Abstract
To investigate whether transcription of hepatitis B virus (HBV) gene occurs in human sperm, total RNA was extracted from sperm of patients with chronic HBV infection (test-1), from donor sperm transfected with a plasmid containing the full-length HBV genome (test-2), and from nontransfected donor sperm (control), used as the template for reverse transcription-polymerase chain reaction (RT-PCR). Positive bands for HBV DNA were observed in the test groups but not in the control. Next, to identify the role of host genes in regulating viral gene transcription in sperm, total RNA was extracted from 2-cell embryos derived from hamster oocytes fertilized in vitro by HBV-transfected (test) or nontransfected (control) human sperm and successively subjected to SMART-PCR, suppression subtractive hybridization, T/A cloning, bacterial amplification, microarray hybridization, sequencing and the Basic Local Alignment Search Tool (BLAST) search to isolate differentially expressed genes. Twenty-nine sequences showing significant identity to five human gene families were identified, with chorionic somatomammotropin hormone 2 (CSH2), eukaryotic translation initiation factor 4 gamma 2 (EIF4G2), pterin-4 alpha-carbinolamine dehydratase 2 (PCBD2), pregnancy-specific beta-1-glycoprotein 4 (PSG4) and titin (TTN) selected to represent target genes. Using real-time quantitative RT-PCR (qRT-PCR), when CSH2 and PCBD2 (or EIF4G2, PSG4 and TTN) were silenced by RNA interference, transcriptional levels of HBV s and x genes significantly decreased (or increased) (P < 0.05). Silencing of a control gene in sperm did not significantly change transcription of HBV s and x genes (P > 0.05). This study provides the first experimental evidence that transcription of HBV genes occurs in human sperm and is regulated by host genes.
Keywords: embryo, hepatitis B virus, host gene, regulation, sperm, transcription
INTRODUCTION
Hepatitis B virus (HBV) can invade the human male germline, and its DNA has been detected at various stages of spermatogenesis in patients with chronic HBV infection.1,2,3 Viral infections may be deleterious to human fertility.4,5,6 Moreover, some studies report that HBV has a negative effect on sperm motility in vivo. The couples in which the male is infected with HBV have a higher risk of a low fertilization rate after in vitro fertilization (IVF).7 Our previous studies found that HBV DNA is able to integrate into the chromosomes of patients' sperm cells and induce chromosomal aberrations and that such HBV DNA-carrying sperm can fertilize oocytes.8 After fertilization, the sperm-introduced HBV genes retain their functions of replication and expression in embryonic cells.9,10 However, it is generally believed that hepatotropism is a prominent feature of HBV infection, and viral infection appears to be restricted to hepatocytes.11 Because sperm cells differ from hepatic cells, whether HBV gene transcription occurs in sperm cells and, in particular, its regulation mechanism has attracted the attention of researchers.
Hepatitis B infection and disease pathogenesis are known to be influenced by a number of factors, including host genetic factors.12 Many cellular proteins that possibly regulate HBV gene transcription in hepatocytes have been identified, including liver-enriched proteins, such as hepatocyte nuclear factor 1 (HNF1), hepatocyte nuclear factor 3 (HNF3), CCAAT/enhancer binding protein (C/EBP), and Krüppel-like factor 15 (KLF15), ubiquitous factors, such as specificity protein 1 (Sp1), regulatory factors X1 (RFX1), nuclear factor-Y (NF-Y), and activator protein-1 (AP1), and members of the nuclear receptor superfamily, such as hepatocyte nuclear factor 4 (HNF4), retinoid X receptor alpha (RXRa), peroxisome proliferator-activated receptor alpha (PPARa), and chicken ovalbumin upstream promoter transcription factor 1 (COUP-TF1).13,14,15,16,17,18 The current study was undertaken to explore whether HBV gene transcription occurs in sperm cells and to isolate and identify host genes that participate in regulating HBV gene transcription in sperm cells, which is critical to reveal the regulation mechanism of viral gene transcription in these cells and to prevent the negative impact of HBV infection on the fertilizing capacity of sperm.
MATERIALS AND METHODS
Semen samples were obtained from healthy donors and patients with chronic HBV infection. Written informed consents were obtained from all study subjects who allowed their sperm samples to be used for research. All protocols used in this study involving human subjects were approved by the Institutional Ethical Review Board of Shantou University Medical College (Approval No. SUMC-2017-04) according to the World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects.19
The animal protocol was designed to minimize pain or discomfort. The animals were acclimatized to laboratory conditions (23°C, 12 h light/12 h dark, 50% humidity, ad libitum access to food and water) for 2 weeks prior to experimentation. All animals were euthanized by barbiturate overdose (intravenous injection, 150 mg kg−1 pentobarbital sodium) for oocyte collection, and then their bodies were subjected to harmless treatment. All procedures involving animals were reviewed and approved by the Medical Animal Care and Welfare Committee at Shantou University Medical College (IACUC protocol No. SUMC2015-152) according to the recommended guide for the care and use of laboratory animals published by the National Research Council (USA).20
Sperm preparation
Semen samples were incubated in a CO2 incubator (37°C, 5% CO2 in air) for 30 min to allow liquefaction. Motile sperm were selected using the swim-up method in Biggers-Whitten-Whittingham (BWW) medium supplemented with 0.3% bovine serum albumin (BSA).21 After washing, these sperm samples were used for subsequent experiments.
