Abstract
Background
Papain-like and legumain-like proteases are proteolytic enzymes which play key roles in plant development, senescence and defense. The activities of proteases in both families could be inhibited by a group of small proteins called cystatin. Cystatin family genes have been well characterized both in tobacco and rice, suggesting their potential roles in seed development. However, their potential targets, papain-like and legumain-like proteases, have not been well characterized in plants, especially in rice, a model plant for cereal biology.
Results
Here, 33 papain-like and 5 legumain-like proteases have been identified in rice genome, respectively. Gene structure, distribution in rice chromosome, and evolutionary relationship to their counterparts in other plants have been well characterized. Comprehensive expression profile analysis revealed that two family genes display divergent expression pattern, which are regulated temporally and spatially during the process of seed development and germination. Our experiments also revealed that the expression of most genes in these two families is sensitively responsive to plant hormones and different abiotic stresses.
Conclusions
Genome-wide identification and comprehensive gene expression pattern analysis of papain-like and legumain-like proteases in rice suggests their multiple and cooperative roles in seed development and response to environmental variations, which provides several useful cues for further in-depth study.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1298-1) contains supplementary material, which is available to authorized users.
Keywords: Papain-like protease, Legumain-like protease, Plant hormones, Stress, Rice
Background
Plant genomes encode hundreds of proteases, which belong to dozens of unrelated families and have been divided into different families and clans on the basis of evolutionary relationships. Among these proteases, papain-like cysteine proteases (PLCPs) in peptidase C1A family and legumain-like cysteine proteases (LLCPs) in peptidase C13 family [1] are known as two specific types of cysteine proteases, whose activities could both be inhibited by a group of small proteins called cystatin [2, 3].
PLCPs contain two domains: an α-helix and β-sheet which delimit a cleft at the surface acting as the substrate-binding groove [4] and a catalytic triad Cys-His-Asn which is highly conserved among different kingdoms. PLCPs are encoded as inactive precursors, which comprise an N-terminal signal peptide, a prodomain and the mature protein. By limited intra- or inter-molecular proteolysis, after cleaving off an inhibitory propeptide in an acidic environment [5], PLCPs become active [6] and function in various physiological processes such as seed germination [7, 8], male organ development [9–12], senescence [13], defense against pathogens [14, 15], and response to insect attack or abiotic stress [16, 17].
LLCPs are a group of Asn-specific proteinases, which were primarily located in the vacuole and responsible for maturation of storage proteins in seeds [18–20]. Considering their intracellular localizations and function, LLCPs are also named ‘vacuolar processing enzymes’ (VPEs) [21]. Similar to PLCPs, VPEs are usually synthesized as inactive precursors composed of a short N- and a much longer C-terminal propeptide flanking the mature enzyme [21, 22]. VPE is usually self-catalytically converted into the mature form at acidic condition by sequential removal of the C-terminal propetide and N-terminal propetide, which is an essential step for enzyme activation [18, 23]. VPEs have been shown to participate in protein processing in several physiological processes, which are responsible for maturation and/or activation of various vacuolar proteins [20, 24]. In addition, several VPEs have also been shown to function in regulating programmed cell death (PCD) both in developmental process and defense responses through their caspase-like activities [25–28].
Rice (Oryza sativa L.) is one of most widely grown crop in the world, which provides main food source for people in Southeast Asia, and has been considered as model species for many basic and applied researches. Great efforts have been made to improve rice yield and resistance to different biotic and abiotic stresses [29–32]. As described above, PLCPs and LLCPs were reported to be involved in seed development and plant defense against different stresses. However, few of them have been well characterized, especially in rice [11, 12, 33]. Thus, genome wide identification and expression analysis of PLCPs and LLCPs is helpful to explore their potential roles in rice seed development, and improve rice yield and resistance to various stresses. Here, 33 PLCPs and 5 LLCPs have been identified and characterized, providing valuable clues to gain insight into their specific physiological roles in the further study.
Results
Identification and cloning of OsCPs and OsVPEs in rice genome
To identify genes encoding PLCPs and LLCPs in rice, tBLASTP program in National Center for Biotechnology Information (NCBI) database was performed using 30 protein sequences of PLCPs in Arabidopsis thaliana identified by Beers et al. [34] and 4 protein sequences of LLCPs in A. thaliana [27], respectively. After removing redundant sequences, candidates with intact open reading frame covering peptidase C1A and peptidase C13 domain were considered as true OsCP and OsVPE in rice. These predicted OsCPs and OsVPEs were further confirmed by PCR using cDNA as templates. Finally, 33 genes (Designated as OsCP1-OsCP33) encoding PLCP and 5 genes (Designated as OsVPE1-OsVPE5) encoding LLCP were identified in rice genome, respectively. The information for each gene in rice was listed in Table 1.
Table 1.
Detailed information of OsCPs and OsVPEs
| Gene | RAP-DB Locus ID | TIGR/MSU Locus ID | Signal peptide | No of amino acid | Mol. Weight (kDa) | PI | Chromosome Location |
|---|---|---|---|---|---|---|---|
| OsCP1 | Os04g0670200 | LOC_Os04g57440 | 21 | 466 | 49,798.7 | 5.02 | Chr4 34,176,732–34,172,494 |
| OsCP2 | Os04g0670500 | LOC_Os04g57490 | 31 | 490 | 52,642.3 | 7.61 | Chr4 34,205,994–34,207,906 |
| OsCP3 | Os11g0255300 | LOC_Os11g14900 | 24 | 378 | 40,933.2 | 5.79 | Chr11 8,382,036–8,379,342 |
| OsCP4 | Os05g0108600 | LOC_Os05g01810 | 22 | 358 | 39,036.6 | 4.96 | Chr5 486,053–488,406 |
| OsCP5 | Os01g0971400 | LOC_Os01g73980 | 27 | 365 | 39,927.8 | 5.06 | Chr1 42,855,657–42,857,466 |
| OsCP6 | Os01g0907600 | LOC_Os01g67980 | 28 | 371 | 40,719.5 | 6.64 | Chr1 39,501,330–39,502,978 |
| OsCP7 | Os02g0715000 | LOC_Os02g48450 | 30 | 366 | 40,599.9 | 8.00 | Chr2 29,667,743–29,669,662 |
| OsCP8 | Os08g0556900 | LOC_Os08g44270 | 30 | 385 | 41,871.8 | 6.85 | Chr8 27,875,142–27,876,599 |
| OsCP9 | Os09g0497500 | LOC_Os09g32230 | 24 | 349 | 37,379.2 | 4.76 | Chr9 19,240,737–19,239,243 |
| OsCP10 | Os05g0508300 | LOC_Os05g43230 | 25 | 450 | 47,620.8 | 5.97 | Chr5 25,153,902–25,157,655 |
| OsCP11 | Os04g0203500 | LOC_Os04g12660 | 23 | 301 | 33,117.8 | 7.78 | Chr4 7,008,959–7,010,088 |
| OsCP12 | Os09g0381400 | LOC_Os09g21370 | 26 | 362 | 38,639.5 | 5.19 | Chr9 12,899,550–12,897,272 |
| OsCP13 | Os06g0582600 | LOC_Os06g38450 | 29 | 357 | 38,509.2 | 7.11 | Chr6 22,779,409–22,781,109 |
| OsCP14 | Os01g0347500 | LOC_Os01g24550 | 26 | 361 | 38,988.9 | 6.38 | Chr1 13,839,339–13,837,906 |
| OsCP15 | Os01g0613500 | LOC_Os01g42780 | 30 | 360 | 37,513.6 | 7.52 | Chr1 24,345,173–24,343,518 |
| OsCP16 | Os01g0330200 | LOC_Os01g22670 | 22 | 357 | 38,565.5 | 6.41 | Chr1 12,747,081–12,745,130 |
| OsCP17 | Os01g0217300 | LOC_Os01g11840 | 28 | 366 | 39,292.3 | 5.75 | Chr1 6,400,890–6,402,066 |
| OsCP18 | Os09g0442300 | LOC_Os09g27030 | 23 | 362 | 39,114.0 | 7.16 | Chr9 16,439,141–16,443,631 |
