Abstract
The World Health Organization has recently introduced molecular prognostic-diagnostic biomarkers in the classification of Central Nervous System (CNS) tumors. In order to characterize subclasses of tumors that cannot find a precise location in the current classification, and, or cannot be tested because of scant material, it is important to find new molecular biomarkers in tissue and, or biological fluid samples. In this study, we identified serum microRNAs that could serve as biomarkers for the diagnosis and prognosis of patients with tumors of glial origin. We retrospectively analyzed microRNA expression in the serum extracellular vesicles of patients with tumors of glial origin. Extracellular vesicles RNA was analyzed by Nanostring. qRT-PCR confirmed 6 overexpressed microRNAs: hsa-miR-4443, hsa-miR-422a, hsa-miR-494-3p, hsa-miR-502-5p, hsa-miR-520f-3p, and hsa-miR-549a. Hsa-miR-4443 was the only microRNA that showed significant differences in most comparisons. In situ hybridization (ISH), confirmed that our signature was mostly expressed in cancer cells.
Importantly, hsa-miR-549a and hsa-miR-502-5p expression predicted prognosis in patients with tumors of glial origin. Although more studies are needed, we demonstrated that serum vesicles microRNA profiles are promising diagnostic and prognostic molecular biomarkers that will find an actual application in the clinical practice of CNS tumors.
Keywords: microRNA, CNS tumors, Serum, Biomarkers, Diagnosis, Prognosis
Highlights
-
•
We identified a microRNA signature in the serum extracellular vesicles of patients with glial tumors.
-
•
The signature was differentially expressed among tumors and controls, showing potentials in the diagnosis of Gliomas.
-
•
Hsa-miR-549a and hsa-miR-502-5p expression predicted prognosis in patients with tumors of glial origin.
Research in context
To find targeted therapies and patients sensitive to specific treatments, we need to better classify different groups of brain tumors identifying novel molecular biomarkers. The most common brain tumors are Gliomas.
In this article, we identified a group of very short genes (microRNAs) in the extra-cellular vesicles of the serum of 8 normal and of 28 patients with gliomas. The expression of such genes differs among the different groups of tumors and normal patients, and can help in the diagnosis of gliomas.
Interestingly, two microRNAs could predict the outcome of the disease and might find future clinical applications.
1. Introduction
In 2016, the World Health Organization (WHO) published a new classification for Central Nervous System (CNS) tumors that, beside deleting and adding histological subgroups, introduced, for the first time, molecular biomarkers that define new prognostic groups (Louis et al., 2016). In attempt to merge the different clinical information contributed by previous classifications, the International Society of Neuropathology (ISN) proposed a “layered” diagnostic path, that includes the histologic classification (layer 2), WHO grading (layer 3), molecular information (layer 4) and an “integrated diagnosis” (layer 1) (Louis et al., 2014).However, because of their extensive biological variability some gliomas cannot be molecularly classified (Masui et al., 2016). Furthermore, a group of glial tumors has not yet found a precise molecular classification: the “Not Otherwise Specified” (NOS) cases include those tumors for which testing cannot be performed due to limited tissue availability, low number of neoplastic cells, or other causes (Louis et al., 2016). Clearly, patients with a non-classified biological variant of gliomas and/or a “NOS”, will not be able to benefit of the molecularly targeted treatment. On the other hand, imaging studies cannot microscopically define clear margin in malignant lesions before and after surgery. Medications affecting Blood Brain Barrier (BBB) permeability, can produce images with falsely reduced tumor burden (“pseudo-response”) (Holdhoff et al., 2013). Treatment of high grade gliomas with radiation and temodar can lead to “pseudo-progression”, causing unnecessary surgery or chemotherapy termination (Santangelo et al., 2017). It is extremely important to find molecular biomarkers aiding in the diagnosis and classification of these tumors on tissue samples and, more importantly, on biological fluids. MicroRNAs are non-coding small RNAs of few nucleotides (Li et al., 2012; Ostrom et al., 2015; Peterson et al., 2015; Skog et al., 2008; Van Deun and Hendrix, 2017) in length that modulate gene expression by regulating mRNA translation/degradation, playing key roles in physiologic and pathologic processes, including cancer (Drusco and Croce, 2017). Moreover, microRNA profiles can classify human cancer, rendering these small molecules suitable biomarkers to classify cancers for diagnostic, prognostic and therapeutic purposes (Lu et al., 2005). MicroRNAs are expressed in all tissues and biological fluids (Drusco et al., 2015; Mitchell et al., 2008; Russo et al., 2017), where they are released either freely as chemically modified molecules, or associated with proteins, or within extracellular vesicles (EVs) (Raposo and Stoorvogel, 2013). Secreted by all type of cells, including cancer cells, EVs are a system of intercellular communication through which proteins and nucleic acids are packed within small membrane spherules and shipped at distant sites (Taylor and Gercel-Taylor, 2008). Thus, microRNA profiling in EVs of patients' biological fluid, may lead to the identification of new diagnostic and prognostic biomarkers. In this study, we identified a microRNA signature of serum EVs of patients with tumors of glial origin that was not only able to differentiate between glioma and normal patients, but also to predict survival.
2. Materials and Methods
2.1. Samples
Serum samples from 9 patients with Oligodendroglioma, 9 patients with Astrocytoma and 10 patients with Glioblastoma were provided by UCSF neurosurgery department (Genomic Shared Resources Core is NIH subsidized Shared Resources CCSG:P30CA016058), while 8 normal controls were provided by Ohio State University, Columbus, OH (Protocol: 2005C0014) (Fig. 1). In situ hybridization FFPE sections of 12 oligodendrogliomas, 10 astrocytomas, 13 glioblastoma samples and 12 non-neoplastic perilesional grey matter specimens were provided by the Regina Elena Institute, Dept. of Pathology, Rome (Protocol: 825/16).Collection of tissue and serum samples has been conducted according to the standards established by the Declaration of Helsinki, and informed consent was obtained according to the approved protocols mentioned above.
