Abstract
A growing appreciation of the metabolic artifacts of cell culture has generated heightened enthusiasm for performing metabolomics on populations of cells purified from tissues and biofluids. Fluorescence activated cell sorting, or FACS, is a widely used experimental approach to purify specific cell types from complex heterogeneous samples. Here we show that FACS introduces oxidative stress and alters the metabolic state of cells. Compared to unsorted controls, astrocytes subjected to FACS prior to metabolomic analysis showed altered ratios of GSSG to GSH, NADPH to NADP+, and NAD+ to NADH. Additionally, a 50% increase in reactive oxygen species was observed in astrocytes subjected to FACS relative to unsorted controls. At a more comprehensive scale, nearly half of the metabolomic features that we profiled by liquid chromatography/mass spectrometry were changed by at least 1.5-fold in intensity due to cell sorting. Some specific metabolites identified to have significantly altered levels as a result of cell sorting included glycogen, nucleosides, amino acids, central carbon metabolites, and acylcarnitines. Although the addition of fetal bovine serum to the cell-sorting buffer decreased oxidative stress and attenuated changes in metabolite concentrations, fetal bovine serum did not preserve the metabolic state of the cells during FACS. We conclude that, irrespective of buffer components and data-normalization strategies we examined, metabolomic results from sorted cells do not accurately reflect physiological conditions prior to sorting.
Highlights
-
•
Subjecting cells to FACS introduces oxidative stress and alters cellular redox state.
-
•
The concentrations of many metabolites change during cell sorting.
-
•
FBS and BSA do not prevent these perturbations during FACS.
-
•
Metabolic changes are non-uniform and cannot be corrected by simple normalization.
1. Introduction
Liquid chromatography/mass spectrometry (LC/MS) and gas chromatography/mass spectrometry (GC/MS) are the most widely used experimental platforms for performing metabolomics [1]. Historically, these technologies have been primarily applied to two types of samples: (i) cells grown in standard monoculture, or (ii) tissues and biofluids harvested from animals and patients. Cell monoculture has some attractive benefits, such as being cost effective and high throughput. Most important to the current work, cell monoculture avoids the challenge of having to resolve metabolites from more than one cell type. As a consequence, the metabolism of cultured cells can be rapidly quenched and their metabolites extracted for profiling without a cell-purification step [2]. Metabolomic analysis of tissues and biofluids by LC/MS or GC/MS, in contrast, is complicated by the presence of multiple cell types. Without a cell-purification step, signal intensities in metabolomic data represent the average concentration of a metabolite from all cell types in the tissue or biofluid and are difficult to interpret in the context of metabolic regulation. Thus, historically, cells in monoculture have been primarily used to study metabolic regulation by LC/MS or GC/MS, whereas whole tissues and biofluids have more frequently been used to screen for biomarkers of disease [3], [4], [5].
A potential complication of studying metabolic regulation in cell culture is that cells are not in their naturally occurring environment, which can introduce non-physiological artifacts in metabolism [6], [7]. Standard cell-culture media, for instance, contains ~10-fold less fatty acids compared to healthy human serum. Proliferating cells have a high demand for fatty acids to support the formation of new membranes. In standard cell-culture media, proliferating cells mostly synthesize fatty acids de novo from glucose [8]. When proliferating cells are cultured in media containing physiological levels of fatty acids, however, they prefer to uptake the fatty acids rather than synthesize them. Although media formulations are emerging that better reflect the composition of human plasma, nutrient availability may not be the only source of metabolic artifacts in cell culture [9]. Despite having access to glutamine, for example, some tumors show minimal utilization of glutamine in vivo. Yet, cell lines derived from these same tumors rely heavily on glutamine in cell culture [10].
With increasing evidence that the metabolism of cells in culture differs from the metabolism of cells in an animal or a patient, there has been heightened enthusiasm to study metabolic regulation in tissues and biofluids with metabolomics. The challenge remains of how to resolve the metabolites of specific cell types within the samples during LC/MS and GC/MS profiling. An experimental strategy that is commonly employed to purify populations of cells from complex samples is fluorescence-activated cell sorting or FACS [11]. One potential workflow is to isolate specific types of cells from complex samples by FACS and subsequently quench their metabolism prior to extracting metabolites for mass spectrometry analysis [12]. While it is provocative to imagine stopping metabolism by enzyme inactivation prior to cell sorting, conventional methods for quenching metabolism are not compatible with FACS. It is therefore important to note that FACS can take up to several hours, depending on experimental conditions, sample type, and number of replicates. Many metabolites turnover on a much faster timescale [13]. By way of illustration, the total pool of ATP can turnover six times per minute in heart tissue [14]. During FACS, cells are transferred to buffers with limited nutrient availability and then subjected to changes in temperature as well as pressure. Here we sought to assess the extent that such environmental perturbations during FACS reprogram cellular metabolism, which has important implications for the physiological relevance of metabolomic data collected from sorted cells.
