Abstract
Background:
Pseudomonas aeruginosa is a biofilm-forming bacterium which can result in serious health problems, particularly in burn patients. Biofilm has been assumed to protect the bacteria from environmental fluctuations such as antimicrobial agent. Mucoid strains generate extensive levels of the alginate exopolysaccharide, which is an important factor of its biofilm.
Materials and Methods:
Totally, 100 isolates of P. aeruginosa has been gathered from wound infections of burn patients. Polymerase chain reaction of exoA gene has been carried out to confirm the bacteriologic identification of isolates. The biofilm-forming capacity has been specified by capsule staining and microtiter plate test as qualitative and quantitative determination, respectively. Antimicrobial susceptibility of the isolates has been specified by disk diffusion method.
Results:
All the isolates carried the exoA gene. The antibiotic resistance was imipenem (90%); levofloxacin (93%); aztreonam (87%); piperacillin-tazobactam (85%); tobramycin (92%); polymyxin b (PB) (2%); and ceftazidime (CAZ) (32%). Totally, multidrug-resistant (MDR) and extended drug-resistant (XDR) isolates were 19% and 75%, respectively. Fortunately, pan drug-resistant (PDR) strain has not been observed. The assessment of biofilm formation has shown that 7% of the isolates were nonbiofilm (N), weak (W) 67%, moderate (M) 22%, and strong (S) 4%.
Conclusions:
As a result, the findings of this survey indicated that PB and CAZ were the most effective antibiotics against P. aeruginosa, which of course indicate a serious problem about the emergence of the PDR strains. There was no relationship between the patterns of biofilm production and antibiotic susceptibility, but high frequency of MDR/XDR and biofilm producer strains has been detected.
Keywords: Alginate, biofilm, burn, multidrug-resistant/extended drug-resistant, Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa is a biofilm-forming opportunistic bacterium, which can make critical health problems, particularly in immunosuppressed hosts such as burn patients, patients suffering from respiratory diseases such as cystic fibrosis (CF), and cancer chemotherapy patients.[1,2] Wound infection by antibiotic-resistant organisms such as P. aeruginosa, Acinetobacter, and Klebsiella should be identified as a potential risk.[3] P. aeruginosa plays a notable role in perilous infections in burn patients. Rapid acquisition of multidrug resistance (MDR) leads to high morbidity and mortality, especially in burn centers.[4,5] P. aeruginosa infections are mostly difficult-to-treat because of the low antibiotic sensitivity and the high rate of the emergence of antimicrobial resistance during the process of the treatment.[6,7] Accumulation of resistance after exposure to various antibiotics and cross-resistance among them may result in MDR, extended drug-resistant (XDR), and pan drug-resistant (PDR).[6,8,9] The rapid emergence of hospital pathogens and antibiotic-resistant organisms necessitate periodic evaluation of bacterial colonization patterns and antibiogram sensitivity in burn wards.[3]
P. aeruginosa is a prevalent biofilm-forming bacterium which hence often used as a model organism in biofilm studies.[10] Biofilm is a complex and compressed microbial community in an exopolysaccharide matrix[11] and allows bacteria to attach to the surfaces protecting it from environmental fluctuations such as antimicrobial agent.[12,13,14,15,16] Mucoid strains of P. aeruginosa produce numerous amounts of the alginate exopolysaccharide which is a significant component of its biofilm.[17] Alginate is a linear heteropolysaccharide formed of D-mannuronic acid and L-glucuronic acid.[18] There are two different hypotheses that antibiotic resistance is different between mucoid and nonmucoid P. aeruginosa strains; first, glycocalyx can act as a barrier to antibiotic diffusion which is related to its polyanionic properties;[19,20,21] the second has revealed that some antibiotics such as tobramycin (TN) can bind to exopolysaccharide and penetrate inside bacteria.[22]
The purpose of the current study was to determine alginate/biofilm production, antibiotic susceptibility pattern, and frequency of MDR and XDR in P. aeruginosa isolated in Imam Musa Kazem Burn Hospital in Isfahan, Iran.
