Abstract
Schwann cells (SCs) contribute to nerve repair following injury; however, the underlying molecular mechanism is poorly understood. MicroRNAs (miRNAs), which are short noncoding RNAs, have been shown to play a role in neuronal disease. In this work, we show that miRNAs regulate the peripheral nerve system by modulating the migration and proliferation of SCs. Thus, miRNAs expressed in peripheral nerves may provide a potential therapeutic target for peripheral nerve injury or repair.
1. Introduction
MicroRNAs (miRNAs) are endogenous small noncoding RNAs that are present as RNA-duplex transcripts of approximately 22 nucleotides. miRNAs regulate mRNAs in eukaryotic cells [1]. In the initial step of processing, primary miRNA transcripts, called pri-miRNAs, are generated in the nucleus with a 5′ cap structure and a 3′ poly-A tail [2, 3]. Next, precursor miRNAs containing a 70-nucleotide stem loop precursor miRNA are generated from the pri-miRNA by microprocessor complexes, such as the nuclear Drosha-DiGeorge syndrome critical region gene 8 (DGCR8) enzyme complex [4]. The pre-miRNA is exported to the cytoplasm via binding with the nuclear export factor, exportin 5. Finally, miRNA maturation is mediated by Dicer, a type-III ribonuclease, which allows the miRNA to form a duplex of 22 nucleotides [5]. The mature miRNA is then loaded onto the RNA-induced silencing complex (RISC), which modulates gene expression by binding via imperfect complementarity to the 3′-untranslated region (UTR) of target mRNAs, resulting in translational repression or degradation of the mRNA [6]. miRNAs are involved in diverse biological functions in aging [7, 8], cancer [9, 10], cell development [11, 12], and neuronal disease [13–15].
The peripheral nervous system, which links the brain and body, is made up of the nerves and ganglia outside the brain and spinal cord. Schwann cells (SCs) are important in the development and maintenance of the peripheral nervous system. In adult peripheral nerves, SCs exist in myelinating and nonmyelinating forms [6]. SCs that arise from the neural crest are surrounded by axons and myelinate large-caliber axons in the peripheral nervous system [16]. Schwann cell dysfunction or injury results in demyelinating neuropathies along with loss of motor and sensory abilities [17]. Mutations in SCs contribute to disease by reduction or gain of function of specific genes, such as peripheral myelin protein-22 (PMP22) or myelin protein zero (MPZ) [18]. There is evidence that misfolded PMP22 or MPZ accumulates within the endoplasmic reticulum of myelinating SCs [19].
Krox20 (also termed early growth response gene: EGR2) is a transcription factor that plays a role in myelin formation during promyelination. SCs are important myelin structural components along with myelin binding protein (MBP) and MPZ [20, 21]. Krox20, MBP, and MPZ levels are increased during development of the sciatic nerve while sex determining region Y-box 2 (SOX2), Notch, and Jun act as negative regulators [22–24].
Following injury by nerve transection or crush, SCs undergo a demyelination process and myelin damage called Wallerian degeneration. Macrophages in peripheral nerves move to the site of damage and are activated to clear the damaged myelin or axonal debris [25]. The clearance of myelin leads to axonal regeneration following peripheral nerve injury. Crush injury and nerve trunk transection are common types of experimentally induced nerve injury [26]. Crush injury results in axonal interruption while maintaining the connective sheaths, which is called axonotmesis, while nerve transection results in disruption of axon connection and nerve loss. Degeneration following nerve transection is more acute than following crush injury [26].
