Abstract
In about half the cases of involuntary childlessness, a male infertility factor is involved. The PIWI-LIKE genes, a subclade of the Argonaute protein family, are involved in RNA silencing and transposon control in the germline. Knockout of murine Piwi-like 1 and 2 homologs results in complete infertility in males. The aim of this study was to analyze whether the mRNA expression of human PIWI-LIKE 1–4 genes is altered in ejaculated spermatozoa of men with impaired sperm characteristics. Ninety male participants were included in the study, among which 47 were with normozoospermia, 36 with impaired semen characteristics according to the World Health Organization (WHO) manual, 5th edition, and 7 with azoospermia serving as negative control for the PIWI-LIKE 1–4 mRNA expression in somatic cells in the ejaculate. PIWI-LIKE 1–4 mRNA expression in the ejaculated spermatozoa of the participants was measured by quantitative real-time PCR. In nonazoospermic men, PIWI-LIKE 1–4 mRNA was measurable in ejaculated spermatozoa in different proportions. PIWI-LIKE 1 (100.0%) and PIWI-LIKE 2 (49.4%) were more frequently expressed than PIWI-LIKE 3 (9.6%) and PIWI-LIKE 4 (15.7%). Furthermore, a decreased PIWI-LIKE 2 mRNA expression showed a significant correlation with a decreased sperm count (P = 0.022) and an increased PIWI-LIKE 1 mRNA expression with a decreased progressive motility (P = 0.048). PIWI-LIKE 1 and PIWI-LIKE 2 mRNA expression exhibited a significant association with impaired sperm characteristics and may be a useful candidate for the evaluation of the impact of PIWI-LIKE 1–4 mRNA expression on male infertility.
Keywords: male infertility, Piwi, PIWI-LIKE genes, sperm quality parameters
INTRODUCTION
Infertility is generally defined as the inability to conceive spontaneously within a period of at least 12 months of unprotected intercourse. In more developed countries, a 12-month prevalence rate of up to 16.7% is reported, setting one of six couples at risk of unwilling childlessness.1 In approximately 40% of cases, male factors contribute to couple infertility. Causes for impaired sperm characteristics as the leading cause for male infertility are diverse, ranging from endocrine disturbances, immunological factors, increased scrotal temperature, urogenital tract infections as well as congenital or acquired urogenital abnormalities and genetic abnormalities (European Association of Urology [EAU] Male Infertility guidelines).2 However, cases of male idiopathic infertility are frequent and molecular data about their aetiology and predictors of impaired sperm characteristics are scarce.3
The members of the PIWI gene family, a subclade of the Argonaute proteins, are defined by their highly conserved P-element induced wimpy testis (PIWI) and Piwi-Argonaute-Zwille (PAZ) domains and their exclusive expression in the germline.4,5 Both domains function in the binding of small RNAs with the PAZ domain ensuring the binding of the 3'OH-overlap of small RNAs and the PIWI domain binding to the 5'-phosphate cap of small RNAs. This binding implicates slicer function and posttranscriptional silencing of target mRNAs.6,7,8 PIWI-LIKE proteins are shown to interact with a subclass of small RNAs called piwi-interacting RNAs (piRNAs), which are longer than other small RNA classes such as microRNAs (26–31 nt instead of 18–25 nt), share a preference for a 5'uridine, and are processed from a few distinct genetic loci.9,10,11 At the cellular level, piRNA/PIWI complexes trigger the silencing of retrotransposons, selfish genetic elements activated for instance by demethylation processes during gametogenesis and able to reintegrate at random loci in the genome.12,13,14 PIWI-LIKE genes also act at the chromatin level and exert epigenetic silencing of distinct genomic areas through regulation of histone K9 methylation.15,16,17,18 Furthermore, in male knockout mice, the loss of murine Piwi-like 1 (Miwi) results in a meiotic arrest at the round spermatid stage,19 while loss of murine Piwi-like 2 (Mili) results in a zygotene-pachytene spermatocytic block,20 resulting in male mice sterility. Taken together, PIWI-LIKE genes play an essential role in male gametogenesis by protecting the differentiating germ cells' genomic stability and the individual's male fertility.