| OsCP19 | Os01g0330300 | LOC_Os01g22680 | 26 | 367 | 39,474.7 | 6.84 | Chr1 12,751,899–12,753,162 |
| OsCP20 | Os02g0469600 | LOC_Os02g27030 | 19 | 373 | 40,591.6 | 6.44 | Chr2 15,891,528–15,895,297 |
| OsCP21 | Os04g0208200 | LOC_Os04g13140 | 26 | 349 | 36,867.3 | 5.10 | Chr4 7,250,358–7,251,846 |
| OsCP22 | Os04g0107700 | LOC_Os04g01710 | 31 | 383 | 41,996.4 | 5.57 | Chr4 472,891–471,100 |
| OsCP23 | Os12g0273800 | LOC_Os12g17540 | 24 | 350 | 37,170.8 | 4.79 | Chr12 10,053,411–10,055,200 |
| OsCP24 | Os09g0562700 | LOC_Os09g38920 | 32 | 382 | 40,882.8 | 6.07 | Chr9 22,353,654–22,351,648 |
| OsCP25 | Os01g0347600 | LOC_Os01g24560 | 29 | 343 | 37,322.2 | 6.90 | Chr1 13,842,272–13,843,749 |
| OsCP26 | Os09g0564600 | LOC_Os09g39110 | 26 | 374 | 41,375.9 | 5.49 | Chr9 22,453,266–22,450,380 |
| OsCP27 | Os01g0613800 | LOC_Os01g42790 | 22 | 359 | 38,250.9 | 7.71 | Chr1 24,353,345–24,351,302 |
| OsCP28 | Os07g0480900 | LOC_Os07g29760 | 22 | 376 | 39,868.1 | 6.39 | Chr7 17,496,502–17,498,225 |
| OsCP29 | Os09g0565100 | LOC_Os09g39170 | 24 | 319 | 34,267.0 | 5.99 | Chr9 22,494,320–22,491,705 |
| OsCP30 | Os04g0311400 | LOC_Os04g24600 | 24 | 381 | 41,555.7 | 6.51 | Chr4 14,129,614–14,132,611 |
| OsCP31 | Os09g0564000 | LOC_Os09g39060 | 19 | 283 | 30,472.5 | 4.93 | Chr9 22,422,558–22,420,898 |
| OsCP32 | Os09g0564200 | LOC_Os09g39070 | 25 | 369 | 39,515.8 | 4.98 | Chr9 22,430,508–22,427,465 |
| OsCP33 | Os05g0310500 | LOC_Os05g24550 | 22 | 434 | 48,152.4 | 7.09 | Chr5 14,161,386–14,167,285 |
| OsVPE1 | Os01g0559600 | LOC_Os01g37910 | 29 | 501 | 54,964.9 | 5.80 | Chr1 21,230,393–21,234,929 |
| OsVPE2 | Os04g0537900 | LOC_Os04g45470 | 23 | 497 | 54,885.8 | 5.14 | Chr4 26,908,679–26,903,866 |
| OsVPE3 | Os02g0644000 | LOC_Os02g43010 | 22 | 496 | 54,847.9 | 5.48 | Chr2 25,895,244–25,890,441 |
| OsVPE4 | Os05g0593900 | LOC_Os05g51570 | 19 | 474 | 51,845.3 | 5.72 | Chr5 29,580,698–29,579,044 |
| OsVPE5 | Os02g0219400 | LOC_Os02g12740 | 35 | 404 | 45,123.2 | 5.70 | Chr2 6,672,950–6,668,822 |
Gene structure analysis of OsCPs and OsVPEs
Intron-exon structure of OsCPs and OsVPEs were determined by comparison of the cDNA sequences with their corresponding genomic sequences. The results revealed that the genes encoding PLCPs could be divided into three groups according to the number of intron (Fig. 1a). The first group consists of OsCPs without any intron. Two OsCPs (OsCP6 and OsCP8) belong to this group. The second group is single-intron gene, and above half of OsCPs (OsCP2, OsCP4, OsCP5, OsCP12, OsCP13, OsCP14, OsCP15, OsCP16, OsCP17, OsCP19, OsCP21, OsCP22, OsCP23, OsCP24, OsCP25, OsCP27, OsCP28 and OsCP31) fall into the second group. The third group is multiple-intron OsCPs, and the remaining OsCPs (OsCP1, OsCP3, OsCP7, OsCP9, OsCP10, OsCP11, OsCP18, OsCP20, OsCP26, OsCP29, OsCP30, OsCP32 and OsCP33) belong to the third group. The number of introns in the third group is divergent, ranging from two to seven (Fig. 1a). As for OsVPEs, most OsVPEs harbored multiple introns with one exception (OsVPE4) (Fig. 1b). However, the length of coding sequences of OsCPs and OsVPEs seems conserved, with 852 to 1473 nucleotides for OsCPs and 1215 to 1506 nucleotides for OsVPEs, indicating that divergent number and length of intron determine the gene size of two families in genome.
Fig. 1.

Genomic structure of OsCPs and OsVPEs in rice. a and b correspond to OsCPs and OsVPEs respectivily
Chromosomal localization and gene duplication analysis
Physical locations of these two families in the rice chromosomes were determined according to their genome sequences. 33 OsCPs were mapped to 10 rice chromosomes with an uneven distribution pattern (Fig. 2). Majority of OsCPs were assigned to chromosome 1, 4 or 9, with 6–9 OsCPs in each chromosome. The distribution of remaining OsCPs was scattered, with one to three genes in each chromosome. As for OsVPEs, they were assigned to four chromosomes (Fig. 2). Chromosome 2 contains two genes (OsVPE3 and OsVPE5), and chromosome 1, 4, 5 harbor one VPE respectively.
Fig. 2.

Localization of OsCPs and OsVPEs on rice chromosomes. Chromosome number was indicated at the top of each chromosome. The size of chromosome was labeled on the left of the figure. Tandem duplicated genes were outlined with red color, and segmental duplicated gene pairs were linked with blue and red lines
Furthermore, gene duplication events of OsCP and OsVPE families during long evolutionary history were also analyzed. Gene pairs separated at most by five intervening genes were considered as tandem duplicates [35]. There are five pairs located in tandem repeats (OsCP1/OsCP2, OsCP14/OsCP25, OsCP16/OsCP19, OsCP15/OsCP27 and OsCP26/OsCP31/OsCP32) in OsCP family (Fig. 2). However, no gene tandem duplication events was found in OsVPE family. At same time, three pairs (OsCP4/OsCP5, OsCP8/OsCP31 and OsCP20/OsCP30) from OsCP family and one pair (OsVPE2/OsVPE3) from OsVPE family were found to be present on the duplicated chromosomal segments, suggesting that OsCPs in rice expanded through both segmental and tandem duplications, but OsVPEs only through segmental duplications.
Ka/Ks ratio, a tool to measure gene divergence, was also calculated in the present study. Ka/Ks < 1 means negative selection; Ka/Ks = 1 means neutral selection, and Ka/Ks > 1 means positive selection [36]. Duplicated pairs (OsCP1/OsCP2, OsCP14/OsCP25, OsCP16/OsCP19, OsCP15/OsCP27 and OsCP26/OsCP31) were suggested to belong to positive selection as indicated from Ka/Ks value, and Ka/Ks ratios of the remaining pairs (OsCP4/OsCP5, OsCP8/OsCP31, OsCP20/OCP30, OsCP26/OsCP32, OsCP31/OsCP32 and OsVPE2/OsVPE3) were < 1, suggesting that negative selection on these duplication events occurred (Table 2). In addition, the dates of gene duplication events were figured out as well based on the proposed divergences of rice from other grasses. Gene duplications among OsCPs probably occurred from 13.28 to 56.21 million years ago, and gene duplications among OsVPEs took place 71.64 million years ago.
Table 2.
Ka/Ks analysis and duplicated date calculation for OsCPs and OsVPEs
| Duplicated pair | Duplicate type | Ka | Ks | Ka/Ks | Positive selection | Date of gene duplication (million years) |
|---|---|---|---|---|---|---|
| OsCP4/OsCP5 | Segmental | 0.1812 | 0.1854 | 0.9773 | No | 14.26 |
| OsCP8/OsCP31 | Segmental | 0.5674 | 0.7307 | 0.7765 | No | 56.21 |
| OsCP20/OCP30 | Segmental | 0.1456 | 0.5487 | 0.2654 | No | 42.21 |
| OsCP1/OsCP2 | Tandem | 0.3532 | 0.1727 | 2.0452 | Yes | 13.28 |
| OsCP14/OsCP25 | Tandem | 0.3036 | 0.2298 | 1.3211 | Yes | 17.68 |
| OsCP16/OsCP19 | Tandem | 0.3724 | 0.3459 | 1.0766 | Yes | 26.61 |
| OsCP15/OsCP27 | Tandem | 0.3776 | 0.3423 | 1.1031 | Yes | 26.33 |
| OsCP26/OsCP31 | Tandem | 0.4620 | 0.3929 | 1.1759 | Yes | 30.22 |
| OsCP26/OsCP32 | Tandem | 0.3422 | 0.6900 | 0.4959 | No | 53.08 |
| OsCP31/OsCP32 | Tandem | 0.2273 | 0.4190 | 0.5425 | No | 32.23 |
| OsVPE2/OsVPE3 | Segmental | 0.1482 | 0.9313 | 0.1591 | No | 71.64 |
Ka/Ks < 1 means negative selection, Ka/Ks = 1 means neutral selection, and Ka/Ks > 1 means positive selection
Protein structure and phylogenetic analysis
To gain insight into potential subcellular location of each OsCP and OsVPE, signal peptide predication of each protein using SignalP 4.1 was firstly carried out [37]. The results revealed that all OsCPs and OsVPEs contain a predicted signal peptide, indicating that all members in these two families could enter the endomembrane system (Figs. 3a and 4a). Subcellular targeting of all members in these two families was also predicted. The results revealed that there are two subcellular targeting sequences in OsCPs. The first is the vacuolar targeting sequence NPIR, which could be detected in the N terminus of OsCP18. The second is the ER targeting sequence, which could be detected both in OsCP3 and OsCP8 (Additional file 1: Figure S1).