Sperm transfection and efficiency
Each sperm sample from a healthy donor was divided into two groups: a test group and a control group. A 100-μl aliquot of a mixture comprised of 1 μl of the plasmid pIRES2-EGFP-HBV (1.5 μg μl−1) containing the full-length HBV genome, 6 μl of Fugene HD, and 93 μl of HEPES-buffered saline was incubated at 28°C for 15 min and then added to the sperm cells in the test group, which was incubated for 1.5 h. The sperm cells in the control group were not transfected.
To measure the transfection efficiency, a Nick Translation kit (Roche, Basel, Switzerland) and Fluorescein-12-dUTP (Thermo Fisher Scientific, Waltham, MA, USA) were used. The plasmid was labeled with fluorescein-12-dUTP by nick translation according to the manufacturer's instructions. Transfected sperm cells were detected by green fluorescence and counted under a fluorescence microscope (Dmil Led, LEICA, Heidelberg, Germany) (Supplementary Figure 1a (227KB, tif) and b (227KB, tif) ). The transfection efficiency (%) was determined by the number of cells showing green fluorescence divided by the total number of cells with and without green fluorescence multiplied by 100. Five high-power fields (×400) per slide were assessed, and the values were averaged.
Identification of transfected and non-transfected sperm cells and sperm-derived embryos. (a) The arrows show three of the transfected sperm cells under a phase-contrast microscope. (b) the same field of view under a fluorescence microscope. (c) The arrows show two 2-cell embryos under a phase-contrast microscope. (d) the arrow shows a transfected two-cell embryo in the same field of view under a fluorescence microscope. Scale bar = 20 μm in (a) and (b), 50 μm in (c) and (d).
Confirmation of HBV gene transcription in sperm cells
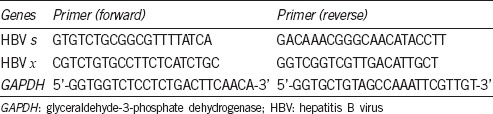
In experiment A, sperm cells in test Group A were obtained from HBV-infected patients, and those in control Group A were from healthy donors. In experiment B, sperm cells were obtained from healthy donors and divided into test Group B or control Group B, which was transfected with a plasmid containing the full-length HBV genome or not transfected, respectively. Total RNA from sperm cells was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and cDNA was synthesized with a PrimeScript™ RT reagent kit and gDNA Eraser (Cat. #RR047A, TaKaRa Biotech, Dalian, China) according to the manufacturer's protocol. Reverse transcription polymerase chain reaction (RT-PCR) was performed in a reaction volume of 20 μl containing cDNA (2 μl), 2× QuantiFast SYBR Green PCR Master Mix (10 μl; Qiagen, Hilden, Germany), forward and reverse primer (10 μmol l−1, 0.2 μl each) for HBV s or HBV x, and RNase-free water (7.6 μl). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was coamplified as an internal control.22 The primer pairs for GAPDH and the HBV x and s genes are shown in Table 1. The cycling conditions were 5 min at 95°C, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. The amplified PCR fragments were subjected to 1.5% agarose gel electrophoresis and stained with ethidium bromide.
Table 1.
The primers were used to amplify the hepatitis B virus s and x genes, and glyceraldehyde-3-phosphate dehydrogenase gene
Isolation and identification of host genes regulating transcription of HBV genes in sperm cells
Oocyte preparation, insemination, and embryo grouping
Oocyte preparation and insemination were performed as described previously.23,24 Briefly, zona-free hamster ova were fertilized by sperm cells from the test and control groups and then incubated in a CO2 incubator (37°C, 5% CO2 in air) for 24 h to obtain two-cell embryos. Two-cell embryos exhibiting green fluorescence, derived from the test group sperm, and those without green fluorescence, derived from the control group sperm, were collected under a fluorescence microscope (Supplementary Figure 1c (227KB, tif) and d (227KB, tif) ). After washing, total RNA from the two-cell embryos was extracted for cDNA library construction by switching mechanism at 5' end of RNA template (SMART) amplification.
SMART amplification and suppression subtractive hybridization (SSH)
SMART amplification and SSH were carried out using SMARTer™ PCR cDNA Synthesis Kit and Clontech PCR-Select™ cDNA Subtraction Kit (Clontech Laboratories, Inc., Mountain View, CA, USA) according to the manufacturer's instructions. Briefly, 3.5 μl (50 ng) total RNA extracted from two-cell embryos of the test and control groups was successively subjected to first-strand cDNA synthesis, amplified by long-distance PCR, purified by column chromatography, digested by Rsa I, diluted to a final concentration of 300 ng μl−1 in 1× Tris-sodium chloride-EDTA (TNE) buffer, and subjected to adaptor ligation. SSH materials consisted of cDNA from the test two-cell embryos as the tester and cDNA from the control two-cell embryos as the driver for forward subtraction, and vice versa for reverse subtraction. After the first and second hybridizations, hybridized samples 1 and 2 were mixed and then incubated at 68°C overnight, followed by a primary PCR and a secondary PCR. After an agarose/ethidium bromide gel analysis, the products were used for DNA ligation. The same procedures listed above were also performed for reverse subtraction and control subtraction.