A clear correlation between the dysregulated expression of human PIWI-LIKE 1 (HIWI) and tumorigenesis and prognosis can be observed in several tumor entities, including seminomas,21 breast cancer,22,23 and soft-tissue sarcoma.24 On the other hand, the data on the impact of human PIWI-LIKE gene mutation or dysregulation on male fertility are scarce. Gu et al.25 identified single-nucleotide polymorphisms in PIWI-LIKE 3 (nonsynonymous) and in PIWI-LIKE 4 (in the 3' untranslated region) to be associated with the occurrence of oligozoospermia. Furthermore, PIWI-LIKE 2 hypermethylation was associated with spermatogenic failure due to germ cell maturation defects in a cohort of 32 patients with azoospermia or severe oligozoospermia.26 Recently, it was shown that single-nucleotide mutations in the D-box domain of PIWI-LIKE 1 prevent ubiquitination and protein decay, resulting in azoospermia probably due to failure in the histone-to-protamine exchange.27 However, no data exist about the predictive potential of human PIWI-LIKE gene expression in ejaculated spermatozoa, which are the cells classically used in andrological practice for the assessment of fertility status and therapy decision.
The aim of this study was to determine whether mRNA expression of the four human PIWI-LIKE orthologs (PIWI-LIKE 1/HWI, PIWI-LIKE 2/HILI, PIWI-LIKE 3/HIWI3, and PIWI-LIKE 4/HIWI2) were detectable in ejaculated spermatozoa of patients. Furthermore, we wanted to evaluate whether these mRNA expressions were associated with impaired clinical semen characteristics.
PATIENTS AND METHODS
Study subjects and ethical approval
Participants were recruited from the couple fertility clinic of the Center for Reproductive Medicine and Andrology, University Hospital Halle (Saale, Germany). Altogether, 90 participants exhibiting different diagnoses of normozoospermia, asthenozoospermia, teratozoospermia, oligozoospermia, and different combinations, as well as azoospermia were enrolled. All patients were examined by an experienced andrologist. A detailed overview of the studied cohort is given in Table 1. The study was approved by the Ethics committee of the Medical Faculty of the Martin-Luther University Halle-Wittenberg. All participants gave written informed consents.
Table 1.
Participants’ clinical characteristics
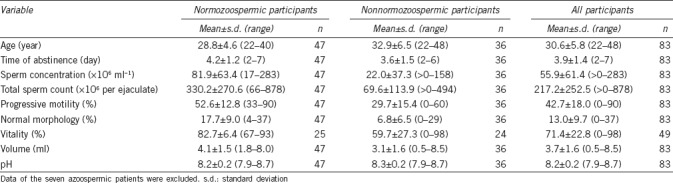
Clinical data assessed for this study contained age, sperm concentration, motility and morphology, occurrence and concentration of round cells, viability, semen volume, and pH. Every ejaculate analysis was conducted in the andrology laboratory of our clinic according to the World Health Organization (WHO) manual, 5th edition.28 The categories for the subdivision of the cohort (oligozoospermia, asthenozoospermia, necrozoospermia, teratozoospermia, and normozoospermia) were applied according to the reference values described in the WHO manual.28 Samples from the seven azoospermic patients were included in this study for the validation of the spermatozoa-specific PIWI-LIKE mRNA detection.
Specimen preparation and RNA isolation
After collection and ejaculate analysis according to the WHO manual, 5th edition, the specimens were centrifuged (1000 g, 10 min) immediately to separate spermatozoa and seminal plasma. The cell pellet was snap frozen and cryopreserved at −80°C until analysis.
Total RNA isolation from ejaculated spermatozoa was performed by the phenol/chloroform method. Briefly, the sperm pellet was dissolved in 1 ml TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and RNA separated by the addition of chloroform (AppliChem, Darmstadt, Germany) and centrifugation. To remove remaining traces of DNA, the solution was treated with DNase I (Qiagen, Hilden, Germany) for 15 min. RNA was precipitated by addition of isopropanol (AppliChem) and incubation at −20°C overnight. RNA pellet was washed twice with ice-cold ethanol and dissolved in 30 μl RNAse-free water. The RNA concentration and purity was assessed by absorption spectrometry in a spectrophotometer.
cDNA synthesis
The cDNA synthesis was carried out with RevertAid H Minus First strand cDNA synthesis kit (ThermoScientific, Waltham, MA, USA) and random primer according to manufacturer's protocol. Then, 1 μg of total RNA was applied for each reaction, yielding a sufficient amount of cDNA for double measurement of each gene.
