Summary
Johnston’s organ is the largest mechanosensory organ in Drosophila. It contributes to hearing, touch, vestibular sensing, proprioception, and wind sensing. In this study, we used in vivo 2-photon calcium imaging and unsupervised image segmentation to map the tuning properties of Johnston’s organ neurons (JONs), at the site where their axons enter the brain. We then applied the same methodology to study two key brain regions that process signals from JONs: the antennal mechanosensory and motor center (AMMC) and the wedge, which is downstream from the AMMC. First, we identified a diversity of JON response types which tile frequency space and form a rough tonotopic map. Some JON response types are direction-selective; others are specialized to encode amplitude modulations over a specific range (dynamic range fractionation). Next, we discovered that both the AMMC and the wedge contain a tonotopic map, with a significant increase in tonotopy – and a narrowing of frequency tuning – at the level of the wedge. Whereas the AMMC tonotopic map is unilateral, the wedge tonotopic map is bilateral. Finally, we identified a subregion of the AMMC/wedge that responds preferentially to the coherent rotation of the two mechanical organs in the same angular direction, indicative of oriented steady air flow (directional wind). Together, these maps reveal the broad organization of the primary and secondary mechanosensory regions of the brain. They provide a framework for future efforts to identify the specific cell types and mechanisms that underlie the hierarchical re-mapping of mechanosensory information in this system.
eTOC blurb
Patella and Wilson show that the primary and secondary mechanosensory regions of the Drosophila brain contain stereotyped and orderly maps of vibration frequency, with different subregions representing distinct features of motion amplitude and phase (direction).
Introduction
Mechanosensation begins with specialized receptor cells that convert mechanical forces into electrical signals. A current challenge is to understand what happens next, as receptor cell signals are translated into patterns of activity in the central nervous system. In vertebrates, the relevant regions of the spinal cord and brainstem are difficult to access, especially in awake organisms. Therefore, organisms with more accessible nervous systems can be especially useful in the study of mechanosensory processing.
In particular, studies of mechanosensory processing in insects have yielded a number of important findings [1–5]. For example, recent studies in crickets have revealed neural mechanisms of acoustic template matching [6, 7]. Recent studies in stick insects have provided insight into the mechanisms of proprioceptive feedback during tactile exploration [8, 9] and the roles of mechanosensory reflexes in voluntary limb movements [10].
The most genetically-tractable insect is currently Drosophila melanogaster. As such, Drosophila has emerged as a useful model for studying the cellular and network mechanisms of mechanosensory processing [5, 11, 12]. The largest mechanosensory organ in Drosophila is Johnston’s organ, which is situated inside the antenna (Figure 1A). This organ contains an array of mechanosensory neurons termed Johnston’s organ neurons [JONs; 13]. JON dendrites are stretched and compressed as the distal antennal segment rotates [14]. The resulting changes in JON firing patterns are conveyed via the antennal nerve into the brain. Experimentally manipulating JONs perturbs a variety of behaviors, including acoustic startle reflexes [15], auditory conditioning [16], social behaviors evoked by courtship song [17–19], locomotor responses to wind [18, 20–22], vestibular sensing [17], somatosensation [23], and proprioceptive feedback during flight [24, 25].
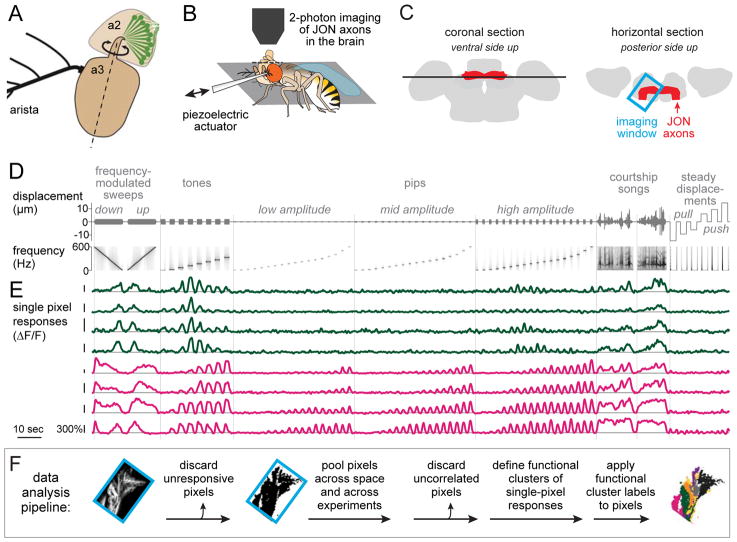
Figure 1. Pipeline for generating functional maps of JON response types.
A. Johnston’s organ neurons (JONs) reside in the second segment of the antenna (a2) and sense rotations of the distal antennal segment (a3) with respect to a2. Dashed line indicates approximate axis of rotation.
B. Experimental setup. The head is rotated 180°, and the proboscis is removed to expose the brain’s ventral side. The displacement of the distal antennal segment is controlled by a piezoelectric actuator attached to the arista.
C. Schematics of the brain showing an optical z-section (black line, left) and an x-y imaging window (blue box, right). JON axons (red) enter from the anterior face of the brain and then bend medially to arborize throughout the brain region called the antennal mechanosensory and motor center (AMMC).
D. Mechanical stimuli, shown in terms of actuator displacement (top) and actuator frequency (bottom). Zero is the normal resting position of the antenna. Positive displacements push the antenna toward the head; negative displacements pull it from the head. For each pixel, responses to different randomly-interleaved trials of the same stimulus are averaged and then concatenated in a fixed sequence.
E. Responses of 8 example pixels, imaged in flies where GCaMPf was expressed in all JONs. Pixels are color-coded according to their functional type, either type vii (dark green) or type x (magenta), as in Figure 2 and Figure 3. Vertical scale bars are 300% ΔF/F.
F. Data analysis pipeline. Image at left shows GCaMP6f-expressing JON axons. Imaging window is oriented as in (C).
Calcium imaging has suggested that there are several JON types, each with distinct tuning properties [17, 18, 26, 27]. However, this may be an underestimate of JON diversity, given that there are almost 500 neurons in Johnston’s organ [13]. Interestingly, neurons homologous to JONs are found in leg mechanosensory organs, and these cells show tremendous physiological diversity [28–31].
JON axons mainly terminate in a brain region termed the antennal mechanosensory and motor center (AMMC). The AMMC contains a range of cell types that respond selectively to specific features of mechanical stimuli [15, 17, 32–38]. However, no studies have undertaken systematic comparisons between JONs and AMMC neurons. Also little is known about the next stage of processing, beyond the AMMC. One region in particular (termed the wedge, or WED, because of its distinctive shape) is a major target of the AMMC, and it receives input from a particularly large diversity of morphological cell types [37], however, its functional organization is largely unexplored.
In this study, our goal was to visualize mechanosensory feature maps in the brain. In order to survey all JON types simultaneously, we used pan-JON calcium imaging; in order to survey all CNS cell types simultaneously, we used pan-neuronal calcium imaging, with the calcium indicator excluded from JONs. Imaging in vivo while delivering a large panel of mechanical stimuli, we found widespread and diverse responses at the level of JONs, the AMMC, and the WED. We used an unsupervised image analysis algorithm to discover a large set of reliably-localized functional response types. These functional maps reveal the broad organization of the primary and secondary mechanosensory regions of the brain. They should guide future work focused on the characterization of specific cell types and the mechanosensory computations they implement.
Results
Mapping peripheral mechanoreceptor responses
Our first goal was to map the representation of mechanical features in JONs. Specifically, we imaged JON axons at the site where they enter the brain and terminate in the AMMC (Figure 1C). We chose to image JON axons – rather than JON somata – in order to directly compare the spatial maps of JON responses with the spatial maps of postsynaptic responses (in AMMC neurons).
In order to displace the antenna in a controlled manner, we attached a piezoelectric actuator to the arista, the rigid branching structure which protrudes from the antenna (Figure 1A,B). We delivered mechanical waveforms that included frequency-modulated (FM) sweeps, narrowband vibrations (“tones”), sustained displacements, and courtship songs (Figure 1D). We used stimulus amplitudes (225 nm - 14 μm) that captured the physiological range of displacements evoked by natural stimuli; these stimuli range from barely-audible sound [15] to high-speed wind [18]. We used frequencies (0 to 600 Hz) that captured the physiological range of antennal movement frequencies [14].
In order to image activity in all JONs simultaneously, we drove expression of GCaMP6f [39] using the pan-JON driver line nan-Gal4 [40]. In separate experiments, we expressed GCaMP6f in subsets of JONs using many sparse and diverse Gal4 lines [13]. Single-pixel responses (Figure 1E) from all genotypes were analyzed together.
The first step in our data analysis pipeline (Figure 1F) was to discard unresponsive pixels. We then pooled pixels across all z-planes, all brains, and all genotypes. We discarded pixels that were functionally uncorrelated with other pixels (i.e., they contained mainly noise), yielding a 102,669×3,549 matrix (pixels×time). We then categorized pixels into response types based on their functional similarities (Figure 2A,B). Finally, pixel categories were projected back into spatial coordinates, allowing us to visualize functional maps in the brain (Figure 2C).
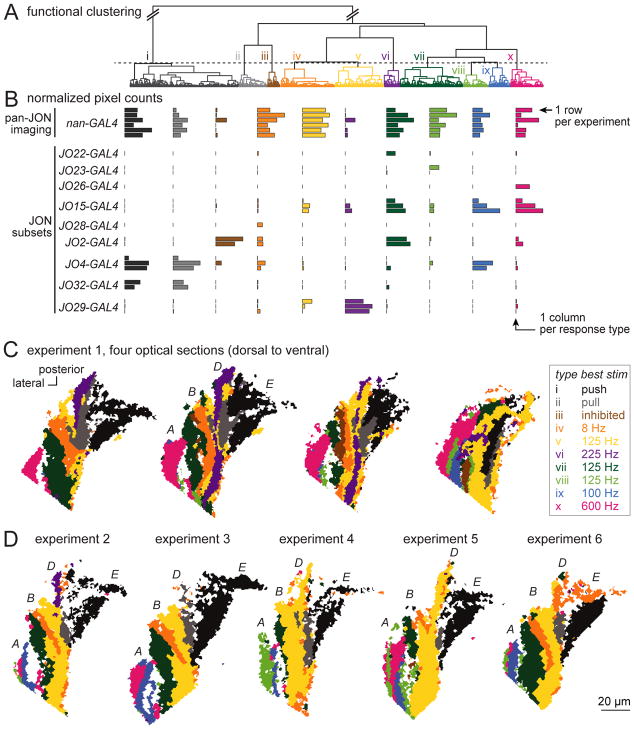
Figure 2. Functional clustering and spatial mapping of JON responses.
A. Functional clustering of the JON dataset. Data were pooled from all pan-JON imaging experiments (using nan-Gal4) and also all experiments imaging from small JON subsets (using the specific Gal4 lines at left). Pixels were hierarchically clustered using Ward’s method. Then, pixels were divided into functional types (i – x) using a fixed threshold (dashed line).
B. Each row is a different experiment, and each column is a different response type. Each bar represents the number of pixels belonging to that type (at the z-level where that type is best represented, normalized to the maximum for that type across brains).