Materials and Methods
Bacterial isolated and identification test
In this cross-sectional study, 100 isolates of P. aeruginosa have been gathered from wound infections of burn patients admitted to Imam Mosa Kazem Burn Hospital in Isfahan, Iran, between March and July 2015. Each isolate has been determined due to the standard bacteriological methods including Gram-staining, growth at 42°C in cetrimide agar, oxidation-fermentation (OF), TSI, and oxidase tests. Furthermore, polymerase chain reaction (PCR) of exoA gene has been carried out to confirm the bacteriologic identification. A 397-bp fragment of the exoA gene has been selected with specified primers (forward: 5’-GACAACGCCCTCAGCATCACCAGC-3,' reverse: 5’-CGCTGGCCCATTCGCTCCAGCGCT-3’).[23] Each PCR reaction was prepared in 20 μL volume include 10 μL the commercial Master Mix (containing Taq DNA polymerase, dNTPs, and MgCl2) (Ampliqon Denmark), 1 μL DNA sample, 0.5 μL of each primer (Metabion, Germany), and 8 μL distilled. Samples were then subjected to one cycle of 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 68°C for 30 s, and 72°C for 45 s and one final cycle of 72°C for 5 min. P. aeruginosa strain ATCC27853 (American Type Culture Collection) was included as the control.
Determination of mucoid strain
Negative stain of capsule
Mucoid strains have been determined by the use of Anthony's capsule staining as the qualitative method.[24] Briefly, for each of 100 isolates, a thin film of skim milk suspension has been prepared and air-dried; the film has been flooded with crystal violet for 60 s. The slide has been gently rinsed with a 20% copper sulfate solution for capsule decolonization. The cell and background have been stained purple, and the capsule appears as a faint blue halo.
Quantification of alginate/biofilm production by microtiter method
The P. aeruginosa isolates have been analyzed to quantify biofilm production using microtiter dish method.[25] In this method, each strain has been grown overnight at 37°C in tryptic soy broth (TSB) including 0.25% glucose. The cultures have been diluted 1:100 in TSB medium. The bacteria suspensions (125 μL) have been aliquoted into a 96-well polystyrene microtiter plate and inoculated for 24 h at 37°C without agitation. The wells have been washed three times with 300 μL distilled water; the attached bacteria have been fixed with absolute methanol for 10 min and finally stained with 125 μL of 0.1% crystal violet solution in water for about 10–15 min. After staining, the wells have been washed three times with distilled water to remove all nonadherent cells. The wells were destained with 125 μL of 30% acetic acid in water. A new sterile flat-bottomed 96-well polystyrene microtiter plate was inoculated with 125 μL destaining solution in each well. The absorbance of the destaining solution has been measured at 570 nm using an ELISA reader (Stat Fax-2100). Every experiment has been carried out in triplicate. As the control, the uninoculated medium was used. According to the optical density of each sample (ODi) and the negative control (ODc), the isolates have been categorized as strong (4× ODc < ODi), moderate (2× ODc < ODi ≤ 4× ODc), weak (ODc < ODi ≤ 2× ODc), or nonproducer of biofilm (ODi < ODc).
Antibiotic susceptibility tests
Agar diffusion methods (Kirby–Bauer method) have been applied to determine the antibiotic susceptibility of isolated bacteria against TN (10 μg), aztreonam (ATM, 30 μg), imipenem (IMI, 10 μg), ceftazidime (CAZ, 30 μg), levofloxacin (LEV, 5 μg), piperacillin-tazobactam (PTZ, 110 μg), and polymyxin B (PB, 300U). MDR, XDR, and PDR strains have been detected according to a new standardized international document.[26] P. aeruginosa (ATCC 27853) was used as a control strain.
Statistical analyses
Statistical Package for Social Sciences software (SPSS Inc. No. 23, Version 23.0. Armonk, NY: IBM Corp.) was used for statistical analyses. Fisher's exact test or Chi-square test was used for the categorical data analysis. P < 0.05 was considered statistically significant.
Results
One hundred P. aeruginosa have been isolated from burn wounds. The isolates were Gram-negative, growing at 42°C, OF, TSI and oxidase-positive. The identification of the isolates has been confirmed by amplification of exoA gene which particularly belongs to P. aeruginosa [Figure 1]. All the isolates carried the exoA gene.
Figure 1.
