., Rostron T., Cerundolo V., Pamer E.G., Abramson S.B., Huttenhower C., Littman D.R. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2 doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E., Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J. Physiol. 2012;590:447–458. doi: 10.1113/jphysiol.2011.219691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen B.E., Nieuwenhuis I.G., Mooij P., Verschoor E.J., Fagrouch Z.C., Kondova I., Boonstra A., Koopman G. Role of microbial translocation in soluble CD14 up-regulation in HIV-, but not in HCV-, infected chimpanzees. J. Gen. Virol. 2016;97:2599–2607. doi: 10.1099/jgv.0.000577. [DOI] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I., Dunham R.M., Iwai S., Maher M.C., Albright R.G., Broadhurst M.J., Hernandez R.D., Lederman M.M., Huang Y., Somsouk M., Deeks S.G., Hunt P.W., Lynch S.V., Mccune J.M. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006438. 193ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., Sinha R., Gilroy E., Gupta K., Baldassano R., Nessel L., Li H., Bushman F.D., Lewis J.D. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., Heath A.C., Warner B., Reeder J., Kuczynski J., Caporaso J.G., Lozupone C.A., Lauber C., Clemente J.C., Knights D., Knight R., Gordon J.I. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary tables