C. Maps of JON response types in the AMMC in one representative experiment. Shown are four optical sections from one experiment in a brain where all JONs expressed GCaMP6f (under nan-Gal4 control). Note that the long axis of each color patch is typically oriented along the anterior-posterior axis, parallel to the long axis of the antennal nerve, as we would expect if each functional type represents a distinct group of JON axons. The four consistently-identifiable major zones of the axon bundle (A, B, D, and E) are labeled in the second map, following the scheme of Kamikouchi et al. (2006). Some functional types are found in more than one zone, consistent with the fact that some JON axons branch and innervate multiple zones [13].
D. Maps from five additional pan-JON imaging experiments, with one optical section shown for each experiment (corresponding to the z-level of the second map in experiment 1).
This approach offers three advantages over traditional image analysis methods. First, we make no assumptions about what mechanical stimulus features might define the functional differences between types. Discovering the relevant mechanical stimulus features was one of our goals. Second, “types” are defined in purely functional terms; the algorithm that categorizes pixels into types has no information about the pixel’s spatial location. Thus, this approach allows us to discover the spatial organization of response types in an unbiased manner. Third, the algorithm is blind to the identity of the brain that contained each pixel. In truth, however, we expect that all brains would contain similar response types. Therefore, we can assess cross-brain similarity as a check on the plausibility of the algorithm’s output.
Using this unsupervised approach, we identified 10 JON response types in total. As expected, each response type was consistently identifiable in every pan-JON imaging experiment (Figure 2B) at a fairly consistent spatial location (Figure 2C,D). There were two exceptions (types iii and vi), which were identifiable in only a subset of pan-JON experiments. However, both types emerged again in sparse genotypes (see below), which supports their validity. If we lowered the threshold for dividing pixels into response types, thereby splitting pixels into finer categories, we found that cross-brain consistency was worse (Figure S1). Thus, the JON response types we have defined are the most finely-split types that are consistently resolvable.
In principle, the response types in the sparse genotypes should simply be subsets of the response types in the broad genotype. This is indeed what we observed (Figure 2B). Moreover, the spatial location of each response type in the sparse genotypes was consistent with its location in pan-JON imaging experiments (Figure S2).
Frequency selectivity and tonotopy in JONs
Two JON response types responded best to steady antennal displacements (Figure 3A, types i and ii). These types responded only weakly to antennal vibrations (Figure 3B,C). Type i was excited when the antenna was pushed toward the head, while type ii was excited when the antenna was pulled away from the head. Both types were localized to the medial edge of the JON axon bundle (Figure 3D). Push responses were medial to pull responses (Figure S3), in agreement with a previous report [18].
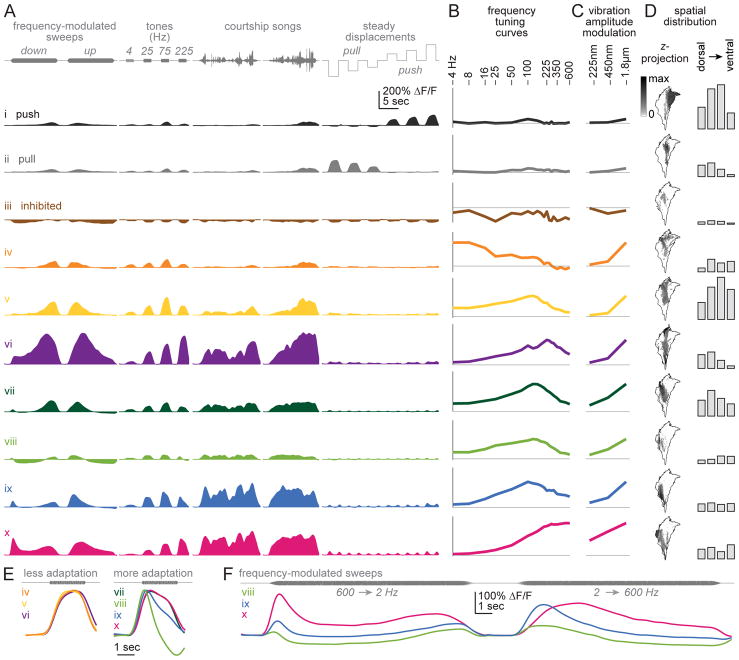
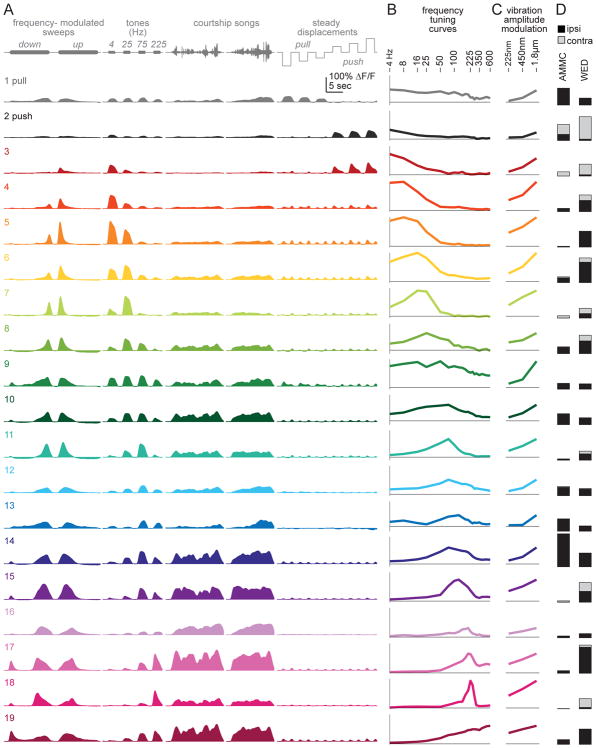
Figure 3. JON response types.
A. Responses to select stimuli for each JON response type, averaged across pixels (24 brains, 102,669 total pixels). Response types are ordered as in the dendrogram in Figure 2A. Types i and ii prefer steady displacements (push/pull). Type iii is inhibited by all stimuli. The remaining types are excited by specific vibration frequencies, and are color-coded according to their center frequency (the center-of-mass of the frequency tuning curve; Figure S4).
B. Vibration frequency tuning curves. Vibration amplitude is 1.800 μm (peak-to-mean). Peak ΔF/F is measured over a 500-msec time window, beginning 100-msec after vibration onset. Horizontal line is zero ΔF/F. The y-axis is scaled in each case to represent 65% of the maximum range of modulation within each type, across all stimuli.
C. Vibration amplitude modulation curves. Shown as in (B) but measured at three stimulus amplitudes, at the best frequency for each type.
D. Left: maps of each type at a z-level near the center of the JON axon bundle. The outline shows the envelope of all registered responsive pixels. Maps are oriented as in Figure 1B. Each gray scale map represents the proportion of responsive pixels assigned to the corresponding type at a particular z-level (second from left in Figure 2C); note that this gray scale is nonlinear to make intermediate values more visible. Right: bar graphs show the number of pixels at four z-levels, pooled across brains.
E. Responses to a sustained 225 Hz tone, enlarged from (A) and normalized to each curve’s maximum.
F. Responses to FM sweeps in select JON types, enlarged from (A). In these JON types, preferred frequency depends strongly on sweep direction. In other words, inverting the direction of the sweep does not invert the response.
Another JON response type (iii) was inhibited by all stimuli (Figure 3A–C). This response type was always localized to the middle of the JON axon bundle (Figure 3D). In pan-JON imaging experiments, it was not always visible, but it was consistently present in a specific Gal4 line (Figure 2B, Figure S2).
All the other JON response types were excited mainly by vibrations (types iv – x). Their best frequencies spanned a wide range, from 4 Hz to 600 Hz (Figure 3A,B). These response types were arranged in a loosely tonotopic spatial order (Figure 2D and Figure 3D), with the representation of high vibration frequencies (>300 Hz) positioned laterally and the representation of lower frequencies more medially. Most medial of all were the JON response types devoted to steady displacements of the antenna (push/pull) – in other words, DC stimuli (~0 Hz). Our results are consistent with previous reports that lateral JON axons prefer high frequencies, while medial JON axons prefer lower frequencies [18, 26, 27].
It is important to note that courtship songs recruit all these JON types (Figure 3A). Thus, there are no specialized “courtship song detectors” at the level of JONs. This is perhaps not surprising, as courtship song is a spectrally broadband stimulus [11].
Amplitude sensitivity and adaptation in JONs
JON response types differed in the range of stimulus amplitudes they were sensitive to. Notably, one type responded robustly to the lowest amplitude we tested (type x, Figure 3C), but increasing stimulus amplitude 8-fold produced only a 2-fold response increase. Thus, this type was nearly saturated at low amplitudes. By contrast, the other vibration-preferring JON types were less sensitive to low amplitudes, but their responses were also less saturated (types iv – ix).
JON response types also differed in their adaptation properties. On one hand, some types showed adaptation to sustained high-frequency vibrations (Figure 3E). Most of these types also showed adaptation during FM sweeps, so their apparent preferred frequency was different during down- and up-modulations (Figure 3F). These response types are likely responsible for the observed adaptation in JON field potentials evoked by sustained vibrations [41, 42]. On the other hand, we found that other JON response types showed little or no adaptation (Figure 3E). These less-adapting types were positioned medially to the more-adapting types (Figure 3D).
In summary, our results indicate considerable specialization among JON response types. Each encodes signals over a specific spectral band. Thus, we can view JONs as a parallel array of bandpass filters, with center frequencies ranging from ~DC (types i–ii) to ≥600 Hz (type x). Some channels are directionally-tuned, meaning that they are sensitive to the phase of antennal motion (push/pull). Moreover, different channels carry amplitude information over distinct dynamic ranges. Strongly-adapting channels should prefer repeated brief bursts of stimulus energy, whereas less-adapting channels should prefer prolonged stimuli.
Mapping mechanosensory responses in central neurons
Our next goal was to map mechanosensory responses in brain regions downstream from JONs in the central nervous system (CNS). In order to image signals from all neurons that are not JONs, we drove GCaMP6f expression using the pan-neuronal Gal4 line nSyb-Gal4 [43] while repressing expression in JONs using pan-JON Gal80. As an alternative approach, in separate experiments, we labeled JONs with a red fluorescent marker (tdTomato) and we discarded the labeled pixels during data analysis. The analyses described below combine data from the two genotypes.
In pilot experiments, we found that reliable responses were generally localized to two brain regions, the AMMC and WED (Figure 4A and 4B). Mechanosensory responses certainly exist outside these regions [35, 44, 45], but they might be undetectable if co-tuned neurons are not co-localized. Also, these responses might depend on behavioral state. Finally, responses in the dorsal part of the brain might be difficult to detect in our experiments, because we chose to image the brain ventral-side up.
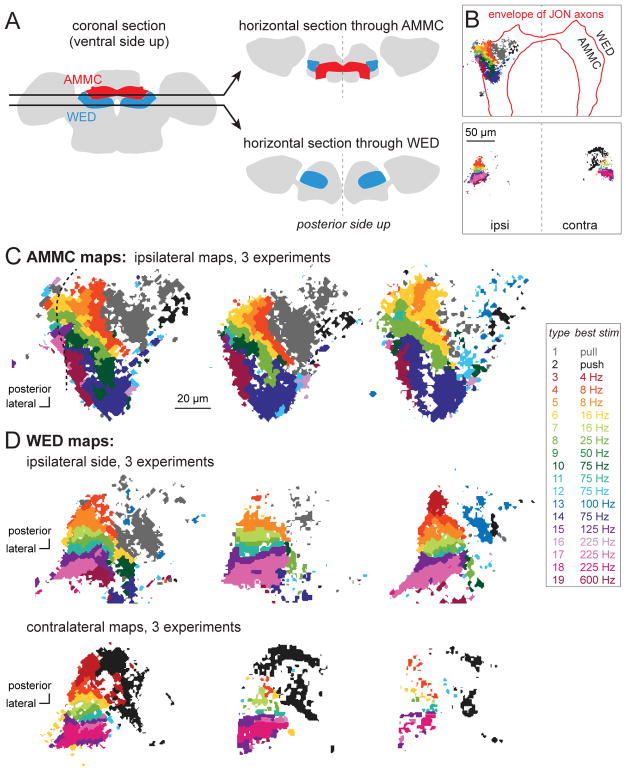
Figure 4. Imaging responses of CNS neurons.