2. The Proliferation of SCs
SC proliferation is regulated by platelet-derived growth factor [27], transforming growth factor (TGF) [28], laminin [29], Notch [24], and neuregulin 1 (NRG-1) [30]. Notch and neuregulin 1 which are axon-associated signals affect SC proliferation in vivo. Notch receptor localizes in SC and Notch ligands exist on axons. Canonical Notch signaling is essential positive regulator of SC proliferation by decreasing the number of SC after Notch inactivation in vivo [24]. NRG-1 is also present on axons and its receptors such as ErbB2 and ErbB3 exist in Schwann cells. NRG treatment stimulates SC proliferation in vitro [30]. TGF-β treatment in Schwann cell enhanced SC proliferation in vitro [31]. D'Antonio et al. showed that type II TGF-β receptor mutant mice reduced SC DNA synthesis [28]. Laminin which is the major component of the basal lamina also contributes to SC proliferation. Yu et al. showed that laminin-null Schwann cells attenuated phosphatidylinositol 3- (PI3-) kinase activity and proliferation of immature SCs [29]. A recent study showed that mTORC1 (mechanistic target of rapamycin) pathway which is a downstream of growth-factor-stimulated phosphatidylinositol 3′ kinase (PI3K)/AKT pathway enhanced proliferation of immature SC as well as the myelin sheath of differentiated SCs [32]. In addition, SC proliferation is important during Wallerian degeneration by increasing SC number in the distal stump [33].
3. The Migration of SCs
After peripheral nerve injury, Schwann cell migrates toward regrowing axons by regulating extracellular matrix (ECM) and ECM substrates such as fibronectin and laminin [34]. Also, extracellular signal-related kinase (ERK) 1/2 and AKT are important players for migration of Schwann cell during nerve degeneration [35]. Chang et al. demonstrated that neuregulin enhanced SC migration through erbB2/3- focal adhesion kinase (FAK) pathway following nerve injury [36]. Bentley and Lee showed that neutrophin receptor p75 mutant mice attenuated SC migration and axon growth [37].
4. miRNAs in the Regeneration of Peripheral Nerves
There is evidence that miRNAs modulate peripheral nerve myelination. Absence of Dicer in sciatic nerve SCs in a mouse model impaired myelin formation and reduced mRNA level of KROX20, a key transcriptional activator of myelin genes [38, 39]. Bremer et al. showed that several miRNAs, including miRNA-34a, miRNA-146, miRNA-30a, miRNA-195, miRNA-140, miRNA-27b, and miRNA-204, were upregulated upon myelination in Dicer mutant mice [39]. Gokey et al. showed that 225 miRNAs were expressed during peripheral myelination by microRNA profiling. miRNA-106a, miRNA-20b, miRNA-338, miRNA-92b, miRNA-19b, miRNA-363, miRNA-350, miRNA-17, and miRNA-340 regulated Sox10, which regulates myelin genes [40].
4.1. Let-7
The lethal-7 (let-7) gene plays a vital role in carcinogenesis. Moreover, several studies have shown that let-7 miRNAs affect neuronal cell fate and regeneration by regulating nerve growth factor (NGF) expression [41]. Li et al. showed that, following sciatic nerve transection, let-7c, -7d, -7e, -7f, -7i, and miRNA-98 levels in the proximal nerve were increased at 1 day after nerve injury and decreased at 4 and 7 days after nerve injury, followed by a rebound at 14 days. A transwell migration assay showed that transfection with let-7d/miRNA-98 mimics significantly suppressed the migratory ability of SCs compared with the control [41]. NGF has been shown to be regulated by let-7 miRNAs through direct binding to the 3′-UTR of NGF mRNA [41]. Thus, let-7 miRNAs affect SC migration and axon outgrowth during regeneration.
4.2. miRNA-1
Expression of miRNA-1 following sciatic nerve crush was drastically decreased at 4, 7, and 14 days after peripheral nerve injury compared with the expression at 0 h, while brain-derived neurotrophic factor (BDNF) expression was enhanced, exhibiting a negative correlation with miRNA-1 [42]. Overexpression of miRNA-1 in primary SCs inhibited the migration and proliferation of SCs. BDNF knockdown by small interfering RNA recapitulated the effects of miR-1 on the proliferation and migration of SCs [42].
4.3. miRNA-9
miRNA-9 has been shown to be an important regulator of SC migration [43], which is critical to the regenerative response of SCs to nerve injury. Overexpression of miRNA-9 mimics attenuated SC migration, whereas inhibition of miRNA-9 enhanced migration by directly targeting collagen triple helix repeat containing protein 1 (CTHRC1) [43].
4.4. miRNA-148-3p
Qian et al. showed that miRNA 148-3p plays a role in the regeneration of peripheral nerves by regulating SC migration [44]. For instance, overexpression of miRNA 148-3p enhanced the migratory ability of SCs while inhibition of miRNA-148-3p attenuated migration in vitro. Furthermore, miRNA-148b-3p enhanced migration via targeting Cullin-associated NEDD8-dissociated protein 1 (Cand1), which negatively regulated the proliferation of lymph node carcinoma cells in the prostate [44].