In this study, we found that subjecting astrocytes to FACS led to oxidative stress and an altered redox state as supported by significant changes in the ratios of NADPH to NADP+ and NAD+ to NADH. In mammalian cells, NAD(H) and NADP(H) are utilized by hundreds of metabolic reactions that span various biochemical functions. Many of these reactions are regulated by the ratios of NADPH to NADP+ and NAD+ to NADH [15]. Hence, it may not be surprising that we also found changes in the concentrations of metabolites involved in many major metabolic pathways as a result of FACS. Our work indicates that metabolomic data from sorted cells do not accurately reflect the native metabolism of cells prior to FACS.
2. Materials and methods
2.1. Tissue culture
Cells were grown in high-glucose Dulbecco's Modified Eagle Media (DMEM) (4.5 g/L glucose) containing 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin at 37 °C with 5% CO2.
2.2. Morphology and viability assay
DI TNC1 astrocytes, plated with the same original seeding density, were left in 1 mL of phosphate buffered saline (PBS) with or without 1% dialyzed FBS (dFBS) at 4 °C for 4 h. Bright field images were taken by using a BioTek Cytation™ 5 Cell Imaging Multi-Mode Reader. Viability and cell number were assessed by using trypan blue and a Nexcelom Cellometer Auto 1000.
2.3. FACS
Cells were resuspended to form a single cell suspension in 1 mL of PBS alone, 1% dFBS in PBS, or 1% bovine serum albumin (BSA) in PBS. The cell suspension was filtered and then sorted by using FACS on a BD FACSAria™ II (nozzle size 85 µM, plate voltage 5000 V, sheath pressure 45 psi, flow rate 1.0, optical path/laser used = 488 nm). Cells were selected with side scatter and forward scatter to collect live, single cells [16], [17]. The population was gated based on the relative size and complexity of the cells using Forward Scatter (FSC) and Side Scatter (SSC) parameters. Doublets were excluded with FSC-Width and SSC-Width (Fig. S1).
2.4. Reactive Oxygen Species (ROS) detection
ROS were detected with the DCFDA/H2DCFDA - Cellular Reactive Oxygen Species Detection Assay Kit from Abcam (Cat. No. ab113851) according to the manufacturer's instructions. In brief, 2’,7’-dichlorofluorescin diacetate (DCFDA) was added to the cells. Oxidation of DCFDA by ROS was monitored by the formation for 2’,7’-dichlorofluoroscein (DCF). Fluorescence of DCF was measured (Ex/Em=485/535 nm).
2.5. Glycogen detection
Glycogen was measured by using a Glycogen Assay Kit from Abcam (Cat. No. 65620) according to the manufacturer's instructions (OxiRed probe, Ex/Em=535/587 nm).
2.6. Assessing NADPH/NADP+
The NADP+/NADPH Quantitation Colorimetric Kit from BioVision (Cat. No. K347) was used according to the manufacturer's instructions. In brief, fluorescence of the NADPH Developer was measured (OD=450 nm), two hours after the addition of the NADP Cycling Mix.
2.7. Hydrogen peroxide treatment
For the H2O2 metabolomic experiments, cells were treated with three mL of PBS +/−1% dFBS and 200 μM H2O2 at 4 °C for 4 h before quenching metabolism with methanol (MeOH).
2.8. LC/MS-based metabolomics
Cells in the rapidly quenched condition had their metabolism quenched quickly with 500 μL MeOH/ 6 × 105 cells after removal from cell-culture plates. Cells in the sorted condition were first subjected to FACS before being quenched with 500 μL MeOH/ 6 × 105 cells. Cells in the delayed-quench condition were left on the benchtop in PBS alone or with 1% dFBS for four hours before being quenched with 500 μL MeOH/ 6 × 105 cells. After extraction, samples were analyzed with hydrophilic interaction liquid chromatography (HILIC) or reversed-phase liquid chromatography (RPLC) coupled to mass spectrometry in negative or positive ionization mode, respectively. All raw data files were converted into mzXML files using msconvert [18]. Data analysis was performed by using either Xcalibur Qual Browser or a combination of in-house software packages implemented in R, which we have described in detail previously [19], [20], [21].