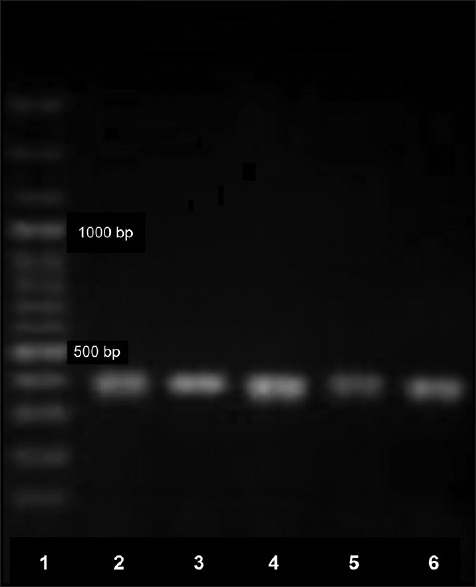
Gel electrophoresis of polymerase chain reaction products of the exoA gene (397 bp) among Pseudomonas aeruginosa isolates. Line 1: Ladder (100 bp), Line 2: Positive control, Line 3–6: Clinical specimens
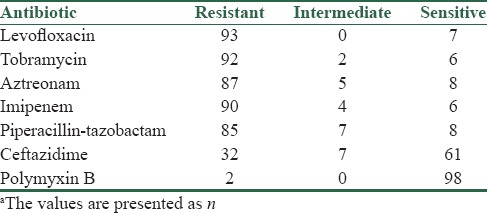
The antibiotic susceptibility patterns of the P. aeruginosa isolates are shown in Table 1. A high rate of resistance has been observed against IMI, TN, and LEV (approximately 90%), ATM (87%), PTZ (85%). The lowest and medium resistances have been observed to PB (2%) and CAZ (32%), respectively. Totally, MDR and XDR isolates were 19% and 75%, respectively. PDR strain has not been observed.
Table 1.
Antibiotic susceptibility patterns of Pseudomonas aeruginosa isolated from burn patients in Isfahan, Irana
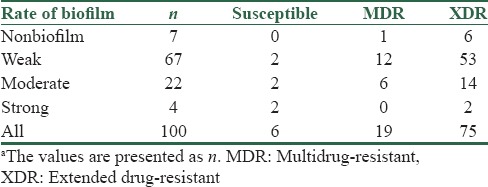
Mucoid strains were determined in all isolates as an alginate producer by capsule stain method (qualitative methods). Ninety-three percent of the isolates indicated biofilm formation. In addition, microtiter method was conducted as a quantitative assessment of biofilm production. The results have been shown that 7% of isolates were nonbiofilm (N), weak (W) 67%, moderate (M) 22%, and strong (S) 4%. The relation between the biofilm formation and the antibiotic susceptibility patterns is shown in Table 2 (P < 0.05).
Table 2.
The relation between biofilm formation and the antibiotic susceptibility patternsa
Discussion
In P. aeruginosa infections, alginate/biofilm production has been measured as an important determinant of pathogenicity.[27] Mucoid strains of P. aeruginosa produce vast levels of the alginate considered the main component of its biofilm.[17] In the present study including P. aeruginosa isolated from burn patients, we assess presence and rate of the alginate as a virulence marker and determine the frequency of MDR and XDR strain.
In this study, 93% of P. aeruginosa isolates have been specified as mucoid type. Frequency of mucoid isolates of P. aeruginosa in our study is consistent with results of some studies that are mentioned below in detail; in Ghanbarzadeh et al.'s study in Iran, 144 isolated from burn patient 92.4% were mucoid; further, in Jabalameli et al.'s study in Iran with 96 sample, the mucoid strains were 96%; further, in India, in a study conducted by Ugargol et al. with overall theme as the characterization of virulence factors, such as alginate in a tertiary care hospital, 96.9% of 250 P. aeruginosa isolates identified as mucoid phenotype; similar proportion has been reported in burns isolates.[2,28,29]
In some surveys, mucoid strains were lower than our result. According to the findings of Kádár et al. in 2010, 23.3% of sixty clinical samples of P. aeruginosa were positive for biofilm formation.[30] In another study performed by Ghadaksaz et al. in Iran, the frequency of biofilm formation was 50.1% among 104 clinical isolates of P. aeruginosa.[31] In a study conducted at Manipal, 68% of P. aeruginosa strains produced alginate.[32] The results mentioned above have shown that mucoid strains have lately increased in comparison with previous studies in Iran and another country.