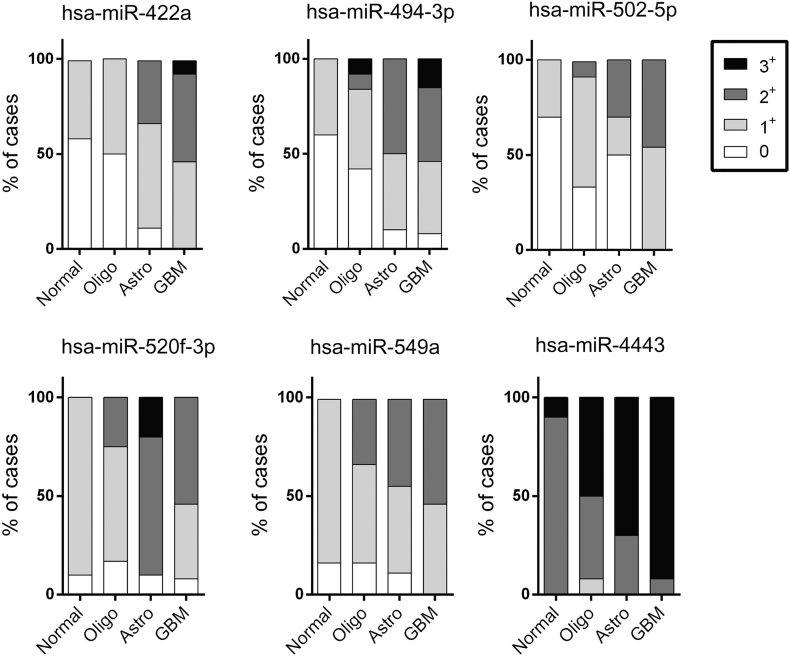
Fig. 1.
In situ hybridization histograms.
ISH was performed on 12 Normal CTRL (perilesional grey matter), 12 Grade II oligodendrogliomas, 10 Grade II astrocytomas and 13 glioblastomas. The intensity of expression of each microRNA of the signature was scored in each group of Glioma with numbers going from 0, in case of absent expression, to 3+ for maximal expression.
2.2. Extracellular Vesicles RNA Extraction
In order to avoid any cellular contamination samples were centrifuged 3 times at 3500 rpm for 20 min. After each centrifugation, supernatant was transferred to a new tube. Extracellular vesicles were extracted from 250μl of serum using Exoquick plus (System Bioscience cat#EXOQ5TM-1) and following manufacturer's instructions. The exosomal pellet was resuspended in 1.5 ml of TRIReagent (SIGMA-ALDRICH cat#T9424-200 ml), and 2 μl of 33.3 attoMole solution of spike-ins (cel-miR-248 and ath-miR159a) were added to each sample. The RNA was then extracted using the NORGEN RNA Clean-up and Concentration kit (cat#23600) and RNA was isolated following manufacturer's instructions.Furthermore, samples were concentrated and cleaned up from any chemical residues by centrifugation through Amicon ultra 0·5 centrifuge filters (SIGMA ALDRICH cat#Z740169-96EA), and then speed-vacued to 10 μl of volume.
2.3. Nanostring nCounter Assay and Data Analysis
A total of 36 samples were processed with NanoString nCounter Human v3 miRNA Expression Assay. A 3 μl volume of RNA suspension was used as input for nCounter miRNA sample preparation reactions according to manufacturer's instructions (NanoStringTechnologies). In this study, we used a volumetric approach rather than fixed concentrations. Extracted RNA was concentrated to a final volume of 10 μl: 3 μl were used for the NanoString array, and 7 μl were diluted 1:7 for RT-PCR validation. NanoString allows to concurrently measure 800 different microRNAs in each sample. Three μl of RNA suspension were annealed with multiplexed DNA tags (miR-tag) and target specific bridges. Mature microRNAs were bound to specific miR-tags using a Ligase enzyme. Tags in excess were removed by an enzyme clean-up step. The tagged microRNAs product was diluted 1:5. Twenty microlitre of reported probes in hybridization buffer and 5 μl of Capture probes, were combined with 5μl of the tagged microRNAs diluted solution. The overnight hybridization (16 to 20 h) at 65 °C allowed to complex sequence specific probes with targets. Probes' excess was removed using two-step magnetic beads based purification on an automated fluidic handling system (nCounter Prep Station), and target/probe complexes were immobilized on the cartridge for data collection. Data collection was carried out on the nCounter Digital Analyzer (NanoString Technologies) following the manufacturer's instructions, to count individual fluorescent barcodes and quantify target RNA molecules present in each sample. For each assay, a high-density scan (600 fields of view) was performed. NanoString raw data was analyzed with nSolver™, a software provided by NanoString Technologies. Negatives were used to perform background subtraction. Positives were used to perform technical normalization to adjust any lane by lane variability due to differences in hybridization, purification or binding. Data analysis was carried out using the software provided by NanoString technologies. Data was normalized by calculating the geometric mean of the top 100 miRNAs in all samples, as recommended by NanoString. P-values were calculated using the LIMMA package (Linear Models for Microarray Data) from the Bioconductor R project. The p-values were adjusted for multiple testing using the Benjamini and Hochberg method to control the False Discovery Rate (FDR) (Drusco et al., 2015). Raw data are available at NCBI GEO: GSE112462.
2.4. Taqman Stem-Loop miRNA RT-qPCR
Expression of mature single miRNAs was assessed in triplicate by the TaqMan Stem-loop miRNAassay (Applied Biosystems, Foster City, CA, USA), and normalized to Cel-miR-248 (Applied Biosystems) in all 36 samples. In this study we used a volumetric approach rather than fixed concentrations. Extracted RNA was concentrated to a final volume of 10 μl: 3 of them were used for the NanoString array, while the remaining 7 μl were diluted 1:7. Reverse transcription was carried out with 1.3 μl of the 1:7 diluted RNA stock. P-Values were calculated by one-tailed t-test. RT-qPCR box plots are represented on Fig. 3 as 2∧−ΔCt relative expression to Cel-miR-248. Means ± standard error of the mean (s.e.m.), *P < 0.05, by two-tailed Student's t-test (Drusco et al., 2015).