Quantitative real-time-PCR
The measurement of the PIWI-LIKE 1–4 mRNA expression in the specimen was quantified by quantitative real-time-PCR. The reaction mix was set up with the HotStart Taq-Polymerase Kit (Qiagen) according to the manufacturer's protocol. Each human PIWI-LIKE gene (PIWI-LIKE 1–4) was amplified by using specific TaqMan primers (PIWI-LIKE 1: Hs_00380290; PIWI-LIKE 2: Hs_01032719; PIWI-LIKE 3: Hs_00908837; PIWI-LIKE 4:Hs_00895218; Applied Biosystems, Darmstadt, Germany). Quantitative real-time PCR measurements applying TaqMan assays were performed with the following program: (1) 95°C for 15 min, (2) 95°C for 20 s, and (3) 60°C for 1 min (repeat cycle 2–3 for 45 times).
The mRNA expression of hypoxanthine phosphoribosyl transferase (HPRT; forward primer: 5'-TTG CTG ACC TGC TGG ATT AC-3'; reverse primer: 5'-CTT GCG ACC TTG ACC ATC TT-3') was measured by using Maxima SYBR Green/ROX Master Mix (ThermoScientific) according to the manufacturer's protocol. HPRT mRNA is the most stable expressed according to free software package NormFinder (available from: http://www.mdl.dk/publicationsnormfinder.htm) and was chosen as the reference gene. The mRNA expression for each gene was calculated by the Δ CT method.29
Statistical analyses
Statistical analyses were performed with SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Bivariate correlation analyses according to Spearman-Rho and nonparametric tests (Mann–Whitney U-Test) were conducted for the evaluation of the association between human PIWI-LIKE 1–4 mRNA expression and sperm characteristics. P < 0.05 was considered statistically significant.
RESULTS
Assessment of PIWI-LIKE mRNA expression in ejaculated spermatozoa
PIWI-LIKE 1–4 mRNA was measurable in ejaculated spermatozoa in different proportions. PIWI-LIKE 1 (100.0% of cases) and PIWI-LIKE 2 (49.4%) were more frequently detectable than PIWI-LIKE 3 (9.6%) and 4 (15.7%). HPRT was expressed stable in every sample and served as internal reference for the standardization of the comparison of the individual PIWI-LIKE 1–4 mRNA expressions. In connection with the use of 1 μg total RNA per se men sample for cDNA synthesis, the application of a reference gene in the calculation of the relative mRNA expression (given as Δ CT value) guarantees the independence of the PIWI-LIKE 1–4 transcript measurement from the number of spermatozoa in the ejaculate sample. On the other hand, in the cellular fraction of ejaculates from seven azoospermic patients analyzed as controls, PIWI-LIKE 1–4 mRNA was not detectable in any sample. Round cells (≥0.1 × 106 ml−1) could be detected in 13 ejaculate samples, while 70 specimens exhibited no accurate determinable round cell contamination (<0.1 × 106 ml−1). The mean concentration of round cells was 0.3 × 106 ml−1 (range: 0–3.8 × 106 ml−1), while the median was 0 ml−1. Furthermore, in nonparametric tests, no correlation between the occurrence of detectable concentrations of round cells in the ejaculate and the expression of either PIWI-LIKE 1–4 mRNA was observed (P ranging from 0.37 to 0.6; Mann–Whitney U-test). A nonparametric bivariate linear correlation analysis was performed to examine whether the mRNA expressions of individual human PIWI-LIKE genes were correlated with each other in human ejaculated spermatozoa. In the ejaculated sperm samples, only the mRNA expression of PIWI-LIKE 1 was correlated to PIWI-LIKE 2 expression (rs = 0.543; P = 2.1 × 10−9).
PIWI-LIKE 1 mRNA is correlated with reduced sperm vitality
Further clinical and biographical data were studied in correlation with the individual PIWI-LIKE gene expression assessing bivariate linear regression analyses. The percentage of nonviable spermatozoa was inversely associated with the expression of the PIWI-LIKE 1 mRNA (rs = −0.34; P = 0.018; n = 49). Additionally, there was a significant positive correlation between increased age of participant and increased PIWI-LIKE 1 (rs = 0.29; P = 0.008; n = 83) or increased PIWI-LIKE 2 mRNA expression (rs = 0.26; P = 0.016; n = 83). Furthermore, PIWI-LIKE 1 mRNA expression was inversely associated with the percentage of motile spermatozoa in the ejaculate (rs = −0.23; P = 0.037), while PIWI-LIKE 2 mRNA expression was associated with sperm concentration (rs = 0.27; P = 0.011) and especially the number of spermatozoa in the ejaculate (rs = 0.31; P = 0.004).