A. Schematic optical sections intersecting the AMMC and WED.
B. Examples of maps at these two z-levels. Pixels that responded to mechanosensory stimuli are color-coded as below. GCaMP6f is expressed under the control of a pan-neuronal promoter, with expression suppressed in JONs (using iav-Gal80). The approximate envelope of JON axons is indicated by a red boundary. Top: most responsive pixels reside in the AMMC (defined as the region inside the red boundary). A few responsive pixels reside in the WED (lateral to the red boundary). Here only the ipsilateral side was imaged. Bottom: at this z-level, the WED was imaged on both ipsi- and contralateral to the stimulated antenna.
C. Maps at the level of the AMMC (3 representative brains). In the case on the left (same as in B), some pixels on the lateral-posterior edge of the map reside outside the boundary of the AMMC; note the discontinuity in the functional map at this location (dashed line).
D. Maps at the level of the WED, imaged on the ipsilateral side (top) or contralateral side (bottom; 3 representative brains each). Contralateral maps are reflected so that lateral is always to the left.
In each experiment, we imaged multiple horizontal planes through the AMMC and WED. We then pooled pixels from AMMC and WED, in all z-planes, and all brains. We used the same data analysis pipeline we devised for the JON data to divide pixels into types.
In total, we identified 19 response types in the CNS. Each response type was found in a fairly consistent location in almost every experiment (Figure 4C,D). If we lowered the threshold for dividing pixels into response types, cross-brain consistency became notably worse. Thus, the 19 response types we define in the CNS are the most finely-split types we can consistently resolve.
Some response types were well-represented in both the AMMC and WED (e.g., type 1). Other types were almost exclusively found in the WED (e.g., type 18). We did not find response types that were exclusive to the AMMC. Figure 5 shows the representation of each type in the AMMC and WED.
Figure 5. CNS response types.
A. Responses to select stimuli for each type, averaged across all pixels belonging to that type (11 brains, 49,608 total pixels). Types are sorted by their center frequency and colored accordingly (Figure S4).
B. Vibration frequency tuning curves. Vibration amplitude is 1.800 μm (peak-to-mean). Peak ΔF/F is measured over a 500-msec time window, beginning 100-msec after vibration onset. The y-axis represents the full range of modulation within each type, across all stimuli.
C. Amplitude modulation curves. Shown as in (B) but measured at three stimulus amplitudes, at the best frequency for each type (Figure S4).
D. Number of pixels belonging to each type at four z-levels, averaged across brains, in the AMMC and WED.
General organization of maps in the AMMC and WED
We found some CNS response types that preferred steady displacements, and other response types that preferred vibrations (Figure 5A,B). Steady displacements were preferred in the medial AMMC and WED, whereas vibrations were preferred in the lateral AMMC and WED (Figure 4C,D). This roughly mirrors the functional divisions of the JON axon bundle.
Within these broad sectors – medial versus lateral – we found multiple functional subregions. The medial AMMC and WED contained subregions that preferred either “push” or “pull” displacements (Figure 4 and Figure 5). Meanwhile, the lateral AMMC and WED contained subregions that preferred different vibration frequencies (Figure 4 and Figure 5). Courtship songs recruited most subregions of the AMMC and WED (Figure 5A).
Frequency selectivity and tonotopy in the AMMC and WED
These data allowed us to compare vibration frequency representations in JONs, the AMMC, and the WED. First, in comparing JONs with the AMMC, we found a shift toward an over-representation of low frequencies in the AMMC. Second, in comparing the AMMC with the WED, we found a shift toward an over-representation of extreme frequencies (both low and high) in the WED (Figure 6A).
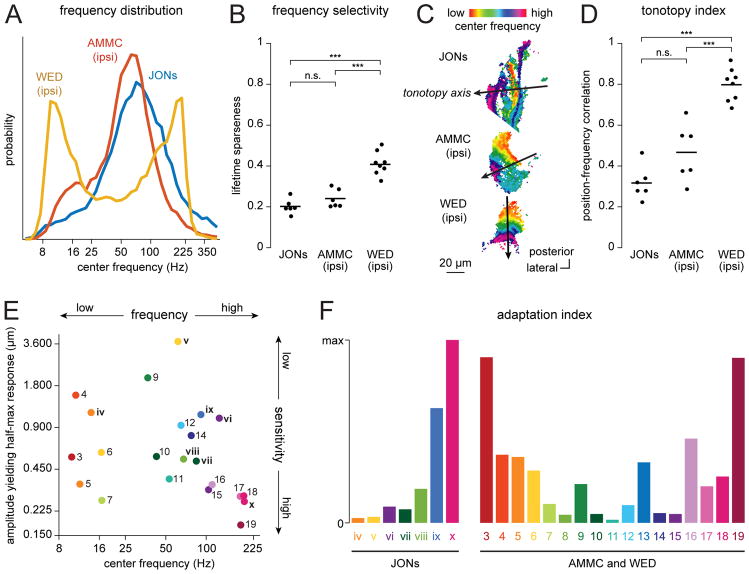
Figure 6. Comparing the vibration-preferring regions in JON, AMMC and WED maps.
A. The distribution of center frequencies of single pixels, aggregated across experiments. The “center frequency” is the center of mass of the frequency tuning curve (Figure S4). Here (and throughout this figure) only vibration-preferring response types are included.
B. Frequency selectivity, defined as the mean lifetime sparseness of all the frequency tuning curves in each map. Each point is a different experiment; lines are averages. One-way ANOVA: p < 0.0001. Tukey-Kramer’s post hoc test (two-sided): ***p<0.0001, n.s. p ≥ 0.05.
C. Representative maps of center frequencies in JONs, AMMC, and WED. The line on each map represents the tonotopy axis. Examples correspond to the second map in Figure 2C (JONs), the second map in Figure 4C (AMMC), and the first map in Figure 4D (ipsilateral WED). All maps are shown in the same orientation.
D. Tonotopy index, defined as the strength of the relationship between center frequency and tonotopy-axis position. Each point is a different experiment; lines are averages (n=6 JON experiments, 6 AMMC experiments, 8 WED experiments). One-way ANOVA: p < 0.0001. Tukey-Kramer’s post hoc test (two-sided): ***p<0.0001, n.s. p ≥ 0.05.
E. Sensitivity versus center frequency, for all response types. The y-axis represents the vibration amplitude eliciting a half-maximum response (which is inversely related to sensitivity). Note that there are high- and low-sensitivity subregions devoted to both high and low vibration frequencies.
F. Adaptation index for each response type. If up- and down-modulated frequency sweeps elicit identical mirror-image responses, the adaptation index is zero. If these responses are very different from mirror images of each other, the adaptation index is large. STAR Methods provides details on the adaptation index, as well as other analyses in this figure.
In addition, we found that overall frequency tuning of individual response types was significantly narrower in the WED than in the AMMC or JONs (Figure 6B). This could arise if individual WED neurons are more narrowly tuned than AMMC neurons or JONs. Alternatively, there might be no difference in frequency selectivity at the single-cell level, but instead WED neurons could simply be more systematically positioned according to frequency. Either way, responses to different frequencies are more spatially segregated in the WED neuropil.
Notably, the tonotopic organization of WED maps was more orderly than that of AMMC and JON maps. To quantify this observation, we identified the main axis of tonotopic organization in each map (Figure 6C). We then projected every pixel position onto that axis. The relationship between axis position and frequency preference is a metric of tonotopic order. This metric was indeed significantly higher in the WED than in the AMMC or JONs (Figure 6D). In other words, there was a significantly stronger relationship between linear position and frequency tuning in this region.
Amplitude sensitivity and adaptation in the AMMC and WED
Overall, the amplitude sensitivity of the AMMC and WED was comparable to that of JONs. Some AMMC and WED response types were particularly sensitive to low-amplitude stimuli. Others were only recruited by high-amplitude stimuli (Figure 5). We found high- and low-sensitivity response types devoted to both high and low vibration frequencies (Figure 6E).
Some AMMC and WED subregions showed strong adaptation, whereas others did not (Figure 5A). Strong adaptation was present in high-frequency-preferring subregions, as well as low-frequency preferring subregions (Figure 6F). This contrasts with the situation in JONs, where only high-frequency-preferring subregions showed strong adaptation.
Unilateral and bilateral maps in the AMMC and WED
The vibration-preferring subregions of the AMMC responded mainly to the ipsilateral antenna, with little or no response to vibrating the contralateral antenna. The situation in the WED was different: here we found consistent and widespread responses to both vibrating either the ipsilateral antenna or the contralateral antenna (Figure 5D). In the WED, the tonotopic maps evoked by ipsi- and contralateral stimuli were similar (Figure 4D). This indicates that each strip in the WED tonotopic map integrates input from similarly-tuned vibration-sensitive channels on the right and left.
Non-vibration-preferring subregions could also show bilateral responses. In particular, responses to contralateral stimulation were found in the subregion recruited by steady ipsilateral “pull”. This subregion is labeled gray in the AMMC maps and the ipsilateral WED maps (Figure 4C and 4D). It straddles the dorsal-ventral division between the AMMC and WED, forming one continuous subregion. Interestingly, the contralateral stimulus that recruited this subregion was not “pull” but “push”. Thus, in the same spatial subregion, the contralateral WED map appears black rather than gray (Figure 4C,D). We wondered whether this subregion might respond best if we presented both of these stimuli simultaneously – i.e., if we pulled the ipsilateral antenna while also pushing the contralateral antenna. Therefore, we set out to test this idea next.
Stimulating both mechanoreceptor organs simultaneously
To pull the ipsilateral antenna (away from the head) while pushing the contralateral antenna (toward the head), we used steady wind directed at the ipsilateral side of the head (Figure 7A, Figure S5). Flipping the orientation of the wind (from ipsi- to contralateral) inverted this pattern of antennal displacements. Next, we found we could emulate a tailwind by delivering ipsi- and contralateral wind at the same time; this pulled both antennae away from the head. Finally, a headwind pushed both antennae toward the head. Thus, we could switch each antenna between two states, yielding four stimulus configurations (Figure 7A, Figure S5).
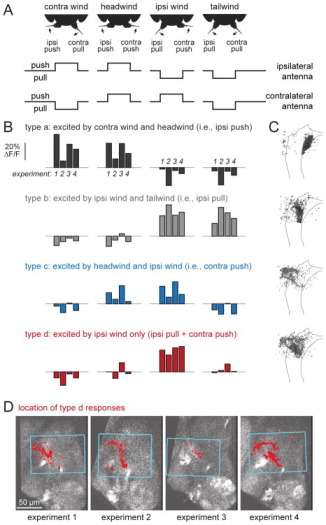
Figure 7. CNS responses to displacement of both antennae (wind stimuli).
A. Four patterns of bilateral antennal displacement. Headwind pushes both antennae toward the head. Tailwind pulls both antennae away from the head. Ipsi wind pulls the ipsi antenna and pushes the contra antenna, while contra wind does the reverse. In all our experiments, ipsi/contra are defined relative to the imaged side; the schematics here show ipsi relative to one side only, but in fact both sides were imaged in all flies and produced comparable results.