Fig. 3.

Schematic diagram of OsCPs and analysis of conserved motifs in OsCPs. a Schematic diagram of OsCPs. b Conservative analysis of the inhibitor domain in OsCPs. c Conservative analysis of the peptdiase C1A domain in OsCPs
Fig. 4.

Schematic diagram of OsVPEs and analysis of conserved motifs in OsVPEs. a Schematic diagram of OsVPEs. b Conservative analysis of peptdiase C13 domain in OsVPEs
Multiple sequence alignment was performed to explore sequence features and to identify functional motifs of two families in rice. As for OsCPs, several typical motifs have been identified. The first, Cathepsin propeptide inhibitor domain was found at the N terminus of all OsCPs except OsCP33 (Fig. 3a and Additional file 1: Figure S1). The motif sequence was ExxxRxxxFxxNxxxI/VxxxN with one mismatch and the most conservative positions in rice are 1, 5 and 12 (Fig. 3b). Instead of “ERFNIN” motif, three OsCPs (OsCP14, OsCP17 and OsCP19) carry a similar “ERWNIR” motif and three OsCPs (OsCP20, OsCP28 and OsCP30) carry a conserved “ERFNAQ” motif just like cathepsin F in animals. Second, catalytic triad (Cys-His-Asn) is conserved in rice PLCPs except OsCP16, in which serine is substituted for cysteine as a nucleophile of enzyme activity, and the amino acids before and after the catalytic triad are also conserved, which could be detected in all OsCPs (Fig. 3c). Third, the active region is highly conserved and rich in polar amino acids. The fourth, a C-terminal extension consisting of a Pro-rich domain followed by a granulin-like domain (Cx5Cx5CCCx7Cx4CCx6CCx5CCx6Cx6C) was detected in three OsCPs (OsCP1, OsCP2 and OsCP10). Granulins are growth hormones that are released upon wounding in the animal kingdom, but this fusion only occurs in plants and is not detected in animals [5, 38]. However, exact roles of granulin domain in OsCP protein are still waiting to be explored in the further study. In contrast to OsCPs, OsVPEs comprise a shorter N-terminal and a much longer C-terminal propeptide (Fig. 4a and Additional file 2: Figure S2). The active region of OsVPEs, rich in polar amino acids, is highly conserved and the catalytic triad (Cys-His-Asn) is also detected in five OsVPEs (Fig. 4b).
To further analyze phylogenetic relationships of OsCPs and OsVPEs to their counterparts from other plants, a total 133 PLCPs from Hordeum vulgare, Zea mays and A. thaliana, and 29 LLCPs from H. vulgare, Z. mays, A. thaliana and Glycine max were used to construct a phylogenetic tree. OsCPs were distributed evenly across evolutionary tree branches. Phylogenetic relationship did not reflect the distinction between monocot and eudicot plants, just like the cystatins in rice (Fig. 5a) [39]. However, LLCPs were divided into the two independent monocots and eudicots, indicating their functional difference between monocots and eudicots LLCPs (Fig. 5b).
Fig. 5.
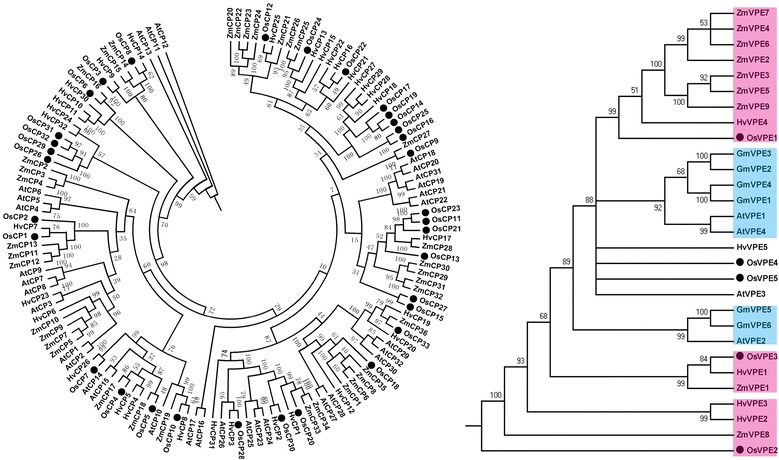
Phylogenetic relationships of papain-like and legumain-like cysteine proteases among rice and other plant species. The tree was calculated with Phylip Ver. 3.68 software using the Protpars method. Rice papain-like and legumain-like cysteine proteases were marked by black dots and plant legumain-like cysteine proteases from monocots and eudicots were shaded in purple and light blue respectively. The numbers at the nodes indicate the bootstrap values
Expression pattern of OsCPs and OsVPEs in different tissues under normal conditions
To assess the potential functions of OsCPs and OsVPEs during rice development, their expression pattern was revealed by two approaches: publicly available expression data and real-time reverse transcription-PCR (RT-qPCR). The expression data of two families from the spatio-temporal gene expression profiles of various tissues/organs at RiceXPro (Rice Expression Profile Database) were obtained and summarized to construct expression profile of OsCPs and OsVPEs. OsCPs display diverse expression patterns as shown in Additional file 3: Figure S3. The transcripts of most OsCPs could be detected in the same tissue, for example, at least 20 OsCPs were detected in roots, 8 genes in flag leaf, 6 genes in palea and lemma, indicating that rice PLCPs may function redundantly in vivo. Two OsCPs (OsCP6 and OsCP28) display a similar expression pattern, which are predominantly expressed in embryo and endosperm, but lower in other tissue tested. Expression pattern of OsCPs and OsVPEs derived from MPSS (massively parallel signature sequencing) were also listed in Additional file 4: Table S4, which is similar to the results from RiceXPro.
To confirm the public data, RT-qPCR was used to construct the expression profile of OsCPs and OsVPEs. cDNAs prepared from different tissues such as leaves, stems, roots, anther, out glume, inner glume and seeds at different developmental stages were chosen as templates for RT-qPCR. Similar to the public data, heatmap analysis based on the relative expression level show that most members of OsCPs display diverse expression pattern (Fig. 6a). Three OsCPs (OsCP1, OsCP20 and OsCP33) were abundantly expressed in each tissue tested. Two OsCPs (OsCP3 and OsCP12) had a similar expression pattern, both preferentially expressed in anther, indicating a potential role in anther development. Generally, VPEs in plants could be separated into two subfamilies: vegetative-type VPEs and seed-type VPEs [40]. However, OsVPEs displayed a rather broad expression profile, which could be detected in both seed and vegetative tissues like HvLeg-2 and HvLeg-4 in barley (Fig. 6b and Additional file 5: Figure S5) [41].
Fig. 6.

Heatmap analysis of OsCP and OsVPE gene expression. (a). Heatmap analysis of OsCPs, (b). Heatmap analysis of OsVPEs. The expression was normalized against OsActin and OsUBC, and data represent the mean with three independent experiments. A red box indicates the higher expression level, whereas the blue box indicates the lower expression level. The scale bar represents the fold change (log2 value)
OsCPs and OsVPEs display dynamic expression pattern during the processes of seed development and germination
Early reports demonstrated that both PLCPs and LLCPs are involved in seed formation and seed germination [7, 8, 27, 33, 42]. To explore potential roles of two family genes in the process of rice seed development, the transcriptional level of each gene in seeds at different developmental stages and different germination stages were comprehensively analyzed. In general, more than half of OsCPs and all the OsVPEs could be detected in seeds at different stages (Fig. 7). Among them, four OsCPs (OsCP1, OsCP8, OsCP20 and OsCP33) and three OsVPEs (OsVPE1, OsVPE2 and OsVPE3) were abundant in seeds. It is common knowledge that seed development in rice consists of the development of embryo and endosperm and the former could be divided into three stages: proembryo development, embryo differentiation and maturation [43]. Seven OsCPs and four OsVPEs were abundant in seeds corresponding to the proembryo developmental stage and nine OsCPs and three OsVPEs could be detected in the seeds (4~ 10 days after pollination, DAP), speculating their roles in organ differentiation of rice embryo development (Fig. 7a). During the process of endosperm development, accumulation of storage compounds is very important and closely related to grain production and quality. Expression pattern analysis revealed that twelve OsCPs (OsCP1, OsCP8, OsCP10, OsCP18, OsCP20, OsCP24, OsCP28, OsCP30, OsCP33, OsVPE1, OsVPE2 and OsVPE3) were strongly expressed in this stage, indicating that they may take part in the processing of storage proteins during storage phase of endosperm development. After 12 days, endosperm cells begin to degrade through PCD, during which the expression levels of five OsCPs and two OsVPEs still kept high, suggesting that these genes perhaps participated in the degradation of endosperm cells.