T/A cloning and bacterial amplification
T/A cloning and amplification of subtraction products were performed using pGEM®-T Easy Vector System (Promega, Madison, WI, USA) according to the manufacturer's instructions. The 174 white (positive) colonies obtained were separately resuspended in 1.5 ml of LB medium with ampicillin (100 μg ml−1) and grown at 37°C for 6 h. PCR amplifications were carried out using 0.3 μl bacterial suspension and Premix Taq™ (TaKaRa Biotech), and 5μl from each PCR reaction was subjected to 1% agarose gel electrophoresis. Positive bands were detected in 152 of 174 colonies.
Microarray hybridization and data analysis
Microarray hybridization and data analysis were performed by TaKaRa Biotech. Briefly, the PCR products for differentially expressed genes from 152 clones, obtained from the subtraction libraries, were purified using 2-propanol precipitation and then spotted in triplicate on TaKaRa glass slides using an Affymetrix Arrayer 417 (TaKaRa Biotech). The PCR products from the forward- and reverse-subtracted libraries (2 μg each) were labeled with Cy3 and Cy5 fluorescence dyes and separately used to probe the glass slides containing the PCR-amplified cDNA. The slides were hybridized overnight at 65°C with labeled purified probes. Quality control of the microarray chips was performed according to TaKaRa's method and standard. Array slides were scanned using an Affymetrix 428 Array Scanner (Affymetrix, Buckinghamshire, UK). The measured intensities are expressed as a ratio of Cy5/Cy3 intensities, which were background-corrected and normalized to the average Cy5/Cy3 ratio. The ratios were log2 transformed, and the following clones were selected for sequencing: (1) clones with a fold change value >2 or <0.5 when compared with the average Cy5/Cy3 intensity ratio, the differential expression of which was statistically significant, were used to select target genes; (2) a clone with a fold change value >0.5 and <2, the differential expression of which was not statistically significant, was chosen as the control.25,26
DNA sequencing and Basic Local Alignment Search Tool (BLAST) analysis
DNA sequencing and analysis were performed by Beijing Genomics Institute Shenzhen Co., Ltd., (Shenzhen, China) using a 3730 XL DNA Analyzer (Applied BioSystems, Foster City, CA, USA). Sequencing reactions were carried out with BigDye v3.1 Mix and POP-7™ polymer (Applied Biosystems). For the acquired sequences, BLAST was used to search for sequences homologous to human genes in the GenBank nucleotide database. Of the sequences, five with statistically significant differential expression were selected as the target genes, and a nonstatistically significant sequence without statistical significance of differential expression was selected as a control.
Identification of target genes regulating transcription of HBV genes in sperm cells
To silence the target and control genes, gene-specific short hairpin (sh) RNA or short interfering (si) RNA were designed and synthesized by Shanghai GenePharma Co., Ltd., or QIAGEN Co., Ltd., (Shanghai, China), respectively (Supplementary Table 1 (363.2KB, tif) ). Each sperm sample from a healthy donor was divided into two groups: a test group, which was cotransfected with plasmids containing the full-length HBV genome and the gene-specific shRNA or siRNA; and a control group, which was transfected with the plasmid containing the full-length HBV genome alone. Real-time qRT-PCR was performed using an ABI 7300 Real-Time PCR System (Applied Biosystems) to compare the transcriptional level of HBV genes in sperm cells of the test and control groups, with GAPDH as an internal control. Total RNA was extracted from plasmid-transfected sperm cells with TRIzol reagent (Life Technologies), and cDNA was synthesized using a PrimeScript™ RT Reagent Kit with gDNA Eraser (Cat. #RR047A, Takara Biotech) according to the manufacturer's protocol. A total volume of 20 μl of reaction mixture contained cDNA (2 μl), 2× QuantiFast SYBR Green PCR Master Mix (10 μl) (Qiagen, Hilden, Germany), forward and reverse primers 10 μmol l−1, 0.2 μl each) for s and x genes and GAPDH (Table 1), and RNase-free water (7.6 μl). Cycling conditions were as follows: 5 min at 95°C; 40 cycles of 15 s at 95°C and 30 s at 60°C; and one cycle of 15 s at 95°C, 2 min at 55°C, and 15 s at 95°C. The data were analyzed quantitatively using the 2−ΔΔCt method.
The gene-specific shRNA and siRNA sequences*
Statistical analysis
Eighteen healthy donors were randomly divided into six groups, and their sperm samples were used to assess the effects of the silencing of five target genes (chorionic somatomammotropin hormone 2 [CSH 2], eukaryotic translation initiation factor 4 gamma 2 [EIF4G2], pterin-4 alpha-carbinolamine dehydratase 2 [PCBD2], pregnancy-specific beta-1-glycoprotein 4 [PSG4], and titin [TTN]) and a control gene (estrogen-related receptor gamma [ESRRG]) on transcription of HBV s and x genes by real-time quantitative (q) RT-PCR. The sperm samples from three donors were individually used for assaying each gene, and each assay was repeated three times. In the 2−ΔΔCt analysis, individual data were converted to a linear form using the 2−Ct calculation27 and then subjected to a paired-sample t-test using SPSS version 16.0 (SPSS Inc., Chicago, IL, USA) to determine a significant difference in average transcription levels of s and x genes between the test and control groups. P < 0.05 was considered statistically significant.