Altered PIWI-LIKE 1 or PIWI-LIKE 2 mRNA expression was associated with sperm characteristics
The mRNA expression of the different human PIWI-LIKE genes in relation to the individual clinical sperm characteristics (concentration, total sperm count, motility, and morphology) was analyzed by applying nonparametric tests.
PIWI-LIKE 2 mRNA expression was decreased in samples exhibiting oligozoospermia (<39 × 106 cells per sample) in comparison with samples with normozoospermia (P = 0.022; Mann–Whitney U-test). The same was true when considering sperm concentration, decreased PIWI-LIKE 2 mRNA was also correlated with lower sperm concentration (<15 × 106 ml−1; P = 0.022; Mann–Whitney U-test). The relationship of the mRNA expression of PIWI-LIKE 1 and PIWI-LIKE 2 and total sperm count is shown in (Figure 1a and b).
Figure 1.
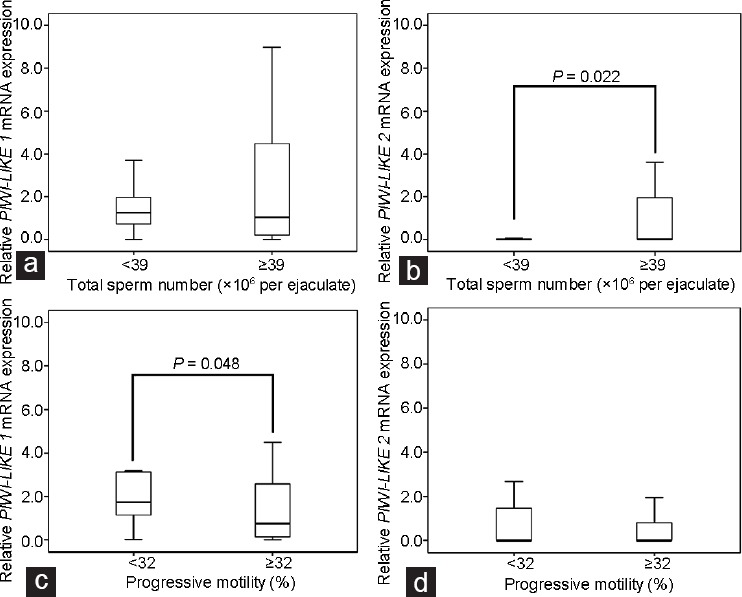
Box-plots comparing the relative mRNA expression of the PIWI-LIKE 1 and PIWI-LIKE 2 (expressed in Δ CT values) in ejaculated spermatozoa stratified to total sperm count and progressive motility. While (a) PIWI-LIKE 1 expression was not associated with sperm count, (b) samples from patients with oligozoospermia (<39 × 106 spermatozoa per ejaculate; n = 26) exhibited a decreased PIWI-LIKE 2 mRNA expression in comparison with normozoospermia (≥39 × 106 spermatozoa per ejaculate; n = 57, P = 0.022, Mann–Whitney U-test). (c) Samples from patients with asthenozoospermia (<32%; n = 23) exhibited an increased PIWI-LIKE 1 mRNA expression compared with normozoospermic patients (≥32%; n = 60; P = 0.048, Mann–Whitney U-test), while (d) PIWI-LIKE 2 mRNA expression was not associated with motility. Note the different y-axis scales indicating the higher expression of PIWI-LIKE 1 mRNA in comparison to PIWI-LIKE 2 mRNA. The boxes designate the distribution of 50% of the measured values; the lines designate the median value. The whiskers define the 2.5% and 97.5% quantiles.
Bearing in mind, that around half the samples had no detectable PIWI-LIKE 2 mRNA expression, we applied additional statistical analyses with the categories PIWI-LIKE 2 mRNA expression “detectable” and “nondetectable.” The results demonstrated that nondetectable PIWI-LIKE 2 mRNA expression was significantly associated with a lower sperm concentration (P = 0.017; Mann–Whitney U-test) and a lower sperm count (P = 0.004; Mann–Whitney U-test).
The association between progressive motility and PIWI-LIKE gene mRNA expression revealed that increased PIWI-LIKE 1 was associated with the occurrence of asthenozoospermia (<32% progressive motile spermatozoa; P = 0.048, Mann–Whitney U-test, (Figure 1c and d). Neither PIWI-LIKE 1 nor PIWI-LIKE 2 mRNA expression was significantly associated with abnormal morphology (<4%; P = 0.24 and P = 0.19; Mann–Whitney U-test).