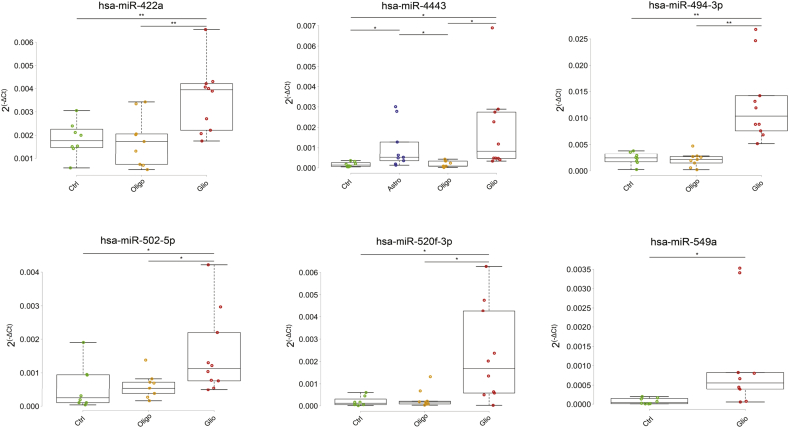
Fig. 3.
RT-PCR boxplots of significant comparisons.
The comparison is represented by the line at the top of the boxplot where each end of the line indicates one group of the comparison. The stars specify the pValue: ** for a p-value < 0.01, * for a p-value < 0.05.
2.5. In Situ Hybridization(ISH)
FFPE sections (Regina Elena Institute, Rome) of primary central nervous system tumors (12 oligodendrogliomas grade II, 10 astrocytomas grade II, 13 glioblastoma multiforme samples) and 12 non-neoplastic perilesional grey matter specimens were stained for the 6 selected miRs. All probes were labeled with 5′-digoxigenin and synthesized by Exiqon (Denmark). In situ hybridization was performed as described, with minor modifications (Drusco et al., 2015). Negative controls included omission of the probe and the use of a scrambled LNA probe; U6 was used as positive control (Exiqon). Slides were counterstained in fast red solution.
2.6. Survival Data Analysis
ISH tissue samples' clinical data were not available. Overall Survival (OS) curves were made analyzing only the clinical data of serum sample patients by using the Kaplan-Meier method. Censoring occurred at the date of the last follow-up or at the date of the death due to any cause. The log-rank test was performed for each comparison between subgroups. OS curves and log-rank test were generated by using ggsurvplot function from survminer R package.
3. Results
This article is the result of a pilot study aimed to find new serum biomarkers in patients with CNS tumors of glial origin. We focused on finding a microRNA signature in serum EVs that could differentiate among these tumors and normal controls. All patients' sera were collected at the time of surgery (Table 1). EVs were extracted from the serum of 9 patients diagnosed with Grade II oligodendroglioma, 9 patients with Grade II astrocytoma, 10 patients with glioblastoma and 8 healthy donors as controls. Patients were arranged into four groups: Normal controls (CTRL), oligodendroglioma (Oligo), astrocyoma (Astro) and glioblastoma (GBM). Samples were processed with the NanoString nCounter Human v3 Expression Assay.In order to reach our goal we created 7 groups: CTRL, GBM, Astro, Oligo, Low grade tumors (Astro and Oligo), High grade tumors (GBM), and Tumors (Astro, Oligo and GBM). We first performed nine comparisons: we compared each of the original 4 patients groups against each other (6 comparisons), CTRL against either High grade tumors or Low grade tumors, High grade tumors against Low grade tumors, and Glial tumors against CTRL (Table 2). Since GBM is the only high grade tumor type of our study, the comparisons CTRL versus GBM, and CTRL versus High grade tumors were considered as one. For each comparison, we then selected significantly differentially expressed microRNAs that showed >35 counts in one of the two compared groups. We finally identified microRNAs that were expressed in more than one group and those that were expressed only in one group. Candidate microRNAs were then selected based on the average number of counts and standard deviation. We also selected few microRNAs that, although not fitting in our selection method, showed interesting count patterns in the initial comparisons but, only one (hsa-miR-502-5p) was confirmed by RT-qPCR. Out of fifteen microRNAs tested, 6 were validated by RT-qPCR: hsa-miR-4443, hsa-miR-422a, hsa-miR-494-3p, hsa-miR-502-5p, hsa-miR-520f-3p, and hsa-miR-549a were significantly differentially expressed among groups, and were all up-regulated (Table 2 & Table2S). Only two microRNA were found significantly down-regulated at NanoString (hsa-miR-144-3p, hsa-miR-19a-3p), but none of them was validated by RT-qPCR. This finding might be due to overly low amounts of down-regulated microRNA in biological fluids. On the other hand, we are comparing two distinct techniques with different sensitivity: one counts single molecules, while the other amplifies an imprecise number of molecules (Bumgarner et al., 2008). Table2 shows the different comparisons with the corresponding p-Values when significant differences were observed with either or both techniques for each microRNA of our signature (fold changes are reported in Table 2S). Both techniques showed significant differences in the following comparisons: CTRL vs Astro for hsa-miR-4443, CTRL vs GBM for hsa-miR-520f-3p and -549a, GBM vs Oligo for hsa-miR-422a, -494-3p and -502-5p, CTRL vs Low Grade for hsa-miR-4443, and in Tumors vs CTRL for hsa-miR-549a and -4443. Significant differences were found only at NanoString between CTRL and Oligo for has-miR-494-3p, -520f-3p, and -549a, between CTRL and Astro for hsa-miR-494-3p, -520f and -549a, between GBM and Astro for hsa-miR-422a, -494-3p and -4443, between CTRL and Low Grade tumors for hsa-miR-494-3p, -520f-3p, and -549a, between Low and High Grade for hsa-miR-422a, -494-3p and a borderline difference for hsa-miR-502-5p, and between Tumors and CTRL for hsa-miR -520f-3p.Since RT-qPCR and in situ hybridization (ISH) are commonly applied technologies in the clinical field for the detection of molecular diagnostic markers, we considered RT-qPCR results