4.5. miRNA-sc8
A transwell migration assay showed that transfection with miRNA-sc8 mimic reduced the migration of SCs, while silencing miRNA-sc8 expression enhanced the proliferation and migration of SCs. In addition, silencing of the epidermal growth factor receptor (EGFR) diminished the positive effects of the miRNA-sc8 inhibitor on SC proliferation and migration [45]. Thus, this evidence suggested that miRNA-sc8 regulates SC proliferation and migration by targeting EGFR.
4.6. miRNA-210
Zhang et al. showed that miRNA-210 modulates peripheral nerve regeneration by enhancing the migration and proliferation of SCs [46]. miRNA-210 levels increased until 14 days after sciatic nerve injury. In addition, overexpression of miRNA-210 enhanced the migration and proliferation of SCs. Moreover, miRNA-210 transfection increased the expression of growth associated protein 43 (GAP43) [46]. Thus, these findings suggested that upregulation of miRNA-210 following sciatic nerve injury may be important for peripheral nerve regeneration.
4.7. miRNA-221/222
Yu et al. performed Agilent miRNA microarray analysis to examine the expression profile of miRNAs in the proximal stump of nerves following sciatic nerve transection [47] and showed that, among 77 miRNAs, miRNA-21, miRNA-31, miRNA-221, miRNA-222, and miRNA-132 levels were enhanced at 1 day after sciatic nerve injury. In situ hybridization revealed that miRNA-221/222 was highly expressed at 4 days after sciatic nerve injury. A transwell migration assay demonstrated that miRNA-221/222 overexpression increased the migratory ability of primary SCs, while miRNA221/22 inhibitor impaired SC migration. miRNA-221/222 modulates longevity assurance homolog 2 (LASS2) in SCs by binding directly to the 3′-UTR of LASS2 mRNA. Thus, sciatic nerve injury enhanced SC migration by regulating miRNA-221/222, which targets LASS2 [47].
4.8. miRNA-sc3
miRNA-sc3 has been shown to be highly expressed in the injured nerve following sciatic nerve transection [48]. Increased expression of miRNA-sc3 promoted the proliferation and migration of primary SCs, while silencing of miRNA-sc3 attenuated proliferation and migration. miRNA-sc3 directly targeted astrotactin 1 (Astn1) and led to translational suppression of Astn1. Thus, these data suggested that miRNA-sc3 modulates the migration and proliferation of primary SCs via Astn1 [48].
4.9. miRNA-132
Yao et al. showed that miRNA-132 expression was increased following sciatic nerve injury. Furthermore, miRNA-132 enhanced the migration of primary SCs. miRNA-132 also enhanced SC migration in hypoxic conditions, while miRNA-132 inhibitor suppressed migration [49]. This study showed that miRNA-132 modulates SC migration by suppressing protein kinase AMP-activated noncatalytic subunit gamma 3 (PRKAG3) expression by binding the 3′-UTR of its mRNA [49].
4.10. miRNA-182
Yu et al. demonstrated that miRNA-182 was highly expressed following sciatic nerve resection by quantitative reverse transcription polymerase chain reaction, in situ hybridization, and miRNA array analysis [50]. Transwell migration and proliferation assays showed that miRNA-182 overexpression significantly attenuated the migration of primary SCs. In addition, miRNA-182 repressed primary SC proliferation and migration through fibroblast growth factor 9 and neurotrimin, implicating miRNAs in peripheral nerve repair [50].
5. MicroRNAs in Peripheral Neuropathy
Damage or dysfunction of the nervous system causes neuropathic pain [51]. While the number of patients experiencing neuropathic pain has increased worldwide [51], the molecular pathogenesis has yet to be elucidated. Therefore, it will be important to understand the molecular mechanism of neuropathic pain and develop novel therapeutic targets. Evidence indicates that miRNAs are involved in neuropathic pain. miRNA-203 attenuated the development of neuropathic pain via regulation of Rap1a expression in neuronal PC12 cells [52]. Expression of miRNA-183 also resulted in suppression of neuropathic pain by repressing the mTOR/VEGF signaling pathway [53]. miRNA-93 was found to attenuate neuropathic pain via modulation of signal transducer and activator of transcription 3 (STAT3) [54]. Recent studies have shown that miRNAs affect peripheral neuropathy.