B. When CNS pixels were divided into types based on their responses to these stimuli, we found four response types that were reliably localized to discrete locations (Figure S5). The first three (types a–c) responded mainly to moving one antenna in a particular direction, and so had similar responses to two of the four stimuli. The last type (type d) responded selectively to pulling the ipsi antenna while also pushing the contra antenna (i.e., the pattern of antennal displacement produced by ipsi wind). The blue box indicates the x-y plane where imaging was performed during wind stimulus presentation.
C. Spatial distribution of these response types, as in Figure 5D projected across z-levels into one horizontal section. The outline delineates the envelope of JON axons, registered across brains, at an intermediate z-level. Gray scale represents the proportion of responsive pixels assigned to the corresponding type. Posterior is up and lateral is left. Type d responses are located dorsal to the other types, and are mainly in the WED (which wraps around the dorsal part of the AMMC).
D. Maps of type d responses from four experiments at a representative z-plane. Posterior is up, lateral is left. Note the fairly consistent location of this response type.
We then imaged responses to these stimuli in the CNS. Again, we expressed GCaMP6f pan-neuronally, while excluding GCaMP6f from JONs. Using our standard data analysis pipeline (see above), we identified several distinct CNS response types. Four types were reliably localized to discrete locations in the brain, and had robust responses to at least one stimulus (Figure S5); we therefore focused on these four subregions.
Among the four wind-responsive subregions we identified, three were sensitive to one antenna only. The first (a) was activated whenever the ipsilateral antenna was pushed, regardless of the state of the contralateral antenna (Figure 7B,C). This subregion mapped to the ipsilateral push zone we identified previously (Figure 4C,D). The second (b) was activated whenever the ipsilateral antenna was pulled, again regardless of the state of the contralateral antenna. This mapped to the ipsilateral pull zone we identified previously. The third (c) responded whenever the contralateral antenna was pushed, regardless of the state of the ipsilateral antenna. This mapped to the contralateral push zone we identified previously.
Interestingly, the fourth subregion (d) was bilateral, meaning that its responses depended on the state of both antennae. This subregion was activated only when the ipsilateral antenna was pulled and the contralateral antenna was pushed (Figure 7B). In other words, it responded only to ipsilateral wind. None of the other wind stimuli produced large or reliable responses. This subregion overlapped with the AMMC/WED border, but it resided mainly in the WED (Figures 7C and 7D), dorsal to the other subregions and positioned at the approximate location we had predicted on the basis of our unilateral stimulus experiments.
In summary, using an independent data set, we confirmed our previous identification of several subregions that detect unilateral antennal displacements; we also identified an additional subregion that detects a particular bilateral antennal configuration. All these subregions detect steady displacements, but they detect different features of those displacements. In short, the medial WED/AMMC contains a detailed representation of the antenna’s steady positional state. This complements the equally detailed representation of the antenna’s movement frequency in the lateral WED/AMMC.
Discussion
Advantages and limitations of neuropil calcium imaging
In this study, we imaged activity in specific neuropil regions in the Drosophila brain. Our focus on neuropil – rather than neural somata – may require explanation for readers unfamiliar with Drosophila neuroanatomy. Briefly, most of the Drosophila brain volume is exclusively neuropil – i.e., axons and dendrites. Axons and dendrites with similar tuning properties are often co-localized, and so pan-neuronal GCaMP imaging can reveal orderly maps of sensory stimulus features in Drosophila brain neuropil [46, 47]. By contrast, neural somata are excluded from the neuropil, and are instead confined to a thin “rind” around the neuropil core; there is no simple rule that relates the location of a neuron’s soma to the location of its axon and dendrites. Thus, similarly-tuned cells often have dissimilar soma locations, and a sensory stimulus typically evokes activity in widely-dispersed somata on the surface of the brain [48]. Our goal was to visualize maps of mechanosensory stimulus features in the brain, and so neuropil imaging was an appropriate choice.
One limitation of neuropil imaging (as compared to somatic imaging) is that we are measuring signals from groups of neurons, not single neurons. Cell-type diversity may disappear due to optical mixing. Alternatively, we might actually overestimate diversity. Consider a hypothetical scenario with only two cell types, occurring in 10 different ratios in 10 different neuropil subregions; our algorithm would identify 10 “response types” because of optical mixing. We can be reasonably certain that we are not overestimating diversity because of optical mixing, because sparse Gal4 lines (where only scattered cells are labeled) collectively revealed the same sort of diversity we saw with a broad Gal4 line (Figure 2A). It seems more likely that we are underestimating diversity, rather than overestimating it.
Calcium imaging also has limited temporal resolution. This is especially relevant to mechanosensation, which is the fastest known sensory modality. Both JONs and AMMC neurons can phase-lock to vibrations as fast as 500 Hz [38, 41]. GCaMP6f signals will reveal only the amplitude modulation envelope of these responses, not their fine structure.
Fundamental features of mechanical stimuli
All the stimuli transduced by Johnston’s organ can be reduced to a single variable – namely, the rotational angle of the distal antennal segment – and its evolution over time. This single variable fully describes the rich content of the natural stimuli that impinge on Johnston’s organ. These include stimuli as diverse as courtship song [19], wind [18, 20, 21], self-generated wingbeat patterns [24, 25], vestibular cues [17], and tactile stimuli [23]. In much the same way, the rich content of human speech and music is also described by a single variable, i.e. the position of our tympanal membrane.
Any one-dimensional time-varying signal can be described in terms of three fundamental features: frequency, amplitude, and phase. Our results show that mechanosensory neurons in this system show specializations for encoding all three of these fundamental features. Collectively, they divide up frequency space, amplitude space, and phase space. Below we discuss each of these features in turn.
Frequency selectivity and tonotopic maps
Almost all of the neural response types we identified were tuned to frequency. Moreover, frequency tuning was consistently related to spatial position. In other words, we found tonotopy at every level of this system, from JONs to AMMC to WED.
Tonotopy may be useful because it allows co-tuned neurons to interact with each other using a minimal expenditure of “wire”. This arrangement should maximize speed and minimize metabolic costs [49]. Tonotopic maps are a prominent feature of vertebrate auditory systems, in peripheral cells and CNS neurons [50]. In insects, tonotopic maps have been described in peripheral cells [51–53], but there is less evidence for tonotopy in CNS neurons. Coarse tonotopy has been reported in some CNS neurons [54, 55], but in other cases there is a lack of tonotopy [56]. Indeed, it has been proposed that the insect CNS generally discards the tonotopic organization of the periphery [3, 57]. Surprisingly, we find that tonotopy is a prominent feature of the AMMC and WED in the Drosophila brain, suggesting there may be more functional similarity than previously suspected in the auditory systems of insects and vertebrates.
The prominence of tonotopy in the AMMC and WED – and the narrowness of frequency tuning in the WED – is also surprising for yet another reason: spectral cues are reportedly irrelevant for determining behavioral responses to courtship song in Drosophila [58–60]. However, courtship behaviors are not the only behaviors that depend on Johnston’s organ. Spectral cues may be important for other Drosophila behaviors that are much less well-studied – e.g., suppression of locomotion by turbulent wind [18], flight steering maneuvers [24], or defensive reactions to predator sounds [61].
Encoding amplitude over a wide dynamic range
We found that different neural channels were specialized to encode stimulus amplitude over different ranges. At one extreme, some channels responded to vibrations as small as 225 nm (Figure 3 and Figure 5). This is close to the smallest vibration amplitude that elicits a detectable electrophysiological [15, 38, 62] or behavioral [16] response.
Interestingly, the most sensitive coding channels were already approaching saturation at low stimulus amplitudes (Figure 3 and Figure 5). These channels should be relatively insensitive to amplitude modulations at high amplitudes. Indeed, when the stimulus is the sound of the fly’s own flight (which is a loud buzz, from the fly’s perspective), high-sensitivity JONs are saturated, and so cannot follow the sound amplitude modulation envelope [25]. This illustrates the need for low-sensitivity channels as well. Accordingly, we found low-sensitivity coding channels which did not saturate at high stimulus amplitudes (Figure 3 and Figure 5). Together, these findings illustrate the principle of “dynamic range fractionation”: different stimulus intensity ranges are allocated to different coding channels. This principle applies to many peripheral sensory cells, including insect auditory receptors [51, 63, 64] and proprioceptors [29].
In other sensory systems, distinct cell types comprise low- and high-threshold subtypes. For example, in the vertebrate somatosensory system, each patch of skin contains low- and high-threshold afferents [65]. In the vertebrate auditory system, each frequency band contains low- and high-threshold auditory fibers [66]. Similarly, in the Drosophila brain, we find that each frequency band contains both high- and low-sensitivity channels (Figure 6E). In other words, these neural channels tile both frequency space and amplitude space.
Phase and direction
Phase is the third fundamental feature of time-varying signals. Like frequency and amplitude, phase is also represented systematically in JONs and downstream CNS neurons. The push/pull channels are a case in point. When the antenna is pushed and pulled, the push/pull channels respond ~180° out of phase (Figure 3 and Figure 5). This is equivalent to saying that these two channels have opposing preferred directions.
Direction-sensitivity has obvious utility in wind sensing. Walking flies use wind direction as a guidance cue [21, 22]. Johnston’s organ is a wind-sensing organ [18, 20], and so it is interesting to compare it with the best-studied insect wind-sensing organ, the cricket cercus. The mechanisms of direction-sensitivity are quite different in the cricket cercus and the fly antenna. The cercus is covered by tiny hairs. Due to the asymmetric structure of the hair socket, each hair has a preferred direction of movement, and wind from different directions will maximally deflect different hairs [67]. Each hair is innervated by one mechanoreceptor neuron which only spikes when the hair is pushed in its preferred direction. By contrast, in the wind-sensing system of Drosophila, there is a single mechanical receiver (i.e., the arista, which is rigidly coupled to the distal antennal segment); this stands in contrast to the many mechanical receivers on the cricket cercus (i.e., the many hairs which move independently). Whereas each receiver in the cricket cercus is innervated by a single neuron, the single receiver in Johnston’s organ is innervated by many neurons (JONs), with some cells having opposing responses to the same receiver movement. Thus, in the cercus peripheral complexity is mechanical, whereas in Johnston’s organ peripheral complexity is neural.
Direction-sensitivity is also potentially useful in sound sensing, because the relative phase (direction) of right and left antennal movements can carry information about the sound source location in the azimuth [68]. Thus direction-sensitivity may exist in vibration-preferring JONs. Indeed, direction-sensitivity does exist in certain vibration-preferring neurons in the AMMC that project to the WED [38]. However, in this study, we could not resolve direction-sensitivity in vibration-preferring JONs or AMMC/WED subregions, because GCaMP signals cannot fluctuate rapidly enough to capture any direction-sensitivity (phase preferences) in vibration responses.
Bilateral integration
Our study provides the first physiological evidence of bilateral integration downstream from Johnston’s organ. Notably, we found that each strip of the WED tonotopic map receives convergent input from both antennae. Within each strip, ipsi- and contralateral frequency preferences are matched.