2.9. Metabolite extraction
Cell pellets were dried on a SpeedVac and subsequently lyophilized. Metabolites were isolated from lyophilized cell pellets by using methanol/acetonitrile/water (2:2:1), adjusted to maintain a ratio of 0.5 mL of solvent per 6 × 105 cells or 1 mL/mg. Following the previously described protocol [22], extracts were dried with a SpeedVac and then reconstituted in 50 μL/6 × 105 cells or 100 μL/mg of acetonitrile/water (1:1) and placed in 4 °C for 1 h. Samples were centrifuged at 14 kRPM and 4 °C for 10 min. Supernatant was transferred to LC/MS vials for analysis.
A more detailed version of materials and methods can be found in the Supplemental information.
3. Results and discussion
To evaluate the effects of FACS on astrocytes, we subjected cells grown in monoculture to three different experimental conditions, which we refer to as rapidly quenched, sorted, or delayed quench. The rapidly quenched condition served as a control, where the metabolism of cultured cells was quickly stopped prior to any analysis. In the sorted condition, cells were subjected to FACS before having their metabolism quenched for subsequent analysis. Lastly, in the delayed-quench condition, cells were left on the benchtop for the same amount of time required to perform FACS before having their metabolism quenched for subsequent analysis. We note that since we started with a monoculture of cells, pure populations of astrocytes were compared in all conditions. We also note that comparison of the sorted and delayed-quench conditions permitted us to isolate the effects of nutrient deprivation from other perturbations associated with FACS.
3.1. FACS alters redox state
Our first objective was to determine the ratio of oxidized glutathione (GSSG) to reduced glutathione (GSH), which is an indicator of cellular oxidative stress [23]. With LC/MS, we determined the relative intensities of GSSG and GSH. For normalization, we set GSSG/GSH to 1 for the rapidly quenched condition and found that the ratio was increased nearly 7 fold in sorted cells (Fig. 1A, S2A-B). As a reference benchmark to help interpret the altered glutathione ratio caused by FACS, we treated astrocytes in FACS buffer with 200 μM hydrogen peroxide for four hours before quenching their metabolism and measuring GSSG/GSH. Compared to untreated cells, astrocytes incubated in hydrogen peroxide showed an approximately 2-fold increase in GSSG/GSH (Fig. 1B). These data support that the oxidative insult imposed by FACS is significant, although we note that some of the effects of hydrogen peroxide may be buffered extracellularly by serum. Interestingly, the ratio of GSSG to GSH was not statistically elevated in the delayed-quench condition relative to the rapidly quenched condition, suggesting that nutrient deprivation alone is not the source of glutathione dysregulation during FACS (Fig. 1A). To test directly whether the level of reactive oxygen species (ROS) changed between the delayed-quench condition and sorted cells, we used the fluorogenic dye DCFDA. Indeed, ROS increased by ~50% in astrocytes subjected to FACS relative to astrocytes in the delayed-quench condition (Fig. 1C).
Fig. 1.
Sorting astrocytes alters their redox state. (A) Relative ratio of GSSG to GSH in rapidly quenched cells, delayed-quench cells, and sorted cells. Ratio was determined by LC/MS. (B) Relative ratio of GSSG to GSH in cells after they were transferred to sorting buffer with or without 200 μM H2O2 for four hours. (C) Comparison of ROS after cells were subjected to either a delayed quench or sorting. (D) Relative ratio of NADPH to NADP+ as determined with a commercial kit in rapidly quenched cells, delayed-quench cells, and sorted cells. (E) Levels of NAD+ and NADH as determined by LC/MS in rapidly quenched cells and sorted cells. NADH was below the limit of detection after sorting. (F) Microscope images of astrocytes in sorting buffer with and without 1% dFBS for four hours demonstrates that dFBS improves cell viability. Quantitation shows that 1% dFBS has a statistically significant effect on cell viability. Data shown are mean values +/- s.d. (n = 3 biological replicates). **p < 0.01, ***p < 0.001; RQ, Rapidly Quenched; DQ, Delayed Quench; dialyzed fetal bovine serum, dFBS.
