Abstract
Background:
Candida dubliniensis is a newly diagnosed species very similar to Candida albicans phenotypically and first discovered in the mouth of people with AIDS in 1995. Among the different phenotypic and genotypic methods, a cost-effective method should be selected which makes it possible to differentiate these similar species.
Materials and Methods:
Polymerase chain reaction (PCR)-restriction fragment length polymorphism with MspI enzyme and the Duplex-PCR method were done by DNA extraction using boiling. The sequencing of the amplified ribosomal region was used to confirm the C. dubliniensis species. Direct examination and colony count of the yeasts were applied for bronchoalveolar lavage (BAL) samples and the growth rate of the yeasts were studied at 45°C. To understand the ability formation of chlamydoconidia in yeast isolates, they were separately cultured on the sunflower seed agar, wheat flour agar, and corn meal agar media.
Results:
Fifty-nine (49.2%) yeast colonies were identified from the total of 120 BAL specimens. Twenty-nine isolated yeasts; including 17 (58.6%) of C. albicans/dubliniensis complex and 12 (41.4%) of nonalbicans isolates produced pseudohypha or blastoconidia in direct smear with a mean colony count of 42000 CFU/mL. C. albicans with the frequency of 15 (42.9%) were the most common isolated yeasts, whereas C. dubliniensis was identified in two nonHIV patients.
Conclusion:
Sequencing of the replicated gene fragment is the best method for identifying the yeasts, but the determination of the species by phenotypic methods such as the creation of chlamydoconidia in sunflower seeds agar and wheat flour agar media can be cost-effective, have sensitivity and acceptable quality.
Keywords: Candida albicans, Candida dubliniensis, genotype, identification, phenotype
Introduction
The genus of candida includes 200 species, of which 30 species have been isolated from human infections, and this number is increasing.[1] Candida is the normal flora of skin and the mucosal surfaces of the body such as the mouth, intestine, vagina, and respiratory system of human body. Under favorable conditions, it proliferates and creates pseudo hypha, capable of causing disease.
In most of the epidemiological studies, Candida albicans have been found to be the dominant source of superficial and systemic infections, other non-C. albicans species are emerging pathogens and can also colonize human mucocutaneous surfaces. Considering the host physiological conditions, having risk factors, underlying diseases, and taking antifungal drugs by the host, these microorganisms are capable contribute to infection.[2] Candida dubliniensis is a newly diagnosed species very similar to C. albicans phenotypically and first discovered in the mouth of people with AIDS in 1995. Both forms a green colony in CHOROMagar Candida, also in serum, they produce germ tube, and in corn meal agar (CMA) or rice medium with Tween 80 and wheat flour agar (WFA) they produce chlamydoconidia. C. dubliniensis forms mycelium and chlamydoconidia in Pal's (sunflower seeds agar) (SFSA) and tobacco agar culture media; thus, these tests can be used to distinguish between both yeasts.[3,4,5,6,7] In a review study conducted at American College of Pathologists, 1046 laboratory samples containing yeasts were sent to the different laboratories to identify the yeasts in the samples. About 39.4% of the laboratories correctly identified the pathogen species as C. dubliniensis and 42.6% of pathogen species, were wrongly identified as C. albicans. In the meantime, the 13 laboratories which used the molecular method diagnosed the agents. The best method for distinguishing these two species is, using of DNA sequencing techniques. DNA probe and the matrix-assisted laser desorption/ionization-time-of-flight methods, which are expensive compared to phenotypic methods.[8] The use of restriction enzymes in the molecular method of polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) is not able to distinguish all yeast species.[9] Using the duplex-PCR method with a pair of specific primers for C. dubliniensis was capable of identifying these organisms in the complex of both species.[10] Recognition of opportunistic species in terms of the prevalence of multiple species in community, changing the physiological behavior and acquiring drug resistance, especially in societies where cancer is growing and organ transplantation is common, is very important.[11,12] Regarding the abundance of C. albicans species in infections and their similarity to C. dubliniensis, as well as the resistance of some species to antifungal drugs, it is important to identify these species in terms of rapid and accurate diagnosis and appropriate treatment and according to the different phenotypic and genotypic methods, the ones should be selected in a way, that is also cost-effective to differentiate between species.
