Abstract
Background
The aim of this study was to examine the expression level of IRRE-like protein 1 (KIRREL) in gastric cancer (GC) and to explore its prognostic significance.
Material/Methods
Bioinformatics methods were used to predict the differential expression levels of KIRREL mRNA in GC and normal gastric tissues by mining cancer-related databases (TCGA and Oncomine). Immunohistochemistry was done to verify the KIRREL protein expression levels in 71 cases of GC tissues combined with matched normal tissues. The relationship between clinicopathologic parameters and KIRREL differential expression levels in GC was investigated by the chi-square test. Kaplan-Meier univariate and Cox multivariate survival analyses were performed to explore the prognostic significance of KIRREL expression in GC patients.
Results
TCGA and GEO data analyses showed that KIRREL mRNA expression level was remarkably higher in GC than that in normal gastric tissues (both P<0.05). KIRREL mRNA levels were dramatically increased from stage I to stage IV (P=0.037). Immunohistochemical results showed that the high positive rate of KIRREL staining in GC was 61.97% (44/71). Moreover, GC patients with KIRREL mRNA or protein high levels had significantly shorter overall survival times than those with KIRREL mRNA or low protein levels (All P<0.05). Additionally, Cox multivariate survival analysis revealed that KIRREL differential expression levels (low vs. high) were the only independent parameter predicting the prognosis of GC patients (P=0.000).
Conclusions
KIRREL was overexpressed in GC and the overexpression of KIRREL could serve as an independent predictor of poor prognosis in GC patients.
MeSH Keywords: Oncogene Proteins, Prognosis, Stomach Neoplasms
Background
Gastric cancer is a malignant disease that seriously threatens human health. Globally, there are 950 000 new cases of gastric cancer and 720 000 deaths every year [1]. Of these, 70% come from Asia, and Chinese patients account for nearly half [2]. Because many gastric cancer patients are diagnosed at the advanced stage, they often cannot receive the best treatment strategy, like receiving the radical gastrectomy. Thus, the overall prognosis of patients remains unfavorable [3–5]. In the field of targeted molecular therapies for gastric cancer, relatively few effective drugs have been demonstrated, such as apatinib [6,7] and Herceptin (for HER-2-positive patients) [8,9]. Therefore, we still need to further explore and develop new molecular targets to improve the therapeutic efficacy and clinical prognosis of patients with gastric cancer.
Kin of IRRE-like protein 1 (KIRREL), also known as NEPH1, is a human protein encoded by the KIRREL gene [10]. KIRREL is a member of the NEPH protein family, which includes KIRREL2 (NEPH3) and KIRREL3 (NEPH2). The cytoplasmic domains of these proteins interact with the C terminus of podocin (NPHS2). Previous studies have shown that KIRREL was expressed in filtration slits of kidney podocytes, cells involved in ensuring size- and charge-selective ultrafiltration of blood [10], which can interact with nephrin (NPHS1) [11,12] and tight junction protein 1 (TJP1) [11,13]. Recently, some studies have found that the abnormal expression of the KIRREL gene was closely related to the occurrence and development, proliferation, and metastasis of drug sensitivity of malignant tumors [14–18]. However, the exact aberrant expression level of KIRREL in gastric cancer and its prognostic significance remain unclear.
Therefore, in order to answer the above questions, the present study was performed. We firstly used bioinformatics methods to predict the differential expression levels of KIRREL mRNA in gastric cancer and normal gastric tissues by mining cancer-related databases (TCGA and Oncomine). Then, immunohistochemistry was used to verify the KIRREL protein expression levels in 71 cases of gastric cancer tissues combined with matched normal tissues, which were collected retrospectively in our hospital. Further, based on the immunohistochemical results, the relationship between clinicopathologic parameters and KIRREL differential expression levels in gastric cancer was investigated by chi-square test. Moreover, Kaplan-Meier univariate and Cox multivariate survival analyses were performed to explore the prognostic significance of KIRREL expression in gastric cancer patients.