The differences of mucoid shift between our result and another study may be related to the site variation of clinical samples, such as ocular infections (no biofilm production), urine (34.2%), chronic rhinosinusitis (28.6%), and CF (33.3%).[2,33,34,35]
Quantitative biofilm determination by the use of the microtiter test showed that 93 strains produced biofilm, which 67 samples were weak biofilm producers, 22 samples were moderate, and four samples were strong. In a study performed by Jabalameli et al., biofilm production has been observed in more than 96% of the isolates which 22.9% were weak biofilm formers, 26% were moderate, and 47% were strong.[29]
According to the earlier studies, it has been thought that antibiotic susceptibility is different between mucoid and nonmucoid P. aeruginosa strains.[1,36,37] In many literature, there are challenges about whether the mucoid phenotype leads to increased resistance. One hypothesis suggests that the glycocalyx usually acts as a polyanionic barrier to antibiotic penetration.[19,20,21] This was refuted by the fact that although some antibiotics such as TN bind to the exopolysaccharide, the resulting reduction in the diffusion coefficient of antibiotic in a biofilm would not be enough to prevent the entry of antibiotics.[38]
According to Table 1 in this study, there was no relationship between the patterns of biofilm production and antibiotic susceptibility. The results of the study performed by Ahangarzadeh et al. showed that the mucoid strains (n = 43) were statistically more resistant to some antibiotic than the nonmucoid strains (n = 90).[18] In two separate studies performed by Ghanbarzadeh et al. and Abidi et al., the statistical analysis showed that biofilm formation in the MDR P. aeruginosa (MDRPA) isolates was higher than that in the non-MDRPA isolates.[28,39]
In contrast, several studies suggest that mucoid strains are more sensitive to some antibiotic than nonmucoid strain.[36,40] For example, in Shawar et al.'s study of inhaled-TN therapy examined the susceptibility of 1240 CF isolates, it found that for all seven antibiotics tested, mucoid strains were more susceptible in comparison with nonmucoid strains.[37] Furthermore, an important finding in Burns et al.'s study was that overall and for each drug tested, mucoid isolates were more susceptible.[41]
P. aeruginosa remains one of the most significant opportunistic causes of nosocomial infections, and it has increased resistance to a range of antimicrobial agents in burn centers.[42] In this study, we also carried out antibiogram test to determine the antibiotic susceptibility pattern. Our results showed that P. aeruginosa isolates were almost resistant to all tested antibiotic, except PB (2%) and CAZ (32%). As a result, MDR and XDR isolates were 19% and 75%, respectively. Because of the XDR isolates are a subset of the MDR isolates; their frequency can be reported as 94% MDR and 85% XDR. Over the recent years, various articles have confirmed an increasing MDR among P. aeruginosa isolated from burn wound infections in Iranian hospitals.[43,44]
According to the results of the study performed by Jabalameli et al., there is a high frequency (>80%) of resistance against all tested antibiotics in our study, except for PB.[29] In a study conducted by Ghanbarzadeh et al. entitled biofilm formation and virulence factors among P. aeruginosa isolated from burn patients, a high rate of resistance has been observed against ATM (86.8%), ciprofloxacin (93.7%), piperacillin (85.4%), amikacin (82%), CAZ (82.6%), and IMI (79.2%). Totally, 93.1% of the isolates were characterized as MDRPA.[28] Ghazi et al. in 2012 investigated antibiotic resistance pattern in clinical isolates of P. aeruginosa and showed that all clinical isolates were resistant to CAZ, PTZ, and cefepime followed by ATM, ticarcillin, amikacin, and TN (96.5%).[45] In a study in 2013 at the Burn Centre of Guilan in the north of Iran, the percentage of resistance to tested antibiotics was as follows: CAZ 57.5%, ciprofloxacin 65%, gentamicin 67.5%, piperacillin 87.5%, amikacin 90%, and IMI 97.5%. Totally, 45.3% were MDR.[46] Yousefi et al. and Shahcheraghi et al. found that 30.1% and 5.46% of the isolates were MDR, respectively.[47,48]
Conclusions
As a result, it can be concluded that the rate of MDR in Iran is higher than other countries. The results of antibiogram showed that PB and CAZ were the most effective drugs against P. aeruginosa in vitro, but the high speed of increased resistance might lead to the emergence of the PDR strains, which is a serious warning to our country. The increased rate of MDR/XDR P. aeruginosa isolates can cause limitations in antibiotic therapy as a final strategy to treat the infections. Therefore, it is important to investigate the antibiotic susceptibility pattern of P. aeruginosa isolates. There was no relationship between the patterns of biofilm production and antibiotic susceptibility, but high frequency of MDR/XDR and biofilm producer strains has been detected.