Materials and Methods
From October 2016 to May 2017, the bronchoalveolar lavage (BAL) samples of 120 patients with the pulmonary disease were collected from the patients referring to the Al-Zahra Hospital, the largest referral center in Isfahan, Iran. After receiving patients' consent forms, and biography, the BAL sample was taken by a pulmonary physician, and a part of the sample was transferred immediately to mycological laboratory for direct examination and culture procedures. To find the amount of yeast colonies in the sample (CFU/mL), 100 μl of the sample was cultured on Sabouraud Dextrose Agar (SDA) (Biolife, Italy) and for microscopic observation the sample was centrifuged for 5 min with the speed of 3000 rpm and two smear was prepared from the sediment using Giemsa staining and 10% KOH. Of 120 BAL specimens, 59 samples (49.2%) showed yeast colonies on culture media, but according to the direct sample and colony count, 30 samples were considered to be normal flora and were not included in the study. Genotypic methods including PCR-RFLP, duplex-PCR, and sequencing of the amplified ribosomal region and the phenotypic methods were carried out on 29 yeast samples.
DNA extraction and polymerase chain reaction-restriction fragment length polymorphism method
DNA of yeasts was extracted using boiling method.[13] Afterward, a reaction with a total volume of 30 μl master mix was prepared as follows: the Premix (AMPLIQON, Denmark) 2 × 15 μl, 0.5 μl of each ITS1 and ITS4 Primers with a concentration of 25 μM, molecular grade water 12 μl and 2 μl DNA. The temperature cycles for amplification were initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 45 s, extension at 72°C for 45 s) and final extension step at 72°C for 5 min. The PCR amplification of the ITS1-5.8S-ITS2 region in ribosomal DNA was carried out by forward (5’-TCCGTAGGTGAACCTGCGG-3’) and reverse (5'TCCTCCGCTTATTGATATGC-3’) primers.[14] 1% gel electrophoresis was used to observe the amplified bands. In PCR-RFLP method, a reaction with the volume of 15 μl containing MspI (EURx, Poland) restriction enzyme 0.5 μl, enzyme buffer 10 × 1.5 μl, molecular grade water 3 μl and 10 μl of PCR product were mixed and incubated at 37°C for 1 h and 30 min. A 1.5% agarose gel was used for electrophoresis.
Duplex-polymerase chain reaction method
Differentiation of C. albicans and C. dubliniensis isolates was performed by duplex PCR using primers targeting sequences in ITS-1 and ITS-2 regions of rDNA as described by Ahmad et al.[10] The sequence of the primers used in the reaction was as follows: CALF (5′-TGGTAAGGCGGGATCGCTT-3′), CALR (5′-GGT CAAAGTTTGAAGATATAC) and CDUF (5′-AA ACTTGTCACGAGATTATTTTT), CDUR (5′-AAA GTTTGAAGAATAAAATGGC-3′). This method was implemented using a total volume of 15 μl for a reaction containing Premix (AMPLIQON, Denmark) 2 × 7.5 μl, 0.5 μl of each CALF, CALR, CDUF, and CDUR primers with a concentration of 20 μM, molecular grade water 4.5 μl, and DNA extracted 1 μl. The temperature cycles for amplification were as follows: initial denaturation step at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 60°C for 45 s, extension at 72°C for 30 s) and final extension step at 72°C for 5 min. 1% gel electrophoresis was used to detect the amplified bands.
Performing the sequencing of rDNA region of Candida dubliniensis isolates
The sequencing of rDNA was used to confirm the results of phenotypic (growth on SFSA culture media) and genotyping methods (the DNA pattern in Duplex-PCR).
The results of sequencing evaluated and compared using of NCBI BLAST searches against fungal sequences existing in DNA databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
To find the growth ability rate of fresh colonies of yeasts, they were cultured on SDA, incubated at 45°C for 48 h. Furthermore, the morphological characteristics of the isolates on different culture media were separately evaluated.