Material and Methods
Bioinformatics prediction
In order to predict the differential expression levels of KIRREL in gastric cancer and normal gastric tissues, TCGA and Oncomine databases were used. We downloaded a total of 408 cases of GC and 211 cases of normal gastric tissues containing KIRREL mRNA expression information from the TCGA database (https://cancergenome.nih.gov/). Then, RStudio (Version 1.1.442), a free and open source data analysis software, was used to analyze the differential expression levels of KIRREL mRNA between the 2 groups and deduce the overall survival (OS) curve. The Oncomine database (https://www.oncomine.org) [19] was also searched to explore the differential expression levels of KIRREL between gastric cancer and normal groups. A total of 9 GEO-sourced datasets were included. Additionally, Kaplan-Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=gastric) [20] was used to draw the OS curve based on the GEO data.
Gastric cancer tissues and relative clinicopathological information of patients
A total of 71 cases of gastric cancer tissues and paired adjacent normal tissues were retrospectively collected to perform the immunohistochemical staining. All the patients had received radical gastrectomy in the Department of General Surgery in our hospital from June 2009 to December 2011. The last follow-up time was July 2017. The pathologic diagnosis of all cases was gastric adenocarcinoma after operation. As shown in Table 1, the relative clinical information of patients, including age, sex, CEA, CA199, and TNM stage, was summarized in detail. This study was approved by the Ethics Committee of our hospital and all patients had signed the informed consent.
Table 1.
Correlation between KIRREL and clinicopathological parameters of gastric cancer patients.
| Clinicopathological parameters | Cases (N) | KIRREL expression levels | χ2 | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | |||||
| ≤60 | 29 | 16 | 13 | 6.114 | 0.013 |
| >60 | 42 | 11 | 31 | ||
| Gender | |||||
| Male | 52 | 20 | 32 | 0.015 | 0.901 |
| Female | 19 | 7 | 12 | ||
| CEA (ng/ml) | |||||
| ≤10 | 34 | 13 | 21 | 0.001 | 0.973 |
| >10 | 37 | 14 | 23 | ||
| CA199 (U/ml) | |||||
| ≤37 | 44 | 16 | 28 | 0.136 | 0.712 |
| >37 | 27 | 11 | 16 | ||
| Tumor location | |||||
| Antrum | 30 | 12 | 18 | 0.086 | 0.770 |
| Other sites | 39 | 15 | 26 | ||
| Tumor size (cm) | |||||
| ≤5 | 38 | 20 | 18 | 7.398 | 0.007 |
| >5 | 33 | 7 | 26 | ||
| Histological differentiation | |||||
| Well/moderate | 40 | 18 | 22 | 1.890 | 0.169 |
| Poor | 31 | 9 | 22 | ||
| T stage | |||||
| T1–2 | 22 | 13 | 9 | 6.001 | 0.014 |
| T3–4 | 49 | 14 | 35 | ||
| N stage | |||||
| N0 | 25 | 13 | 12 | 3.196 | 0.074 |
| N1–3 | 46 | 14 | 32 | ||
| TNM stage | |||||
| I–II | 32 | 17 | 15 | 5.634 | 0.018 |
| III–IV | 39 | 10 | 29 | ||
Immunohistochemical staining and interpretation of results
Based on the protocol manual, immunohistochemical staining was done to examine the differential expression levels of KIRREL in 71 cases of gastric cancer and paired normal tissues. KIRREL monoclonal antibody (ab82804, Abcam, UK) was used with a working concentration of 1: 100. The results of immunohistochemistry were independently judged and interpreted by 2 double-blinded pathologists. According to the previous literature reports [21,22], the immunoreaction score (IRS) of each slice was calculated, which was produced by staining intensity (SI) × number of stained cells (PP). The range of IRS score was 0–12. If the IRS score was greater than 4, KIRREL expression was considered to be high and IRS score ≤4 indicated KIRREL expression was low.
Statistical analysis
SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA) was used to statistically analyze the experimental data. Quantitative data was expressed as mean and standard deviation. TCGA and Oncomine data were analyzed by the independent-samples t test to compare the differential expression levels of KIRREL mRNA between gastric cancer and normal gastric tissue group. Chi-square test or Fisher’s exact test was employed to analyze the relationship between KIRREL differential expression levels and clinicopathological parameters of gastric cancer patients. Kaplan-Meier univariate and Cox multivariate survival analyses were performed to the assess the prognostic significance of KIRREL in gastric cancer patients. The difference was considered statistically significant when the P value was less than 0.05.