One potential function of bilateral integration is sound localization. In vertebrates [50] and large insects [1–4], lateralized sound produces a detectable difference in the amplitude and/or timing of sound pressure cues at the two auditory organs. However, as body size decreases, sound pressure differences become difficult to resolve. For example, in the fly Ormia ochracea, the two auditory organs are only ~500 μm apart; this species has evolved mechanisms for amplifying left/right differences in sound pressure [69–71]. Drosophila melanogaster is even smaller than Ormia ochracea, meaning left/right differences in sound pressure are correspondingly smaller as well. Accordingly, Drosophila has evolved an auditory organ that does not sense sound pressure: instead, the distal antennal segment blows back and forth with air particle velocity fluctuations (like a flag), rather than expanding and compressing with air pressure fluctuations (like a balloon) [14, 72]. Thus, each antenna has intrinsic direction-sensitivity [68]. Because the two aristae are positioned at different angles (Figure 7A), they have different preferred directions. Bilateral comparisons would still be needed for true directional hearing, because one organ alone could not tell the difference between a quiet sound coming from a preferred direction and a loud sound coming from a nonpreferred direction. The key point is that each organ is inherently directional, so there is no need for them to be separated by a large distance [73].
In crickets, bilateral integration for sound localization occurs in cells directly postsynaptic to peripheral auditory afferents. These cells receive antagonistic input from ipsi- and contralateral auditory organs [74]. By contrast, in Drosophila, bilaterality does not seem to emerge in cells directly postsynaptic to JONs (AMMC neurons). Instead, we find the first evidence for bilaterality in the WED.
In addition to finding bilaterality in vibration-preferring subregions, we also found bilaterality in one subregion of the CNS that preferred steady antennal displacements. This subregion spans the border between the AMMC and WED. This subregion is particularly interesting because it has antagonistic directional preferences for ipsi- and contralateral displacements: it responds best when the ipsilateral antenna is pulled while the contralateral antenna is pushed. This pattern of bilateral antagonism confers selectivity for wind directed at the ipsilateral side of the head.
Of course, bilateral integration does not necessarily involve left/right antagonism. Instead, excitatory signals from the two auditory organs may simply be added together. This sort of bilateral pooling could improve the accuracy of behavioral decisions based on the temporal or spectral features of sound stimuli [7].
From maps to cells
Drosophila neurobiologists refer to the little-studied regions of the fly brain as “terra incognita” [75]. New tools have recently opened these brain regions to functional characterization. Like explorers in an unknown land, Drosophila neurobiologists are now facing the task of map-making.
Here we illustrate a general approach to map-making which makes no assumptions about the scale or shape of functional compartments, or the functional properties that distinguish them. This approach yielded fine-grained maps of mechanosensory feature representations. Maps like these will complement new bioinformatic tools that allow researchers to search genetic driver lines using fine-grained anatomical criteria. Together, these tools will enable detailed investigations of specific cell types and the neural computations they implement.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for resources and information should be directed to Rachel Wilson (rachel_wilson@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Flies were raised on standard cornmeal-agar based medium and kept on a 12-hr light/12-hr dark cycle at 25°C. All experiments were performed on female flies 1–2 days post-eclosion. Genotypes were as follows:
Figures 1, 2, 3, 6 (pan-JON): 20XUAS-IVS-GCaMP6f/nan-Gal4;+/+
Figures 4, 5, 6, 7 (pan-neuronal, excluding JONs): 20XUAS-IVS-GCaMP6f/8XLexAop2-IVS-Gal80-WPRE;nSyb-Gal4/iav-LexA
Figures 5, 6 (pan-neuronal, masking JONs): 20XUAS-IVS-GCaMP6f/LexAop-myr-tdTom;nSyb-Gal4/iav-LexA
-
Figure 2B (JON subsets): JO2-Gal4/+; 20XUAS-IVS-GCaMP6f/+; +/+
20XUAS-IVS-GCaMP6f/JO4-Gal4; +/+
20XUAS-IVS-GCaMP6f/+; JO15-Gal4
20XUAS-IVS-GCaMP6f/JO22-Gal4; +/+
20XUAS-IVS-GCaMP6f/+; JO23-Gal4/+
20XUAS-IVS-GCaMP6f/JO26-Gal4; +/+
20XUAS-IVS-GCaMP6f/+; JO28-Gal4/+
JO29-Gal4/+; 20XUAS-IVS-GCaMP6f/+; /+
20XUAS-IVS-GCaMP6f/JO32-Gal4; +/+
Transgenic stocks were previously described as follows: 20XUAS-IVS-GCaMP6f [39], nan-Gal4 [40], 8XLexAop2-IVS-GAL80-WPRE [76, 77], nSyb-GAL4 a.k.a. GMR57C10-Gal4 [43], iav-LexA [78], and LexAop-myr-tdTom [79]. Specific JON driver lines (JO2-Gal4, JO4-Gal4, JO15-Gal4, JO22-Gal4, JO23-Gal4, JO26-Gal4, JO28-Gal4, JO29-Gal4, JO32-Gal4) are described in [13]. The sources of these stocks are listed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental Models: Organisms/Strains | ||
| Drosophila: 20XUAS-IVS-GCaMP6f in attP40 | Bloomington Drosophila Stock Center (BDSC) | RRID: BDSC_42747 |
| Drosophila: 8XLexAop2-IVS-GAL80- WPRE in su(Hw)attP5 | BDSC | RRID: BDSC_32216 |
| Drosophila: LexAoP-myr-tdTomato in su(Hw)attP5 | BDSC | RRID: BDSC_56142 |
| Drosophila: nSyb-Gal4 a.k.a. GMR57C10-Gal4 (in attP2 | BDSC | RRID: BDSC_69152 |
| Drosophila: nan-GAL4 | BDSC | RRID: BDSC_24903 |
| Drosophila: iav-LexA in VK00013 | BDSC | RRID: BDSC_52246 |
| Drosophila: NP1046(JO2)-GAL4 | KYOTO Stock Center (DGRC) at Kyoto Institute of Technology | DGRC_103-867 |
| Drosophila: NP6303(JO4)-GAL4 | DGRC | DGRC_113-902 |
| Drosophila: JO15-GAL4 | BDSC | RRID: BDSC_6753 |
| Drosophila: NP1346(JO22)-GAL4 | DGRC | DGRC_112-636 |
| Drosophila: NP3595(JO23)-GAL4 | DGRC | DGRC_113-359 |
| Drosophila: NP1109(JO26)-GAL4 | DGRC | DGRC_112-511 |
| Drosophila: NP0383(JO28)-GAL4 | DGRC | DGRC_112-162 |
| Drosophila: NP3259(JO29)-GAL4 | DGRC | DGRC_113-185 |
| Drosophila: NP7067(JO32)-GAL4 | DGRC | DGRC_105-355 |
| Software and Algorithms | ||
| MATLAB 2016a | Mathworks (http://www.mathworks.com/) | |
| Scanimage 3.8 | Pologruto et al., 2003 | |
| ImageJ 1.48v | NIH (https://imagej.nih.gov/ij) | |
| Efficient subpixel image registration | (https://www.mathworks.com/matlabcentral/fileexchange/18401-efficient-subpixel-image-registration-by-cross-correlation) | |
METHOD DETAILS
Dissection
Each fly was anesthetized by briefly cooling it on ice. It was immobilized by wedging its dorsal thorax in the hole of a thin sheet of stainless steel foil (0.001 inch thick). The hole was machined with a small end mill on a commercial Computer Numerical Control (CNC) machine. The thin steel foil was bent to accommodate the preparation, and it was fixed in an aperture in a rigid plastic platform. The fly’s head was gently turned through a 180° angle so that its ventral side was accessible from above the platform, while the antennae were free to vibrate beneath the platform. The head, thorax, tarsi and wings were then fastened to the foil with wax to prevent motion. Next, the first and second segments of the antennae (a1 and a2) were glued to the head with a small drop of UV-cured epoxy (Kemxert Corp.) to prevent active movement of the antennae. A drop of saline was then added to the recording chamber on the upper side of the foil. The proboscis and adjacent cuticle, along with the medial parts of the eyes, were removed to expose the ventral brain. The rostral part of the esophagus, tracheas, and excessive adipose tissue were removed to increase optical clarity. Finally, the muscles of the frontal pulsatile organ [muscle 16; 80] were severed to reduce brain motion.
Piezoelectric stimulation of the antenna (Figures 1–6)
A closed-loop piezo-controlled linear actuator with 30 μm travel range (Physik Instrumente) was mounted on a micro-manipulator (Sutter Instruments). A custom-milled attachment linearly coupled the piezo actuator to a sharpened tungsten filament, which was then attached to the arista. To increase the stability of the tungsten filament, it was inserted into a glass pipette filled with epoxy and rigidly coupled to the custom-milled attachment, with the tungsten extending 1 mm past the end of the glass. The tungsten filament was oriented 5° above horizontal, approximately perpendicular to the axis of antennal rotation, and the platform was rotated horizontally so to place the right arista perpendicular to the tungsten filament in the transversal plane. The tip of the tungsten filament was coated with a thin layer of flange sealant (Loctite 515) and was brought in contact with the right arista, consistently in the same location across flies. Two cameras (FLIR Firefly USB equipped with InfiniStix 44 mm/3.00× lenses and 2× DL Tube modules) were used to precisely position the filament and arista relative to each other. One camera’s viewing direction was parallel to the plane of the arista, and the other camera’s viewing angle was orthogonal to the arista. The two cameras were used to verify that actuator commands were producing purely rotational movements of the arista.
The stimulus set included 6 classes of stimuli: frequency-modulated (FM) sweeps, narrowband vibrations (“tones”), pips, pips-with-bias, courtship songs, and steady displacements. Each class included multiple stimuli, for a total of 96 stimuli, described as follows. (Amplitudes are always reported as mean-to-peak amplitudes. Positive displacements push the antenna toward the head, while negative displacements pull it away.) FM sweeps had frequencies linearly sweeping from 2 to 600 Hz (or from 600 Hz to 2 Hz) over 12 s, with an amplitude of 1.8 μm. Tones were 2 s in duration, and had frequencies of 4, 25, 75, 125, 175, 225, 275, 325 Hz; tones were presented at two different amplitudes (450 nm and 1.8 μm). The four lowest-frequency tones were presented in both phases. Pips were 1-s vibrations that were sinusoidally amplitude-modulated at 2 Hz, with carrier frequencies of 8, 16, 25, 50, 75, 100, 125, 150, 175, 200, 225, 250, 275, 300, 350, 400, 500, and 600 Hz; pip amplitudes were 225 nm, 450 nm, and 1.8 μm. Pips-with-bias were the same, except that there was a steady offset of ± 9.246 μm in the antenna’s position during the pip (beginning 1.25 s before pip onset); pips-with-bias had carrier frequencies of 8, 50, 150, 250, 350 Hz, and the pip amplitude (relative to the bias) was 1.8 μm. Courtship songs were two low-pass filtered extracts (12.8 s and 15.8 s) of Drosophila melanogaster courtship song [81]. Steady displacements were 2.5 s in duration, with amplitudes of −14.250, −9.246, −6.000, +6.000, +9.246, and +14.250 μm.
Directional air flow stimulation of both antennae (Figure 7)
To deliver controlled air flow (“wind”), we built a module that could be coupled to the platform holding the fly. This module was an acrylic mount for three 18G steel syringe needles, arranged radially around the fly’s head. The needles were spaced 50° apart, with the middle needle pointing directly at the anterior face of the head. The tip of each needle was positioned 5 mm from the head. The needles were placed in narrow grooves in the acrylic and fixed with dental wax.