Production of chlamydoconidia on three different culture media; corn meal agar with tween 80, wheat flour agar and sunflower seed agar
The medium of CMA (QUELAB, Canada) with 1% tween 80 was prepared according to the instructions with a pH of 6 ± 0.2. To prepare 1 l of WFA medium, 32 g of wheat flour and 12 g of agar is required as reported before.[7] First, the wheat flour boiled in 500 ml of distilled water for 15 min, then the agar was boiled and added to the wheat flour, and the pH was adjusted to 5.5–6. The SFSA was prepared as reference, doing by Al Mosaid et al.[3] In brief, 50 g of sunflower seeds were milled for 5 min and boiled with 1 l of distilled water for 30 min. Then, they were passed through the sterilized gas. Furthermore, 1 g of KH2po4, 1 g of glucose, 1 g of creatinine, and 15 g of agar were added, and the result was adjusted to 1 l culture media with pH 5.5. The fresh colonies were cultured on three mentioned media and incubated at 28°C, for 72 h, and production of chlamydoconidia was examined. For all phenotypic tests, negative control (C. glabrata) and positive controls (C. albicans and C. dubliniensis) were considered.
Results
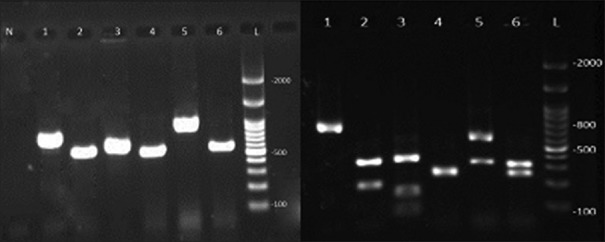
Following PCR amplification, the ITSI-5.8S-ITSII PCR products sizes were observed about 881 bp, 526 bp, 720 bp, 510 bp, 607 bp, and 537 bp in different isolates.[9] Six restriction fragments were visualized and compared by 2% agarose gel electrophoresis after treating with MspI with fragment sizes as about (320,561 bp), (186,340 bp), (720 bp), (250, 260) (82, 155, 370 bp), and (239, 298 bp).[9]
According to the amplification of target fragments, six species of Candida were identified [Figure 1]. Discriminate between C. albicans and C. dubliniensis using duplex-PCR, displayed in Figure 2.
Figure 1.
The left image shows the polymerase chain reaction product and the right image is the bands after adjacency with the MspI enzyme in various candida species isolated from the bronchoalveolar lavage samples. L is a marker of 100 bp. Bands number 1–6 are Candida kefyr, Candida tropicalis, Candida guilliermondii, Candida krusei, Candida glabrata and Candida albicans and N is negative control
Figure 2.
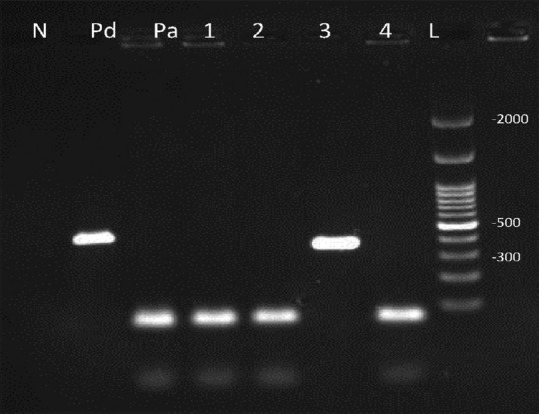
Duplex-polymerase chain reaction products: 1, 2, 4 were identified Candida albicans with 100 bp bands and number 3 Candida dubliniensis with 325 bp. Pa: Positive control (Candida albicans), Pd: Positive control (Candida dubliniensis) N: Negative control and L is a 100 bp marker
In the present study from the 29 studied BAL samples, 15 showed pseudohypha and blastoconidia in Giemsa staining and KOH preparation, meanwhile, in 14 rest BAL samples, no pseudohypha, but many blastoconidia were observed. Of 29 yeast samples, 17 isolates (58.6%) C. albicans/dubliniensis complex, 12 isolates (41.4%) of nonalbicans were identified. Furthermore, six samples were mixed and contained two different strains of Candida. In total, as shown in Table 1, seven different species of Candida were distinguished in BAL specimens as follows: C. dubliniensis two (5.6%), C. albicans 15 (42.9%), C. glabrata 10 (28.7%), C. tropicalis 2 (5.6%), C. kefyr 1 (2.9%), C. krusei 4 (11.4%), and C. guilliermondii one (2.9%). The growth rate of yeasts at 45°C on SDA and formation of chlamydoconidia structures on various culture media, including CMA with Tween 80, WFA and SFSA are presented in Table 2.
Table 1.