Fig. 7.

Dynamic changes of the expression level of OsCPs and OsVPEs during the processes of seed development and germination. (a). Overview of the expression of OsCPs and OsVPEs during the processes of embryo and endosperm development. (b-c). Dynamic changes of the expression level of OsCPs during the processes of seed development (b) and germination (c). (d-e). Dynamic changes of the expression level of OsVPEs during the processes of seed development (c) and germination (d). The expression was normalized against OsActin and OsUBC. The data represent fold change (log2 value) and bars indicate the standard deviation of with three independent repetitions
From the view of dynamic change of the expression pattern, OsCPs could be grouped into four classes (Fig. 7b). The expression level of the first group is relatively stable and shows no visible change during the whole process of seed formation. Four OsCP genes (OsCP1, OsCP3, OsCP8 and OsCP33) fall into this group. The second group consists of three OsCPs (OsCP4, OsCP5 and OsCP12) and the transcripts of them could only be detected in the seeds before 7 DAP, suggesting an important role in early seed development. The third group is that their expression peak is at seeds 7–9 DAP. OsCP2, OsCP13, OsCP28 and OsCP29 belong to this group. The last group contains the rest of OsCPs whose expression levels show dynamic changes during the process of seed development in rice. As for OsVPE genes, the transcriptional level of OsVPE4 decreases gradually during the processes of seed formation, and the other members exhibited relatively stable expression patterns (Fig. 7d).
In the process of seed germination, most of OsCPs and all the OsVPEs could be detected and the striking feature is that the expression level of all the members of OsVPEs decreased remarkably in the process of seed germination (Fig. 7E). Several OsCP genes (OsCP1, OsCP6, OsCP10, OsCP11, OsCP15, OsCP28, OsCP29 and OsCP33) showed similar expression pattern as OsVPEs (Fig. 7c). The transcriptional level of the rest OsCPs expressed in the germinating seeds varied significantly during the process of seed germination, except OsCP20, which showed an abundant and relatively stable expression pattern.
Differential responses of OsCPs to hormone and stress treatments
A remarkable feature of PLCPs from plants is that the transcription of them is regulated by different hormones and various stresses [17, 44, 45], and thus function in different physiological processes. To gain insight into their potential roles in response to various hormones and different severe environments, their relative transcriptional levels in seedlings after different hormones treatments (NAA, KT, ABA, GA3 and JA) and abiotic treatments (cold, drought and salt) were investigated by RT-qPCR. Based on the relative expression level of each OsCP, histograms were created (Fig. 8) and overview of OsCPs in response to different hormones and abiotic stresses was listed in Table 3.
Fig. 8.
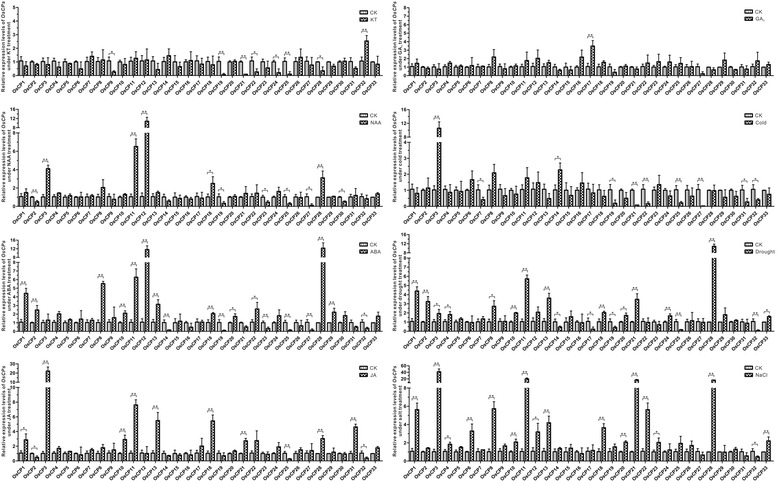
Expression levels of OsCPs in seedlings under different hormone and abiotic treatments. The expression was normalized against OsActin and OsUBC. The data represent the relative expression level compared with that in seedlings under normal growth conditions and bars indicate the standard deviation of with three independent repetitions. ‘*’ and ‘**’ indicate statistical difference compared to the WT (t-test, p < 0.05 or 0.01, respectively)
Table 3.
Overview of OsCPs and OsVPEs in response to different hormones and abiotic stresses
| Name | Hormone | Abiotic stress | ||||||
|---|---|---|---|---|---|---|---|---|
| KT | NAA | ABA | GA | JA | Cold | Drought | Salt | |
| OsCP1 | No | No | ++ | No | + | No | ++ | ++ |
| OsCP2 | No | – | + | No | – | No | + | No |
| OsCP3 | No | ++ | No | No | ++++ | +++ | No | +++++ |
| OsCP4 | No | No | No | No | No | No | No | No |
| OsCP5 | No | No | No | No | No | No | No | No |
| OsCP6 | No | No | No | No | No | No | No | + |
| OsCP7 | No | No | No | No | No | – | No | No |
| OsCP8 | No | No | ++ | No | No | No | + | ++ |
| OsCP9 | – | No | No | No | No | No | No | No |
| OsCP10 | No | No | + | No | + | No | + | + |
| OsCP11 | No | ++ | ++ | No | ++ | No | ++ | +++ |
| OsCP12 | No | +++ | +++ | No | No | No | No | + |
| OsCP13 | No | No | + | No | ++ | No | + | ++ |
| OsCP14 | No | No | – | No | No | + | – | No |
| OsCP15 | No | No | No | No | No | No | No | No |
| OsCP16 | No | No | No | No | No | No | No | No |
| OsCP17 | No | No | No | + | No | No | – | No |
| OsCP18 | No | + | + | No | ++ | No | + | + |
| OsCP19 | – | – | – | No | No | – | – | No |
| OsCP20 | No | No | No | No | No | No | No | + |
| OsCP21 | – | No | No | No | + | – | + | ++++ |
| OsCP22 | – | No | + | No | No | – | No | ++ |
| OsCP23 | No | – | – | No | No | No | No | + |
| OsCP24 | – | No | No | No | No | No | No | No |
| OsCP25 | – | – | – | No | – | – | – | No |
| OsCP26 | No | No | No | No | No | No | No | No |
| OsCP27 | No | – | – | No | No | – | No | No |
| OsCP28 | – | + | +++ | No | + | No | +++ | ++++ |
| OsCP29 | No | No | + | No | No | No | No | No |
| OsCP30 | No | – | No | No | No | No | No | No |
| OsCP31 | No | No | No | No | ++ | – | No | No |
| OsCP32 | + | No | – | No | – | – | – | – |
| OsCP33 | No | No | No | No | No | No | No | + |
| OsVPE1 | No | No | + | No | No | No | + | ++ |
| OsVPE2 | No | No | + | No | + | No | + | ++ |
| OsVPE3 | No | + | ++++ | No | + | No | ++ | ++++ |
| OsVPE4 | No | No | No | No | – | No | No | No |
| OsVPE5 | No | No | No | No | No | No | No | No |
The expression was normalized against OsActin and OsUBC and data represent the mean with three independent experiments. ‘+’ or ‘-’ represents that the expression level was up-regulated or down-regulated respectively. One ‘+’ or ‘-’ means > 2 fold change; two ‘+’ or ‘-’ mean > 4 fold change; three ‘+’ or ‘-’ mean > 8 fold change; four ‘+’ or ‘-’ mean > 16 fold change; five ‘+’ or ‘-’ mean > 32 fold change
Twenty four OsCPs except OsCP4, OsCP5, OsCP6, OsCP7, OsCP15, OsCP16, OsCP20, OsCP26 and OsCP33 are response to at least one hormone treatment (Table 3 and Fig. 8). However no OsCP was commonly regulated by five hormones tested. Generally, these OsCPs display variable responses to different stresses. Only OsCP17 is responsible to GA3. After KT treatment, the expression levels of OsCPs were commonly down-regulated significantly (< 2 fold) apart from OsCP32. For the other three hormone treatments, OsCPs exhibited differential expression pattern. The expression level of OsCP3 (> 16 fold) increased significantly after JA treatment. Whereas the expression level of OsCP27 (< 16 fold) decreased significantly after NAA treatment. Microarray data of 33 OsCPs in 7-day-old seedlings subjected to six hormones (ABA, GA3, Auxin, Brassinosteroid, Cytokinin and JA) were also extracted from the Rice Expression Profile Database (Additional file 6: Figure S6). Consistent with our results, OsCPs were more sensitive to ABA and JA among six plant hormone treatments.