Given that glutathione homeostasis is tightly coupled to the overall redox state of the cell, we next aimed to determine whether cell sorting also affects the ratio of NADPH to NADP+ and NAD+ to NADH. We found that when astrocytes were sorted, their NADPH/NADP+ shifted towards the reduced state. While cells experiencing a delayed-quench did show an increased ratio, the ratio was twice as high after FACS (Fig. 1D). It is intriguing that the GSSG to GSH balance shifts in the oxidative direction, whereas the NADPH to NADP+ balance shifts in the reductive direction. One possible rationalization for this difference is that as cells transition from proliferating in cell culture to the stressful environment of FACS, their metabolism shifts from a state of anabolism to catabolism. Such a metabolic transition decreases reductive biosynthetic reactions utilizing NADPH. Thus, although NADPH consumption by glutathione reductase may increase, this change in NADPH flux might be smaller than that resulting from decreased reductive biosynthesis. We also observed changes in the NADH and NAD+ levels. Here we show LC/MS data for each metabolite because the concentration of NADH was below the detection limit in sorted cells, which is consistent with a shift towards a catabolic state (Fig. 1E).
In some cell-sorting experiments, serum (e.g., FBS) is added to the cell-sorting buffer. Although the addition of serum during FACS is often to introduce proteins that reduce non-specific binding of antibodies and protect against apoptosis [24], we wished to explore whether serum can also minimize redox perturbations due to sorting. In principle, including high concentrations of serum in the sorting buffer creates an environment that more closely resembles conventional cell-culture conditions, but there are limitations to such a strategy. First, it is generally recommended that dialyzed serum be used when sorting to minimize the concentration of cations such as calcium and magnesium. These ions promote cell to cell adhesion and clumping, which adversely affect FACS performance [25], [26], [27]. Second, high concentrations of serum in the sorting buffer may result in high concentrations of protein. Since the FACS sheath fluid is usually protein free, too much serum protein can theoretically create a Schlieren effect and result in distortions in light scattering that may affect data quality [28]. Thus, in our experiments, we tested the effects of 1% dFBS, which is commonly used in FACS. Initially, we evaluated astrocytes incubated in buffer with and without 1% dFBS for four hours on the bench top. Including dFBS in the buffer decreased morphological changes characteristic of apoptosis and improved overall cell viability (Fig. 1F). The addition of 1% dFBS also helped preserve the redox state in sorted cells. Strikingly, the ratio of GSSG to GSH was not statistically different in astrocytes sorted with 1% dFBS compared to unsorted controls that were rapidly quenched prior to analysis (Fig. 1A). We observed similar protective effects from dFBS when astrocytes in buffer were challenged with hydrogen peroxide (Fig. 1B). Interestingly, however, 1% dFBS only lessened changes in NADPH/NADP+ and ROS due to sorting (Fig. 1C-D). It also did not prevent disruption of the NAD+/NADH balance during FACS (Fig. 1E).
3.2. FACS alters metabolite concentrations
The redox state of the cell plays a critical role in regulating various reactions at the center of metabolism such as glycolysis, the pentose phosphate pathway, and the tricarboxylic acid (TCA) cycle [15]. We predicted that subjecting cells to FACS not only alters their redox state, but also changes the concentrations of many metabolites. As a general indicator of metabolic perturbation, we first measured the relative intensities of ADP and ATP by LC/MS. For comparison, we set ADP/ATP to 1 for the rapidly quenched condition and determined that this ratio increased over 4 fold when astrocytes were sorted (Fig. 2A). By adding 1% dFBS to the sorting buffer, we were able to mitigate the change in ADP/ATP as a result of FACS to ~2 fold (Fig. 2A, S2C-D). When we stressed cells in sorting buffer with hydrogen peroxide instead of FACS, we observed consistent alterations in ADP/ATP (Fig. 2B).
Fig. 2.
Sorting cells causes widespread alterations in metabolism. (A) Ratio of ADP to ATP in rapidly quenched and sorted cells. (B) Ratio of ADP to ATP in cells after they were transferred to sorting buffer with or without 200 μM H2O2 for four hours. Data shown (A-B) are mean values +/- s.d. (n = 3 biological replicates). (C-F) Scatter plots displaying signals detected by untargeted metabolomics after filtering to remove isotopes, adducts, etc. For each signal, fold changes were calculated from the mean of rapidly quenched cells versus sorted cells (n = 3 biological replicates). Data were transformed by log2() for display. Signals above the y = log2(1.5) line were altered by a fold change ≥1.5. (C) Comparison of sorted cells to rapidly quenched cells without 1% dFBS when using a HILIC separation and negative-ionization mode. (D) Comparison of sorted cells to rapidly quenched cells without 1% dFBS when using a RPLC separation and positive-ionization mode. (E) Same as (C), but with 1% dFBS. (F) Same as (D), but with 1% dFBS. **p < 0.01.