5.1. miRNA-146a
miRNA-146a plays a role in diabetic peripheral neuropathy [55, 56]. miRNA-146a in sciatic nerve tissue was attenuated in mice with type 2 diabetes and miRNA-146a levels in plasma and sciatic nerve tissue were enhanced following administration of miRNA-146a mimics in diabetic mice. Additionally, miRNA-146a mimics enhanced the axonal diameter and myelin thickness of sciatic nerves [56].
6. The Role of microRNAs in SC Myelination
A recent study showed that Dicer, which is a player for miRNA biogenesis, is important for SC myelination. For instance, Dicer deficient SCs diminished their ability to myelinate and microarray analysis of miRNA from Dicer deficient nerves showed that 109 miRNAs were significantly up- or downregulated. miRNA-1224, miRNA-9, and miRNA-455 were reduced upon myelination and downregulated following Dicer mutant from SCs [39]. Yun et al. showed that miRNA 138 showed a developmental increase between P2 and P21 while it repressed Ccnd1, Sox2, or c-jun which are negative regulators of myelination implying that miRNA138 might be essential for SC differentiation [57]. Also, the expression of miRNA-193, miRNA-222, miRNA-129, miRNA-145, and miR-29a was significantly downregulated in inhibition of Dicer [58]. Gökbuget et al. showed that let-7 miRNAs were significantly upregulated during myelination and inversely correlated to the level of lin28 homolog B (Lin28B) which is a microRNA regulator. Lin28B expression leads to impaired onset of SC myelination and let-7 miRNAs enhanced expression of Krox 20, which is a key transcription factor for myelination, via suppression of Notch signaling [59]. These results imply that the Lin28/let-7 regulatory axis may be an important player during remyelination.
7. MicroRNAs in Demyelination Diseases
Though several studies reported that miRNAs expressed from active and inactive demyelinating lesions [60] and interleukin 17- (IL-17-) producing T helper cells from peripheral blood cells [61–63] from patients with multiple sclerosis which is a chronic disease of the central nervous system, a few studies for miRNA expression in demyelination disease of peripheral nervous system have been studied. Guillain–Barré syndrome (GBS) which is an acute inflammatory demyelinating polyneuropathy affects the peripheral nervous system [64]. Lv et al. showed that has-miR-4717-5p and has-miR-642b-5p were upregulated in the GBS patients [65]. Also, low level of miRNA155 was expressed in peripheral blood mononuclear cells from GBS patients and silencing of miR155 enhanced the production of Th1-type cytokines such as TNF-α, IL-1β, IFN-γ, and IL-12 in vitro [66]. To identify miRNAs in demyelinating diseases may be important for developing new biomarkers for demyelinating diseases.
8. Conclusion
MiRNAs affect proliferation, migration, and myelination for SCs by regulating their targets in normal state. As shown in Figure 1, alterations in miRNA expression and their target may lead to dysfunction for SC differentiation or nerve regeneration. Therefore, it is essential to identify miRNA expression during development and in disease states which may provide important information for therapeutic and diagnostic use. Also, due to stable expression of miRNA during the tissue sampling [67], miRNA is suggested as a useful tool for biomarker.
Figure 1.