Total air flow into the system was controlled by a flow meter. After the flow meter was a 2-way relief valve which was normally open (i.e., venting to the room), and so normally diverting a portion of the input air flow away from the needles. The remaining air flow was sent in parallel to the three needles, with a 3-way valve positioned in front of each needle. Each 3-way valve normally vented to the room. All valves were controlled by an Arduino Uno board in serial communication with the acquisition computer. Switching the middle 3-way valve pushed both antennae toward the head. Switching the left or right 3-way valve pulled the ipsilateral antenna away from the head and pushed the contralateral antenna toward the head. Switching both the right and left 3-way valves (while also closing the normally open relief valve to keep total airflow constant) pulled both antennae away from the head.
We monitored the movement of the two antennae using two cameras placed parallel to the two aristae. At the beginning of each experiment, we manually adjusted the position of the wind-delivery module relative to the fly until each stimulus produced the desired movement of the antennae. Occasionally we also made small manual adjustments of the individual needles. We used the two video cameras to visualize both antennae during all four air flow configurations. Antennal movements were checked at the beginning and end of each experiment (Figure S5A). Stimulus-induced displacements were generally stable across the entire experiment, except for 2 flies that were excluded from further analysis. To ensure that all “push” displacements were similar (regardless of how they were produced), and to ensure that all “pull” displacements were also similar (again regardless of how they were produced), an independent observer (who was blind to the results of each experiment) checked all the video images. We retained the 5 experiments where this blinded observer judged that stimulus control was good; 11 experiments were excluded. We also excluded one experiment where the signal-to-noise ratio of the GCaMP6f fluorescence was particularly poor, leaving 4 flies in our final data set.
Two-photon calcium imaging
Images were acquired on a custom-built two-photon microscope using ScanImage 3.8 software [82]. The system was equipped with a mode-locked Ti:Sapphire laser (Mai Tai DeepSee, Newport Spectra-Physics) tuned to 925 nm, a water immersion objective (XLUMPlanFL N 20×/1.00 W, Olympus), and separate fluorescence detection channels for GCaMP6f emission and tdTomato emission (R9110, Hamamatsu).
Immediately before beginning to record stimulus-evoked fluorescence changes, an anatomical z-stack of the resting GCaMP6f fluorescence was acquired at high resolution for the entire field of view. For this anatomical stack, frames (512×512 pixels, ~15 repetitions per z-level) were acquired at 0.98 Hz, sampling a volume of the ventral brain (generally 150 μm in depth, by 5 μm steps).
Acquisition of stimulus-evoked fluorescence changes was performed in frame scan mode. Several acquisition runs were performed per fly at different xyz coordinates identified from the anatomical stack. Each acquisition run was performed at a fixed z level and was divided into discrete trials, where each trial contained a randomly-selected stimulus. Each trial comprised a number of frames ranging from 41 to 204, depending on the duration of the stimulus. Stimuli were drawn without replacement until each stimulus was repeated 3 times. Frames were 128×86 pixels in size and imaged at 11.63 Hz, scanning from the top left of each frame to bottom right. One fly in Figure 7 was imaged at 10 Hz (128×100 pixels per frame) and later upsampled to 11.63 Hz. Four flies in Figure S3 were imaged at 12.5 (128×80 pixels) and later downsampled to 11.63 Hz. Between consecutive trials there was a gap extending for 65% the duration of the preceding stimulus.
During the entire experiment, the brain was perfused constantly (at ~4 ml/min) with saline solution that was continuously bubbled with 95% O2, 5% CO2. The saline was held at a constant temperature of 22°C. We observed spontaneous and evoked neural activity throughout the course of the experiment.
In the JON imaging experiments, the boundaries of the imaging volume were chosen to encompass all significantly responsive pixels; this included all regions where any resting GCaMP6f expression was observed. The imaging volume therefore included the entire AMMC, as well as the portion of the WED where a small group of JONs terminate [branch "AD"; 13]. The imaging volume also included the portion of the gnathal ganglion where a few JONs terminate [branch "AV"; 13]; however, we did not reliably observe significant responses in the gnathal ganglion, so the gnathal ganglion does not appear in our JON maps, although the more proximal parts of the AV axons may contribute to our AMMC maps. While most JONs terminate strictly ipsilaterally, a small number of JONs extend to the contralateral side of the brain [branch “EDC-c”; 13]; we did not reliably observe significant responses on the contralateral side, so it does not appear in our JON maps.
In CNS imaging experiments, the boundaries of the z-stack were again chosen to encompass all significantly responsive pixels. These pixels were limited to the AMMC and WED. There are certainly regions outside the AMMC and WED that contain neurons responsive to Johnston’s organ stimulation; however, they did not appear as significantly responsive in our experiments. One key limitation was that regions in the more dorsal part of the brain (i.e., deeper regions in our inverted-brain preparation) were subject to more light scattering. Therefore, we might not detect deep (dorsal) regions even if they were responsive. A second consideration is that neurons in regions outside the AMMC and WED might not be spatially organized according to their mechanosensory tuning properties, and so their responses might not be detectable in our pan-neuronal imaging experiments. Notably, we did not find significant responses in the gnathal ganglion, although this region was very accessible in our preparation, and it receives a direct JON projection [13, 23].
Data pre-processing
In each experiment, data were split in green/red channels when applicable, and saved into movies using ImageJ 1.48v (NIH). Each movie was then loaded into MATLAB 2016a, which was used for all subsequent analysis. Pixels whose raw fluorescence intensity exceeded the 99.998th percentile were set to the 99.998th percentile value; this was done to reduce the impact of “noise pixels” on the subsequent analysis, and it had no consequences for signal-pixels. In order to align data in the x-y plane on a frame-by-frame basis, each movie was first low-pass filtered with a Gaussian kernel (SD=0.5×0.5×3) and further smoothed in z with a moving average filter (15 frames). The magnitude of a Sobel gradient operator was calculated for each frame, and its frame-by-frame registration shifts were computed using efficient subpixel motion registration [83]. This procedure was generally successfully applied on data where only the green channel (GCaMP6f fluorescence) was available. When the red channel (tdTomato) was available, we calculated the registration shifts on the red channel. Subsequently, these registration shifts were applied to the raw data.
Registered movies were downsampled in x-y to 70% of the original pixel number (bilinear interpolation), and temporally smoothed with a Gaussian kernel (sigma = 3).
Next, responses to different trials of the same stimulus were averaged. For each pixel and each stimulus, ΔF/F0 was computed, where F0 was calculated over an extended baseline window including the baseline of the current trial, and that of the 3 preceding and 3 following trials. A trial-averaged ΔF/F0 series was then calculated for each stimulus type.
Then, we discarded nonresponsive pixels. Pixels were considered responsive if they fluctuated significantly in response to at least one stimulus. For each pixel and for every stimulus, the distribution of fluorescence values over a fixed stimulus window was compared to the distribution of fluorescence values in the extended baseline window described above using a t-test. The stimulus time window began 0.8 s after stimulus onset and ended 0.1 s after stimulus offset for the piezo-stimulation experiments; it began 0.2 s after stimulus onset and ended 0.2 s after stimulus offset for the wind experiments. Each pixel was deemed responsive or unresponsive based on a threshold p value (adjusted for false discovery rate) which was determined separately for each experiment, based on the expectation that GCaMP-negative pixels should be unresponsive. We also discarded pixels whose mean intensity projection was close to zero.
We grouped the imaging data into three main data sets (JON, piezo-CNS, wind-CNS). Within each dataset, we pooled responsive pixels from all experiments and z-levels. In the CNS datasets where JONs expressed tdTomato, we removed all tdTomato-positive pixels.
Clustering pixels by functional similarity
Our unsupervised, function-based analysis approach is similar to that employed by several previous pan-neuronal calcium imaging studies of the Drosophila brain [46, 47]. First, we concatenated each pixel’s trial-averaged responses to all the stimuli to obtain a response vector r(t). For the piezo-stimulation experiments (Figures 1–6), we took the window from stimulus onset to 0.7 s after stimulus offset, for each stimulus. Each vector r(t) contained 3549 time points. For the wind-stimulation experiments (Figure 7), we took the window starting 1.6 s after stimulus onset and ending 0.4 s before stimulus offset. Each vector r(t) contained 92 time points.
We next z-scored each pixel so that its response vector r(t) had zero mean and unit SD, and we used a Euclidean distance metric to hierarchically agglomerate functionally similar pixels using the “average-linkage method”. We discarded clusters of pixels which showed a high degree of dissimilarity from the majority of the dataset, because the fluorescence fluctuations in these pixels appeared to be mainly noise. We established the threshold for dissimilarity by visual inspection of the dendrogram branching pattern and the corresponding single-pixel response profiles. With this procedure, we discarded 6% of the responsive pixels in the JON dataset and 17% in the piezo-CNS dataset. We did not apply the pixel-exclusion procedure to the wind-CNS dataset; instead, the “noise pixels” are mainly aggregated in types e-h of that data set (Figure S5). The final sizes of the data matrices were 102669 × 3549 (JON data set), 49608 × 3549 (piezo-CNS data set), and 4496× 92 (wind-CNS data set); matrices are arranged as (pixels × time).
Finally, we re-computed the hierarchical agglomerative clustering, this time using “Ward’s method”, which produces more uniform group sizes than the “average-linkage method” does. We selected a threshold for dividing pixels into types so as to create as many types as possible, while also satisfying two criteria: (1) we assumed that spatial maps should be as similar across flies as possible; (2) we assumed that pixels belonging to the same functional type should be spatially contiguous. Thus, we progressively lowered the threshold for splitting pixels into types until the quality of the map began to decline markedly (according to these two criteria), and then fixed the threshold just above that point (Figure S1).
The thresholds used to cut pixels into clusters were different for JON experiments and CNS experiments. These thresholds are defined in terms of the Euclidean distance between the z-scored response vectors of the pixels. However, the choice of thresholds was governed by the same rules in both cases: we aimed to create as many types as possible, while also maintaining cross-brain consistency, and also maintaining spatial contiguity within each type.
Anatomical registration and drawing regional boundaries
Anatomical registration was performed for three purposes: (1) for mapping tdTomato-positive pixels onto brains where tdTomato was not used to label JONs, (2) for dividing pixels into AMMC versus WED domains, and (3) for generating example display images. Anatomical registration was not used for clustering pixels by functional similarity, because the clustering was purely based on r(t), without regard to space. In order to perform anatomical registration, we semi-automatically registered the mean projection of each x-y imaging window to the anatomical z-stack we acquired at the beginning of that experiment, by applying a non-reflective similarity transformation calculated over manually selected control points. Each brain (i.e., each anatomical z-stack) was then registered to a template z-stack semi-automatically (as above) in x-y and manually in z.
We defined the AMMC as the region delimited by the envelope of the JON axon bundle [84]; the only exception was a small dorsolateral branch of the JON axon bundle which was assigned to the WED based on anatomical and functional landmarks; the localization of this small branch to the WED was confirmed by a previous study [which called it branch "AD"; 13]. The boundary of the AMMC was drawn on the basis of tdTomato fluorescence in brains where tdTomato was expressed in all JONs. In brains where this marker was not used, the boundary of the AMMC was mapped by registering a tdTomato-positive sample onto that brain.
We defined the WED as the region immediately lateral and dorsal to the AMMC. We did not employ the term “saddle” [which has been recently proposed as a term to be applied to the region interposed between the AMMC and WED; 84] because we did not find evidence of a separate functional map interposed between the AMMC map and the WED map.
Responses in the AMMC were resolvable in experiments where JON GCaMP expression was suppressed using Gal80. Responses in the WED were resolvable in all CNS imaging experiments.