Financial support and sponsorship
This study was supported by Isfahan University of Medical Science, Grant no. 394656.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Rezaee MA, Behzadiyan-Nejad Q, Owlia P, Pirayeh SN. In vitro activity of imipenem and ceftazidim against mucoid and non-mucoid strains of Pseudomonas aeruginosa isolated from patients in Iran. Arch Iran Med. 2002;4:251–4. [Google Scholar]
- 2.Ugargol AR, Srikanth N, Shilpa K, Patil S. Characterisation and detection of virulence factors, alginate and phospholipase 'C' in Pseudomonas aeruginosa in a Tertiary Care Hospital. Int J Health Sci Res. 2014;4:82–7. [Google Scholar]
- 3.Rezaei E, Safari H, Naderinasab M, Aliakbarian H. Common pathogens in burn wound and changes in their drug sensitivity. Burns. 2011;37:805–7. doi: 10.1016/j.burns.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Japoni A, Alborzi A, Kalani M, Nasiri J, Hayati M, Farshad S. Susceptibility patterns and cross-resistance of antibiotics against Pseudomonas aeruginosa isolated from burn patients in the South of Iran. Burns. 2006;32:343–7. doi: 10.1016/j.burns.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Moghoofei M, Fazeli H, Poursina F, Nasr Esfahani B, Moghim S, Vaez H, et al. Morphological and bactericidal effects of amikacin, meropenem and imipenem on Pseudomonas aeruginosa. Jundishapur J Microbiol. 2015;8:e25250. doi: 10.5812/jjm.25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50:43–8. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaez H, Faghri J, Isfahani BN, Moghim S, Yadegari S, Fazeli H, et al. Efflux pump regulatory genes mutations in multidrug resistance Pseudomonas aeruginosa isolated from wound infections in Isfahan hospitals. Adv Biomed Res. 2014;3:117. doi: 10.4103/2277-9175.133183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shu JC, Chia JH, Siu LK, Kuo AJ, Huang SH, Su LH, et al. Interplay between mutational and horizontally acquired resistance mechanisms and its association with carbapenem resistance amongst extensively drug-resistant Pseudomonas aeruginosa (XDR-PA) Int J Antimicrob Agents. 2012;39:217–22. doi: 10.1016/j.ijantimicag.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbin C, Maley M, Harkness J, Benn R, Malouf M, Glanville A, et al. The impact of pan-resistant bacterial pathogens on survival after lung transplantation in cystic fibrosis: Results from a single large referral centre. J Hosp Infect. 2004;56:277–82. doi: 10.1016/j.jhin.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Hammond AA, Miller KG, Kruczek CJ, Dertien J, Colmer-Hamood JA, Griswold JA, et al. An in vitro biofilm model to examine the effect of antibiotic ointments on biofilms produced by burn wound bacterial isolates. Burns. 2011;37:312–21. doi: 10.1016/j.burns.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrd MS, Pang B, Hong W, Waligora EA, Juneau RA, Armbruster CE, et al. Direct evaluation of Pseudomonas aeruginosa biofilm mediators in a chronic infection model. Infect Immun. 2011;79:3087–95. doi: 10.1128/IAI.00057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy P, Brammah S, Wills E. Burns, biofilm and a new appraisal of burn wound sepsis. Burns. 2010;36:49–56. doi: 10.1016/j.burns.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 13.Charlesworth CJ, Saran VV, Volpiana LK, Woods HL. The role of biofilm structure in the mechanism of gentamicin and ciprofloxacin antibiotic resistance in P. aeruginosa PAO1 biofilms. J Exp Microbiol Immunol. 2008;12:27–33. [Google Scholar]
- 14.Cotton LA, Graham RJ, Lee RJ. The role of alginate in P. aeruginosa PAO1 biofilm structural resistance to gentamicin and ciprofloxacin. J Exp Microbiol Immunol. 2009;13:58–62. [Google Scholar]