Apart from nine OsCP genes (OsCP4, OsCP5, OsCP9, OsCP15, OsCP16, OsCP24, OsCP26, OsCP29 and OsCP30), other OsCPs are responsive to different stress treatments (Table 3 and Fig. 8). However, only two OsCPs (OsCP21 and OsCP32) show response to all three stresses (> 2 fold). OsCP32 was always down-regulated by three different stress treatments, which indicated a common role of OsCP32 in cold, drought and salt stress resistance. However other OsCPs were differentially regulated by different stress. In responsible to cold treatment, two OsCPs (OsCP3 and OsCP14) were up-regulated (> 2-fold change) and eight OsCPs (OsCP7, OsCP19, OsCP21, OsCP22, OsCP25, OsCP27, OsCP31 and OsCP32) were down-regulated (> 2-fold change). Notably, for rice PLCPs, the degree of response to cold stress varied significantly. In response to drought treatment, nine OsCPs (OsCP1, OsCP2, OsCP8, OsCP10, OsCP11, OsCP13, OsCP18, OsCP21 and OsCP28) were up-regulated and five OsCPs (OsCP14, OsCP17, OsCP19, OsCP25 and OsCP32) were down-regulated. In responsible to salt treatment, almost half of OsCPs were up-regulated and only OsCP32 was down-regulated. The expression data of OsCPs from MPSS database under abiotic stress treatments were summarized in Additional file 7: Table S7. Three OsCPs (OsCP1, OsCP8 and OsCP25) showed a similar response to drought stress and the expression level of OsCP1 was up-regulated after salt treatment which was also detected in present study. Apart from this, potential binding motifs in the promoters of these proteases have been screened, and the results were listed in Additional file 8: Table S8.
Differential responses of OsVPEs to hormone and stress treatments
Similar to PLCPs, the expression level of VPEs in plants also increased in the process of senescence [46], wounding [47], pathogen infection [26, 28] and abiotic stresses [46, 47]. To verify whether VPEs in rice display similar responses, the relative expression level of each OsVPE was quantified after different treatments. Generally, all OsVPEs except OsVPE5 are response to one or several plant hormones or abiotic stresses (Table 3 and Fig. 9). However, different treatments have diverse effects on the change of expression levels of OsVPEs. For hormone treatment, no OsVPE show response to KT and GA3. Only one OsVPE3 was up-regulated by NAA treatment (Fig. 9a). In contrast, the expression of most OsVPEs was regulated by ABA and JA. The expression level of three OsVPEs (OsVPE1, OsVPE2 and OsVPE3) increased significantly in seedlings after ABA treatment. JA has the same effect on OsVPE2 and OsVPE3, but reverse effect on OsVPE4 expression. For stress treatments, no OsVPE shows response to cold treatment. However, three OsVPEs (OsVPE1, OsVPE2 and OsVPE3) were commonly regulated (> 2 fold change) by salt and drought treatment, indicating their common roles in tolerance to salt and drought stresses (Fig. 9b).
Fig. 9.
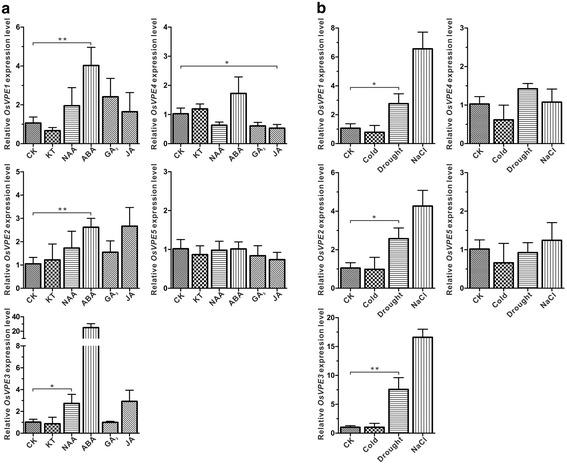
Expression levels of OsVPEs in seedlings under different hormone and abiotic treatments. The expression was normalized against OsActin and OsUBC. The data represent the relative expression level compared with that in seedlings under normal growth conditions and bars indicate the standard deviation of with three independent repetitions. ‘*’ and ‘**’ indicate statistical difference compared to the WT (t-test, p < 0.05 or 0.01, respectively)
Discussion
Characteristics of the PLCPs in rice
As described above, several typical conserved motifs for PLCPs have been identified in most OsCPs. The catalytic triad (Cys-His-Asn) is essentially responsible for proteolytic activity of PLCPs [48], and this central typical motif could be detected in all of OsCPs except OsCP16, in which serine is substituted for cysteine. “ERFNIN” motif provides the core structure of the auto-inhibitory prodomain in most rice PLCPs [49] and the other OsCPs carry the similar ERFNAQ or ERWNIR motif instead. Generally, PLCPs could be divided into four major groups: cathepsin B-, F-, H-, and L-like proteases according to the motif in N-terminal pre-domain and their closest counterparts in animals and ERFNIN motif is typical for cathepsin L- and H-like proteases, but not for cathepsin B-like proteases [50, 51]. ERFNAQ motif is a marker motif for cathepsin F-like proteases [50]. According to this principle, one cysteine protease OsCP33 was grouped into cathepsin B-like protease, since no typical motif could be detected in pre-domain. Three OsCPs (OsCP20, OsCP28 and OsCP30) with an ERFNAQ motif in their proregions fall into cathepsin F-like protease. And typical ERFNIN motif could be detected in the remaining OsCPs through the alignment of protein sequences. However, differences in their substrates and physiological roles of papain-like proteases in different groups are still largely unknown. Granulin-like domain (Cx5Cx5CCCx7Cx4CCx6CCx5CCx6Cx6C), which may serve to regulate thiolprotease activity in plants, was also detected in the C-terminal of several rice PLCPs (OsCP1, OsCP2 and OsCP10). Although the fusion of a granulin domain in the C-terminal of PLCPs has been observed in several plants, but the exact roles of this domain in PLCPs needs to be further studied in the future.
Potential roles of OsCPs in seed development
During past decades, PLCPs were reported to play essential roles in different developmental processes, especially in various types of PCD in different tissues. NtCP14, a papain-like protease with a granulin domain in the C terminal, was approved as a key protease in triggering PCD of suspensor in early embryogenesis. Overexpression of NtCP14 could induce precocious cell death of basal cell lineages of the embryo [52]. Besides its role in PCD of suspensor cell, PLCPs were also associated with the development of inner integument. In Brassica napus, BnCysP1 encoding a papain-like protease was reported to be responsible for PCD of the inner integument [53]. In the present study, our expression pattern analysis results revealed that the transcripts of all OsCPs could be detected in seeds, but the expression levels of them display dynamic changes during the process of seed development in rice, indicating potential roles of PLCPs in seed development, potentially in PCD of endosperm, which are worthy to be explored in the future study. Another striking feature of the expression profile is that the expression level of most OsCPs display striking changes during the process of seed germination, indicating potential roles in regulating seed germination. In barley, two cathepsin L-like proteases, HvPap-6 and HvPap-10, could degrade B, C, and D hordeins stored in the endosperm of barley seeds, which is critical for successful seed germination [7]. Similarly, a gibberellin-inducible cysteine proteinase named gliadain, could digest the storage protein gliadin into low molecular mass peptide almost specifically in wheat for seed germination [8].
There are also some examples indicating that PLCPs are regulated not only at the transcriptional level, but also at the level of protease activity. In barley, the transcripts of a cathepsin B-like cysteine protease (CatB) increased upon germination in the aleurone, leading to the increase of CatB activities in the process of seed germination [44]. Similarly, cathepsin L-like peptidases have also been shown to be involved in the mobilization of hordeins in the barley seed but this process could be partially inhibited by barley cystatins [7]. In the present study, the transcripts of four OsCPs (OsCP1, OsCP6, OsCP18 and OsCP20) are abundant during the first three days of seed germination and decreased later. Consistent with this result, the expression levels of most cystatin genes were higher in seeds at early stages and then decreased dramatically upon seed germination [39]. Hence, the balance between cystatins and PLCPs seems important for seed germination.