as reference. ISH further confirmed our findings and provided information on the localization, type of cell producing the tested microRNA and, as for most ISH diagnostic and prognostic biological markers, its quantitative evaluation (Fig. 1, Fig. 2).We performed RT-qPCR on total RNA of serum EVs, and ISH on tissue samples coming from a different set of patients with tumors of glial origin for every identified microRNA. At RT-qPCR, every single microRNA of our signature was able to significantly differentiate between CTRL and GBM (Table2 & Fig. 3), while none was able to distinguish between CTRL and Oligo, GBM and Astro, and Low versus High grade (Table 2 & Fig. 3). ISH histograms representing the different degree of expression of each microRNA tested on tissue samples are shown in Fig. 1. The most representative ISH images of each group of tumors for every microRNA of our signature are displayed in Fig. 2. Tissue staining showed that every probe was able to differentiate CTRL from GBM. With the exception of miR-4443, that was able to differentiate between CTRL and Oligo, all microRNAs showed overlapping coloration intensities in the two groups. Only miR-4443 was significantly up-regulated in Astro compared to CTRL at RT-qPCR and NanoString (Table 2 & Fig. 3). At ISH, Astrocytomas were staining darker than CTRL in most of the cases. With the exception of miR-549a, all the remaining microRNAs tested by RT-qPCR were able to significantly differentiate between GBM and Oligo (Table 2 & Fig. 3). As shown in Fig. 1, tissue staining strongly confirmed this finding for miR-422a and miR-4443, while miR-494-3p, -502-5p and -520f-3p expressions levels between the two groups were overlapping. Although with borderline p-Values, RT-qPCR showed that miR-4443 was able to distinguish between Oligo and Astro and between Low Grade tumors and CTRLs (Table 2). The latter comparison was also significant at NanoString with a p-value = 0.003, suggesting that this difference could become more reliable using a larger number of samples. Fig. 1 shows that miR-4443 and miR549a tissue expression gradually increased throughout the four groups. A similar pattern of expression was observed with all the microRNAs of our signature with the exception of miR-520f-3p, which instead showed the higher variability of expression in Astrocytomas.
Table 1.
Glial tumor patients' clinical and molecular data. In the table are reported the age and gender of each patient, the number of days from the diagnosis to the time of the last follow up, the prognosis at the time of the last follow-up, and the molecular findings. Empty boxes correspond to those cases in which the biomarker was not tested.
| Patients' clinical & molecular data | ||||||||
|---|---|---|---|---|---|---|---|---|
| Oligodendroglioma (WHO gr2) | Age | Sex | Time to follow up | Status | IDH 1 | ATRX | 1p19q | Integrated diagnosis |
| 1 | 57 | M | 2225 | DEAD | 1 | 1 | Astrocytoma,IDH mutant | |
| 2 | 47 | M | 4697 | ALIVE | 1 | 0 | Oligodendroglioma,IDH mutant, NOS | |
| 3 | 39 | F | 4725 | ALIVE | 1 | 0 | 1 | Oligodendroglioma,IDH mutant, 1p19q codeleted |
| 4 | 31 | M | 3907 | ALIVE | 1 | 1 | Oligodendroglioma,IDH mutant, NOS | |
| 5 | 47 | F | 3182 | ALIVE | 1 | 1 | Astrocytoma,IDH mutant | |
| 6 | 39 | F | 3354 | ALIVE | 1 | 1 | Oligodendroglioma,IDH mutant, 1p19q codeleted | |
| 7 | 48 | F | 3001 | ALIVE | 1 | 1 | Oligodendroglioma,IDH mutant, 1p19q codeleted | |
| 8 | 62 | F | 1687 | DEAD | 1 | 1 | Oligodendroglioma,IDH mutant, 1p19q codeleted | |
| 9 | 74 | M | 1242 | DEAD | 1 | 1 | Oligodendroglioma,IDH mutant, 1p19q codeleted | |
| Astrocytoma (WHO gr2) |
Age | Sex | Time to follow up | Status | IDH 1 | ATRX | 1p19q | Integrated diagnosis |
| 1 | 48 | F | 537 | DEAD | 0 | 0 | Astrocytoma, NOT IDH mutant R132H | |
| 2 | 33 | M | 2216 | DEAD | NA | NA | Astrocytoma, NOS | |
| 3 | 29 | F | 4109 | ALIVE | 0 | 0 | Astrocytoma, NOT IDH mutant R132H | |
| 4 | 34 | M | 4565 | DEAD | 1 | 1 | Astrocytoma, IDH mutant | |
| 5 | 31 | F | 4151 | ALIVE | 1 | 0 | Astrocytoma, IDH mutant,NOS | |
| 6 | 46 | M | 3133 | ALIVE | 1 | 1 | Oligodendroglioma, IDH mutant, 1p/19q codeleted | |
| 7 | 42 | F | 3211 | ALIVE | 1 | 1 | Oligodendroglioma, IDH mutant, 1p/19q codeleted | |
| 8 | 47 | M | 2285 | ALIVE | 1 | 1 | Astrocytoma, IDH mutant | |
| 9 | 48 | F | 2250 | ALIVE | 0 | 0 | Astrocytoma, NOT IDH mutant R132H | |
| GBM (WHO gr4) |
Age | Sex | Time to follow up | Status | IDH 1 | ATRX | 1p19q | Integrated diagnosis |
| 1 | 37 | M | 3076 | ALIVE | 1 | 1 | GBM, IDH mutant | |
| 2 | 57 | M | 537 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 3 | 59 | M | 212 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 4 | 59 | F | 2297 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 5 | 57 | M | 4 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 6 | 63 | F | 247 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 7 | 77 | M | 136 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 8 | 65 | M | 386 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 9 | 43 | F | 247 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
| 10 | 46 | M | 392 | DEAD | 0 | GBM, NOT IDH mutant R132H | ||
Table 2.