To obtain a global perspective of metabolic dysregulation, we next applied untargeted metabolomics (Fig. 2C-F, S3). Metabolites extracted from cells were analyzed using two different chromatographic methods to increase metabolome coverage, HILIC and RPLC. About 800 signals (also known as metabolomic features) were found in the HILIC data after filtering isotopes, adducts, etc [21]. A comparison of the rapidly quenched and sorted astrocytes revealed that nearly 50% of the HILIC signals were altered by at least 1.5 fold. We note that, based on analyses of technical and biological variability, fold changes of 1.5 or higher are commonly used as thresholds of significance in metabolomics [29]. The number of altered signals was not substantially reduced when dFBS was added, but the overall magnitude of the fold changes did decrease as illustrated by the scatter plots in Fig. 2C and E. Comparable results were obtained using the RPLC method, however, the effects of adding 1% dFBS were larger. Only 38% of the signals were altered by more than 1.5 fold when dFBS was included in the FACS buffer, whereas 46% were altered by more than 1.5 fold when it was omitted (Fig. 2D and F). These results suggest that hydrophobic metabolites are more sensitive to the addition of dFBS than hydrophilic metabolites. Notably, many metabolomic signals did not change, or changed in opposite directions. This indicates that the observed metabolite changes were not a result of systematic experimental error.
It is important to acknowledge that metabolomic comparisons based on unidentified LC/MS signals alone can be misleading. First, some data sets are characterized by a high frequency of artifacts and contaminants [30]. Second, many metabolites appear as a variable number of LC/MS signals, depending on their tendencies to oligomerize, fragment, etc. [21]. As such, the number of altered metabolomic signals does not necessarily correlate with the magnitude of metabolic perturbation. A more accurate consideration of metabolism requires examining structurally identified compounds. We focused on pathways in central carbon metabolism since they are frequently investigated in metabolic studies. Intriguingly, most metabolites we identified decreased in concentration as a result of FACS. These included amino acids and acylcarnitines as well as intermediates from glycolysis, the pentose phosphate pathway, and the TCA cycle (Fig. 3, S4 and S5). Other metabolites, such as the nucleosides uridine and inosine, increased in concentration due to cell sorting (Fig. 3D, S4F). We also point out that the changes observed after sorting cells were different from those observed after a delayed-quench (Fig. S6), indicating that the metabolic perturbations caused by FACS are not merely a result of nutrient deprivation. Although the addition of 1% dFBS to the sorting buffer helped mitigate metabolic changes, in most cases metabolite concentrations were still altered by several fold.
Fig. 3.
Some representative metabolites whose relative concentrations change due to sorting. (A-E) Metabolite levels were quantified by LC/MS. (F) Glycogen levels were measured with a commercial kit. Data shown are mean values +/- s.d. (n = 3 biological replicates). *p < 0.05, **p < 0.01, ***p < 0.001; RQ, Rapidly Quenched; dFBS, dialyzed fetal bovine serum; lysoPC, lysophosphatidylcholine.
Since nutrients are limited during FACS and our delayed-quench condition, we next considered sources of biomass from which carbon may be derived. Glycogen in the brain is predominantly localized to astrocytes, where it is broken down during periods of hypoglycemia [31]. Using the OxiRed dye, we found glycogen storages to be severely depleted in astrocytes after cell sorting, even after addition of 1% dFBS (Fig. 3F). After a delayed quench, in contrast, glycogen was only decreased by 15%. This difference in glycogen utilization suggests that carbon is uniquely mobilized as a result of FACS, potentially in response to oxidative stress.
3.3. Neither dFBS nor BSA prevents metabolic alterations
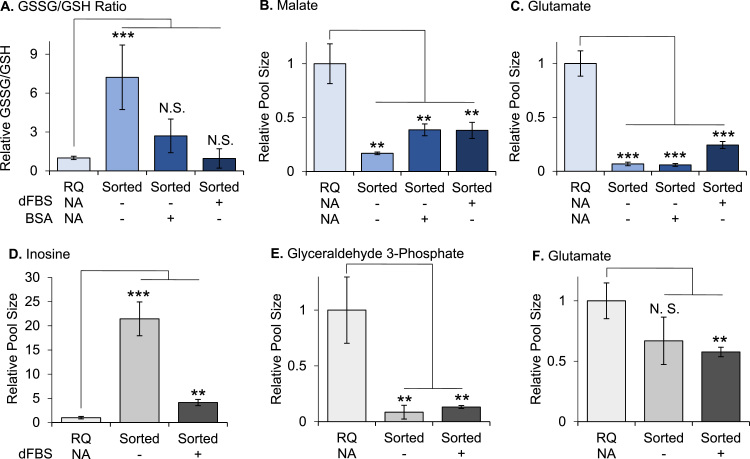
Instead of adding 1% dFBS to the sorting buffer, it is also common to add 1% BSA. Since much of the total protein content in serum is albumin, which has antioxidant properties, we sought to assess whether BSA helps limit metabolic alterations to the same extent as dFBS during sorting [32], [33], [34]. As expected, the metabolic effects of including BSA in the sorting buffer were generally similar to those of including dFBS (Fig. 4A-C). We do note, however, that dFBS might be slightly more effective at reducing oxidative stress and metabolic perturbations during FACS compared to BSA, possibly due to the presence of other unique antioxidant proteins in serum such as transferrin and ceruloplasmin [35], [36], [37]. Indeed, complementing sample buffers with antioxidants during purification procedures has been shown to improve metabolic function previously [38].