RESULTS
Transfection efficiency, and confirmation of the transcription of HBV genes in sperm cells
The transfection efficiency for a plasmid containing the full-length HBV genome into sperm cells was 39.51% ± 2.72%.
Based on RT-PCR, bands for GAPDH were observed in both experiments A and B, either in the test groups or the control groups. In contrast, positive bands for HBV s and x genes were observed only in test Group A and B and were not observed in control Group A and B or the minus reverse transcriptase (–RT) and minus template (–T) controls. The product sizes for GAPDH, HBV s and x genes were 127 bp, 95 bp, and 170 bp, respectively (Figure 1).
Figure 1.
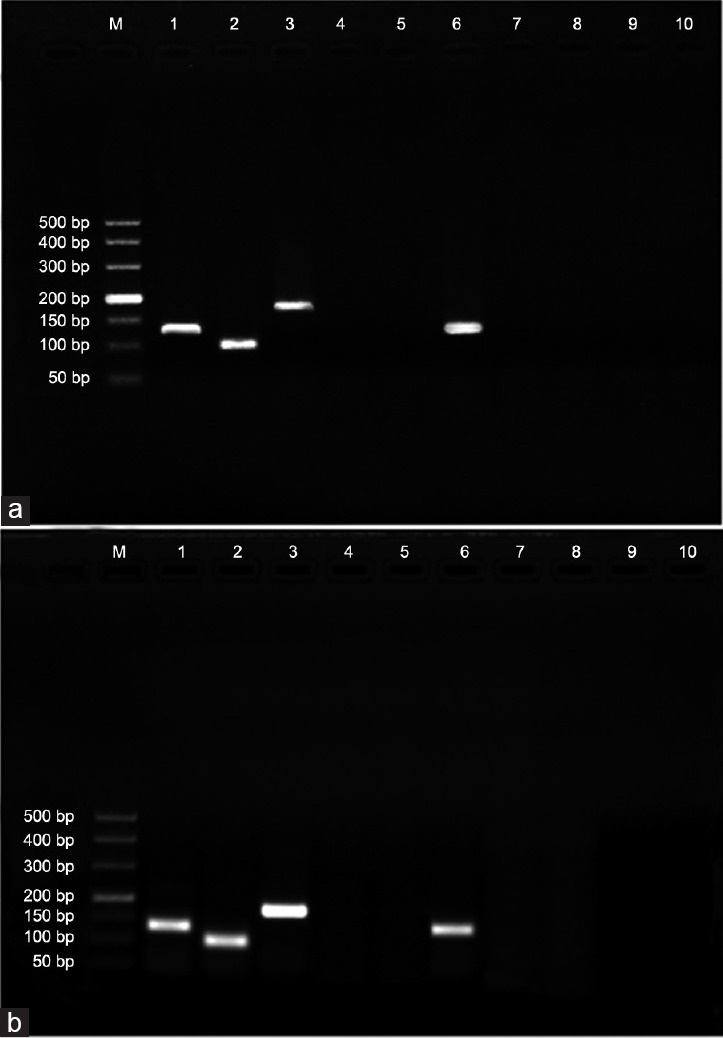
Confirmation of HBV s and x gene transcription in sperm cells. (a) Sperm cells in test group A were obtained from patients with chronic HBV infection, and those in control Group A were obtained from healthy donors. (b) Sperm cells were obtained from healthy donors and then divided into test Group B, which were transfected with a plasmid containing the full-length HBV genome, and control Group B, which were nontransfected. M: marker DL 50~500; Lane 1: GAPDH in test Group A and B; Lane 2: s gene in test Group A and B; Lane 3: x gene in test Group A and B; Lanes 4 and 9: minus reverse transcriptase control (–RT); Lanes 5 and 10: minus template control (–T). Lane 6: GAPDH in control Group A and B; Lane 7: amplification of s gene in control Group A and B; Lane 8: amplification of x gene in control Group A and B. Positive bands for GAPDH were observed in test Group A and B as well as in control Group A and B; positive bands for s and x genes were observed only in test Group A and B and not in control Group A and B or in –RT and –T, suggesting that s and x gene transcriptional activity occurs in sperm cells. GAPDH: glyceraldehyde-3-phosphate dehydrogenase; HBV: hepatitis B virus.
Isolation and identification of host genes regulating transcription of HBV genes in sperm cells
SMART amplification, SSH, T/A cloning, and bacterial amplification
Total RNA from two-cell embryos in the test and control groups was used as the template in SMART amplification. Control mouse liver total RNA (positive group), deionized H2O (negative group), and the primers (3' SMART CDS Primer II A and 5' PCR Primer II A) were provided by the kit. Dispersed positive bands were detected in the test, control, and positive groups but not in the negative group, indicating successful amplification of cDNA of two-cell embryos derived from transfected sperm or nontransfected sperm (Supplementary Figure 2 (166.4KB, tif) ). After SSH, T/A cloning and bacterial amplification, 174 white (positive) colonies from subtracted libraries (123 from the forward and 51 from the reverse SSH libraries) were amplified by PCR, resulting in 152 positive insert-containing clones (103 from the forward- and 49 from the reverse-subtracted libraries). The insert sizes ranged from 500 to 1500 bp, and the majority of which were approximately 1000 bp, in agreement with the statistical prediction of Rsa I digestion.