Nanostring and RT-qPCR significant comparisons. RT = RT-PCR, NS = NanoString Assay, n.s. = Not Significant. Nanostring and RT-PCR significant pValues are showed for each tested microRNA and comparison. P-Values are written in bold for those microRNAs that showed a significant differential expression in the corresponding comparison. Bold boxes frame those p-Values that were found significant with both technologies (RT & NS). All microRNAs were found up-regulated in significant comparisons. Comparison boxes that did not show any significant difference with both techniques were left empty.
| Nanostring and RT-qPCR Significant comparisons | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Values |
||||||||||||
| miR-422a | miR-494-3p | miR-502-5p | miR-520f | miR-549a | miR-4443 | |||||||
| Comparisons | RT | NS | RT | NS | RT | NS | RT | NS | RT | NS | RT | NS |
| CTRL vs OLIGO | n.s. | 0.017 | n.s. | 0.014 | n.s. | 0.007 | ||||||
| CTRL vs ASTRO | n.s. | 0.022 | n.s. | 0.001 | n.s. | 0.001 | 0.045 | 0.0006 | ||||
| CTRL vs GBM | 0.005 | n.s. | 0.001 | n.s. | 0.045 | n.s. | 0.012 | 0.001 | 0.040 | 0.042 | 0.032 | n.s. |
| GBM vs OLIGO | 0.006 | 0.016 | 0.001 | 0.038 | 0.040 | 0.046 | 0.017 | 0.033 | n.s. | |||
| GBM vs ASTRO | n.s. | 0.010 | n.s. | 0.048 | n.s. | 0.020 | ||||||
| OLIGO vs ASTRO | 0.048 | n.s. | ||||||||||
| CTRL vs LOW GRADE | n.s. | 0.008 | n.s. | 0.001 | n.s. | 0.001 | 0.049 | 0.003 | ||||
| LOW vs HIGH GRADE | n.s. | 0.004 | n.s. | 0.018 | n.s. | 0.049 | ||||||
| TUMORS vs CTRL | n.s. | 0.0004 | 0.017 | 0.002 | 0.005 | 0.012 | ||||||
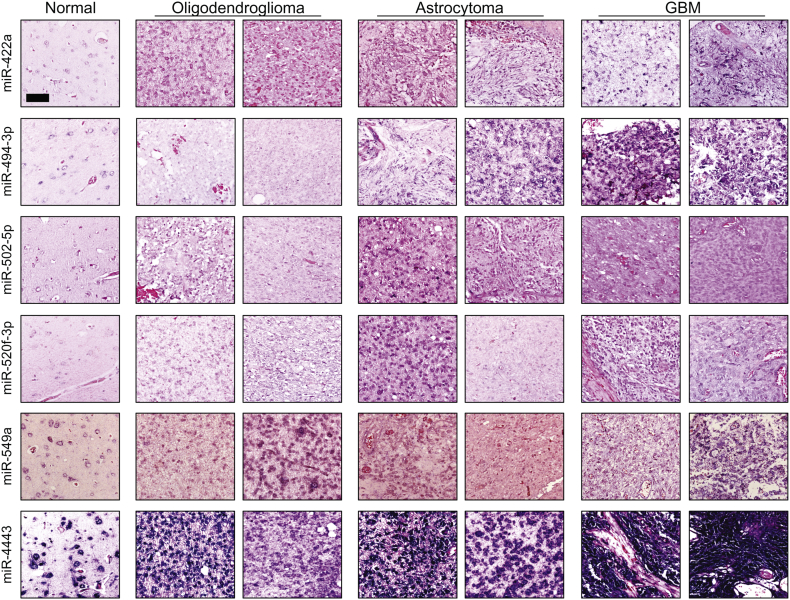
Fig. 2.
Representative in situ hybridization images.
In Situ Hybridization images of the most two representative tissue sections in different type of Gliomas for each microRNA of the signature. Normal perilesional tissue pictures of every tested microRNA are instead in singleton.
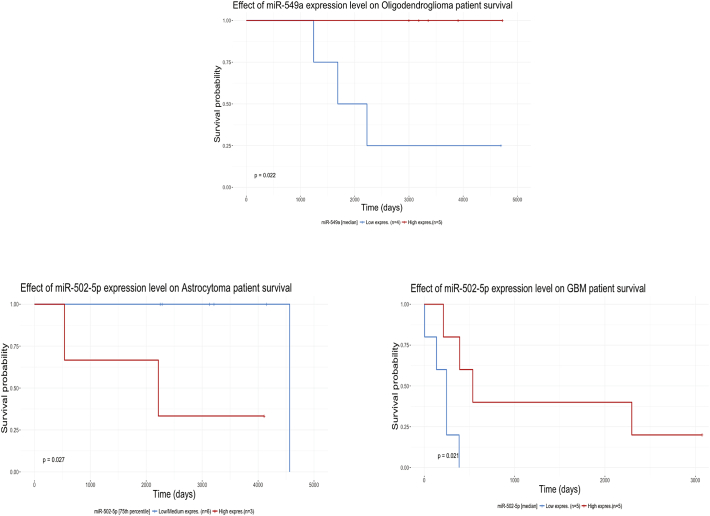
Only miR-4443 and miR-549a were significantly differentiating between Glial Tumors and CTRL at both RT-qPCR and NanoString. ISH staining showed stronger differences of miR-4443 expression in glial tumors compared to control, but confirmed miR-549 ability of differentiating between Glial Tumors and CTRL in half of the cases. Although ISH was confirmatory of RT-qPCR results, our aim was to find diagnostic microRNAs in serum vesicles of patients with glial tumors. ISH role was to understand what type of cells was actually producing the tested microRNA, and to identify those microRNA that could lead us to new therapeutic targets. Fig. 3 boxplots represent RT-qPCR significant findings for each microRNA of our signature: all of them were able to differentiate between a Glioblastoma and a Normal patient, but only miR-549a and miR-4443 were able to distinguish between the serum of patients with Glial Tumors and Normal healthy patients. We built survival curves to investigate if any of those microRNAs could be of any prognostic significance. Survival rates were evaluated as days from the diagnosis to the last follow-up. Using the median as threshold, we observed that higher levels of expression of miR-549a (p value = 0.022) and miR-502-5p (p value = 0.021) were significantly associated with a prolonged survival of patients with Oligodendroglioma and Glioblastoma respectively (Fig. 4). When we used the 75 percentile as threshold, miR-502-5p overexpression was linked to increased mortality in patients with Astrocytoma (Fig. 4). Only one young patient with low expression of miR-502-5p died after >4000 days from diagnosis. We did not have enough information and, or follow-up serum samples to explain this event. MiR-502-5p expression levels might have changed during the course of the disease. ISH patients' clinical information were not available to validate serum EVs patients' data. More studies with follow-up samples are needed in order to strengthen these findings.