Results
KIRREL was highly expressed in gastric cancer
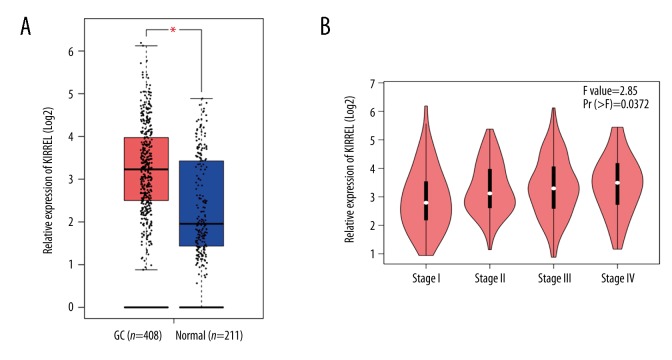
We initially used bioinformatics mining to explore the differential expression levels of KIRREL mRNA between gastric cancer and normal tissues based on TCGA and GEO (from Oncomine database) data. Results of TCGA data analysis showed that KIRREL mRNA expression level was remarkably higher in gastric cancer than that in normal gastric tissues (P<0.05, Figure 1A). Based on the different TNM stage (I vs. II vs. III vs. IV), KIRREL mRNA levels were dramatically elevated from stage I to stage IV (P=0.037, Figure 1B). Additionally, there were 9 GEO-sourced datasets mined from the Oncomine database, and meta-analysis results showed that KIRREL mRNA levels were significantly higher in gastric cancer than those in normal gastric tissues (P=0.017, Figure 2).
Figure 1.
Overexpression of KIRREL mRNA in gastric cancer predicted by TCGA data. (A) KIRREL mRNA levels in gastric cancer vs. normal gastric tissue. (B) Based on the different TNM stage (I vs. II vs. III vs. IV), KIRREL mRNA levels were dramatically elevated from stage I to stage IV.
Figure 2.
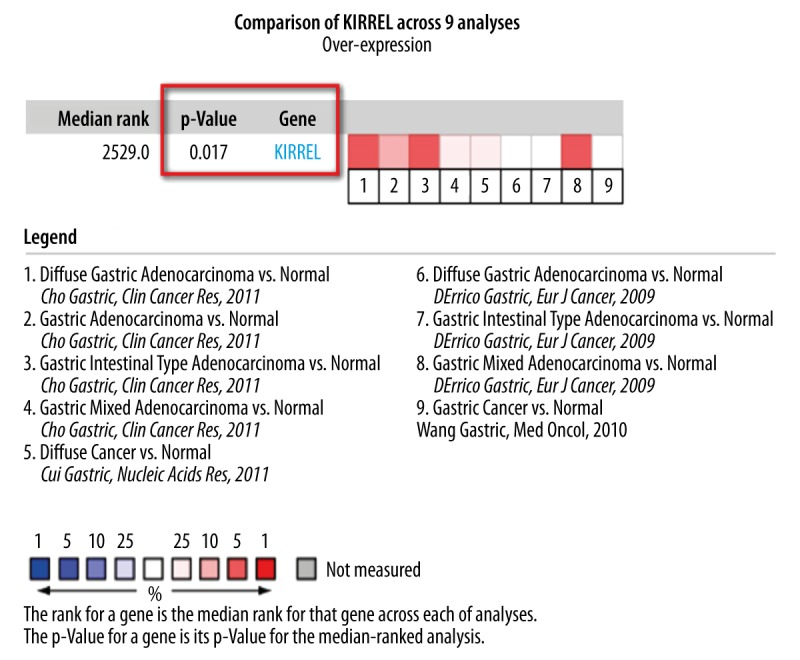
Meta-analysis of the 9 datasets on KIRREL mRNA levels in gastric cancer vs. normal gastric tissue searched by Oncomine database. The GSE number of each dataset were: Cho Gastric, 4 datasets, GEO: GSE13861; Cui Gastric, 1 dataset, GEO: GSE27342; DErrico Gastric, 3 datasets, GEO: GSE13911; Wang Gastric, 1 dataset, GEO: GSE19826.