SMART amplification of cDNA. M: marker; T: the test group; C: the control group; +: the positive group; -: the negative group.
Microarray analysis
In this study, the 2-fold average ratio of Cy5/Cy3 intensities was 5.7400, and the 0.5-fold average ratio of Cy5/Cy3 intensities was 1.4349. Both are the thresholds for determining whether gene differential expression is statistically significant. Of 152 positive insert-containing clones, 29 showed fold change values >2 or <0.5, and 123 exhibited fold change values >0.5 and <2. These 29 clones and one of the 123 clones were selected for sequencing (Supplementary Table 2 (2.9MB, tif) ).
The sequence homology analysis of the differentially expressed genes and a control gene*
DNA sequencing and BLAST analysis
For the acquired sequences, BLAST analysis revealed 29 sequences showing significant identity to five human gene families (Supplementary Table 2 (2.9MB, tif) ). Of these, five representative genes (one from each family, identity: ≥96%, E-value: 0.0)28 were selected as target genes (Supplementary Table 3 (201.9KB, tif) ; the selection criteria are shown in (Supplementary Table 2 (2.9MB, tif) ). The target genes were CSH2, EIF4G2, PCBD2, PSG4, and TTN. A control sequence showed significant identity to the ESRRG family (identity: 99%, E-value: 0.0) (Supplementary Table 3 (201.9KB, tif) ).
The host target genes and a control gene searched by basic local alignment search tool
Identification of target genes regulating transcription of HBV genes in sperm cells
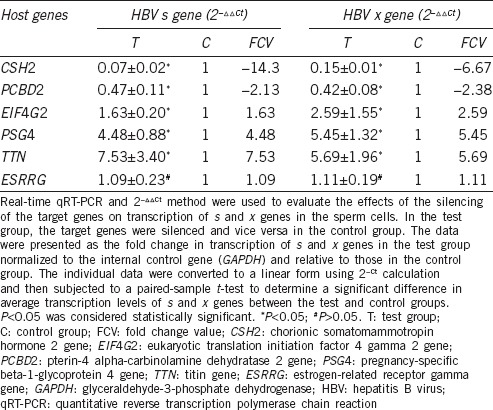
The transcriptional levels of s and x genes in sperm cells after the silencing of the target genes and a control gene by interference (RNAi) are shown in Table 2. When CSH2 and PCBD2 were silenced, s and x gene expression in sperm cells was downregulated between 2.13 to 14.3 folds (P < 0.05). When EIF4G2, PSG4, and TTN were silenced, s and x gene expression in sperm cells was upregulated between 1.63 and 7.53 folds (P < 0.05). There was no significant difference in the levels of s and x genes transcription in sperm cells silenced for ESRRG compared to nonsilenced sperm cells (P > 0.05) (Table 2).
Table 2.
Effect of silencing the target genes and a control gene on transcriptional levels of s and x genes in the sperm cells
DISCUSSION
In this study, RT-PCR detected expression of HBV s and x genes in patient sperm cells but no expression in control sperm cells, suggesting that transcription of HBV genes occurs in sperm cells. Positive bands were observed for donor sperm transfected with a plasmid containing the full-length HBV genome; such transcription should not occur prior to the elongation stage of spermatids but should occur in mature sperm cells.
Whether sperm cells express genes has long been a contentious issue. It was previously generally accepted that RNA was lost or degraded during spermiogenesis, when nuclear condensation is accompanied by the loss of most of the cytoplasm (and cytoplasmic RNA) and the sperm cells are transcriptionally silent cells. This is based on the fact that nucleosomal histones, present in spermatogonia, are replaced by protamine in elongating spermatids and sperm cells and that these proteins are responsible for highly stable chromatin condensation, leading to a sperm genome that is transcriptionally inactive.29,30 However, an increasing number of studies have reported that a complex population of RNAs exists in human ejaculate sperm31 that is unequivocally spermatozoal in origin.32 Sperm RNA, previously thought to be left-overs from spermatogenesis, likely is active in both the sperm cell itself and, perhaps more intriguingly, once it gains access to the ooplasm.33 Moreover, some studies have reported a variety of unsuspected metabolic functions in the nuclei of sperm cells, in contrast to the traditional view that these cells are metabolically inert.34 Indeed, efficient machinery is present in sperm cells that can transcribe, splice, and reverse-transcribe exogenous DNA molecules.34,35,36 The possible mechanism of transcriptional activity occurring in sperm cells might be due to a histone-to-protamine exchange process that appears to be incomplete in sperm cells. Although the majority of DNA is packaged by protamine, part of it (up to 15%) comprises histone-bound chromatin scattered throughout the paternal genome, largely at gene promoter regions,37,38 which maintains some features of active chromatin, mainly the presence of acetylated histones.38,39,40 Therefore, detectable levels of transcription in sperm cells persist,41 and transcriptional activity is carried out in the head and mid-piece regions of human sperm cells.42
In this study, RT-PCR showed the transcriptional activity of HBV genes in sperm cells. In addition to the presence of efficient machinery that can transcribe, splice, and reverse-transcribe exogenous DNA molecules,34,35,36 another reason for the occurrence of transcriptional activity in sperm cells might be that after integrating into the sperm genome, the HBV genome may not undergo the same chromatin compaction as the vast majority of genes in sperm cells. RNA polymerase II catalyzes transcription of DNA to synthesize mRNA precursors and most small nuclear (sn) RNAs and microRNAs. In the HBV life cycle, after entry into liver cells, the HBV genome produces viral mRNA using cellular RNA polymerase II transcriptional machinery.43 As RNA polymerase II levels are particularly high in round spermatid nuclei,44 we speculate that s and x genes may be transcribed using the RNA polymerase II transcriptional machinery stored in the sperm nucleus.