Demographic information and results of colony count and direct smear of yeasts isolated from patients with pulmonary symptoms
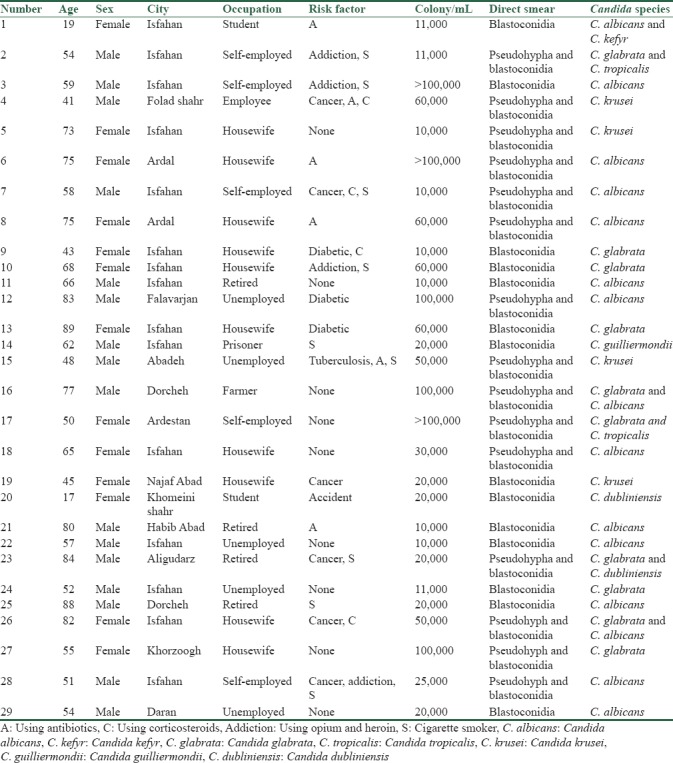
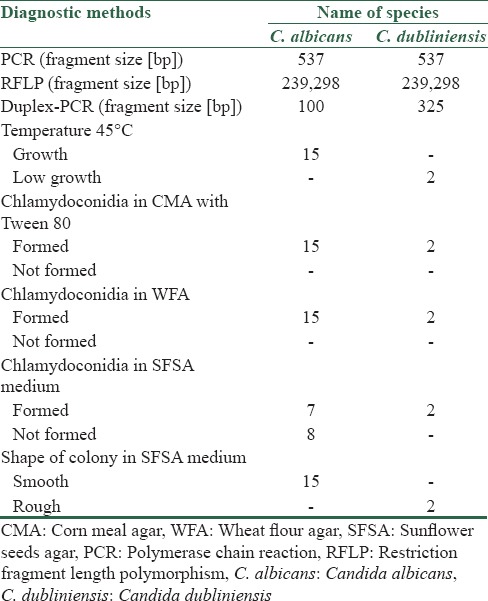
Table 2.
Differentiation of Candida albicans and Candida dubliniensis using genotypic and phenotypic methods
Discussion
From the total of 120 BAL specimens, in 59 samples (49.2%) yeast colonies grew on SDA, which were considered to be normal flora, colonization, or infection. The BAL samples of 29 patients with qualified inclusion criteria included in the study. The most common predisposing factors in this research were pulmonary symptoms notably (asthma, cough, sputum, and hemoptysis) with the frequency of 26 (89.7%) patients. Of the 17 identified BAL Candida, the most frequent was the C. albicans/dubliniensis complex which was differentiated by genotypic and phenotypic methods. In two separate studies, C. albicans, showed the highest frequency in pulmonary samples of people with different risk factors or immunosuppressed and anatomical disease.[15,16] In the present study also C. albicans was the common fungus in the BAL samples of patients with pulmonary symptoms. It was demonstrated as pseudohypha and blastoconidia in direct smear of 15(51.7%) specimens with the mean of colony count of (42,000/mL) in the BAL samples. That is notable, the normal flora of the C. albicans colonies with colony count of <10,000 colonies/ml of BAL and also no formation of pseudohypha and blastoconidia in direct smear is not accounted at this project.