In JON imaging experiments, we were able to reliably identify four major zones in the JON axon bundle (A, B, D, and E). The fifth reported major zone (C) could not be reliably identified, probably because it is relatively small; it is visible in high-resolution confocal z-stacks of fixed tissue [13], but not in our lower-resolution images. Our functional maps of the JON axon bundle are largely consistent with previous descriptions of the JON map in the brain [18, 26, 27], except that that the “pull-preferring” subregion in our JON maps (the subregion in gray) was typically localized to the most medial part of the axon bundle, immediately adjacent to the “push”-preferring subregion (black). Previously, “pull” responses were attributed to the C zone, which would situate the “pull” subregion between the B and D branches, i.e. slightly more lateral relative to its location in our maps [18].
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed using ImageJ (Version 1.48v) and custom software written in MATLAB (Version R2016a). Portions of this research were conducted on the Orchestra High Performance Computer Cluster at Harvard Medical School. This NIH-supported shared facility consists of thousands of processing cores and terabytes of associated storage and is partially provided through grant NCRR 1S10RR028832-01.
No statistical methods were used to predetermine sample sizes in advance, but sample sizes are similar to those reported in other studies in the field. Data collection and analysis were generally not performed blind to the conditions of the experiments, except when specifically noted above. Some experiments were excluded from the final wind-CNS data set based solely on the accuracy of antennal displacements and the overall signal-to-noise ratio of the recording (as described above) by an observer blind to the results of the experiment. In the JON data set and the piezo-CNS data set, a relatively small number of recordings were discarded prior to any analysis due to excessive drift and/or poor signal-to-noise ratio.
The analyses in Figure 6 focused only on pixels belonging to vibration-preferring response types in the piezoelectric-stimulation experiments (JON and CNS). Several of these analyses use the metric we call “center frequency”, which we define as the center-of-mass of the frequency tuning curve (Figure S4). In Figure 6A, we computed a histogram of frequency preferences by aggregating the center frequency of all pixels in all experiments. When we repeated this analysis using “peak frequency” instead of center frequency, our conclusions were unchanged. In Figure 6B, we measured “frequency selectivity” as follows: for each response type and each experiment, we computed a frequency tuning curve (averaged over pixels); we then log-interpolated the tuning curves and measured their lifetime sparseness [85], and finally averaged these values within each map (JONs, AMMC, WED) for each experiment, weighted according to the relative representation of each response type within each map. In Figure 6C, the axis of tonotopy was computed as the spatial axis in each map (each xy imaging plane) that minimizes the variance in center frequency as a function of axis position. (In principle, the true axis of tonotopy might have resided outside the xy plane – e.g., the tonotopy axis might actually have been the z-axis; however, we found that maps we obtained at adjacent z-depths were quite similar, with only gradual changes in tonotopic maps as we traversed the z-axis in 4–10 μm increments.) In Figure 6D, we computed a “tonotopy index” as follows: for each experiment and each map (JONs, AMMC, WED), we measured each pixel’s center frequency and its projected position on the tonotopy axis. We then measured the correlation between the vector of center frequencies ordered by frequency and the same values ordered by axis position. If axis position varied perfectly monotonically with frequency, then the tonotopy index would be one. When we repeated this analysis using “peak frequency” instead of center frequency, our conclusions were unchanged. In Figure 6E, we identified the vibration amplitude eliciting a half-maximum response as follows: we fit sigmoidal functions to the amplitude-response curves (Figures 3C and 5C) and we took the vibration amplitude of the fitted function at half-maximum. In Figure 6F, we computed an “adaptation index” as follows: for each response type (using data from Figures 3A and 5A), we compared responses to frequency up-modulations and down-modulations, after first inverting the down-modulation response and shifting it slightly to maximize its similarity to the up-modulation response. The adaptation index was taken as the sum of the squared difference between the up-response and the (inverted) down-response. Thus, if up- and down-modulation were to elicit symmetrical responses, the adaptation index would be zero.
DATA AND SOFTWARE AVAILABILITY
Data and software may be obtained upon request.
Supplementary Material
Highlights.
Primary and secondary mechanosensory regions of the fly brain contain tonotopic maps
The secondary map is bilateral; it has more orderly tonotopy and narrower tuning
Different subregions encode vibrations over different amplitude ranges
One subregion integrates bilateral signals to specifically detect ipsilateral wind
Acknowledgments
We thank Matt Pecot for fly stocks, and Mehmet Fi3ek for building the 2-photon microscope. Members of the Wilson lab provided advice, as well as comments on the manuscript. Ofer Mazor and Pavel Gorelik provided engineering support via the Harvard Medical School Research Instrumentation Core Facility. This work was funded by grant R01 NS101157 from the NIH. P.P. was supported by the Armenise-Harvard Foundation. R.I.W. is an HHMI Investigator.
Footnotes
Author contributions
P.P. performed the experiments and analyzed the data. P.P. and R.I.W. designed the study and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hennig RM, Franz A, Stumpner A. Processing of auditory information in insects. Microsc Res Tech. 2004;63:351–374. doi: 10.1002/jemt.20052. [DOI] [PubMed] [Google Scholar]
- 2.Romer H. Directional hearing: from biophysical binaural cues to directional hearing outdoors. J Comp Physiol [A] 2015;201:87–97. doi: 10.1007/s00359-014-0939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Göpfert MC, Hennig RM. Hearing in insects. Annu Rev Entomol. 2016;61:257–276. doi: 10.1146/annurev-ento-010715-023631. [DOI] [PubMed] [Google Scholar]
- 4.Hedwig B. Sequential filtering processes shape feature detection in crickets: A framework for song pattern recognition. Front Physiol. 2016;7:46. doi: 10.3389/fphys.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuthill JC, Wilson RI. Mechanosensation and adaptive motor control in insects. Curr Biol. 2016;26:R1022–1038. doi: 10.1016/j.cub.2016.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schöneich S, Kostarakos K, Hedwig B. An auditory feature detection circuit for sound pattern recognition. Sci Adv. 2015;1:e1500325. doi: 10.1126/sciadv.1500325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostarakos K, Hedwig B. Calling song recognition in female crickets: temporal tuning of identified brain neurons matches behavior. J Neurosci. 2012;32:9601–9612. doi: 10.1523/JNEUROSCI.1170-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg E, Buschges A, Schmidt J. Single perturbations cause sustained changes in searching behavior in stick insects. J Exp Biol. 2013;216:1064–1074. doi: 10.1242/jeb.076406. [DOI] [PubMed] [Google Scholar]
- 9.Berg EM, Hooper SL, Schmidt J, Buschges A. A leg-local neural mechanism mediates the decision to search in stick insects. Curr Biol. 2015;25:2012–2017. doi: 10.1016/j.cub.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Hellekes K, Blincow E, Hoffmann J, Buschges A. Control of reflex reversal in stick insect walking: effects of intersegmental signals, changes in direction, and optomotor-induced turning. J Neurophysiol. 2012;107:239–249. doi: 10.1152/jn.00718.2011. [DOI] [PubMed] [Google Scholar]
- 11.Coen P, Murthy M. Singing on the fly: sensorimotor integration and acoustic communication in Drosophila. Curr Opin Neurobiol. 2016;38:38–45. doi: 10.1016/j.conb.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa Y, Kamikouchi A. Auditory system of fruit flies. Hear Res. 2016;338:1–8. doi: 10.1016/j.heares.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 13.Kamikouchi A, Shimada T, Ito K. Comprehensive classification of the auditory sensory projections in the brain of the fruit fly Drosophila melanogaster. J Comp Neurol. 2006;499:317–356. doi: 10.1002/cne.21075. [DOI] [PubMed] [Google Scholar]
- 14.Göpfert MC, Robert D. The mechanical basis of Drosophila audition. J Exp Biol. 2002;205:1199–1208. doi: 10.1242/jeb.205.9.1199. [DOI] [PubMed] [Google Scholar]
- 15.Lehnert BP, Baker AE, Gaudry Q, Chiang AS, Wilson RI. Distinct roles of TRP channels in auditory transduction and amplification in Drosophila. Neuron. 2013;77:115–128. doi: 10.1016/j.neuron.2012.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menda G, Bar HY, Arthur BJ, Rivlin PK, Wyttenbach RA, Strawderman RL, Hoy RR. Classical conditioning through auditory stimuli in Drosophila: methods and models. J Exp Biol. 2011;214:2864–2870. doi: 10.1242/jeb.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamikouchi A, Inagaki HK, Effertz T, Hendrich O, Fiala A, Göpfert MC, Ito K. The neural basis of Drosophila gravity-sensing and hearing. Nature. 2009;458:165–171. doi: 10.1038/nature07810. [DOI] [PubMed] [Google Scholar]
- 18.Yorozu S, Wong A, Fischer BJ, Dankert H, Kernan MJ, Kamikouchi A, Ito K, Anderson DJ. Distinct sensory representations of wind and near-field sound in the Drosophila brain. Nature. 2009;458:201–205. doi: 10.1038/nature07843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coen P, Clemens J, Weinstein AJ, Pacheco DA, Deng Y, Murthy M. Dynamic sensory cues shape song structure in Drosophila. Nature. 2014;507:233–237. doi: 10.1038/nature13131. [DOI] [PubMed] [Google Scholar]
- 20.Budick SA, Reiser MB, Dickinson MH. The role of visual and mechanosensory cues in structuring forward flight in Drosophila melanogaster. J Exp Biol. 2007;210:4092–4103. doi: 10.1242/jeb.006502. [DOI] [PubMed] [Google Scholar]
- 21.Bell JS, Wilson RI. Behavior reveals selective summation and max pooling among olfactory processing channels. Neuron. 2016;91:425–438. doi: 10.1016/j.neuron.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steck K, Veit D, Grandy R, Badia SB, Mathews Z, Verschure P, Hansson BS, Knaden M. A high-throughput behavioral paradigm for Drosophila olfaction - The Flywalk. Sci Rep. 2012;2:361. doi: 10.1038/srep00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hampel S, Franconville R, Simpson JH, Seeds AM. A neural command circuit for grooming movement control. eLife. 2015;4:e08758. doi: 10.7554/eLife.08758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mamiya A, Straw AD, Tomasson E, Dickinson MH. Active and passive antennal movements during visually guided steering in flying Drosophila. J Neurosci. 2011;31:6900–6914. doi: 10.1523/JNEUROSCI.0498-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mamiya A, Dickinson MH. Antennal mechanosensory neurons mediate wing motor reflexes in flying Drosophila. J Neurosci. 2015;35:7977–7991. doi: 10.1523/JNEUROSCI.0034-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuo E, Yamada D, Ishikawa Y, Asai T, Ishimoto H, Kamikouchi A. Identification of novel vibration- and deflection-sensitive neuronal subgroups in Johnston's organ of the fruit fly. Front Physiol. 2014;5:179. doi: 10.3389/fphys.2014.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa Y, Okamoto N, Nakamura M, Kim H, Kamikouchi A. Anatomic and physiologic heterogeneity of subgroup-A auditory sensory neurons in fruit flies. Front Neural Circuits. 2017;11:46. doi: 10.3389/fncir.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zill SN. Plasticity and proprioception in insects. I Responses and cellular properties of individual receptors of the locust metathoracic femoral chordotonal organ. J Exp Biol. 1985;116:435–461. doi: 10.1242/jeb.116.1.435. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann T, Koch UT, Bässler U. Physiology of the femoral chordotonal organ in the stick insect, Cuniculina impigra. J Exp Biol. 1985;114:207–223. [Google Scholar]