We found CSH2, PCBD2, EIF 4G2, PSG4, and TTN to be involved in transcriptional regulation of HBV s and x genes in sperm cells. CSH2 is located on chromosome 17q23.3 and encodes a protein that plays an important role in growth control and utilizes multiple transcription initiation sites.45 PCBD2, which is located at chromosome 5q31.1, codes for a protein involved in tetrahydrobiopterin biosynthesis.45 Silencing of CSH2 and PCBD2 by RNAi decreased the levels of s and x gene transcription in sperm cells, indicating that both genes upregulated transcription of these HBV genes. At chromosome 11p15.4, EIF4G2 encodes for a protein that functions as a general repressor of translation by forming translationally inactive complexes,45 and PSG4 at chromosome 19q13.31 codes pregnancy-specific glycoprotein (PSG), which may play a role in regulation of the innate immune system.45 TTN (chromosome 2q31.2) encodes a large abundant protein of striated muscle, which appears to play a role in chromosome condensation and segregation during mitosis.45 When EIF4G2, PSG4, and TTN were silenced by RNAi, the transcriptional levels of s and x genes in sperm cells increased, suggesting that these genes downregulate s and x gene transcription. These results suggest that certain host genes participate in regulating HBV gene transcription in sperm cells.
However, the interplay between the virus and host cells is complex, and it remains largely unknown how these genes interact with each other or act independently to allow differential transcription of HBV genes. Some clues may help in understanding their interaction. First, certain previously identified host HBV-regulatory genes share high similarity in function and signaling pathways with genes (identified herein) that regulate HBV gene transcription. For example, Sp1 is a ubiquitous factor that binds to GC-rich motifs in many promoters, can regulate transcription of HBV genes,15 and is involved in many cellular processes, including cell differentiation, growth, and apoptosis. The protein encoded by CSH 2 also plays an important role in growth control and provides avenues for developmental regulation and tissue specificity. Both genes function in the prolactin signaling pathway, suggesting possible regulation of HBV gene transcription by a similar mechanism. Next, some host genes might indirectly participate in regulating HBV gene transcription through general activation of transcription regulators or factors. For example, HNF1 is a liver-enriched transcription factor that can regulate transcription of HBV genes.16 PCBD2, identified herein, regulates dimerization of HNF1α and enhances its transcriptional activity.
Development of the human embryo begins with the fusion of sperm and egg, and sperm quality can affect embryogenesis from a very early stage.46 Our previous studies demonstrated that the HBV S protein can cause a series of apoptotic events resulting in reduced sperm motility, loss of sperm membrane integrity, sperm dysfunction, decreased fertility, and sperm death;47,48,49 HBV infection can induce various chromosomal aberrations in sperm, including aneuploidy, acentric fragment and deletion, ring chromosome, triradial, dicentric chromosomes and pulverization.8 Nonetheless, sperm with these chromosomal aberrations can achieve normal fertilization and introduce these aberrations into the embryo, which may increase the risk of abortion, stillbirth, or birth defects.8 Therefore, understanding HBV genes transcription and their regulatory mechanism in sperm cells is critical for further exploring the mechanisms of transcriptional regulation of viral genes in sperm cells and for preventing viral threat to sperm fertilizing capacity and early embryonic development.
CONCLUSIONS
This study, for the first time, provides experimental evidence that transcription of HBV genes occurs in human sperm cells and is regulated by host genes. The findings are significant for both medically oriented infertility research and more basic viral biology questions.
AUTHOR CONTRIBUTIONS
THH and YZ carried out the study design and execution, analysis and interpretation, manuscript preparation, and critical discussion. DLL, MMMA, PHL, XLZ, QDX, XQX, TTH, ZWH, JHH, and LX performed the experiments and analyzed the data. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENT
This work was supported by the National Natural Science Foundation of China (No. 30972526) and by the Applied Basic Research Programs of Sichuan Province (No. 2014JY0110). The authors thank Prof. Stanley Lin for his assistance in revising the final draft of the manuscript and editing for English grammar and syntax.