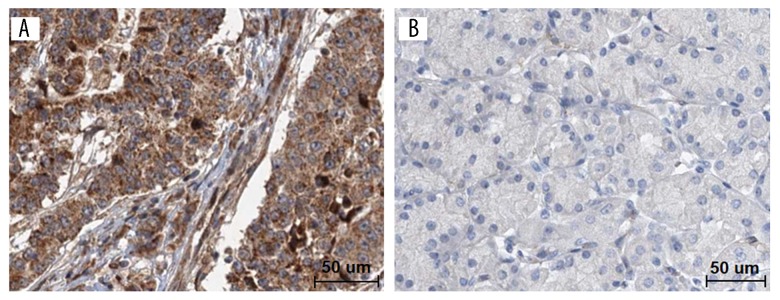
To verify the above predictive results, immunohistochemical staining was done to examine the expression levels of KIRREL protein in 71 cases of gastric cancer and paired normal tissues (Figure 3). As shown in Figure 3A, KIRREL was positively stained in cytomembrane and cytoplasm of gastric cancer. The high positive rate of KIRREL staining in gastric cancer was 61.97% (44/71). The expression levels of KIRREL protein were significantly higher in gastric cancer than those in paired normal gastric tissues (χ2=7.213, P=0.007, Table 2).
Figure 3.
KIRREL protein levels in 71 cases of gastric cancer and paired adjacent normal tissues by immunohistochemistry. (A) High expression of KIRREL in gastric cancer. (B) Low expression of KIRREL in paired normal tissue. Bar=50 um.
Table 2.
Overexpression of KIRREL in 71 cases of gastric cancer tissues.
| Tissues | KIRREL expression levels (Cases, %) | χ2 | P | |
|---|---|---|---|---|
| Low | High | |||
| Gastric cancer | 27 (38.03) | 44 (61.97) | 7.213 | 0.007 |
| Matched normal tissues | 43 (60.56) | 28 (39.44) | ||
Correlation between KIRREL and clinicopathological parameters of gastric cancer patients
To explore the correlation between KIRREL and clinicopathological parameters of gastric cancer patients, chi-square test was performed under KIRREL protein differential expression levels (low vs. high) and different subgroups of each parameter. As shown in Table 1, the differential expression levels of KIRREL protein (low vs. high) were significantly correlated with the age (≤60 vs. >60 years), tumor size (≤5 vs. >5 cm), T stage (T1–2 vs. T3–4), and TNM stage (I–II vs. III–IV) of gastric cancer patients (All P<0.05).
Prognostic significance of KIRREL in gastric cancer patients
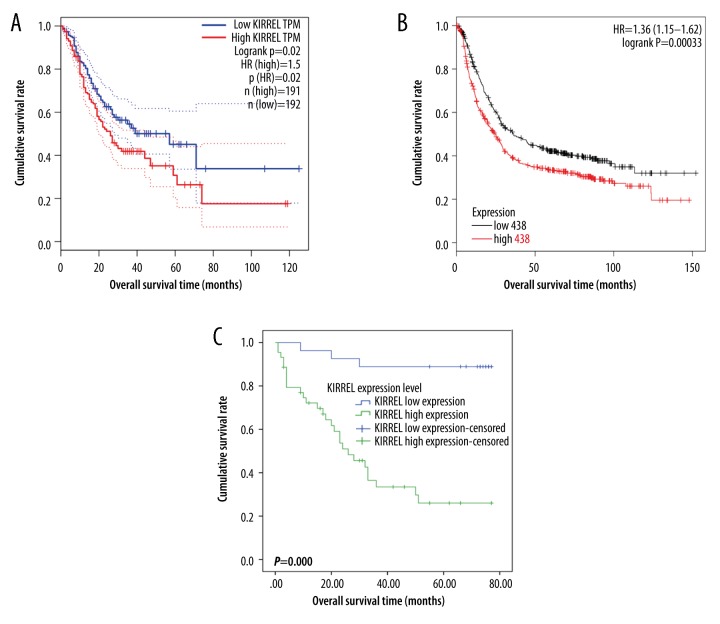
Lastly, the prognostic significance of KIRREL in gastric cancer patients was explored. By mining the TCGA database (Figure 4A) and using the Kaplan-Meier Plotter online tool (Figure 4B), we deduced that gastric cancer patients with KIRREL mRNA high levels had significantly decreased overall survival time than those with low KIRREL mRNA levels (All P values <0.05). Then, to test the above predictive findings, immunohistochemistry was carried out and the Kaplan-Meier univariate survival analysis revealed that gastric cancer patients with high levels of KIRREL protein had significantly shorter overall survival times than those with low levels of KIRREL protein (P=0.000, Figure 4C). Additionally, KIRREL levels, tumor size, histological differentiation, T stage, N stage, and TNM stage were all the statistically significant factors influencing the survival time of gastric cancer patients as determined by log-rank univariate survival analysis (Table 3). Subsequently, these 6 indicators were included in the Cox multivariate survival analysis. The results showed that KIRREL differential expression level (low vs. high) was the only independent parameter to predict the prognosis of gastric cancer patients (P=0.000, Table 4).