Relationship between papain-like and legumain-like cysteine proteases
Papain-like and LLCPs are two important proteolytic enzymes in two subfamilies of cysteine proteases in the Merops protease database [1], which are usually synthesized as inactive poenzyme and use a catalytic Cys as a nucleophile during proteolysis. Auto-inhibitory domain in the N-terminal of PLCPs will be processed to generate a mature form in an acid condition [54]. In contrast, autocatalytic activation of the LLCPs needs two sequential steps by cleavage of the C- and N-terminal propeptides [23]. Although many distinctions between two families in protein structure and biochemical properties exist, the activities of both of them could be inhibited by a group of small proteins called cystatins [2, 3], which spontaneously raises questions about whether papain-like and LLCPs are cross-linked in same biological process.
Previous work has proved that both papain-like and LLCPs participated in hypersensitive response (HR) [15, 26, 28] and seed germination [7, 8, 19]. During the hypersensitive response (HR), the transcriptional level of a papain-like protease called NbCathB was quickly induced, which is critical for HR. When the activities was blocked by treatment with protease inhibitors or downregulation of NbCathB, the HR induced by two distinct nonhost bacterial pathogens (Erwinia amylovora and Pseudomonas syringae pv. Tomato) was prevented [15]. Similarly roles of LLCPs in HR have also been found. Silencing of VPEs in N. benthamiana could abolish the hypersensitive cell death triggered by tobacco mosaic virus (TMV) [26]. In addition, both papain-like and LLCPs are presumed to be responsible for the mobilization of the storage proteins during the process of seed germination. The storage protein-phaseolin in common bean could not be degraded either by papain-like protease CPPh1 or by legumain-like proteases LLP, but only be degraded by papain-like protease CPPh1 and legumain-like proteases LLP in a synergetic way [55]. Furthermore, VmPE-1 had a potential to process the papain-like proteinase designated SH-EP to its intermediate in vitro, which had a major role in the degradation of seed storage protein in Vigna mungo [42]. All these data implied that papain-like and LLCPs may be linked together in many physiological processes. In the present study, some papain-like and legumain-like family genes were found to have a similar expression pattern, for example, OsCP10/OsCP18/OsVPE4 during the process of seed formation and OsCP1/OsCP6/OsVPE2 during the process of seed germination. Furthermore, the expression of some papain-like and legumain-like family genes (OsCP8/OsVPE1, OsCP1/OsCP13/OsVPE2 and OsCP11/OsCP18/OsCP28/OsVPE3) are commonly regulated by hormones and different abiotic stresses, suggesting their potential cooperative roles in plant development and stress environments.
Conclusions
In the present study, 33 OsCPs encoding PLCPs and 5 OsVPEs encoding LLCPs were identified in rice genome respectively. Systematic analysis of OsCP and OsVPE family genes including gene structure, chromosomal distribution, gene duplication, phylogenetic relationship, sequence characteristics and expression pattern analysis were performed. Comprehensive expression profile analysis of both families during the whole process of seed development and germination was also carried out, suggesting their potential roles in seed development and germination. RT-qPCR analysis during diverse stress environments revealed that most of them were regulated by plant hormones and in response to different stress treatments including cold, drought and salt stress. This work suggests their common roles in seed development and stress tolerance, which provides potential clues for further in-depth study of the selected genes in two families.
Methods
Identification of OsCPs and OsVPEs in rice genome
To identify OsCPs and OsVPEs in O. sativa, tBLASTP program of the National Center for Biotechnology Information (NCBI) in the rice protein database (http://www.ncbi.nlm.nih.gov/) with AtCP and AtVPE protein sequences of A. thaliana was performed. Returned nucleotide sequences were considered as OsCP and OsVPE candidates. After removing the redundant genes, deduced protein sequences of all putative OsCP and OsVPE were used to perform BLASTP program, and the sequences with intact peptidase C1A and peptidase C13 domain were considered as true OsCPs and OsVPEs in O. sativa. Corresponding full-length cDNAs were downloaded from Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE).
Analysis of genomic structure and chromosomal localization
Exon-intron organization was determined by the alignment of their coding sequence to their corresponding genome full-length sequence. Diagrams were drawn with Gene Structure Display Server (GSDS: http://gsds.cbi.pku.edu.cn/). OsCPs and OsVPEs were positioned on the rice chromosomes using BLASTN at the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/).
Gene duplication and duplication date calculation
Genes on the duplicated chromosomal segments were identified using Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) with the maximum distance permitted between collinear gene pairs of 500 kb. Homologous genes separated by five genes at most were regarded as tandem duplicated genes. Calculation of the duplication dates was according to the previous methods [39].
Protein sequence alignment and phylogenetic analysis
Multiple sequence alignments of amino acid sequences were performed using Clustal X ver. 1.81 with the default multiple alignment parameters. Phylogenetic tree was generated with Phylip Ver. 3.68 using the Protpars method. Protein sequences of papain-like and LLCPs from A. thaliana, H. vulgare and Z. mays were used in this analysis, and their accession numbers are listed in Additional file 9: Table S9 and Additional file 10: Table S10.
Digital expression analysis of OsCPs and OsVPEs
Expression profile data from rice microarrays are available in the Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp), which is a repository of gene expression profiles derived from microarray analysis of tissues/organs encompassing the entire life cycle of the rice plant under natural and plant hormone-treated conditions. The expression results of OsCPs and OsVPEs were summarized in Additional file 6: Figure S6 and Additional file 11: Figure S11.
Rice MPSS database (http://mpss.udel.edu/rice/mpss_index.php) was searched to obtain the expression levels of OsCPs and OsVPEs. The criteria was that the signature must be unique in the genome (hits = 1) and a perfect match (100% identity over the tag length). TPM (tags per million) means the normalized abundance of the signatures, which is the best estimate of the expression level for a given gene. The expression data under normal conditions were listed in Additional file 4: Table S4. The expression data from rice plants under abiotic stress treatments were summarized in Additional file 7: Table S7.
Plant materials and the methods for various treatments
Oryza sativa L. japonica cv. Nipponbare was grown in the greenhouse at Wuhan University with temperature difference between day and night (30/26 °C) under a photoperiod of 16 h light and 8 h dark. For the expression pattern analyses of OsCPs and OsVPEs under normal conditions, total RNA was extracted from root, stem and leaf of 5-day-old seedlings growing on 1/2 MS solid medium and other different tissues including anther, out glume, inner glume, seeds at different developmental stages.
For various treatments, seeds were sterilized according to previous protocol [39] and then inoculated on 1/2 MS solid medium contain 1% sucrose. For Abscisic acid (ABA), Gibberellin acid (GA3), 1-Naphthaleneacetic acid (NAA), Kinetin (KT), Jasmonic acid (JA) treatment, 5-day-old seedlings were cultured in 1/2 MS liquid medium containing 25 μM ABA, 5 μM GA3, 5 μΜ NAA, 5 μM KT and 50 μM JA for 12 h, respectively [35]. For salt stress, 5-day-old seedlings were transferred into 1/2 MS liquid medium containing 400 mM NaCl; for drought stress, seedlings were dried between folds of the sterile filter paper; for cold stress, the culture plates containing the seedlings were kept at 4 °C. All the treatments were for 12 h.
cDNA synthesis and RT-qPCR
Total RNA of different tissues were isolated with Trizol reagent according to the manufacturer’s instructions (Life Technology,USA). The residual genomic DNA was removed by RNase-free DNase I (Promega, USA). First-strand cDNA was synthesized using M-MLV reverse transcriptase following the manufacturer’s instructions (Invitrogen, USA). RT-qPCR was introduced for OsCPs and OsVPEs expression analysis according to the protocol described previously [56]. Five house-keeping genes including ACTIN, eEF-1a, UBC, UBQ5 and GAPDH were chosen as internal reference genes for RT-qPCR. The stability of five reference genes in different tissues was evaluated using geNorm (Version 3.5). Two most stable reference genes ACTIN and UBC were chosen for the calculating normalization factors for different tissues. Thus, the relative expression level of each gene in different tissues was calculated according to the previous protocol [56]. Gene-specific primers were listed in Additional file 12: Table S12.