Fig. 4.
Attempts to preserve metabolism during sorting with dFBS and BSA as well as strategies to infer metabolites levels prior to FACS with data normalization failed. (A) Relative ratio of GSSG to GSH in rapidly quenched and sorted cells ±1% dFBS or 1% BSA, as determined by LC/MS. (B-C) Relative levels of malate and glutamate in rapidly quenched and sorted cells ±1% dFBS or 1% BSA, as determined by LC/MS. (D-F) Normalizing LC/MS data by the median metabolite signal intensity in each sample does not correct for metabolic alterations that result from sorting. See Fig. 3D, B, and Fig. S4B for comparisons of inosine, glyceraldehyde 3-phosphate, and glutamate before data normalization, respectively. Data shown are mean values +/- s.d. (n = 3 biological replicates). **p < 0.01, ***p < 0.001; RQ, Rapidly quenched; dFBS, dialyzed fetal bovine serum.
3.4. Metabolite alterations cannot be corrected by data normalization
Most identified metabolites had decreased concentrations after sorting. It may therefore be tempting to imagine a correction strategy to normalize the data so that relative metabolite concentrations before FACS could be estimated. Here we evaluated normalizing signal intensities from the metabolomic data by the median signal intensity of each sample. In most cases, data normalization was unable to correct for metabolic alterations associated with FACS and, in some cases, it actually led to artificial amplification of these metabolic changes (Fig. 4D-F, S7). We reason that such a normalization strategy fails because each metabolite is altered by a different fold change due to FACS and because of the non-linear relationship between mass spectrometry signals and concentrations.
4. Conclusions
It is appealing to consider applying FACS to isolate pure populations of cells from complex animal samples for mass spectrometry-based metabolomics, so that metabolite concentrations can be quantified from cells in their naturally occurring environments. Here we show, however, that sorting astrocytes significantly disrupts their metabolism. Although here we focused on one cell type (astrocytes), caution should be taken when using metabolomics to analyze other sorted cells as well. Our results show that FACS introduces oxidative stress, alters cellular redox state, and changes the intensity of nearly half of the metabolomic signals detected. Specifically, we found alterations in the concentrations of amino acids, acylcarnitines, nucleosides, glycogen, as well as intermediates from glycolysis, the pentose phosphate pathway, and the TCA cycle. Adding dFBS or BSA to the cell-sorting buffer improved results, but did not prevent metabolic perturbations during FACS. Thus, we conclude that the metabolism of cells in their naturally occurring environments is not accurately represented by metabolomic data after samples have been subjected to sorting.