In a study in Japan, which focused on cases of postmortem bronchopulmonary specimens, the frequency Candida was significantly higher (34.7%) than other organs.[17] In a study was done in India on patients with pulmonary tuberculosis, C. albicans with the frequency of 50% was a dominant comparison to the other Candida species.[15] In similar researches done in different regions of Iran on the BAL and the other pulmonary samples, the most prevalent organism isolated from cultivating BAL samples were C. albicans with different but about similar percent of the organism (33.3%), (39.4%), (35.3%).[18,19] Also in a study on colonization of respiratory fungi done in Babol and Sari, the most common isolated fungus was Candida (64.7%).[20] However in the present study if we account all of the isolated Candida from the BAL samples, it was 59 (49.2%) and show this amount is less than in the center of Iran compared to the north. Candida is the normal flora of the mouth and it generally isolated from the sputum of 20%–50% of healthy individuals.[21]
C. dubliniensis is a newly diagnosed species very similar to C. albicans phenotypically. It has a high prevalence in people with AIDS, and this frequency is increasing in people with other predisposing factors.[22,23,24,25,26] Identification and differentiation of opportunistic yeasts in immunocompromised individuals and also the appearance of resistant isolates to antifungal drugs are particular importance in identifying these yeasts.[27] Most of the studies done on the pulmonary specimens in the world and in Iran, did not mention about the differentiation of these two yeasts C. albicans and C. dubliniensis and have not mentioned anything about the colony count or observation of fungal elements in direct examination.
In the present study, two samples were confirmed to be C. dubliniensis, one was recognized in BAL sample of a 17-year-old girl with a history of accident and with no other predisposing factors and the other was recognized in a 84-year-old male with a history of prostate surgery due to prostate cancer and tobacco smoker. Therefore, we found C. dubliniensis in nonAIDS individuals, and it is important to differentiate it from the other yeast in nonAIDS patients. Although Candida are considered to be normal lung flora and do not have clinical diagnostic value except in biopsy specimens, however, due to the fact that all 29 isolates from the BAL sample in the present study were found to be highly abundant colonies in culture (more than 10,000/mL of BAL) and also observation of pseudohypha and blastoconidia or just abundant blastoconidia in direct smears cannot be ignored with the presence of this yeast population in the lung. Even if this yeast population been colonized or formed biofilm in the lung, we prepossessed to the pulmonary physicians to this point because of the presence of Candida mycelium are considered to be pathogenic in biopsy specimens.[28] Of course, the invasiveness method of the biopsy specimen should also be considered. As in candidal vaginitis or other candida infections, observing the pseudohypha and blastoconidia in the specimens suggests a vaginal candidiasis and causes complications and symptoms, such as itching and burning and sometimes even allergic symptoms the abundant presence of these fungal elements in BAL samples should not be ignored.[29,30]
According to Morrell et al., delay in diagnosis and treatment of invasive candidiasis is considered a risk factor for mortality.[31] To perform diagnostic tests, the specificity of the tests should also be considered. In a study done on 58 diabetic patients and 48 control groups using PCR-RFLP despite using Msp1 and HinfI out of the 32 isolated Candida spp. in DM patients, 25 (43.1%) were identified as Candida albicans, whereas C. dubliniensis, was not isolated from any of the study groups. Msp1 restriction enzyme cannot differentiate the two species so using this enzymes not specific to discriminates C. dubliniensis from the other species.[32] Of course, there are other molecular tests such as multiplex-PCR, nested-PCR, real-time PCR, etc., which are not cost-effective due to the expensive materials and devices they need.[33,34]
Phenotypic methods such as producing chlamydoconidia on the CMA with tween 80, WFA and SFSA medium, sugar assimilation and fermentation tests, germ tube production, have different cost and accuracy for differentiation of the yeasts.[6]
In Sullivan's study the formation of chlamydoconidia, was induced on rice agar and this medium was able to differentiate C. albicans from C. dubliniensis.[35] In Pal's Agar medium all of 128 C. dubliniensis isolates produced a hyphal fringe with rough colonies. In contrast, none of the 124 C. albicans isolates tested produced soft colonies with hyphal fringe and none of them produced chlamydospores on Pal's agar, whereas 120 of 128 of the C. dubliniensis isolates tested (93.75%) produced chlamydospores within 48–72 h.[3] According to this study, all 15 isolates of C. albicans produced soft milky colonies on SFSA culture media, and both of C. dubliniensis formed rough milky colonies with chained chlamydoconidia [Figure 3].
Figure 3.