DISCUSSION
In this study, we evaluated the mRNA expression of the spermatogenesis-relevant genes PIWI-LIKE 1–4 in ejaculated spermatozoa of patients referred to our center for couple infertility. To the best of our knowledge, the analysis of mRNA expression of all four PIWI-LIKE genes in ejaculated spermatozoa has not been published. Heyn et al.26 analyzed PIWI-LIKE 2 transcript levels in spermatozoa of normozoospermic individuals in comparison to testicular samples with patients with spermatogenic arrest. In our study, we detected a significant correlation between the expression of PIWI-LIKE 1 and PIWI-LIKE 2 mRNA, suggesting an overlapping expression pattern of these genes in sperm development as it is described for the homologous Piwi-like 1 (Miwi) and Piwi-like 2 (Mili) genes in mice.30 Specifically, the expression of both murine gene homologs is detected in late stages of spermatogenesis, with Miwi expressed only in the mid-pachytene spermatocytes and later stages.19,31 Most recently, Hempfling et al.32 reported an almost ubiquitous expression of PIWI-LIKE 1 and PIWI-LIKE 2 in testicular samples of patients with normozoospermia or hypospermatogenesis. In contrast, PIWI-LIKE 4 was only analyzed in samples of patients with spermatogenic arrest and showed a high expression in these samples, while PIWI-LIKE 3 was not detectable in any testicular sample.32
Intriguingly, increased PIWI-LIKE 1 and also PIWI-LIKE 2 mRNA expression was significantly correlated with elevated participants' age. An association between PIWI-LIKE gene expression and increasing age has not been described before and may be interesting in the context of aging males' protection of their genetic material against transposons activation, environmental stressors, and oxidative radicals. Furthermore, especially PIWI-LIKE 2 mRNA expression seems to play an essential role in these processes, leading to a worsened prognosis for soft-tissue sarcoma patients with a decreased mRNA expression of either PIWI-LIKE 2 or PIWI-LIKE 4.33 However, as age was only a secondary endpoint of this study and the age range was rather small, general conclusions on an age-dependence of PIWI-LIKE 1–4 mRNA expression in spermatozoa are not possible yet.
Analogous to the detrimental effect of a decreased PIWI-LIKE 2 mRNA expression in tumors on patients' outcome, a decreased expression of PIWI-LIKE 2 mRNA in our cohort was associated with a decreased total sperm count (P = 0.022). This result is in line with the results of Heyn et al.,26 showing a decreased PIWI-LIKE 2 transcript level due to PIWI-LIKE 2 promoter hypermethylation in patients' samples exhibiting different stages of spermatogenic failure. The application of the same amount of total RNA (1 μg) for cDNA synthesis of each patient's sample and the usage of a reference gene (HPRT) for the normalization of the gene expression both ruled out a bias in the measured PIWI-LIKE 2 mRNA expression, thus additionally strengthening this result. Furthermore, a significant association between an increased PIWI-LIKE 1 mRNA expression and a decreased percentage of progressive motile spermatozoa could be demonstrated in our study cohort, which may indicate the need for a PIWI-LIKE gene silencing in proper sperm development.
The protein expression of PIWI-LIKE 1 and 2 in human ejaculated spermatozoa is still rarely studied; however, recently Hempfling et al.32 demonstrated with immunofluorescence in testicular biopsies that PIWI-LIKE 1 protein is detectable in spermatocytes and spermatids, but not in elongated spermatids. This is a strong hint that the protein expression of the PIWI-LIKE genes in human spermatozoa resembles that detected in murine spermatozoa, where PIWI-LIKE 2 and 1 protein expression is diminished in late phases of spermatogenesis.30 Therefore, we would expect no detectable PIWI-LIKE 1 or 2 protein expression in normal sperm samples. From the recent literature, one could speculate that mammalian PIWI-LIKE 1 or 2 mRNA (or potentially protein expression) is a surrogate for proper sperm development, maturation, and better genomic integrity rather than a functional regulator of sperm activity.
In summary, we measured mRNA expression of the four human PIWI-LIKE genes in a cohort of 83 men with different semen status. PIWI-LIKE 1 and PIWI-LIKE 2 mRNA expressions were especially identified as putative new targets for further studies on the association between sperm-bound mRNA transcriptome and male fertility impairments. These interactions should be addressed in broader studies applying more holistic techniques such as microarrays and RNA-seq.