Acknowledgements
We thank the Flow Cytometry & Fluorescence Activated Cell Sorting Core at Washington University School of Medicine in St. Louis for help with cell sorting. G.J.P. received financial support for this work from NIH grants R35ES028365 and R21CA191097, as well as the Alfred P. Sloan Foundation, the Pew Scholars Program in the Biomedical Sciences, and the Edward Mallinckrodt, Jr., Foundation.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.03.004.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Zamboni N., Saghatelian A., Patti G.J. Defining the metabolome: size, flux, and regulation. Mol. Cell. 2015;58:699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villas-Bôas S.G., Bruheim P. Cold glycerol–saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal. Biochem. 2007;370:87–97. doi: 10.1016/j.ab.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Mayers J.R., Vander Heiden M.G. Nature and nurture: what determines tumor metabolic phenotypes? Cancer Res. 2017;77:3131–3134. doi: 10.1158/0008-5472.CAN-17-0165. [DOI] [PubMed] [Google Scholar]
- 4.Zanetti K.A., Mette E., Maruvada P., Milner J., Moore S.C., Nicastro H.L., Ross S.A., Sampson J.N., Verma M., L J.S. The future of metabolomic profiling in population-based research: opportunities and challenges. J. Anal. Bioanal. Tech. 2014;5:1–4. [Google Scholar]
- 5.Ng P.C., Lam H.S. Biomarkers in neonatology: the next generation of tests. Neonatology. 2012;102:145–151. doi: 10.1159/000338587. [DOI] [PubMed] [Google Scholar]
- 6.Mayers J.R., Vander Heiden M.G. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem. Sci. 2015;40:130–140. doi: 10.1016/j.tibs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamphorst J.J., Nofal M., Commisso C., Hackett S.R., Lu W., Grabocka E., Vander Heiden M.G., Miller G., Drebin J.A., Bar-Sagi D., Thompson C.B., Rabinowitz J.D. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao C.-H., Fowle-Grider R., Mahieu N.G., Liu G.-Y., Chen Y.-J., Wang R., Singh M., Potter G.S., Gross R.W., Schaefer J., Johnson S.L., Patti G.J. Exogenous fatty acids are the preferred source of membrane lipids in proliferating fibroblasts. Cell Chem. Biol. 2016;23:483–493. doi: 10.1016/j.chembiol.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantor J.R., Abu-Remaileh M., Kanarek N., Freinkman E., Gao X., Louissaint A., Lewis C.A., Sabatini D.M. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell. 2017;169:258–272. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson S.M., Papagiannakopoulos T., Olenchock B.A., Heyman J.E., Keibler M.A., Luengo A., Bauer M.R., Jha A.K., O’Brien J.P., Pierce K.A., Gui D.Y., Sullivan L.B., Wasylenko T.M., Subbaraj L., Chin C.R., Stephanopolous G., Mott B.T., Jacks T., Clish C.B., Van Der Heiden M.G. Environment impacts the metabolic dependencies of ras-driven non-small cell lung cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonner W.A., Hulett H.R., Sweet R.G., Herzenberg L.A. Fluorescence activated cell sorting. Rev. Sci. Instrum. 1972;43:404–409. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]
- 12.Roci I., Gallart-Ayala H., Schmidt A., Watrous J., Jain M., Wheelock C.E., Nilsson R. Metabolite profiling and stable isotope tracing in sorted subpopulations of mammalian cells. Anal. Chem. 2016;88:2707–2713. doi: 10.1021/acs.analchem.5b04071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villas-Bas S.G. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2006. Sampling and Sample Preparation, in: Metabolome Anal. An Introd; pp. 39–82. [Google Scholar]
- 14.Jacobus W.E. Respiratory control and the integration of heart high-energy phosphate metabolism by mitochondrial creatine kinase. Annu. Rev. Physiol. 1985;47:707–725. doi: 10.1146/annurev.ph.47.030185.003423. [DOI] [PubMed] [Google Scholar]
- 15.Gelman S.J., Naser F., Mahieu N.G., Mckenzie L.D., Dunn G.P., Chheda M.G., Correspondence G.J.P., Patti G.J. Consumption of NADPH for 2-HG synthesis increases pentose phosphate pathway flux and sensitizes cells to oxidative stress. Cell Rep. 2018;22:512–522. doi: 10.1016/j.celrep.2017.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loken M.R., Sweet R.G., Herzenberg L.A. Cell discrimination by multiangle light scattering. J. Histochem. Cytochem. 1976;24:284–291. doi: 10.1177/24.1.1254923. [DOI] [PubMed] [Google Scholar]
- 17.Benson M.C., McDougal D.C., Coffey D.S. The application of perpendicular and forward light scatter to assess nuclear and cellular morphology. Cytometry. 1984;5:515–522. doi: 10.1002/cyto.990050513. [DOI] [PubMed] [Google Scholar]