Supplementary information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Hadchouel M, Scotto J, Huret JL, Molinie C, Villa E, et al. Presence of HBV DNA in spermatozoa: a possible vertical transmission of HBV via the germ line. J Med Virol. 1985;16:61–6. doi: 10.1002/jmv.1890160109. [DOI] [PubMed] [Google Scholar]
- 2.Davison F, Alexander GJ, Trowbridge R, Fagan EA, Williams R. Detection of hepatitis B virus DNA in spermatozoa, urine, saliva and leucocytes, of chronic HBsAg carriers. A lack of relationship with serum markers of replication. J Hepatol. 1987;4:37–44. doi: 10.1016/s0168-8278(87)80007-7. [DOI] [PubMed] [Google Scholar]
- 3.Lang ZW. [Distribution of hepatitis B virus in testicle tissue in patients with hepatitis B infection] Chin Med J Peking (Zhonghua yi xue za zhi) 1993;73:329–31. 379 [Article in Chinese] [PubMed] [Google Scholar]
- 4.Durazzo M, Premoli A, Di Bisceglie C, Bertagna A, Faga E, et al. Alterations of seminal and hormonal parameters: an extrahepatic manifestation of HCV infection? World J Gastroenterol. 2006;12:3073–6. doi: 10.3748/wjg.v12.i19.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vicari E, Arcoria D, Di Mauro C, Noto R, Noto Z, et al. Sperm output in patients with primary infertility and hepatitis B or C virus; negative influence of HBV infection during concomitant varicocele. Minerva Med. 2006;97:65–77. [PubMed] [Google Scholar]
- 6.Moretti E, Federico MG, Giannerini V, Collodel G. Sperm ultrastructure and meiotic segregation in a group of patients with chronic hepatitis B and C. Andrologia. 2008;40:173–8. doi: 10.1111/j.1439-0272.2007.00818.x. [DOI] [PubMed] [Google Scholar]
- 7.Oger P, Yazbeck C, Gervais A, Dorphin B, Gout C, et al. Adverse effects of hepatitis B virus on sperm motility and fertilization ability during IVF. Reprod Biomed Online. 2011;23:207–12. doi: 10.1016/j.rbmo.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Huang JM, Huang TH, Qiu HY, Fang XW, Zhuang TG, et al. Effects of hepatitis B virus infection on human sperm chromosomes. World J Gastroenterol. 2003;9:736–40. doi: 10.3748/wjg.v9.i4.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali BA, Huang TH, Salem HH, Xie QD. Expression of hepatitis B virus genes in early embryonic cells originated from hamster ova and human spermatozoa transfected with the complete viral genome. Asian J Androl. 2006;8:273–9. doi: 10.1111/j.1745-7262.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- 10.Ali BA, Huang TH, Xie QD. Detection and expression of hepatitis B virus X gene in one and two-cell embryos from golden hamster oocytes in vitro fertilized with human spermatozoa carrying HBV DNA. Mol Reprod Dev. 2005;70:30–6. doi: 10.1002/mrd.20185. [DOI] [PubMed] [Google Scholar]
- 11.Tang H, McLachlan A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci U S A. 2001;98:1841–6. doi: 10.1073/pnas.041479698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennig BJ, Hall AJ. Host genetic factors in hepatitis B infection, liver cancer and vaccination response: a review with a focus on Africa. Sci Total Environ. 2012;423:202–9. doi: 10.1016/j.scitotenv.2010.09.036. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Xie Y, Wu X, Kong Y, Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371–8. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology. 1991;183:825–9. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 15.Raney AK, Le HB, McLachlan A. Regulation of transcription from the hepatitis B virus major surface antigen promoter by the Sp1 transcription factor. J Virol. 1992;66:6912–21. doi: 10.1128/jvi.66.12.6912-6921.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang WX, Li M, Wu X, Wang Y, Li ZP. HNF1 is critical for the liver-specific function of HBV enhancer II. Res Virol. 1998;149:99–108. doi: 10.1016/s0923-2516(98)80085-x. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Mertz JE. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol. 1997;71:9366–74. doi: 10.1128/jvi.71.12.9366-9374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Tan T, Tian Y, Zheng B, Ou JH, et al. Kruppel-like factor 15 activates hepatitis B virus gene expression and replication. Hepatology. 2011;54:109–21. doi: 10.1002/hep.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–4. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 20.National Research Council. 8th ed. Washington DC: National Academies Press; 2011. Guide for the Care and Use of Laboratory Animals. [PubMed] [Google Scholar]
- 21.World Health Organization. 5th ed. Geneva: WHO Press; 2010. Direct swim-up. In: WHO editor. WHO Laboratory Manual for the Examination and Processing of Human Semen; p. 164. [Google Scholar]
- 22.Mori R, Wang Q, Danenberg KD, Pinski JK, Danenberg PV. Both beta-actin and GAPDH are useful reference genes for normalization of quantitative RT-PCR in human FFPE tissue samples of prostate cancer. Prostate. 2008;68:1555–60. doi: 10.1002/pros.20815. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization. Zona-free hamster oocyte penetration test. In: WHO editor. 5th ed. Geneva: WHO Press; 2010. WHO Laboratory Manual for the Examination and Processing of Human Semen; p. 152. [Google Scholar]
- 24.Ahmed MM, Huang TH, Xie QD. An improved experimental model for studying vertical transmission of hepatitis B virus via human spermatozoa. J Virol Methods. 2008;151:116–20. doi: 10.1016/j.jviromet.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 25.Farztdinov V, McDyer F. Distributional fold change test – A statistical approach for detecting differential expression in microarray experiments. Algorithms Mol Biol. 2012;7:29. doi: 10.1186/1748-7188-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, et al. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002:3, research0062.1–research00621.2. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Fassler J, Cooper P BLAST Glossary. Created; 14 July. 2011. [Last accessed on 2017 Mar 09]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK62051/pdf/Bookshelf_NBK62051.pdf .