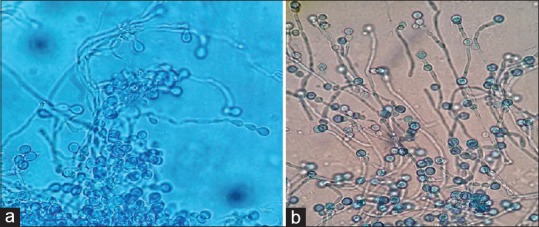
Chlamydoconidia formation of Candida dubliniensis. (a) On sunflower seeds agar medium, (b) On wheat flour agar medium ×400
Contrary to other studies that indicate that C. dubliniensis does not grow at a temperature of 45°C, both isolates of C. dubliniensis showed slight growth on the SDA medium at 45°C after 48 h., but all of 15 C. albicans and also the other mentioned none-albicans isolates at the present study grew well on the SDA medium at 45°C after 48 h.
Cereals such as wheat, corn, and rice are rich carbon and due to the abundance of this element in the body of yeasts, this factor is essential for the growth of microorganisms, including Candida.[36] In addition to the biochemical composition of the culture medium, the temperature, pH, and the physiology of microorganisms are also important in consuming the material from the environment. Therefore, the composition of the medium can change the conditions of the growth of a particular species and create special morphological structures, such as the production of chlamydoconidia or the creation of hyphae in the candida species. The two WFA and CMA with Tween 80 culture media exhibited the same specificity and sensitivity in the differentiation of the C. albicans/dubliniensis complex. It should be noted that the WFA medium for the first time in this study has been applied to this number of yeasts (29 isolates) detected from the clinical specimens. WFA without adding the Tween 80, is quite doing similar to the CMA medium with the Tween 80, which is cheaper and easier to use than the commercial CMA Tween 80 medium.[37] In general, it can be concluded that each isolate has particular conditions that respond differently depending on its growth environment and should be considered in the experiments. C. dubliniensis species are more resistant to fluconazole because these isolates are usually detected from the specimens of AIDS patients who have received fluconazole treatment. However in general, according to numerous studies, and considering all aspects whether the infection is sporadic or epidemic, it can be said that determining the species in cases of an epidemic of the disease is much more important than the sporadic ones.[8] Filamentous growth is an essential virulence trait of the human pathogenic yeasts within the genus Candida, and the greater propensity of C. albicans to form hyphae has been proposed to account for the greater virulence of this species relative C. dubliniensis.[38] Analysis of C. dubliniensis blastoconidia in the stomach and kidney of infected mice revealed that they grow predominantly in the yeast phase, whereas C. albicans could be recovered in both the yeast and hyphal phases.[39]
Conclusion
By conducting chlamydoconidia tests on CMA tween 80 and WFA media, the C. albicans/dubliniensis complex can be distinguished from other Candida species. The SFSA medium was completely consistent with the duplex-PCR molecular test, and it was possible to differentiate between the two species of C. albicans and C. dubliniensis. Also, the temperature of 45°C did not completely differentiate between the two species of C. albicans and C. dubliniensis, but to some extent, it has been helpful. Therefore, it is not possible to determine the definite identity of microorganism by applying just a single method.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent forms. In the form the patient(s) has/have given his/her/their consent for his/her/their images and other clinical information to be reported in the journal. The patients understand that their names and initials will not be published and due efforts will be made to conceal their identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
This study was financially supported by Isfahan University of Medical Sciences (Grant No: 395780).
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We would like to thank the Isfahan University of Medical Sciences for financial support. Also personals, cooperation of Reference Pulmonary Clinic Centre in Alzahra Hospital is appreciated.