- 18.Chambers M.C., MacLean B., Burke R., Amodei D., Ruderman D.L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T.A., Brusniak M.Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S.L., Nuwaysir L.M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E.W., Moritz R.L., Katz J.E., Agus D.B., MacCoss M., Tabb D.L., Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahieu N.G., Spalding J.L., Gelman S.J., Patti G.J. Defining and detecting complex peak relationships in mass spectral data: the Mz.unity algorithm. Anal. Chem. 2016;88:9037–9046. doi: 10.1021/acs.analchem.6b01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahieu N.G., Spalding J.L., Patti G.J. Warpgroup: increased precision of metabolomic data processing by consensus integration bound analysis. Bioinformatics. 2015;32:btv564. doi: 10.1093/bioinformatics/btv564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahieu N.G., Patti G.J. Systems-level annotation of a metabolomics data set reduces 25,000 features to fewer than 1000 unique metabolites. Anal. Chem. 2017;89:10397–10406. doi: 10.1021/acs.analchem.7b02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y.-J., Huang X., Mahieu N.G., Cho K., Schaefer J., Patti G.J. Differential incorporation of glucose into biomass during warburg metabolism. Biochemistry. 2014;53:4755–4757. doi: 10.1021/bi500763u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asensi M., Sastre J., V Pallardo F., Lloret A., Lehner M., Garcia-de-la Asuncion J., Viña J. Ratio of reduced to oxidized glutathione as indicator of oxidative stress status and DNA damage. Methods Enzymol. 1999;299:267–276. doi: 10.1016/s0076-6879(99)99026-2. 〈http://www.ncbi.nlm.nih.gov/pubmed/9916205〉 (Accessed 5 February 2018) [DOI] [PubMed] [Google Scholar]
- 24.Davey H.M., Kell D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol. Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attramadal A. The effect of divalent cations on cell adhesion. J. Periodontal Res. 1969;4:281–285. doi: 10.1111/j.1600-0765.1969.tb01980.x. [DOI] [PubMed] [Google Scholar]
- 26.Hornby J.E. Measurements of cell adhesion. Development. 1973;30 [Google Scholar]
- 27.Brennan J.K., Mansky J., Roberts G., Lichtman M.A. Improved methods for reducing calcium and magnesium concentrations in tissue culture medium: application to studies of lymphoblast proliferation in vitro. In Vitro. 1975;11:354–360. doi: 10.1007/BF02616371. [DOI] [PubMed] [Google Scholar]
- 28.Settles L., Hackett G.S., Miller E.B., Weinstein J.D. Begell House, Inc.; 1995. Full-Scale Schlieren Flow Visualization, in: Flow Vis. VII; pp. 2–13. [Google Scholar]
- 29.Crews B., Wikoff W.R., Patti G.J., Woo H.-K., Kalisiak E., Heideker J., Siuzdak G. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Anal. Chem. 2009;81:8538–8544. doi: 10.1021/ac9014947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahieu N.G., Huang X., Chen Y.-J., Patti G.J. Credentialing features: a platform to benchmark and optimize untargeted metabolomic methods. Anal. Chem. 2014;86:9583–9589. doi: 10.1021/ac503092d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown A.M., Ransom B.R. Astrocyte glycogen and brain energy metabolism. Glia. 2007;55:1263–1271. doi: 10.1002/glia.20557. [DOI] [PubMed] [Google Scholar]
- 32.Bourdon E., Blache D. The importance of proteins in defense against oxidation. Antioxid. Redox Signal. 2001;3:293–311. doi: 10.1089/152308601300185241. [DOI] [PubMed] [Google Scholar]
- 33.Roche M., Rondeau P., Singh N.R., Tarnus E., Bourdon E. The antioxidant properties of serum albumin. FEBS Lett. 2008;582:1783–1787. doi: 10.1016/j.febslet.2008.04.057. [DOI] [PubMed] [Google Scholar]
- 34.Halliwell B. Albumin-An important extracellular antioxidant? Biochem. Pharmacol. 1988;37:569–571. doi: 10.1016/0006-2952(88)90126-8. [DOI] [PubMed] [Google Scholar]
- 35.Taysi S., Akcay F., Uslu C., Dogru Y., Gulcin I. Trace elements and some extracellular antioxidant protein levels in serum of patients with laryngeal cancer. Biol. Trace Elem. Res. 2003;91:11–18. doi: 10.1385/BTER:91:1:11. [DOI] [PubMed] [Google Scholar]
- 36.Pacht E.R., Davis W.B. Role of transferrin and ceruloplasmin in antioxidant activity of lung epithelial lining fluid. J. Appl. Physiol. 1988;64:2092–2099. doi: 10.1152/jappl.1988.64.5.2092. [DOI] [PubMed] [Google Scholar]
- 37.Gutteridge J.M.C. Antioxidant properties of the proteins caeruloplasmin, albumin and transferrin. A study of their activity in serum and synovial fluid from patients with rheumatoid arthritis. Biochim. Biophys. Acta - Protein Struct. Mol. Enzymol. 1986;869:119–127. doi: 10.1016/0167-4838(86)90286-4. [DOI] [PubMed] [Google Scholar]
- 38.Brewer G., Jones T., Wallimann T., Schlattner U. Higher respiratory rates and improved creatine stimulation in brain mitochondria isolated with anti-oxidants. Mitochondrion. 2004;4:49–57. doi: 10.1016/j.mito.2004.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material