Figure 4.
Overall survival (OS) curves of gastric cancer patients based on the differential expression levels KIRREL mRNA or protein (low vs. high). (A) OS curve based on TCGA data; (B) OS curve based on GEO data (analyzed in Kaplan-Meier Plotter website); (C) OS curve based on the immunohistochemical data.
Table 3.
Kaplan-Meier univariate survival analysis of KIRREL and other clinicopathological parameters in gastric cancer patients.
| Clinicopathological parameters | Mean survival time (months) | 95% CI | P value |
|---|---|---|---|
| KIRREL levels | |||
| Low | 70.630 | 63.748–77.511 | 0.000 |
| High | 34.907 | 26.001–43.812 | |
| Age (years) | |||
| ≤60 | 51.862 | 41.117–62.607 | 0.678 |
| >60 | 48.478 | 38.583–58.373 | |
| Gender | |||
| Male | 49.938 | 41.512–58.364 | 0.965 |
| Female | 49.485 | 34.937–64.032 | |
| CEA (ng/ml) | |||
| ≤10 | 53.364 | 43.084–63.644 | 0.363 |
| >10 | 46.560 | 36.324–56.797 | |
| CA199 (U/ml) | |||
| ≤37 | 48.706 | 39.047–58.365 | 0.754 |
| >37 | 51.824 | 40.864–62.783 | |
| Tumor location | |||
| Antrum | 52.858 | 41.392–64.324 | 0.420 |
| Other sites | 47.725 | 38.344–57.106 | |
| Tumor size (cm) | |||
| ≤5 | 61.286 | 52.272–70.301 | 0.000 |
| >5 | 35.336 | 25.623–45.048 | |
| Histological differentiation | |||
| Well/moderate | 56.106 | 47.039–65.173 | 0.048 |
| Poor | 41.709 | 30.392–53.026 | |
| T stage | |||
| T1–2 | 67.477 | 58.545–76.410 | 0.002 |
| T3–4 | 41.437 | 32.506–50.369 | |
| N stage | |||
| N0 | 63.561 | 53.703–73.420 | 0.008 |
| N1–3 | 42.075 | 32.851–51.299 | |
| TNM stage | |||
| I–II | 66.234 | 58.174–74.295 | 0.000 |
| III–IV | 36.256 | 26.695–45.818 | |
Table 4.
Cox multivariate analysis of KIRREL and other clinicopathological parameters in gastric cancer patients.
| Covariates | HR | 95% CI for HR | P value |
|---|---|---|---|
| KIRREL expression levels (low vs. high) | 7.918 | 2.317–27.060 | 0.001 |
| Histological differentiation (well/moderate vs. poor) | 1.325 | 0.634–2.769 | 0.455 |
| Tumor size (≤5 vs. >5 cm) | 1.367 | 0.545–3.431 | 0.505 |
| T stage (T1–2 vs. T3–4) | 1.759 | 0.515–6.004 | 0.367 |
| N stage (N0 vs. N1–3) | 1.558 | 0.583–4.168 | 0.377 |
| TNM stage (I–II vs. III–IV) | 2.256 | 0.802–6.350 | 0.123 |
Discussion
There have been few studies on the KIRREL gene in tumors. Existing studies have revealed that KIRREL gene is abnormally expressed in some malignancies. For example, Hu et al. [14] performed integrative analysis to mine the GEO microarray data and found a total of 2325 differentially expressed genes (DEGs) involved in pancreatic cancer. Further, gene coexpression network analysis identified that KCTD10, KIRREL, DPP10, and UNC80 were the hub genes associated with the pancreatic carcinogenesis. Johnson et al. [15] revealed that KIRREL serves as a novel substrate specifically cleaved by RHBDL2, affecting epithelial homeostasis. Wang et al. [16] explored the CpG methylation patterns associated with gene expression variation in osteosarcoma, showing that several differentially methylated sites were associated with upregulation of SEZ6L2, KIRREL, CEP72, and CDK4 and might have a key role in the pathogenesis of osteosarcomas through promotion of cell proliferation and metastasis. Additionally, KIRREL was found to be associated with the resistance of molecularly-targeted drugs in lung cancer [17] and wild-type gastrointestinal stromal tumors (GIST) [18]. However, the expression levels of KIRREL in gastric cancer and its prognostic significance have not been explored in detail.