References
- 1.Miceli MH, Díaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11:142–51. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 2.Wu JQ, Zhu LP, Ou XT, Xu B, Hu XP, Wang X, et al. Epidemiology and risk factors for non-Candida albicans candidemia in non-neutropenic patients at a Chinese teaching hospital. Med Mycol. 2011;49:552–5. doi: 10.3109/13693786.2010.541948. [DOI] [PubMed] [Google Scholar]
- 3.Al Mosaid A, Sullivan DJ, Coleman DC. Differentiation of Candida dubliniensis from Candida albicans on Pal's agar. J Clin Microbiol. 2003;41:4787–9. doi: 10.1128/JCM.41.10.4787-4789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan ZU, Ahmad S, Mokaddas E, Chandy R. Simplified sunflower (Helianthus annuus) seed agar for differentiation of Candida dubliniensis from Candida albicans. Clin Microbiol Infect. 2004;10:590–2. doi: 10.1111/j.1469-0691.2004.00923.x. [DOI] [PubMed] [Google Scholar]
- 5.Livério HO, Ruiz LDS, Freitas RS, Nishikaku A, Souza AC, Paula CR, et al. Phenotypic and genotypic detection of Candida albicans and Candida dubliniensis strains isolated from oral mucosa of AIDS pediatric patients. Rev Inst Med Trop Sao Paulo. 2017;59:e14. doi: 10.1590/S1678-9946201759014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neppelenbroek KH, Seó RS, Urban VM, Silva S, Dovigo LN, Jorge JH, et al. Identification of Candida species in the clinical laboratory: A review of conventional, commercial, and molecular techniques. Oral Dis. 2014;20:329–44. doi: 10.1111/odi.12123. [DOI] [PubMed] [Google Scholar]
- 7.Dehghan P, Shadzi S, Zadeh AH. Growth rate and identification of yeasts in three different media: Flour, sprout of cereals and commercial media. J Res Med Sci. 2002;7:1–5. [Google Scholar]
- 8.Brandt ME, Lockhart SR. Recent taxonomic developments with Candida and other opportunistic yeasts. Curr Fungal Infect Rep. 2012;6:170–7. doi: 10.1007/s12281-012-0094-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammadi R, Mirhendi H, Rezaei-Matehkolaei A, Ghahri M, Shidfar MR, Jalalizand N, et al. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Med Mycol. 2013;51:657–63. doi: 10.3109/13693786.2013.770603. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad S, Khan Z, Asadzadeh M, Theyyathel A, Chandy R. Performance comparison of phenotypic and molecular methods for detection and differentiation of Candida albicans and Candida dubliniensis. BMC Infect Dis. 2012;12:230. doi: 10.1186/1471-2334-12-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer J, van Koningsbruggen-Rietschel S, Rietschel E, Vehreschild MJ, Wisplinghoff H, Krönke M, et al. Prevalence and molecular characterization of azole resistance in Aspergillus spp. Isolates from German cystic fibrosis patients. J Antimicrob Chemother. 2014;69:1533–6. doi: 10.1093/jac/dku009. [DOI] [PubMed] [Google Scholar]
- 12.Holding KJ, Dworkin MS, Wan PC, Hanson DL, Klevens RM, Jones JL, et al. Aspergillosis among people infected with human immunodeficiency virus: Incidence and survival. Adult and adolescent spectrum of HIV disease project. Clin Infect Dis. 2000;31:1253–7. doi: 10.1086/317452. [DOI] [PubMed] [Google Scholar]
- 13.Silva GA, Bernardi TL, Schaker PD, Menegotto M, Valente P. Rapid yeast DNA extraction by boiling and freeze-thawing without using chemical reagents and DNA purification. Braz Arch Biol Technol. 2012;55:319–27. [Google Scholar]
- 14.Korabecna M. The variability in the fungal ribosomal DNA (ITS1, ITS2, and 5.8 S rRNA gene): Its biological meaning and application in medical mycology. Commun Curr Res Educ Top Trends Appl Microbiol. 2007;2:783–7. [Google Scholar]
- 15.Kali A, Charles MP, Noyal MJ, Sivaraman U, Kumar S, Easow JM, et al. Prevalence of candida co-infection in patients with pulmonary tuberculosis. Australas Med J. 2013;6:387–91. doi: 10.4066/AMJ.2013.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chadeganipour M, Shadzi S, Dehghan P, Bijary J. The incidence of opportunistic fungi in patients suspected of tuberculosis. Mycoses. 2000;43:269–72. doi: 10.1046/j.1439-0507.2000.00572.x. [DOI] [PubMed] [Google Scholar]
- 17.Yamazaki T, Kume H, Murase S, Yamashita E, Arisawa M. Epidemiology of visceral mycoses: Analysis of data in annual of the pathological autopsy cases in Japan. J Clin Microbiol. 1999;37:1732–8. doi: 10.1128/jcm.37.6.1732-1738.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajialiakbar V, Emami M, Eskandari A. Frequency of fungal pathogens in bronchoalveolar lavage and biopsy specimens of patients hospitalized in Chamran and Amir al momenin Hospitals. EBNESINA. 2007;10:21–5. [Google Scholar]