- 30.Hofmann T, Koch UT. Acceleration receptors in the femoral chordotonal organ of the stick insect, Cuniculina impigra. J Exp Biol. 1985;114:225–237. [Google Scholar]
- 31.Matheson T. Responses and locations of neurones in the locust metathoracic femoral chordotonal organ. J Comp Physiol [A] 1990;166:915–927. [Google Scholar]
- 32.Lai JS, Lo SJ, Dickson BJ, Chiang AS. Auditory circuit in the Drosophila brain. Proc Natl Acad Sci U S A. 2012;109:2607–2612. doi: 10.1073/pnas.1117307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tootoonian S, Coen P, Kawai R, Murthy M. Neural representations of courtship song in the Drosophila brain. J Neurosci. 2012;32:787–798. doi: 10.1523/JNEUROSCI.5104-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaughan AG, Zhou C, Manoli DS, Baker BS. Neural pathways for the detection and discrimination of conspecific song in D. melanogaster. Curr Biol. 2014;24:1039–1049. doi: 10.1016/j.cub.2014.03.048. [DOI] [PubMed] [Google Scholar]
- 35.Clemens J, Girardin CC, Coen P, Guan XJ, Dickson BJ, Murthy M. Connecting neural codes with behavior in the auditory system of Drosophila. Neuron. 2015;89:629–644. doi: 10.1016/j.neuron.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang AEB, Vaughan AG, Wilson RI. A mechanosensory circuit that mixes opponent channels to produce selectivity for complex stimulus features. Neuron. 2016;92:888–901. doi: 10.1016/j.neuron.2016.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuo E, Seki H, Asai T, Morimoto T, Miyakawa H, Ito K, Kamikouchi A. Organization of projection neurons and local neurons of the primary auditory center in the fruit fly Drosophila melanogaster. J Comp Neurol. 2016;524:1099–1164. doi: 10.1002/cne.23955. [DOI] [PubMed] [Google Scholar]
- 38.Azevedo AW, Wilson RI. Active mechanisms of vibration encoding and frequency filtering in central mechanosensory neurons. Neuron. 2017;96:446–460. e449. doi: 10.1016/j.neuron.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Chung YD, Park DY, Choi S, Shin DW, Soh H, Lee HW, Son W, Yim J, Park CS, et al. A TRPV family ion channel required for hearing in Drosophila. Nature. 2003;424:81–84. doi: 10.1038/nature01733. [DOI] [PubMed] [Google Scholar]
- 41.Eberl DF, Hardy RW, Kernan MJ. Genetically similar transduction mechanisms for touch and hearing in Drosophila. J Neurosci. 2000;20:5981–5988. doi: 10.1523/JNEUROSCI.20-16-05981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemens J, Ozeri-Engelhard N, Murthy M. Fast intensity adaptation enhances the encoding of sound in Drosophila. Nat Commun. 2018;9:134. doi: 10.1038/s41467-017-02453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou C, Pan Y, Robinett CC, Meissner GW, Baker BS. Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron. 2014;83:149–163. doi: 10.1016/j.neuron.2014.05.038. [DOI] [PubMed] [Google Scholar]
- 45.Zhou C, Franconville R, Vaughan AG, Robinett CC, Jayaraman V, Baker BS. Central neural circuitry mediating courtship song perception in male Drosophila. eLife. 2015:4. doi: 10.7554/eLife.08477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strother JA, Nern A, Reiser MB. Direct observation of ON and OFF pathways in the Drosophila visual system. Curr Biol. 2014;24:976–983. doi: 10.1016/j.cub.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 47.Weir PT, Dickinson MH. Functional divisions for visual processing in the central brain of flying Drosophila. Proc Natl Acad Sci USA. 2015;112:E5523–5532. doi: 10.1073/pnas.1514415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris DT, Kallman BR, Mullaney BC, Scott K. Representations of taste modality in the Drosophila brain. Neuron. 2015;86:1449–1460. doi: 10.1016/j.neuron.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaas JH. Topographic maps are fundamental to sensory processing. Brain Res Bull. 1997;44:107–112. doi: 10.1016/s0361-9230(97)00094-4. [DOI] [PubMed] [Google Scholar]
- 50.Schnupp J, Nelken I, King A. Auditory Neuroscience: Making Sense of Sound. Cambridge, Massachusetts: MIT Press; 2011. [Google Scholar]
- 51.Oldfield BP. Central projections of primary auditory fibers in Tettigoniidae (Orthoptera: Ensifera) J Comp Physiol [A] 1983;151:389–395. [Google Scholar]
- 52.Römer H. Tonotopic organization of the auditory neuropile in the bushcricket Tettigonia viridissima. Nature. 1983:306. [Google Scholar]
- 53.Stolting H, Stumpner A. Tonotopic organization of auditory receptors of the bushcricket pholidoptera griseoaptera (Tettigoniidae, decticinae) Cell Tissue Res. 1998;294:377–386. doi: 10.1007/s004410051187. [DOI] [PubMed] [Google Scholar]
- 54.Römer H. Anatomical representation of frequency and intensity in the auditory system of orthoptera. In: Kalmring K, Elsner N, editors. Acoustic and Vibration Communication in Insects. Berlin: Paul Parey; 1985. pp. 25–32. [Google Scholar]
- 55.Atkins G, Pollack GS. Correlations between structure, topographic arrangement, and spectral sensitivity of sound-sensitive interneurons in crickets. J Comp Neurol. 1987;266:398–412. doi: 10.1002/cne.902660307. [DOI] [PubMed] [Google Scholar]
- 56.Stumpner A, Molina J. Diversity of intersegmental auditory neurons in a bush cricket. J Comp Physiol [A] 2006;192:1359–1376. doi: 10.1007/s00359-006-0164-z. [DOI] [PubMed] [Google Scholar]
- 57.Hildebrandt KJ. Neural maps in insect versus vertebrate auditory systems. Curr Opin Neurobiol. 2014;24:82–87. doi: 10.1016/j.conb.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 58.Bennet-Clark HC, Ewing AW. Pulse interval as a critical parameter in the courtship song of D. melanogaster. Anim Behav. 1969;17:755–759. [Google Scholar]
- 59.Rybak F, Aubin T, Moulin B, Jallon JM. Acoustic communication in Drosophila melanogaster courtship: Are pulse- and sine-song frequencies important for courtship success? Can J Zool. 2002;80:987–996. [Google Scholar]
- 60.Talyn BC, Dowse HB. The role of courtship song in sexual selection and species recognition by female Drosophila melanogaster. Anim Behav. 2004;68:1165–1180. [Google Scholar]
- 61.Zhang W, Yan Z, Jan LY, Jan YN. Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae. Proc Natl Acad Sci U S A. 2013;110:13612–13617. doi: 10.1073/pnas.1312477110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Effertz T, Wiek R, Göpfert MC. NompC TRP channel is essential for Drosophila sound receptor function. Curr Biol. 2011;21:592–597. doi: 10.1016/j.cub.2011.02.048. [DOI] [PubMed] [Google Scholar]
- 63.Kanou M, Shimozawa T. Varieties of filiform hairs - range fractionation by sensory afferents and cercal interneurons of a cricket. J Comp Physiol [A] 1984;155:485–493. [Google Scholar]
- 64.Oldfield BP, Kleindienst HU, Huber F. Physiology and tonotopic organization of auditory receptors in the cricket Gryllus bimaculatus DeGeer. J Comp Physiol [A] 1986;159:457–464. doi: 10.1007/BF00604165. [DOI] [PubMed] [Google Scholar]
- 65.Blake DT, Johnson KO, Hsiao SS. Monkey cutaneous SAI and RA responses to raised and depressed scanned patterns: effects of width, height, orientation, and a raised surround. J Neurophysiol. 1997;78:2503–2517. doi: 10.1152/jn.1997.78.5.2503. [DOI] [PubMed] [Google Scholar]
- 66.Liberman MC. Auditory-nerve response from cats raised in a low-noise chamber. J Acoust Soc Am. 1978;63:442–455. doi: 10.1121/1.381736. [DOI] [PubMed] [Google Scholar]
- 67.Gnatzy W, Tautz J. Ultrastructure and mechanical properties of an insect mechanoreceptor: stimulus-transmitting structures and sensory apparatus of the cercal filiform hairs of Gryllus. Cell Tissue Res. 1980;213:441–463. doi: 10.1007/BF00237890. [DOI] [PubMed] [Google Scholar]
- 68.Morley EL, Steinmann T, Casas J, Robert D. Directional cues in Drosophila melanogaster audition: structure of acoustic flow and inter-antennal velocity differences. J Exp Biol. 2012;215:2405–2413. doi: 10.1242/jeb.068940. [DOI] [PubMed] [Google Scholar]
- 69.Miles RN, Robert D, Hoy RR. Mechanically coupled ears for directional hearing in the parasitoid fly Ormia ochracea. J Acoust Soc Am. 1995;98:3059–3070. doi: 10.1121/1.413830. [DOI] [PubMed] [Google Scholar]
- 70.Robert D, Miles RN, Hoy RR. Tympanal mechanics in the parasitoid Ormia ochracea: intertympanal coupling during mechanical vibration. J Comp Physiol [A] 1998;183:443–452. [Google Scholar]
- 71.Mason AC, Oshinsky ML, Hoy RR. Hyperacute directional hearing in a microscale auditory system. Nature. 2001;410:686–690. doi: 10.1038/35070564. [DOI] [PubMed] [Google Scholar]
- 72.Robert D, Hoy RR. Auditory systems in insects. In: North G, editor. Invertebrate Neurobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 155–184. [Google Scholar]
- 73.Michelsen A. Hearing and sound communication in small animals: evolutionary adaptations to the laws of physics. In: Webster DB, Fay RR, Popper AN, editors. The Evolutionary Biology of Hearing. New York: Springer-Verlag; 1992. pp. 61–77. [Google Scholar]
- 74.Selverston AI, Kleindienst HU, Huber F. Synaptic connectivity between cricket auditory interneurons as studied by selective photoinactivation. J Neurosci. 1985;5:1283–1292. doi: 10.1523/JNEUROSCI.05-05-01283.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panser K, Tirian L, Schulze F, Villalba S, Jefferis G, Buhler K, Straw AD. Automatic segmentation of Drosophila neural compartments using GAL4 expression data reveals novel visual pathways. Curr Biol. 2016;26:1943–1954. doi: 10.1016/j.cub.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pfeiffer B, Ngo TTB, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfeiffer B. Personal communication to FlyBase. (2010.11.11) Pfeiffer insertions. 2010 http://flybase.org/reports/FBrf0212441.
- 78.Shearin HK, Dvarishkis AR, Kozeluh CD, Stowers RS. Expansion of the gateway multisite recombination cloning toolkit. PLoS One. 2013;8:e77724. doi: 10.1371/journal.pone.0077724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y, Akin O, Nern A, Tsui CY, Pecot MY, Zipursky SL. Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron. 2014;81:280–293. doi: 10.1016/j.neuron.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller A. The internal anatomy and histology of the imago of Drosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. Cold Spring Harbor Laboratory Press; 1994. pp. 420–534. [Google Scholar]
- 81.Arthur BJ, Sunayama-Morita T, Coen P, Murthy M, Stern DL. Multi-channel acoustic recording and automated analysis of Drosophila courtship songs. BMC Biol. 2013;11:11. doi: 10.1186/1741-7007-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33:156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 84.Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, et al. A systematic nomenclature for the insect brain. Neuron. 2014;81:755–765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 85.Willmore B, Tolhurst DJ. Characterizing the sparseness of neural codes. Network. 2001;12:255–270. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.