- 15.Lawley R, Curtis L, Davis J. The food safety hazard guidebook: Royal Society of Chemistry. 2012 [Google Scholar]
- 16.Hengzhuang W, Song Z, Ciofu O, Onsøyen E, Rye PD, Høiby N. OligoG CF-5/20 Disruption of mucoid Pseudomonas aeruginosa biofilm in a murine lung infection model. Antimicrob Agents Chemother. 2016;60:2620–6. doi: 10.1128/AAC.01721-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–26. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 18.Ahangarzadeh RM, Behzadiannezhad Q, NAJJAR PS, Oulia P. Higher aminoglycoside resistance in mucoid Pseudomonas aeruginosa than in non-mucoid strains. Arch Iranian Med. 2002;5:108–10. [Google Scholar]
- 19.Slack MP, Nichols WW. Antibiotic penetration through bacterial capsules and exopolysaccharides. J Antimicrob Chemother. 1982;10:368–72. doi: 10.1093/jac/10.5.368. [DOI] [PubMed] [Google Scholar]
- 20.Costerton JW, Irvin RT, Cheng KJ. The bacterial glycocalyx in nature and disease. Annu Rev Microbiol. 1981;35:299–324. doi: 10.1146/annurev.mi.35.100181.001503. [DOI] [PubMed] [Google Scholar]
- 21.Diaz E, Haaf H, Lai A, Yadana J. Role of alginate in gentamicin antibiotic susceptibility during the early stages of Pseudomonas aeruginosa PAO1 biofilm establishment. J Exp Microbiol Immunol. 2011;15:71–8. [Google Scholar]
- 22.DeVries CA, Ohman DE. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in ALGT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–87. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan AA, Cerniglia CE. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl Environ Microbiol. 1994;60:3739–45. doi: 10.1128/aem.60.10.3739-3745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prescott LM, Harley JP, Klein D. Microbiology. Dubuque, Iowa: Wm C C Brown; 1996. [Google Scholar]
- 25.O'Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011 doi: 10.3791/2437. pii: 2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 27.Choy MH, Stapleton F, Willcox MD, Zhu H. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens- and non-contact lens-related keratitis. J Med Microbiol. 2008;57(Pt 12):1539–46. doi: 10.1099/jmm.0.2008/003723-0. [DOI] [PubMed] [Google Scholar]
- 28.Ghanbarzadeh Corehtash Z, Khorshidi A, Firoozeh F, Akbari H, Mahmoudi Aznaveh A. Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J Microbiol. 2015;8:e22345. doi: 10.5812/jjm.22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jabalameli F, Mirsalehian A, Khoramian B, Aligholi M, Khoramrooz SS, Asadollahi P, et al. Evaluation of biofilm production and characterization of genes encoding type III secretion system among Pseudomonas aeruginosa isolated from burn patients. Burns. 2012;38:1192–7. doi: 10.1016/j.burns.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 30.Kádár B, Szász M, Kristóf K, Pesti N, Krizsán G, Szentandrássy J, et al. In vitro activity of clarithromycin in combination with other antimicrobial agents against biofilm-forming Pseudomonas aeruginosa strains. Acta Microbiol Immunol Hung. 2010;57:235–45. doi: 10.1556/AMicr.57.2010.3.8. [DOI] [PubMed] [Google Scholar]
- 31.Ghadaksaz A, Fooladi AA, Hosseini HM, Amin M. The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J Appl Biomed. 2015;13:61–8. [Google Scholar]
- 32.Prasad SV, Ballal M, Shivananda PG. Slime production a virulence marker in Pseudomonas aeruginosa strains isolated from clinical and environmental specimens: A comparative study of two methods. Indian J Pathol Microbiol. 2009;52:191–3. doi: 10.4103/0377-4929.48914. [DOI] [PubMed] [Google Scholar]
- 33.Prince AA, Steiger JD, Khalid AN, Dogrhamji L, Reger C, Eau Claire S, et al. Prevalence of biofilm-forming bacteria in chronic rhinosinusitis. Am J Rhinol. 2008;22:239–45. doi: 10.2500/ajr.2008.22.3180. [DOI] [PubMed] [Google Scholar]