In this study, we initially used bioinformatics mining method to explore the differential expression levels of KIRREL mRNA between gastric cancer and normal tissues based on TCGA and GEO (from Oncomine database) data. Results of both TCGA and GEO data mining showed that KIRREL mRNA expression levels were remarkably higher in gastric cancer than that in normal gastric tissues. Based on the different TNM stage (I vs. II vs. III vs. IV), KIRREL mRNA levels were dramatically elevated from stage I to stage IV. Further, to verify the above predictive results, immunohistochemical staining was done to examine the expression levels of KIRREL protein in 71 cases of gastric cancer and paired normal tissues. Consistent with the predictive results, the high positive rate of KIRREL staining in gastric cancer was 61.97% and the expression levels of KIRREL protein were significantly higher in gastric cancer than those in paired normal gastric tissues. All these findings suggest that KIRREL serves as an oncogene in advancing the occurrence and development of gastric cancer.
We also explored and demonstrated the correlation between KIRREL and clinicopathological parameters of gastric cancer patients and its prognostic significance. Chi-square testing revealed that the differential expression levels of KIRREL protein (low vs. high) were significantly correlated with the age, tumor size, T stage, and TNM stage of gastric cancer patients. By mining the TCGA database and using the Kaplan-Meier Plotter online tool, we found that gastric cancer patients with high KIRREL mRNA levels had significantly decreased overall survival time compared to those with low KIRREL mRNA levels. Moreover, Kaplan-Meier univariate survival analysis based on the immunohistochemical data revealed that gastric cancer patients with high levels of KIRREL protein had significantly shorter overall survival time than those with low levels of KIRREL protein. Additionally, Cox multivariate survival analysis revealed that KIRREL differential expression levels (low vs. high) were the only independent parameter to predict the prognosis of gastric cancer patients. Taken together, these results suggest that high KIRREL expression could be used as an independent predictor to indicate the poor prognosis of gastric cancer patients and KIRREL might be one of the pivotal target genes involved in the growth and metastasis of gastric cancer.
Our study has certain limitations. Firstly, small gastric cancer tissue samples were collected retrospectively, which might lead to biased statistical results to some extent. Secondly, because most of the retrospectively collected GC patients lost the time of disease-free survival (DFS) information, we could not perform the relative statistical analysis. Thirdly, only immunohistochemical staining, a semi-quantitative method, was used in our validation experiment and some other quantitative methods like Western blot or qRT-PCR will be performed in our future research. Lastly, the exact biological function of KIRREL on gastric cancer and its detailed molecular regulation mechanisms were not assessed in the present study. These deficiencies will be improved and studied in our future experiments.
Conclusions
This preliminary study confirmed that KIRREL was overexpressed in gastric cancer and high KIRREL expression could be used as an independent parameter to predict the poor prognosis of gastric cancer patients. In future, KIRREL may become a new therapeutic target for the treatment of gastric cancer.
Acknowledgements
We sincerely thank Dr. Shan Huang and Dr. Rong Qin (pathologists, Department of Pathology, the Second Affiliated Hospital of Anhui Medical University, Hefei, China) for their kind help with the pathology work.
Footnotes
Source of support: Departmental sources
Conflicts of interest
None.