Additional files
Figure S1. Multiple protein sequences alignment of rice papain-like cysteine proteases. The inhibitor domain and peptidase C1A domain was shaded in red and black respectively. The granulin domain was marked with black boxes. The similar amino acid residues were marked in blue and the identical acid residues were boxed in purple. Red and black dots indicated the catalytic triad and the retention signal in ER respectively. The ‘NPIR’ was shaded in a black ellipse. (DOCX 868 kb)
Figure S2. Multiple protein sequences alignment of rice legumain-like cysteine proteases. The peptidase C13 domain was shaded. The similar amino acid residues were marked in blue and the identical acid residues were boxed in purple. Red dots indicate the catalytic triad. (DOCX 481 kb)
Figure S3. Expression heatmap of OsCPs in different tissues under normal conditions. (DOCX 2617 kb)
Table S4. Expression abundance of OsCPs and OsVPEs in various tissues under normal conditions. (DOCX 20 kb)
Figure S5. Expression heatmap of OsVPEs in different tissues under normal conditions. (DOCX 263 kb)
Figure S6. Expression profile of OsCPs in the shoots and roots under different plant hormones treatments. (DOCX 482 kb)
Table S7. Expression change of OsCPs and OsVPEs under stress treatments. The date was from MPSS: gene analysis (http://mpss.udel.edu/rice/GeneQuery.php). The experiment materials were 14-day-old seedlings. Salt treatment: 250 mM NACL for 24 h; drought treatment: stressed in drought for 5d; cold treatment: 4 °C for 24 h. Compared with normal condition, the expression fold change > 2 or < 0.5 were indicated in red or blue respectively. (DOCX 16 kb)
Table S8. Overview of cis-elements in the promoters of OsCPs and OsVPEs. ABRE: ABA-responsive element; LTRE: Low-temperature-responsive element; DRE: Dethydration-responsive-element; T/G-box: DNA-binding motif of MYC2 (the key transcriptional activator of jasmonate responses). The number of predicted cis-elements were presented by the number of “+”. (DOCX 17 kb)
Table S9. Papain-like Cysteine Proteases in three plant species. (DOCX 18 kb)
Table S10. Legumain-like Cysteine Proteases in four plant species. (DOCX 16 kb)
Figure S11. Expression profile of OsVPEs in the shoots and roots under different plant hormones treatments. (DOCX 214 kb)
Table S12. Primers used in this study. (DOCX 2617 kb)
Acknowledgements
We would like to thank the reviewers and editors for their careful reading and helpful comments on this manuscript.
Funding
This project was supported by the National Natural Science Fund of China (31600244 and 31400171). XMZ, the funder of NSFC31600244, performed some experiments. PZ, the funder of NSFC31400171, designed the experiments and wrote the paper.
Availability of data and materials
Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp) and Rice MPSS database (http://mpss.udel.edu/rice/mpss_index.php) were used for expression pattern analysis. cDNA and genome sequence for each gene were downloaded from Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE). Protein sequence and phylogeny data, including alignments, have been deposited in the TreeBASE repository (http://purl.org/phylo/treebase/phylows/study/TB2:S22378).
Abbreviations
- ABA
Abscisic acid
- GA3
Gibberellin acid
- HR
Hypersensitive response
- JA
Jasmonic acid
- Ka
Nonsynonymous
- Ks
Synonymous
- KT
Kinetin
- LLCPs
Legumain-like cysteine proteases
- MPSS
Massively parallel signature sequencing
- NAA
1-Naphthaleneacetic acid
- PCD
Programmed cell death
- PLCPs
Papain-like cysteine proteases
- RiceXPro
Rice Expression Profile Database
- RT–qPCR
Quantitative real-time reverse transcription-PCR
- VIGS
Virus-induced gene silencing
- VPE
Vacuolar processing enzyme
Authors’ contributions
MXS contributed all reagents and materials used in the experiments. WW, PZ, MXS designed the experiments and wrote the paper. WW, PZ, XMZ, HXX and WYM performed the experiments. WW and PZ analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interest
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1298-1) contains supplementary material, which is available to authorized users.
References
- 1.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38(Database issue):D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai S, Watanabe H, Kondo H, Emori Y, Abe K. Papain-inhibitory activity of oryzacystatin, a rice seed cysteine proteinase inhibitor, depends on the central Gln-Val-Val-ala-Gly region conserved among cystatin superfamily members. J Biochem. 1991;109(2):294–298. [PubMed] [Google Scholar]
- 3.Alvarez-Fernandez M, Barrett AJ, Gerhartz B, Dando PM, Ni J, Abrahamson M. Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J Biol Chem. 1999;274(27):19195–19203. doi: 10.1074/jbc.274.27.19195. [DOI] [PubMed] [Google Scholar]
- 4.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20(17):4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stuber K, Colby T, Overkleeft HS, Bogyo M, Van der Hoorn RA. Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 2012;158(4):1583–1599. doi: 10.1104/pp.112.194001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiederanders B, Kaulmann G, Schilling K. Functions of propeptide parts in cysteine proteases. Curr Protein Pept Sc. 2003;4(5):309–326. doi: 10.2174/1389203033487081. [DOI] [PubMed] [Google Scholar]
- 7.Martinez M, Cambra I, Carrillo L, Diaz-Mendoza M, Diaz I. Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 2009;151(3):1531–1545. doi: 10.1104/pp.109.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiyosaki T, Matsumoto I, Asakura T, Funaki J, Kuroda M, Misaka T, Arai S, Abe K. Gliadain, a gibberellin-inducible cysteine proteinase occurring in germinating seeds of wheat, Triticum aestivum L., specifically digests gliadin and is regulated by intrinsic cystatins. FEBS J. 2007;274(8):1908–1917. doi: 10.1111/j.1742-4658.2007.05749.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XM, Wang Y, Lv XM, Li H, Sun P, Lu H, Li FL. NtCP56, a new cysteine protease in Nicotiana tabacum L., involved in pollen grain development. J Exp Bot. 2009;60(6):1569–1577. doi: 10.1093/jxb/erp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell. 2014;26(7):2939–2961. doi: 10.1105/tpc.114.127282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18(11):2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Jung KH, An G, Chung YY. Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant Mol Biol. 2004;54(5):755–765. doi: 10.1023/B:PLAN.0000040904.15329.29. [DOI] [PubMed] [Google Scholar]
- 13.Ueda T, Seo S, Ohashi Y, Hashimoto J. Circadian and senescence-enhanced expression of a tobacco cysteine protease gene. Plant Mol Biol. 2000;44(5):649–657. doi: 10.1023/A:1026546004942. [DOI] [PubMed] [Google Scholar]
- 14.Kruger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science. 2002;296(5568):744–747. doi: 10.1126/science.1069288. [DOI] [PubMed] [Google Scholar]
- 15.Gilroy EM, Hein I, van der Hoorn R, Boevink PC, Venter E, McLellan H, Kaffarnik F, Hrubikova K, Shaw J, Holeva M, et al. Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 2007;52(1):1–13. doi: 10.1111/j.1365-313X.2007.03226.x. [DOI] [PubMed] [Google Scholar]
- 16.Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell. 2000;12(7):1031–1040. doi: 10.1105/tpc.12.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanna-Chopra R, Srivalli B, Ahlawat YS. Drought induces many forms of cysteine proteases not observed during natural senescence. Biochem Bioph Res Co. 1999;255(2):324–327. doi: 10.1006/bbrc.1999.0195. [DOI] [PubMed] [Google Scholar]
- 18.Hiraiwa N, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS Lett. 1999;447(2–3):213–216. doi: 10.1016/S0014-5793(99)00286-0. [DOI] [PubMed] [Google Scholar]
- 19.Hara-Nishimura I, Inoue K, Nishimura M. A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett. 1991;294(1–2):89–93. doi: 10.1016/0014-5793(91)81349-D. [DOI] [PubMed] [Google Scholar]
- 20.Hara-Nishimura I. Vacuolar processing enzyme responsible for maturation of vacuolar proteins. Seikagaku. The Journal of Japanese Biochemical Society. 1995;67(5):372–377. [PubMed] [Google Scholar]
- 21.Muntz K, Shutov AD. Legumains and their functions in plants. Trends Plant Sci 2002; 7(8):340-344. [DOI] [PubMed]
- 22.Hara-Nishimura I, Takeuchi Y, Nishimura M. Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell. 1993;5(11):1651–1659. doi: 10.1105/tpc.5.11.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroyanagi M, Nishimura M, Hara-Nishimura I. Activation of Arabidopsis vacuolar processing enzyme by self-catalytic removal of an auto-inhibitory domain of the C-terminal propeptide. Plant Cell Physiol. 2002;43(2):143–151. doi: 10.1093/pcp/pcf035. [DOI] [PubMed] [Google Scholar]
- 24.Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, et al. Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem. 2003;278(34):32292–32299. doi: 10.1074/jbc.M305740200. [DOI] [PubMed] [Google Scholar]
- 25.Hatsugai N, Kuroyanagi M, Nishimura M, Hara-Nishimura I. A cellular suicide strategy of plants: vacuole-mediated cell death. Apoptosis. 2006;11(6):905–911. doi: 10.1007/s10495-006-6601-1. [DOI] [PubMed] [Google Scholar]