AUTHOR CONTRIBUTIONS
MG performed the data analysis and revised the manuscript; CM and LM carried out the clinical sample processing and the qPCR measurements; TG and HMB conceived the study design, prepared and revised the manuscript. All authors read and approved the final version of the manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENTS
The authors would like to thank Constanze Kloß for providing excellent technical assistance and performing the semen analysis. Ivan Hoffmann, Dr. Petra Kaltwaßer, and Dr. Solveig Köller are thanked for the recruitment of participants for this study. The study was sponsored in part by a grant from the Wilhelm-Roux program of the Medical Faculty of the Martin Luther University Halle-Wittenberg to TG (FKZ 25/43). The funding source played no role in the study design and data evaluation.
REFERENCES
- 1.Boivin J, Bunting L, Collins JA, Nygren KG. International estimates of infertility prevalence and treatment-seeking. Potential need and demand for infertility medical care. Hum Reprod. 2007;22:1506–12. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- 2.Jungwirth A, Giwercman A, Tournaye H, Diemer T, Kopa Z, et al. European association of urology guidelines on male infertility. The 2012 update. Eur Urol. 2012;62:324–32. doi: 10.1016/j.eururo.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 3.Gianotten J, Lombardi MP, Zwinderman AH, Lilford RJ, van der Veen F. Idiopathic impaired spermatogenesis. Genetic epidemiology is unlikely to provide a short-cut to better understanding. Hum Reprod Update. 2004;10:533–9. doi: 10.1093/humupd/dmh045. [DOI] [PubMed] [Google Scholar]
- 4.Cox DN, Chao A, Baker J, Chang L, Qiao D, et al. A novel class of evolutionarily conserved genes defined by Piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–27. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins. The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–2. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 6.Yan KS, Yan S, Farooq A, Han A, Zeng L, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–74. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 7.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–37. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, et al. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–94. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 10.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 11.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs. The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 12.Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–9. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 14.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–9. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–72. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 16.Sugimoto K, Kage H, Aki N, Sano A, Kitagawa H, et al. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem Biophys Res Commun. 2007;359:497–502. doi: 10.1016/j.bbrc.2007.05.136. [DOI] [PubMed] [Google Scholar]
- 17.Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, et al. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–9. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50:601–8. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Deng W, Lin H. Miwi, a murine homolog of Piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–30. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 20.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, et al. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–19. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of Hiwi, a human member of the Piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–99. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Ren Y, Xu H, Pang D, Duan C, et al. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22:217–23. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan P, Ghosh S, Graham K, Mackey JR, Kovalchuk O, et al. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget. 2016;7:37944–56. doi: 10.18632/oncotarget.9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taubert H, Greither T, Kaushal D, Würl P, Bache M, et al. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene. 2007;26:1098–100. doi: 10.1038/sj.onc.1209880. [DOI] [PubMed] [Google Scholar]
- 25.Gu A, Ji G, Shi X, Long Y, Xia Y, et al. Genetic variants in Piwi-interacting RNA pathway genes confer susceptibility to spermatogenic failure in a Chinese population. Hum Reprod. 2010;25:2955–61. doi: 10.1093/humrep/deq274. [DOI] [PubMed] [Google Scholar]
- 26.Heyn H, Ferreira HJ, Bassas L, Bonache S, Sayols S, et al. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS One. 2012;7:e47892. doi: 10.1371/journal.pone.0047892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gou LT, Kang JY, Dai P, Wang X, Li F, et al. Ubiquitination-deficient mutations in human Piwi cause male infertility by impairing histone-to-protamine exchange during spermiogenesis. Cell. 2017;169:1090–104e13. doi: 10.1016/j.cell.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. 5th ed. Geneva: WHO Press; 2010. WHO Laboratory Manual for the Examination and Processing of Human Semen. [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Bak CW, Yoon TK, Choi Y. Functions of PIWI proteins in spermatogenesis. Clin Exp Reprod Med. 2011;38:61–7. doi: 10.5653/cerm.2011.38.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–81. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hempfling AL, Lim SL, Adelson D, Evans J, O'Connor AE, et al. Expression patterns of HENMT1 and PIWIL1 in human testis – Implications for transposon expression. Reproduction. 2017;154:363–74. doi: 10.1530/REP-16-0586. [DOI] [PubMed] [Google Scholar]
- 33.Greither T, Koser F, Kappler M, Bache M, Lautenschläger C, et al. Expression of human Piwi-like genes is associated with prognosis for soft tissue sarcoma patients. BMC Cancer. 2012;12:272. doi: 10.1186/1471-2407-12-272. [DOI] [PMC free article] [PubMed] [Google Scholar]