References
- 1.Globocan 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012 [EB/OL] WHO: International Agency for Research on Cancer; [Accessed 8 April 2016]. http://globocan.iarc.if/Default.aspx. [Google Scholar]
- 2.Shen L, Shan YS, Hu HM, et al. Management of gastric cancer in Asia: Resource-stratified guidelines. Lancet Oncol. 2013;14:e535–47. doi: 10.1016/S1470-2045(13)70436-4. [DOI] [PubMed] [Google Scholar]
- 3.Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39:1010428317714626. doi: 10.1177/1010428317714626. [DOI] [PubMed] [Google Scholar]
- 4.Ajani JA, Lee J, Sano T, et al. Gastric adenocarcinoma. Nat Rev Dis Primers. 2017;3:17036. doi: 10.1038/nrdp.2017.36. [DOI] [PubMed] [Google Scholar]
- 5.Yang K, Hu JK. Gastric cancer treatment: Similarity and difference between China and Korea. Transl Gastroenterol Hepatol. 2017;2:36. doi: 10.21037/tgh.2017.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Qin S, Xu J, et al. Apatinib for chemotherapy-refractory advanced metastatic gastric cancer: Results from a randomized, placebo-controlled, parallel-arm, phase II trial. J Clin Oncol. 2013;31:3219–25. doi: 10.1200/JCO.2013.48.8585. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Qin S, Xu J, et al. Randomized, double-blind, placebo-controlled phase III trial of apatinib in patients with chemotherapy-refractory advanced or metastatic adenocarcinoma of the stomach or gastroesophageal junction. J Clin Oncol. 2016;34:1448–54. doi: 10.1200/JCO.2015.63.5995. [DOI] [PubMed] [Google Scholar]
- 8.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 9.Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet. 2016;388:2654–64. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 10.Sellin L, Huber TB, Gerke P, et al. NEPH1 defines a novel family of podocin interacting proteins. FASEB J. 2003;17:115–17. doi: 10.1096/fj.02-0242fje. [DOI] [PubMed] [Google Scholar]
- 11.Liu G, Kaw B, Kurfis J, et al. Neph1 and nephrin interaction in the slit diaphragm is an important determinant of glomerular permeability. J Clin Invest. 2003;112:209–21. doi: 10.1172/JCI18242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerke P, Huber TB, Sellin L, et al. Homodimerization and heterodimerization of the glomerular podocyte proteins nephrin and NEPH1. J Am Soc Nephrol. 2003;14:918–26. doi: 10.1097/01.asn.0000057853.05686.89. [DOI] [PubMed] [Google Scholar]
- 13.Huber TB, Schmidts M, Gerke P, et al. The carboxyl terminus of Neph family members binds to the PDZ domain protein zonula occludens-1. J Biol Chem. 2003;278:13417–21. doi: 10.1074/jbc.C200678200. [DOI] [PubMed] [Google Scholar]
- 14.Hu B, Shi C, Jiang HX, et al. Identification of novel therapeutic target genes and pathway in pancreatic cancer by integrative analysis. Medicine (Baltimore) 2017;96:e8261. doi: 10.1097/MD.0000000000008261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson N, Březinová J, Stephens E, et al. Quantitative proteomics screen identifies a substrate repertoire of rhomboid protease RHBDL2 in human cells and implicates it in epithelial homeostasis. Sci Rep. 2017;7:7283. doi: 10.1038/s41598-017-07556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q. CpG methylation patterns are associated with gene expression variation in osteosarcoma. Mol Med Rep. 2017;16:901–7. doi: 10.3892/mmr.2017.6635. [DOI] [PubMed] [Google Scholar]
- 17.Gimenez-Xavier P, Pros E, Bonastre E, et al. Genomic and molecular screenings identify different mechanisms for acquired resistance to MET inhibitors in lung cancer cells. Mol Cancer Ther. 2017;16:1366–76. doi: 10.1158/1535-7163.MCT-17-0104. [DOI] [PubMed] [Google Scholar]
- 18.Beadling C, Patterson J, Justusson E, et al. Gene expression of the IGF pathway family distinguishes subsets of gastrointestinal stromal tumors wild type for KIT and PDGFRA. Cancer Med. 2013;2:21–31. doi: 10.1002/cam4.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–33. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Zhang M, Peng Y, et al. Ubiquitin associated protein 2-like (UBAP2L) overexpression in patients with hepatocellular carcinoma and its clinical significance. Med Sci Monit. 2017;23:4779–88. doi: 10.12659/MSM.907071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu PL, He YF, Yao HH, et al. Martrilin-3 (MATN3) overexpression in gastric adenocarcinoma and its prognostic significance. Med Sci Monit. 2018;24:348–55. doi: 10.12659/MSM.908447. [DOI] [PMC free article] [PubMed] [Google Scholar]