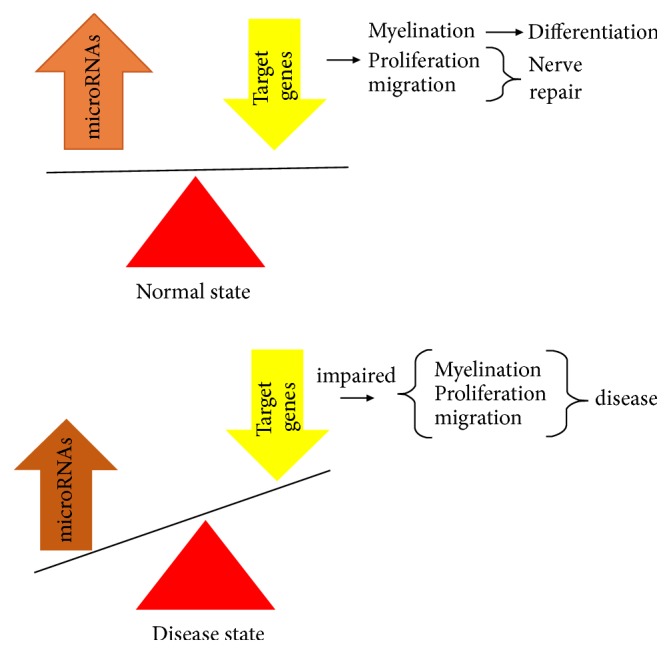
Alternation of microRNA expression and their target contribute to impaired onset of SC differentiation or nerve regeneration.
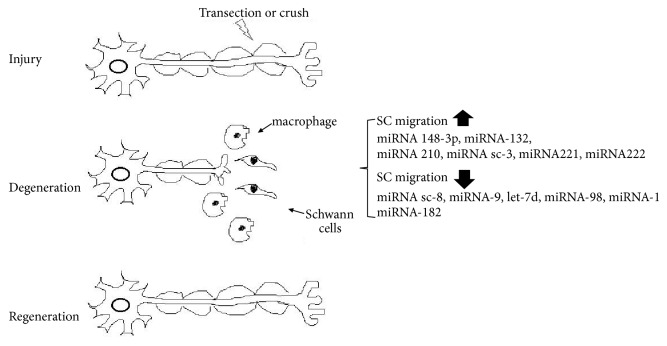
Taken together, these studies demonstrate the importance of miRNAs in peripheral nerves. SC migration plays an important role in peripheral nerve regeneration following injury. As shown in Figure 2, miRNA-148-3p, miRNA-210, miRNA221/222, and miRNA-sc3 enhanced the migration of SCs while miRNA-sc8, miRNA-9, let-7d, miRNA98, miRNA-1, and miRNA-98 attenuated migration. The miRNAs in these studies contributed to peripheral nerve regeneration. Therefore, the regulation of miRNAs in peripheral nerves may provide a potential therapeutic target for peripheral nerve injury or neuropathy.
Figure 2.
MicroRNAs regulate SC migration during nerve regeneration: miRNA 148-3p, miRNA-132, miRNA 210, miRNA sc-3, miRNA221, and miRNA222 increased the ability of SC migration while miRNA sc-8, miRNA-9, let-7d, miRNA-98, and miRNA-1miRNA-182 downregulated the ability of SC migration.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by Basic Research Program (2015R1C1A1A02036842) and Korea government (MSIT) (no. 2016R1A5A2007009).
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Rana T. M. Illuminating the silence: understanding the structure and function of small RNAs. 2007;8(1):23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 2.Cai X., Hagedorn C. H., Cullen B. R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha M., Kim V. N. Regulation of microRNA biogenesis. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y., Ahn C., Han J., et al. The nuclear RNase III Drosha initiates microRNA processing. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Kim V. N., Han J., Siomi M. C. Biogenesis of small RNAs in animals. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Griffin J. W., Thompson W. J. Biology and pathology of nonmyelinating schwann cells. 2008;56(14):1518–1531. doi: 10.1002/glia.20778. [DOI] [PubMed] [Google Scholar]
- 7.Park S., Kang S., Min K. H., Woo Hwang K., Min H. Age-associated changes in MicroRNA expression in bone marrow derived dendritic cells. 2013;42(3):179–190. doi: 10.3109/08820139.2012.717328. [DOI] [PubMed] [Google Scholar]
- 8.Munk R., Panda A. C., Grammatikakis I., Gorospe M., Abdelmohsen K. Senescence-Associated MicroRNAs. 2017;334:177–205. doi: 10.1016/bs.ircmb.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yonemori K., Kurahara H., Maemura K., Natsugoe S. MicroRNA in pancreatic cancer. 2017;62(1):33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Duarte R. J., Cazares-Ordonez V., Avila-Chavez E. The microRNA biogenesis machinery: regulation by steroid hormones and alterations in cancer. 2014;66:460–464. [PubMed] [Google Scholar]
- 11.Raghuwanshi S., Gutti U., Kandi R., Gutti R. K. MicroRNA-9 promotes cell proliferation by regulating RUNX1 expression in human megakaryocyte development. 2017 doi: 10.1111/cpr.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuziwara C. S., Kimura E. T. MicroRNAs in thyroid development, function and tumorigenesis. 2017;456:44–50. doi: 10.1016/j.mce.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Fiore R., Siegel G., Schratt G. MicroRNA function in neuronal development, plasticity and disease. 2008;1779(8):471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Li S., Lv X., Zhai K., et al. MicroRNA-7 inhibits neuronal apoptosis in a cellular Parkinson’s disease model by targeting Bax and Sirt2. 2016;8(2):993–1004. [PMC free article] [PubMed] [Google Scholar]