- 26.Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;305(5685):855–858. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- 27.Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I. A vacuolar processing enzyme, deltaVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell. 2005;17(3):876–887. doi: 10.1105/tpc.104.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroyanagi M, Yamada K, Hatsugai N, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J Biol Chem. 2005;280(38):32914–32920. doi: 10.1074/jbc.M504476200. [DOI] [PubMed] [Google Scholar]
- 29.Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153(1):185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, et al. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci U S A. 2009;106(52):22163–22168. doi: 10.1073/pnas.0912139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng H, Liu H, Li X, Xiao J, Wang S. A CCCH-type zinc finger nucleic acid-binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol. 2012;158(2):876–889. doi: 10.1104/pp.111.191379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo YS, Chern M, Bartley LE, Han M, Jung KH, Lee I, Walia H, Richter T, Xu X, Cao P, et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011;7(4):e1002020. doi: 10.1371/journal.pgen.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Zhu S, Liu S, Jiang L, Chen L, Ren Y, Han X, Liu F, Ji S, Liu X, et al. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 2009;58(4):606–617. doi: 10.1111/j.1365-313X.2009.03801.x. [DOI] [PubMed] [Google Scholar]
- 34.Beers EP, Jones AM, Dickerman AW. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry. 2004;65(1):43–58. doi: 10.1016/j.phytochem.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Xia K, Liu T, Ouyang J, Wang R, Fan T, Zhang M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.) DNA Res. 2011;18(5):363–377. doi: 10.1093/dnares/dsr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18(9):486. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 37.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 38.Bateman A, Bennett HP. The granulin gene family: from cancer to dementia. BioEssays. 2009;31(11):1245–1254. doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Zhao P, Zhou XM, Xiong HX, Sun MX. Genome-wide identification and characterization of cystatin family genes in rice (Oryza sativa L.) Plant Cell Rep. 2015;34(9):1579–1592. doi: 10.1007/s00299-015-1810-0. [DOI] [PubMed] [Google Scholar]
- 40.Hara-Nishimura I, Kinoshita T, Hiraiwa N, Nishimura M. Vacuolar processing enzymes in protein-storage vacuoles and lytic vacuoles. J Plant Physiol. 1998;152(6):668–674. doi: 10.1016/S0176-1617(98)80028-X. [DOI] [Google Scholar]
- 41.Julian I, Gandullo J, Santos-Silva LK, Diaz I, Martinez M. Phylogenetically distant barley legumains have a role in both seed and vegetative tissues. J Exp Bot. 2013;64(10):2929–2941. doi: 10.1093/jxb/ert132. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto T, Minamikawa T. Molecular cloning and characterization of Vigna mungo processing enzyme 1 (VmPE-1), an asparaginyl endopeptidase possibly involved in post-translational processing of a vacuolar cysteine endopeptidase (SH-EP) Plant Mol Biol. 1999;39(1):63–73. doi: 10.1023/A:1006170518002. [DOI] [PubMed] [Google Scholar]
- 43.Deng ZY, Gong CY, Wang T. Use of proteomics to understand seed development in rice. Proteomics. 2013;13(12–13):1784–1800. doi: 10.1002/pmic.201200389. [DOI] [PubMed] [Google Scholar]
- 44.Martinez M, Rubio-Somoza I, Carbonero P, Diaz I. A cathepsin B-like cysteine protease gene from Hordeum vulgare (gene CatB) induced by GA in aleurone cells is under circadian control in leaves. J Exp Bot. 2003;54(384):951–959. doi: 10.1093/jxb/erg099. [DOI] [PubMed] [Google Scholar]
- 45.Esteban-Garcia B, Garrido-Cardenas JA, Alonso DL, Garcia-Maroto F. A distinct subfamily of papain-like cystein proteinases regulated by senescence and stresses in Glycine max. J Plant Physiol. 2010;167(13):1101–1108. doi: 10.1016/j.jplph.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 1999;19(1):43–53. doi: 10.1046/j.1365-313X.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamada K, Nishimura M, Hara-Nishimura I. The slow wound-response of gammaVPE is regulated by endogenous salicylic acid in Arabidopsis. Planta. 2004;218(4):599–605. doi: 10.1007/s00425-003-1131-3. [DOI] [PubMed] [Google Scholar]
- 48.Kamphuis IG, Drenth J, Baker EN. Thiol proteases. Comparative studies based on the high-resolution structures of papain and actinidin, and on amino acid sequence information for cathepsins B and H, and stem bromelain. J Mol Biol. 1985;182(2):317–329. doi: 10.1016/0022-2836(85)90348-1. [DOI] [PubMed] [Google Scholar]
- 49.Karrer KM, Peiffer SL, Ditomas ME. Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci U S A. 1993;90(7):3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kramer L, Turk D, Turk B. The future of cysteine cathepsins in disease management. Trends Pharmacol Sci. 2017;38(10):873–898. doi: 10.1016/j.tips.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Coulombe R, Grochulski P, Sivaraman J, Menard R, Mort JS, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15(20):5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao P, Zhou XM, Zhang LY, Wang W, Ma LG, Yang LB, Peng XB, Bozhkov PV, Sun MX. A bipartite molecular module controls cell death activation in the basal cell lineage of plant embryos. PLoS Biol. 2013;11(9):e1001655. doi: 10.1371/journal.pbio.1001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan LL, Xia Q, Qiu X, Selvaraj G. Early stages of seed development in Brassica napus: a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J. 2002;30(1):1–10. doi: 10.1046/j.1365-313X.2002.01262.x. [DOI] [PubMed] [Google Scholar]
- 54.Grudkowska M, Zagdanska B. Multifunctional role of plant cysteine proteinases. Acta Biochim Pol. 2004;51(3):609–624. [PubMed] [Google Scholar]
- 55.Zakharov A, Carchilan M, Stepurina T, Rotari V, Wilson K, Vaintraub I. A comparative study of the role of the major proteinases of germinated common bean (Phaseolus vulgaris L.) and soybean (Glycine max (L.) Merrill) seeds in the degradation of their storage proteins. J Exp Bot. 2004;55(406):2241–2249. doi: 10.1093/jxb/erh247. [DOI] [PubMed] [Google Scholar]
- 56.Ma L, Xin H, Qu L, Zhao J, Yang L, Zhao P, Sun M. Transcription profile analysis reveals that zygotic division results in uneven distribution of specific transcripts in apical/basal cells of tobacco. PLoS One. 2011;6(1):e15971. doi: 10.1371/journal.pone.0015971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Multiple protein sequences alignment of rice papain-like cysteine proteases. The inhibitor domain and peptidase C1A domain was shaded in red and black respectively. The granulin domain was marked with black boxes. The similar amino acid residues were marked in blue and the identical acid residues were boxed in purple. Red and black dots indicated the catalytic triad and the retention signal in ER respectively. The ‘NPIR’ was shaded in a black ellipse. (DOCX 868 kb)
Figure S2. Multiple protein sequences alignment of rice legumain-like cysteine proteases. The peptidase C13 domain was shaded. The similar amino acid residues were marked in blue and the identical acid residues were boxed in purple. Red dots indicate the catalytic triad. (DOCX 481 kb)
Figure S3. Expression heatmap of OsCPs in different tissues under normal conditions. (DOCX 2617 kb)
Table S4. Expression abundance of OsCPs and OsVPEs in various tissues under normal conditions. (DOCX 20 kb)
Figure S5. Expression heatmap of OsVPEs in different tissues under normal conditions. (DOCX 263 kb)
Figure S6. Expression profile of OsCPs in the shoots and roots under different plant hormones treatments. (DOCX 482 kb)
Table S7. Expression change of OsCPs and OsVPEs under stress treatments. The date was from MPSS: gene analysis (http://mpss.udel.edu/rice/GeneQuery.php). The experiment materials were 14-day-old seedlings. Salt treatment: 250 mM NACL for 24 h; drought treatment: stressed in drought for 5d; cold treatment: 4 °C for 24 h. Compared with normal condition, the expression fold change > 2 or < 0.5 were indicated in red or blue respectively. (DOCX 16 kb)
Table S8. Overview of cis-elements in the promoters of OsCPs and OsVPEs. ABRE: ABA-responsive element; LTRE: Low-temperature-responsive element; DRE: Dethydration-responsive-element; T/G-box: DNA-binding motif of MYC2 (the key transcriptional activator of jasmonate responses). The number of predicted cis-elements were presented by the number of “+”. (DOCX 17 kb)
Table S9. Papain-like Cysteine Proteases in three plant species. (DOCX 18 kb)
Table S10. Legumain-like Cysteine Proteases in four plant species. (DOCX 16 kb)
Figure S11. Expression profile of OsVPEs in the shoots and roots under different plant hormones treatments. (DOCX 214 kb)
Table S12. Primers used in this study. (DOCX 2617 kb)
Data Availability Statement
Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp) and Rice MPSS database (http://mpss.udel.edu/rice/mpss_index.php) were used for expression pattern analysis. cDNA and genome sequence for each gene were downloaded from Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE). Protein sequence and phylogeny data, including alignments, have been deposited in the TreeBASE repository (http://purl.org/phylo/treebase/phylows/study/TB2:S22378).