Fig. 4.
Survival curves.
Kaplan-Meier Survival curves for hsa-miR-549a and hsa-miR-502-5p expressed in serum extracellular vesicles of patients with Oligodendroglioma and, Astrocytoma and Glioblastoma respectively. The red line indicates the overexpression, while the blue line the down-regulation of the tested microRNA.
4. Discussion
The molecular characterization of tumors has become an essential diagnostic step for the management of several cancers. As a result, the concept of personalized therapy has entered in the clinical oncologic practice, where teams of physicians are focused in targeting mutations and treat the specific tumor, rather than addressing a generic disease. Moreover, the discovery of microRNAs has disclosed the complex network of pathways regulating cellular physiologic and pathologic processes, and has given new hopes for the classification and the diagnostic and prognostic evaluation of cancer patients. Glioblastoma tissue signatures have shown to differentiate glioblastoma subclasses and have been connected to signaling pathways involved in cell growth (Kim et al., 2011). MicroRNAs have been found in all kind of biological fluids, including serum and plasma, where they freely circulate, or within extracellular vesicles. Within the CNS, extracellular vesicles are released by neurons, oligodendroglial cell, and microglia (Fauré et al., 2006; Krämer-Albers et al., 2007; Lachenal et al., 2011), and by tumor cells including gliomas (Skog et al., 2008). Gliomas are the most common primary tumor of the CNS, representing the 31% of all brain tumors, and the 81% of brain malignant tumors (Ostrom et al., 2015). The heterogeneity of gliomas accounts for their different prognostic behavior that can't be always predicted by current biomarkers. In an effort to find new molecular biomarkers, we profiled microRNAs in the serum extracellular vesicles of patients with tumor of glial origin. Extracellular vesicles were extracted from serum with the Exoquick polymeric precipitation (Peterson et al., 2015) which according to Van Deun et al. leads to a concomitant isolation of non-exosomal AGO2-bound microRNAs (Van Deun and Hendrix, 2017). Instead Li et al. showed that AGO2-microRNAs complexes are also present in extracellular vesicles (Li et al., 2012). Clearly, the extracellular vesicles topic will remain opened until the scientific community has definitive findings and standardized sorting methods. The signature included 6 up-regulated microRNAs: miR-422a, -494-3p, -4443, -502-5p, -520f-3p, and -549a. All microRNAs significantly differentiated between GBM and Normal controls, but only miR-4443 was able to differentiate Astrocytomas from controls and Oligodendrogliomas. MicroRNA-4443 and -549a could distinguish between tumors of glial origin and normal controls. Interestingly, down-regulation of miR-549a was also associated with poor survival in patients with Oligodendrogliomas (Fig. 4 p value = 0.022). All Oligodendroglioma patients were carrying IDH mutation, 5 had an associated 1p/19q codeletion, and 2 were NOS. None of these molecular markers could be connected with the prognosis. Patients with miR-549a up-regulation were not living >2225 days from diagnosis. While miR-4443 seems to induce epirubicin resistance in breast tumors (Chen et al., 2016), there is no reported involvement of miR-549a in cancer. ISH demonstrated that our signature was mainly expressed by cancer cells, and showed, with the exception of miR-520f-3p, that at different degrees of intensity, the expression of all probes was gradually increasing from controls to Oligodendrogliomas, from the latter to Astrocytomas, reaching the highest staining in Glioblastomas. Compared to the other groups, miR-520f-3p was mostly expressed in Astrocytomas showing the two highest levels of expression. This microRNA is down-regulated in Glioblastoma microvascular proliferation (Xu and Li, 2016). The proneural subtype of Gliablastoma is characterized by microvascular proliferation (Barciszewska, 2016) and, thus, responds to antiangiogenic drugs. We found that patients with high serum vesicles levels of miR-520f-3p showed an almost significant (p value = 0.089) increase in survival compared to those in which the microRNA was down-regulated (Fig. 1S). It is possible that different levels of expression of miR-520f-3p might differentiate among different Astrocytomas prognostic and, or therapeutic groups: those that won't, and those that will inevitably progress to Glioblastoma, those that will respond and those that will not respond to antiangiogenic therapy. Compared to the other groups, serum extracellular vesicle microRNA expression showed the highest variability in Astrocytoma. Overall, Astrocytoma patients behaved more similarly to Glioblastoma patients rather than Controls. There was no significant differential expression when comparing the Astrocytoma against the Glioblastoma group, but, although with borderline pValue (p value = 0.045), miR-4443 could significantly distinguish between Astrocytoma and Controls. Oligodendroglioma extracellular vesicles samples behavior was mostly similar to Normal controls. MiR-422a, -494-3p, and 502-5p were