- 15.O’Toole S. M., Ferrer M. M., Mekonnen J., et al. Dicer maintains the identity and function of proprioceptive sensory neurons. 2017;117(3):1057–1069. doi: 10.1152/jn.00763.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunge R. P. Expanding roles for the Schwann cell: ensheathment, myelination, trophism and regeneration. 1993;3(5):805–809. doi: 10.1016/0959-4388(93)90157-T. [DOI] [PubMed] [Google Scholar]
- 17.Scherer S. S., Wrabetz L. Molecular mechanisms of inherited demyelinating neuropathies. 2008;56(14):1578–1589. doi: 10.1002/glia.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jerath N. U., Shy M. E. Hereditary motor and sensory neuropathies: Understanding molecular pathogenesis could lead to future treatment strategies. 2015;1852(4):667–678. doi: 10.1016/j.bbadis.2014.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Colby J., Nicholson R., Dickson K. M., et al. Pmp22 carrying the trembler or trembler-J mutation is intracellularly retained in myelinating Schwann cells. 2000;7(6):561–573. doi: 10.1006/nbdi.2000.0323. [DOI] [PubMed] [Google Scholar]
- 20.Jessen K. R., Mirsky R. The origin and development of glial cells in peripheral nerves. 2005;6(9):671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 21.Topilko P., Schneider-Maunoury S., Levi G., et al. Krox-20 controls myelination in the peripheral nervous system. 1994;371(6500):796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 22.Le N., Nagarajan R., Wang J. Y. T., Araki T., Schmidt R. E., Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. 2005;102(7):2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkinson D. B., Bhaskaran A., Arthur-Farraj P., et al. c-Jun is a negative regulator of myelination. 2008;181(4):625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodhoo A., Alonso M. B. D., Droggiti A., et al. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. 2009;12(7):839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry V. H., Brown M. C. Role of macrophages in peripheral nerve degeneration and repair. 1992;14(6):401–406. doi: 10.1002/bies.950140610. [DOI] [PubMed] [Google Scholar]
- 26.Dubový P. Wallerian degeneration and peripheral nerve conditions for both axonal regeneration and neuropathic pain induction. 2011;193(4):267–275. doi: 10.1016/j.aanat.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Eccleston P. A. Regulation of Schwann cell proliferation: Mechanisms involved in peripheral nerve development. 1992;199(1):1–9. doi: 10.1016/0014-4827(92)90455-H. [DOI] [PubMed] [Google Scholar]
- 28.D'Antonio M., Michalovich D., Paterson M., et al. Gene profiling and bioinformatic analysis of Schwann cell embryonic development and myelination. 2006;53(5):501–515. doi: 10.1002/glia.20309. [DOI] [PubMed] [Google Scholar]
- 29.Yu W.-M., Feltri M. L., Wrabetz L., Strickland S., Chen Z.-L. Schwann cell-specific ablation of laminin γ1 causes apoptosis and prevents proliferation. 2005;25(18):4463–4472. doi: 10.1523/jneurosci.5032-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levi A. D. O., Bunge R. P., Lofgren J. A., et al. The influence of heregulins on human Schwann cell proliferation. 1995;15(2):1329–1340. doi: 10.1523/JNEUROSCI.15-02-01329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridley A. J., Davis J. B., .Stroobant P., Land H. Transforming growth factors-β1 and β2 are mitogens for rat Schwann cells. 1989;109(6):3419–3424. doi: 10.1083/jcb.109.6.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beirowski B., Wong K. M., Babetto E., Milbrandt J. MTORC1 promotes proliferation of immature Schwann cells and myelin growth of differentiated Schwann cells. 2017;114(21):E4261–E4270. doi: 10.1073/pnas.1620761114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas G. A. Quantitative histology of Wallerian degeneration; nuclear population in two nerves of different fibre spectrum. 1948;82(3):135–145. [PubMed] [Google Scholar]