- 19.Djahromi S, Khaksar A. Fungal isolates of the respiratory tract. Mycoses. 2002;45:6. [Google Scholar]
- 20.Khodavaisy S, Alialy M, Mahdavi OS, Habibi MR, Amri P, Monadi M, et al. The study on fungal colonization of respiratory tract in patients admitted to intensive care units of Sari and Babol hospitals. Med J Mashad Univ Med Sci. 2011:177–184. [Google Scholar]
- 21.BAUM GL. The significance of Candida albicans in human sputum. N Engl J Med. 1960;263:70–3. doi: 10.1056/NEJM196007142630204. [DOI] [PubMed] [Google Scholar]
- 22.Haghdoost AA, Mostafavi E, Mirzazadeh A, Navadeh S, Feizzadeh A, Fahimfar N, et al. Modelling of HIV/AIDS in Iran up to 2014. J AIDS HIV Res. 2011;3:231–9. [Google Scholar]
- 23.Chavasco JK, Paula CR, Hirata MH, Aleva NA, Melo CE, Gambale W, et al. Molecular identification of Candida dubliniensis isolated from oral lesions of HIV-positive and HIV-negative patients in São paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2006;48:21–6. doi: 10.1590/s0036-46652006000100005. [DOI] [PubMed] [Google Scholar]
- 24.Meis JF, Ruhnke M, De Pauw BE, Odds FC, Siegert W, Verweij PE, et al. Candida dubliniensis candidemia in patients with chemotherapy-induced neutropenia and bone marrow transplantation. Emerg Infect Dis. 1999;5:150–3. doi: 10.3201/eid0501.990119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman DC, Sullivan DJ, Bennett DE, Moran GP, Barry HJ, Shanley DB, et al. Candidiasis: The emergence of a novel species, Candida dubliniensis. AIDS. 1997;11:557–67. doi: 10.1097/00002030-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan DJ, Moran GP, Coleman DC. Candida dubliniensis: Ten years on. FEMS Microbiol Lett. 2005;253:9–17. doi: 10.1016/j.femsle.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC, et al. Hidden killers: Human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 28.Masur H, Rosen PP, Armstrong D. Pulmonary disease caused by Candida species. Am J Med. 1977;63:914–25. doi: 10.1016/0002-9343(77)90546-0. [DOI] [PubMed] [Google Scholar]
- 29.Peters BM, Yano J, Noverr MC, Fidel PL., Jr Candida vaginitis: When opportunism knocks, the host responds. PLoS Pathog. 2014;10:e1003965. doi: 10.1371/journal.ppat.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hainer BL, Gibson MV. Vaginitis: Diagnosis and treatment. Am Fam Physician. 2011;83:807–15. [PubMed] [Google Scholar]
- 31.Morrell M, Fraser VJ, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for hospital mortality. Antimicrob Agents Chemother. 2005;49:3640–5. doi: 10.1128/AAC.49.9.3640-3645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammadi F, Javaheri MR, Nekoeian S, Dehghan P. Identification of Candida species in the oral cavity of diabetic patients. Curr Med Mycol. 2016;2:1–7. doi: 10.18869/acadpub.cmm.2.2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arvanitis M, Anagnostou T, Fuchs BB, Caliendo AM, Mylonakis E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin Microbiol Rev. 2014;27:490–526. doi: 10.1128/CMR.00091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin JH, Ranken R, Sefers SE, Lovari R, Quinn CD, Meng S, et al. Detection, identification, and distribution of fungi in bronchoalveolar lavage specimens by use of multilocus PCR coupled with electrospray ionization/mass spectrometry. J Clin Microbiol. 2013;51:136–41. doi: 10.1128/JCM.01907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. Nov: Phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141(Pt 7):1507–21. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 36.Moore-Landecker E. Fundamentals of the Fungi. USA: Prentice Hall; 1996. [Google Scholar]
- 37.Atlas RM. Handbook Of Microbiological Media. USA: CRC press; 2010. [Google Scholar]
- 38.Caplice N, Moran GP. Candida albicans exhibits enhanced alkaline and temperature induction of Efg1-regulated transcripts relative to Candida dubliniensis. Genom Data. 2015;6:130–5. doi: 10.1016/j.gdata.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stokes C, Moran GP, Spiering MJ, Cole GT, Coleman DC, Sullivan DJ, et al. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet Biol. 2007;44:920–31. doi: 10.1016/j.fgb.2006.11.014. [DOI] [PubMed] [Google Scholar]