differentiating Controls and Oligodendrogliomas from Glioblastoma patients. In the comparison CTRL vs OLIGO by RT-qPCR, we could not validate any microRNA that showed significant differences at Nanostring. MicroRNA-422a suppresses cell proliferation, migration and invasion in Gliomas through targeting of insulin-like growth factor1 (IGF1) (Wang et al., 2017a), and its loss is associated with carcinogenesis in Glioblastoma (Liang et al., 2016). In our survival study, down-regulation of miR-422a in Glioblastoma patient was almost significantly associated with patients with a poor prognosis (Fig. 1S p value = 0.09). MiR-494-3p inhibits invasion and proliferation while promoting apoptosis in glioma cell lines through PTEN/AKT pathway (Li et al., 2015), and it is also associated with the progression from Glioma grade III to grade IV (Barciszewska, 2016). Interestingly, miR-502-5p could predict survival in Astrocytoma and Glioblastoma patients. Down-regulation of the microRNA was associated with a favorable prognosis in Astrocytoma (p value = 0.027) with an expectancy of more of 4000 days of survival (Fig. 4). Only one patient with low levels of miR-502-5p and an IDH mutant phenotype died at 4565 days from diagnosis, while patients with high expression did not survive more that 2216 days from the diagnosis. Due to the lack of follow-up samples we could not verify the level of expression of miR-502-5p at the time of death. The samples of this study were collected at the time of diagnosis (time 0). Conversely, in Glioblastoma patients miR-502-5p overexpression was associated with a longer survival (Fig. 4 p value = 0.021). Patients with low levels of the microRNA were not surviving >537 days, while two patients with high expression of miR-502-5p survived >2000 days. One patient was IDH negative and the one with longer survival was instead an IDH mutant phenotype. There are no literature reports of an involvement of miR-502-5p in CNS tumors, but it has been found up-regulated in the serum of patients with subarachnoid hemorrhage (SAH) (Lai et al., 2017). Other Authors have conducted studies on serum, plasma, and whole blood samples of Glioblastoma or Low-grade glioma patients to isolate freely circulating, cellular and exosomal microRNA signatures (Gozé et al., 2017; Regazzo et al., 2016; Roth et al., 2011; Wang et al., 2017b). Although all signatures showed promising results, they could not correlate to survival. More Glioma cases and their follow-up samples need to be tested to validate the strength of our signature. However, we demonstrated that extracellular vesicle microRNAs can realistically find a clinical application as diagnostic and prognostic molecular biomarkers in tumors of glial origin.
The following are the supplementary data related to this article.
Nanostring and RT-qPCR fold changes in significant comparisons. RT = RT-PCR, NS = NanoString Assay, n.s. = Not Significant. Nanostring and RT-PCR fold changes for significant pValues are showed for each tested microRNA and comparison. Fold changes are written in bold for those microRNAs that showed a significant differential expression in the corresponding comparison. Bold boxes frame those fold changes that showed significant p-values with both technologies (RT & NS). Comparison boxes that did not show any significant difference with both techniques were left empty.
Supplementary survival curves. Kaplan-Meier Survival curves for hsa-miR-520f-3p and hsa-miR-422a expressed in serum extracellular vesicles of patients with Astrocytoma. The red line indicates the overexpression, while the blue line the down-regulation of the tested microRNA.
Funding
UCSF NIH/NCI P50 CA097257 for biospecimens and annotation. The study was financed by the NIH/NCI Grant 1 R35 CA 197706-01.This manuscript has not been published previously in print or electronic format and is not under consideration by another publication or electronic medium.
Conflict of Interest
All authors do not own, receive an income, or have been financed from a private or public company.
Author Contributions
Drusco A: Wrote the manuscript, study design and, supervised and participated to experiments.
Fadda P: qRT-PCR statistical analysis, experiments set-up.
Nigita G: Nanostring and survival statistical analysis.
Fassan M, Gardiman MP, Sacchi D: In situ hybridization set up, execution, interpretation, figures.
Carosi M, Pescarmona E, Casini B: In situ hybridization samples' diagnosis and FFPE slides.
Calore F: Provided microvesicle extraction set-up protocol.
Antenucci A: Provided serum samples treatment protocol.
Bottoni A, Kelani H: Microvesicles extraction.
Di Leva G, Zanesi N: Manuscript editing and experiments strategies/trouble shooting.
Berger MS: Provided serum samples.
Croce CM: Study design, manuscript editing of the article, financial support.
Aknowledgement
We would like to thank Joanna Phillips and the UCSF Brain Tumor SPORE Tissue Core (NIH/NCI P50 CA097257) for biospecimens and annotation. The CCC - Genomics Shared Resource Core is NCI subsidized Shared Resource (CCSG: P30CA016058).
Contributor Information
Drusco A, Email: adrusco@gmail.com.
Croce CM, Email: carlo.croce@osumc.edu.