- 29.Grootegoed JA, Siep M, Baarends WM. Molecular and cellular mechanisms in spermatogenesis. Baillieres Best Pract Res Clin Endocrinol Metab. 2000;14:331–43. doi: 10.1053/beem.2000.0083. [DOI] [PubMed] [Google Scholar]
- 30.Lambard S, Galeraud-Denis I, Martin G, Levy R, Chocat A, et al. Analysis and significance of mRNA in human ejaculated sperm from normozoospermic donors: relationship to sperm motility and capacitation. Mol Hum Reprod. 2004;10:535–41. doi: 10.1093/molehr/gah064. [DOI] [PubMed] [Google Scholar]
- 31.Miller D, Briggs D, Snowden H, Hamlington J, Rollinson S, et al. A complex population of RNAs exists in human ejaculate spermatozoa: implications for understanding molecular aspects of spermiogenesis. Gene. 1999;237:385–92. doi: 10.1016/s0378-1119(99)00324-8. [DOI] [PubMed] [Google Scholar]
- 32.Miller D. Sperm RNA: reading the hidden message. In: Rousseaux S, Khochbin S, editors. Epigenetics and Human Reproduction. Berlin Heidelberg: Springer; 2011. pp. 329–53. [Google Scholar]
- 33.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 34.Pittoggi C, Beraldi R, Sciamanna I, Barberi L, Giordano R, et al. Generation of biologically active retro-genes upon interaction of mouse spermatozoa with exogenous DNA. Mol Reprod Dev. 2006;73:1239–46. doi: 10.1002/mrd.20550. [DOI] [PubMed] [Google Scholar]
- 35.Sciamanna I, Vitullo P, Curatolo A, Spadafora C. Retrotransposons, reverse transcriptase and the genesis of new genetic information. Gene. 2009;448:180–6. doi: 10.1016/j.gene.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Spadafora C. Sperm-mediated ‘reverse’ gene transfer: a role of reverse transcriptase in the generation of new genetic information. Hum Reprod. 2008;23:735–40. doi: 10.1093/humrep/dem425. [DOI] [PubMed] [Google Scholar]
- 37.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, et al. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–49. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–6. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–6. [PubMed] [Google Scholar]
- 40.Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts. DNA, histones, protamines and epigenetics? Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- 41.Miteva K, Valkov N, Goncharova-Peinoval J, Kovachev K, Zlatarev S, et al. Electron microscopic data for the presence of post-meiotic gene expression in isolated ram sperm chromatin. Cytobios. 1995;83:85–90. [PubMed] [Google Scholar]
- 42.Naz RK. Effect of actinomycin D and cycloheximide on human sperm function. Arch Androl. 1998;41:135–42. doi: 10.3109/01485019808987955. [DOI] [PubMed] [Google Scholar]
- 43.Uprichard SL, Wieland SF, Althage A, Chisari FV. Transcriptional and posttranscriptional control of hepatitis B virus gene expression. Proc Natl Acad Sci U S A. 2003;100:1310–5. doi: 10.1073/pnas.252773599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt EE, Schibler U. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development. 1995;121:2373–83. doi: 10.1242/dev.121.8.2373. [DOI] [PubMed] [Google Scholar]
- 45.The Human Gene Database, Gene Cards. [Last accessed on 2017 Mar 06]. Available from: http://www.genecards.org .
- 46.Loutradi KE, Tarlatzis BC, Goulis DG, Zepiridis L, Pagou T, et al. The effects of sperm quality on embryo development after intracytoplasmic sperm injection. J Assist Reprod Genet. 2006;23:69–74. doi: 10.1007/s10815-006-9022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, Zhong Y, Fang X, Xie Q, Kang X, et al. Hepatitis B virus s protein enhances sperm apoptosis and reduces sperm fertilizing capacity in vitro . PLoS One. 2013;8:e68688. doi: 10.1371/journal.pone.0068688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang X, Xie Q, Zhou X, Li F, Huang J, et al. Effects of hepatitis B virus S protein exposure on sperm membrane integrity and functions. PLoS One. 2012;7:e33471. doi: 10.1371/journal.pone.0033471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou XL, Sun PN, Huang TH, Xie QD, Kang XJ, et al. Effects of hepatitis B virus S protein on human sperm function. Hum Reprod. 2009;24:1575–83. doi: 10.1093/humrep/dep050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of transfected and non-transfected sperm cells and sperm-derived embryos. (a) The arrows show three of the transfected sperm cells under a phase-contrast microscope. (b) the same field of view under a fluorescence microscope. (c) The arrows show two 2-cell embryos under a phase-contrast microscope. (d) the arrow shows a transfected two-cell embryo in the same field of view under a fluorescence microscope. Scale bar = 20 μm in (a) and (b), 50 μm in (c) and (d).
The gene-specific shRNA and siRNA sequences*
SMART amplification of cDNA. M: marker; T: the test group; C: the control group; +: the positive group; -: the negative group.
The sequence homology analysis of the differentially expressed genes and a control gene*
The host target genes and a control gene searched by basic local alignment search tool