- 34.Chernousov M. A., Yu W., Chen Z., Carey D. J., Strickland S. Regulation of Schwann cell function by the extracellular matrix. 2008;56(14):1498–1507. doi: 10.1002/glia.20740. [DOI] [PubMed] [Google Scholar]
- 35.Yu H., Zhu L., Li C., et al. ERK1/2 and AKT are vital factors in regulation of the migration of rat schwann cells. 2015;77(4):427–432. doi: 10.1292/jvms.14-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang H.-M., Shyu M.-K., Tseng G.-F., et al. Neuregulin Facilitates Nerve Regeneration by Speeding Schwann Cell Migration via ErbB2/3-Dependent FAK Pathway. 2013;8(1) doi: 10.1371/journal.pone.0053444.e53444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentley C. A., Lee K.-F. p75 Is important for axon growth and Schwann cell migration during development. 2000;20(20):7706–7715. doi: 10.1523/JNEUROSCI.20-20-07706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira J. A., Baumann R., Norrmén C., et al. Dicer in Schwann cells is required for myelination and axonal integrity. 2010;30(19):6763–6775. doi: 10.1523/jneurosci.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bremer J., O'Connor T., Tiberi C., Rehrauer H., Weis J., Aguzzi A. Ablation of dicer from murine Schwann cells increases their proliferation while blocking myelination. 2010;5(8) doi: 10.1371/journal.pone.0012450.e12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gokey N. G., Srinivasan R., Lopez-Anido C., Krueger C., Svaren J. Developmental regulation of microRNA expression in Schwann cells. 2012;32(2):558–568. doi: 10.1128/MCB.06270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S., Wang X., Gu Y., et al. Let-7 microRNAs regenerate peripheral nerve regeneration by targeting nerve growth factor. 2015;23(3):423–433. doi: 10.1038/mt.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi S., Yuan Y., Chen Q., et al. Regulation of Schwann cell proliferation and migration by MIR-1 targeting brain-derived neurotrophic factor after peripheral nerve injury. 2016;6 doi: 10.1038/srep29121.29121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou S., Gao R., Hu W., et al. MiR-9 inhibits schwann cell migration by targeting cthrc1 following sciatic nerve injury. 2014;127(5):967–976. doi: 10.1242/jcs.131672. [DOI] [PubMed] [Google Scholar]
- 44.Qian T.-M., Zhao L.-L., Wang J., et al. miR-148b-3p promotes migration of schwann cells by targeting cullin-associated and neddylationdissociated 1. 2016;11(6):1001–1005. doi: 10.4103/1673-5374.184504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu Y., Chen C., Yi S., et al. MIR-sc8 inhibits schwann cell proliferation and migration by targeting EGFR. 2015;10(12) doi: 10.1371/journal.pone.0145185.e0145185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Gong X., Qiu J., Zhang Y., Gong F. MicroRNA-210 contributes to peripheral nerve regeneration through promoting the proliferation and migration of Schwann cells. 2017;14(4):2809–2816. doi: 10.3892/etm.2017.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu B., Zhou S., Wang Y., et al. Mir-221 and mir-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury. 2012;125(11):2675–2683. doi: 10.1242/jcs.098996. [DOI] [PubMed] [Google Scholar]
- 48.Yi S., Wang S., Zhao Q., et al. MiR-sc3, a novel microRNA, promotes schwann cell proliferation and migration by targeting astn1. 2016;25(5, article no. 10):973–982. doi: 10.3727/096368916X690520. [DOI] [PubMed] [Google Scholar]
- 49.Yao C., Shi X., Zhang Z., et al. Hypoxia-Induced Upregulation of miR-132 Promotes Schwann Cell Migration After Sciatic Nerve Injury by Targeting PRKAG3. 2016;53(8):5129–5139. doi: 10.1007/s12035-015-9449-y. [DOI] [PubMed] [Google Scholar]
- 50.Yu B., Qian T., Wang Y., et al. MiR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. 2012;40(20):10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neville A., Peleg R., Singer Y., Sherf M., Shvartzman P. Chronic pain: A population-based study. 2008;10(10):676–680. [PubMed] [Google Scholar]