- 34.Coban AY, Ciftci A, Onuk EE, Erturan Z, Tanriverdi Cayci Y, Durupinar B. Investigation of biofilm formation and relationship with genotype and antibiotic susceptibility of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis. Mikrobiyol Bul. 2009;43:563–73. [PubMed] [Google Scholar]
- 35.Hou W, Sun X, Wang Z, Zhang Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections biofilm-forming capacity of human flora bacteria. Invest Ophthalmol Vis Sci. 2012;53:5624–31. doi: 10.1167/iovs.11-9114. [DOI] [PubMed] [Google Scholar]
- 36.Srifuengfung S, Tiensasitorn C, Yungyuen T, Dhiraputra C. Prevalence and antimicrobial susceptibility of Pseudomonas aeruginosa mucoid and non-mucoid type. Southeast Asian J Trop Med Public Health. 2004;35:893–6. [PubMed] [Google Scholar]
- 37.Shawar RM, MacLeod DL, Garber RL, Burns JL, Stapp JR, Clausen CR, et al. Activities of tobramycin and six other antibiotics against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother. 1999;43:2877–80. doi: 10.1128/aac.43.12.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols WW, Dorrington SM, Slack MP, Walmsley HL. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother. 1988;32:518–23. doi: 10.1128/aac.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abidi SH, Sherwani SK, Siddiqui TR, Bashir A, Kazmi SU. Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 2013;13:57. doi: 10.1186/1471-2415-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owlia P, Nosrati R, Alaghehbandan R, Lari AR. Antimicrobial susceptibility differences among mucoid and non-mucoid Pseudomonas aeruginosa isolates. GMS hygiene and infection control. 2014;9(2) doi: 10.3205/dgkh000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burns JL, Saiman L, Whittier S, Larone D, Krzewinski J, Liu Z, et al. Comparison of agar diffusion methodologies for antimicrobial susceptibility testing of Pseudomonas aeruginosa isolates from cystic fibrosis patients. J Clin Microbiol. 2000;38:1818–22. doi: 10.1128/jcm.38.5.1818-1822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev. 2006;19:403–34. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shahcheraghi F, Feizabadi MM, Yamin V, Abiri R, Abedian Z. Serovar determination, drug resistance patterns and plasmid profiles of Pseudomonas aeruginosa isolated from burn patients at two hospitals of Tehran (IRAN) Burns. 2003;29:547–51. doi: 10.1016/s0305-4179(03)00142-6. [DOI] [PubMed] [Google Scholar]
- 44.Nikbin VS, Abdi-Ali A, Feizabadi MM, Gharavi S. Pulsed field gel electrophoresis & plasmid profile of Pseudomonas aeruginosa at two hospitals in Tehran, Iran. Indian J Med Res. 2007;126:146–51. [PubMed] [Google Scholar]
- 45.Ghazi M, Khanbabaee G, Fallah F, Kazemi B, Mahmoudi S, Navidnia M, et al. Emergence of Pseudomonas aeruginosa cross-infection in children with cystic fibrosis attending an Iranian referral pediatric center. Iran J Microbiol. 2012;4:124–9. [PMC free article] [PubMed] [Google Scholar]
- 46.Nikokar I, Tishayar A, Flakiyan Z, Alijani K, Rehana-Banisaeed S, Hossinpour M, et al. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbiol. 2013;5:36–41. [PMC free article] [PubMed] [Google Scholar]
- 47.Yousefi S, Nahaei M, Farajnia S, Ghojazadeh M, Akhi M, Sharifi Y, et al. Class 1 integron and imipenem resistance in clinical isolates of Pseudomonas aeruginosa: Prevalence and antibiotic susceptibility. Iran J Microbiol. 2010;2:115–21. [PMC free article] [PubMed] [Google Scholar]
- 48.Shahcheraghi F, Badmasti F, Feizabadi MM. Molecular characterization of class 1 integrons in MDR Pseudomonas aeruginosa isolated from clinical settings in Iran, Tehran. FEMS Immunol Med Microbiol. 2010;58:421–5. doi: 10.1111/j.1574-695X.2009.00636.x. [DOI] [PubMed] [Google Scholar]