References
- Barciszewska A.M. MicroRNAs as efficient biomarkers in high-grade gliomas. Folia Neuropathol. 2016;54(4):369–374. doi: 10.5114/fn.2016.64812. [DOI] [PubMed] [Google Scholar]
- Bumgarner R.E., Birditt B., Dahl T. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008 Mar;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhong S.L., Lu P. mi R-4443 participates in the malignancy of breast cancer. PLoS One. 2016 Aug 9;11(8) doi: 10.1371/journal.pone.0160780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusco A., Croce C.M. MicroRNAs and Cancer: a long story for short RNAs. Adv. Cancer Res. 2017;135:1–24. doi: 10.1016/bs.acr.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Drusco A., Bottoni A., Laganà A. A differentially expressed set of microRNAs in cerebrospinal fluid (CSF) can diagnose CNS malignancies. Oncotarget. 2015 Aug 28;6(25):20829–20839. doi: 10.18632/oncotarget.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauré J., Lachenal G., Court M. Exosomes are released by cultured cortical neurons. Mol. Cell. Neurosci. 2006 Apr;31(4):642–648. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Gozé C., Reynes C., Forestier L., Sabatier R., Duffau H. Pilot study of whole blood microRNAs as potential tools for diffuse low-grade gliomas. Cell. Mol. Neurobiol. 2017 Aug 16 doi: 10.1007/s10571-017-0536-7. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Holdhoff M., Yovino S.G., Boadu O., Grossman S.A. Blood-based biomarkers for malignant gliomas. J. Neuro-Oncol. 2013 Jul;113(3):345–352. doi: 10.1007/s11060-013-1144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.M., Huang W., Park R., Park P.J., Johnson M.D. A developmental taxonomy of glioblastoma defined and maintained by microRNAs. Cancer Res. 2011 May 1;71(9):3387–3399. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer-Albers E.M., Bretz N., Tenzer S. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clin. Appl. 2007 Nov;1(11):1446–1461. doi: 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- Lachenal G., Pernet-Gallay K., Chivet M. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 2011 Feb;46(2):409–418. doi: 10.1016/j.mcn.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Lai N.S., Zhang J.Q., Qin F.Y., Sheng B., Fang X.G., Li Z.B. Serum microRNA are non-invasive biomarkers for the presence and progression of subarachnoid haemorrhage. Biosci. Rep. 2017 Feb 23;37(1) doi: 10.1042/BSR20160480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhu D., Huang L. Argonaute 2 complexes selectively protect circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.T., Wang H.Z., Wu Z.W. MiR-494-3p regulates cellular proliferation, invasion, migration, and apoptosis by PTEN/AKT signaling in human glioblastoma cells. Cell. Mol. Neurobiol. 2015 Jul;35(5):679–687. doi: 10.1007/s10571-015-0163-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Wang R., Jin Y. MiR-422a acts as a tumor suppressor in glioblastoma by targeting PIK3CA. Am. J. Cancer Res. 2016 Aug 1;6(8):1695–1707. [PMC free article] [PubMed] [Google Scholar]
- Louis D.N., Perry A., Burger P. International society of neuropathology—Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014 Sep;24(5):429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D.N., Perry A., Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016 Jun;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A. MicroRNA expression profiles classify human cancers. Nature. 2005 Jun 9;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Masui K., Mischel P.S., Reifenberger G. Molecular classification of gliomas. Handb. Clin. Neurol. 2016;134:97–120. doi: 10.1016/B978-0-12-802997-8.00006-2. [DOI] [PubMed] [Google Scholar]
- Mitchell P.S., Parkin R.K., Kroh E.M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 2008 Jul 29;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom Q.T., Gittleman H., Stetson L., Virk S.M., Barnholtz-Sloan J.S. Epidemiology of gliomas. In: Raizer J., Parsa A., editors. Current Understanding and Treatment of Gliomas. Cancer Treatment and Research, vol 163. Springer International Publishing; Switzerland: 2015. [DOI] [PubMed] [Google Scholar]
- Peterson M.F., Otoc N., Sethi J.K., Gupta A., Antes T.J. Integrated systems for exosome investigation. Methods. 2015 Oct 1;87:31–45. doi: 10.1016/j.ymeth.2015.04.015. [DOI] [PubMed] [Google Scholar]
- Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013 Feb 18;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regazzo G., Terrenato I., Spagnuolo M. A restricted signature of serum mi RNAs distinguishes glioblastoma from lower grade gliomas. J. Exp. Clin. Cancer Res. 2016 Jul 30;35(1):124. doi: 10.1186/s13046-016-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth P., Wischhusen J., Happold C. A specific mi RNA signature in the peripheral blood of glioblastoma patients. A. J. Neurochem. 2011 Aug;118(3):449–457. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- Russo F., Di Bella S., Vannini F. mi Randola 2017: a curated knowledge base of non-invasive biomarkers. Nucleic Acids Res. 2017 Sep:25. doi: 10.1093/nar/gkx854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo A., Tamanini A., Cabrini G., Dechecchi M.C. Circulating microRNA as emerging non-invasive biomarkers for gliomas. Ann Transl. Med. 2017 Jul;5(13):277. doi: 10.21037/atm.2017.06.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J., Würdinger T., van Rijn S. Glioblastoma microvesicles transport RNA and proteins that promote tomour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008 Dec;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008 Jul;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Van Deun J., Hendrix A. EV-TRACK consortium. Is your article EV-TRACKed? J. Extracell Vesic. 2017 Nov 10;6(1):1379835. doi: 10.1080/20013078.2017.1379835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Tang C., Na M. MiR-422a inhibits glioma proliferation and invasion by targeting IGF1 and IGF1R. Oncol. Res. 2017 Jn 26;25(2):187–194. doi: 10.3727/096504016X14732772150389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Q., Zhang M.Y., Deng M.L., Weng N.Q., Wang H.Y., Wu S.X. Low serum levels of mi R-485-3p predicts poor survival in patients with glioblastoma. PLoS One. 2017 Sept 20;12(9) doi: 10.1371/journal.pone.0184969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Li J.Y. Differential expression of PDGFRB and EGFR in microvascular proliferation in glioblastoma. Tumour Biol. 2016 Aug;37(8):10577–10586. doi: 10.1007/s13277-016-4968-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nanostring and RT-qPCR fold changes in significant comparisons. RT = RT-PCR, NS = NanoString Assay, n.s. = Not Significant. Nanostring and RT-PCR fold changes for significant pValues are showed for each tested microRNA and comparison. Fold changes are written in bold for those microRNAs that showed a significant differential expression in the corresponding comparison. Bold boxes frame those fold changes that showed significant p-values with both technologies (RT & NS). Comparison boxes that did not show any significant difference with both techniques were left empty.
Supplementary survival curves. Kaplan-Meier Survival curves for hsa-miR-520f-3p and hsa-miR-422a expressed in serum extracellular vesicles of patients with Astrocytoma. The red line indicates the overexpression, while the blue line the down-regulation of the tested microRNA.