- 52.Li H., Huang Y., Ma C., Yu X., Zhiyong Z., Shen L. MiR-203 involves in neuropathic pain development and represses rap1a expression in nerve growth factor differentiated neuronal PC12 cells. 2015;31(1):36–43. doi: 10.1097/AJP.0000000000000070. [DOI] [PubMed] [Google Scholar]
- 53.Xie X., Ma L., Xi K., Fan D., Zhang W. MicroRNA-183 suppresses neuropathic pain and expression of AMPA receptors by targeting mTOR/VEGF signaling pathway. 2017;41(1):181–192. doi: 10.1159/000455987. [DOI] [PubMed] [Google Scholar]
- 54.Yan X.-T., Ji L.-J., Wang Z., et al. MicroRNA-93 alleviates neuropathic pain through targeting signal transducer and activator of transcription 3. 2017;46:156–162. doi: 10.1016/j.intimp.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 55.Feng Y., Chen L., Luo Q., Wu M., Chen Y., Shi X. Involvement of microRNA-146a in diabetic peripheral neuropathy through the regulation of inflammation. 2018;Volume 12:171–177. doi: 10.2147/DDDT.S157109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X. S., Fan B., Szalad A., et al. MicroRNA-146a mimics reduce the peripheral neuropathy in type 2 diabetic mice. 2017;66(12):3111–3121. doi: 10.2337/db16-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun B., Anderegg A., Menichella D., Wrabetz L., Feltri M. L., Awatramani R. MicroRNA-deficient Schwann cells display congenital hypomyelination. 2010;30(22):7722–7728. doi: 10.1523/JNEUROSCI.0876-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verrier J. D., Semple-Rowland S., Madorsky I., Papin J. E., Notterpek L. Reduction of dicer impairs Schwann cell differentiation and myelination. 2010;88(12):2558–2568. doi: 10.1002/jnr.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gökbuget D., Pereira J. A., Bachofner S., et al. The Lin28/let-7 axis is critical for myelination in the peripheral nervous system. 2015;6, article 8584 doi: 10.1038/ncomms9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Junker A., Krumbholz M., Eisele S., et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. 2009;132(12):3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 61.Qiao Y., Ma C., Nie S., et al. Percutaneous treatment with drug-eluting stent vs bypass surgery in patients suffering from chronic stable angina with multivessel disease involving significant proximal stenosis in left anterior descending artery. 2009;73(10):1848–1855. doi: 10.1253/circj.CJ-08-1060. [DOI] [PubMed] [Google Scholar]
- 62.Keller A., Leidinger P., Lange J., et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. 2009;4(10) doi: 10.1371/journal.pone.0007440.e7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Otaegui D., Baranzini S. E., Armañanzas R., et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. 2009;4(7) doi: 10.1371/journal.pone.0006309.e6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hughes R. A. C., Hadden R. D. M., Gregson N. A., Smith K. J. Pathogenesis of Guillain-Barre syndrome. 1999;100(1-2):74–97. doi: 10.1016/S0165-5728(99)00195-2. [DOI] [PubMed] [Google Scholar]
- 65.Lv Z., Shi Q., Huang W., et al. MicroRNA expression profiling in Guillain-Barré syndrome. 2016;301:12–15. doi: 10.1016/j.jneuroim.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y.-Z., Feng X.-G., Shi Q.-G., et al. Silencing of miR155 promotes the production of inflammatory mediators in guillain-barré syndrome in vitro. 2013;36(2):337–345. doi: 10.1007/s10753-012-9551-5. [DOI] [PubMed] [Google Scholar]
- 67.Jung M., Schaefer A., Steiner I., et al. Robust MicroRNA stability in degraded RNA preparations from human tissue and cell samples. 2010;56(6):998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]