Abstract
The endonuclease G gene (Endog), which codes for a mitochondrial nuclease, was identified as a determinant of cardiac hypertrophy. How ENDOG controls cardiomyocyte growth is still unknown. Thus, we aimed at finding the link between ENDOG activity and cardiomyocyte growth. Endog deficiency induced reactive oxygen species (ROS) accumulation and abnormal growth in neonatal rodent cardiomyocytes, altering the AKT-GSK3β and Class-II histone deacethylases (HDAC) signal transduction pathways. These effects were blocked by ROS scavengers. Lack of ENDOG reduced mitochondrial DNA (mtDNA) replication independently of ROS accumulation. Because mtDNA encodes several subunits of the mitochondrial electron transport chain, whose activity is an important source of cellular ROS, we investigated whether Endog deficiency compromised the expression and activity of the respiratory chain complexes but found no changes in these parameters nor in ATP content. MtDNA also codes for humanin, a micropeptide with possible metabolic functions. Nanomolar concentrations of synthetic humanin restored normal ROS levels and cell size in Endog-deficient cardiomyocytes. These results support the involvement of redox signaling in the control of cardiomyocyte growth by ENDOG and suggest a pathway relating mtDNA content to the regulation of cell growth probably involving humanin, which prevents reactive oxygen radicals accumulation and hypertrophy induced by Endog deficiency.
Keywords: cardiac hypertrophy, ENDOG, mitochondrial DNA, humanin
Highlights
-
•
Endog deficiency induces cardiomyocyte hypertrophy and superoxide accumulation.
-
•
Hypertrophy induced by Endog deficiency requires ROS accumulation.
-
•
ENDOG is involved in mtDNA replication independently of ROS accumulation.
-
•
Electron Transport Chain activity is not affected by Endog deficiency.
-
•
Humanin restricts ROS accumulation and blocks cardiomyocyte growth.
1. Introduction
The mitochondrial endonuclease G (Endog) gene was identified as a blood pressure-independent determinant of cardiac hypertrophy [1]. Reduced Endog expression was associated with abnormal cardiomyocyte growth and increased reactive oxygen species (ROS) abundance in the heart of rodents in vivo and Endog silencing was sufficient to induce cardiomyocyte growth and to increase ROS production in vitro [1]. The involvement of redox signaling in mediating cardiac hypertrophy induced by several neurohormonal stimuli has been previously reported [2]. ROS and the transduction signaling mediated by the protein kinase AKT/PKB have been involved in hypertension-associated cardiac hypertrophy [3], [4], [5], and oxidation of several Cysteine residues of the Class-II HDAC4 induces its nuclear export, derrepressing positive mediators of cardiomyocyte growth [6]. These results suggested that increased ROS abundance in the heart of Endog null mice (Endog-/-) could mediate cardiomyocyte growth, but the possibility was not yet assessed. Identifying the mechanisms relating Endog deficiency with hypertrophy is important due to its potential clinical relevance. ENDOG-like activity was first identified in extracts of bovine heart mitochondria [7] and, despite further work characterizing its activity in vitro as a DNA/RNA endonuclease, its biological function is presently unknown. Biochemical studies showed that ENDOG cleaves mitochondrial RNA and DNA (mtDNA), suggesting a potential role in mtDNA replication [8]. However, the role of ENDOG in mtDNA biology has never been assessed in organelle or in cells. On the other hand, ENDOG was shown to induce genomic DNA fragmentation upon its nuclear translocation during caspase-independent cell death [9], in particular in cardiomyocytes [10], which repress apoptotic gene expression during differentiation [11]. The fact that the adult myocardium silences the expression of apoptotic genes but keeps the expression of Endog [10], coupled to its identification as a candidate gene for the control of cardiac hypertrophy whose deficiency induces ROS accumulation, prompted us to further investigate these events. Here, we addressed the question whether ROS are involved in cardiomyocyte growth induced by Endog deficiency and which are the mechanisms involved, using in vivo and in vitro models.
2. Materials and Methods
2.1. Animal experimentation, primary cardiomyocyte and fibroblast cultures
The investigation with experimental animals was approved by the Experimental Animal Ethic Committee of the University of Lleida and conforms to the Guide for the Care and Use of Laboratory Animals, 8th Edition, published in 2011 by the US National Institutes of Health. The EndoG mice (Mus musculus) line has a C57BL/6 J background and has been crossed in our lab for several years [1]. Cardiomyocytes from Endog+/+ and Endog-/- hearts were obtained as in [12] and either stained for α-actinin to measure size [13] or cultured and treated as described in the figure legends. Rat (Rattus norvegicus) neonatal cardiomyocytes were obtained from the ventricles of 1 to 4-day-old Sprague-Dawley pups and cultured as previously described [11]. Four to six microscopic images per condition (genotype, gene silencing and drug treatment) were recorded at a resolution of 4080×3072 pixels with an Olympus IX71 fluorescence microscope and U-RFL-T power supply system (20× objective and 10× ocular magnifications plus 1.5× optical enhancement). Cross-sectional areas of approximately 100 cells / condition were measured using the Image-J software (NIH). In this setting, 1 µm2 occupies 225 pixels. Cardiomyocyte cross-sectional areas were transformed from pixels to µm2. Primary mouse embryonic fibroblasts (MEF) were prepared by mincing E14.5 embryos from EndoG mice, treating them with trypsin for 20 to 30 minutes, and plating in complete DMEM media supplemented with 10% FBS and antibiotics. The cells were split at 1:2–1:3 ratios when freshly confluent, passaged two or three times to obtain a morphologically homogenous culture, and then frozen or expanded for further studies. Primary cell cultures were treated with N-acetyl-L-cysteine (NAC, Sigma-Aldrich; A7250), a ROS scavenger. NAC was dissolved in PBS or complete DMEM media to produce a 612 mM stock solution. Following filter sterilization (0.2 μm pore-size filters), NAC was added to the cell media at a concentration of 0.2 mM during the time required according to the experimental design. NAC stocks were made fresh. L-Glutathione reduced (GSH, Sigma-Aldrich; G4251) was stored at 163 mM and prepared at a concentration of 10 mM in culture medium at the time of the experiment, as previously described [14]. MitoTEMPO (Sigma-Aldrich; SML0737) was stored at 20 mM and prepared in water and added to the culture at a final concentration of 25 µM [15]. Lyophilized Humanin peptide, sequence MAPRGFSCLLLLTGEIDLPVK (custom synthesized, purity > 90%; GenScript, USA) was reconstituted in water to obtain a 2 mM stock solution that was subsequently dissolved in culture media to obtain the final working concentrations.
2.2. Endog gene silencing
Endog gene silencing in cardiomyocytes was achieved by transduction of lentiviral particles containing either of two independent Endog-specific small hairpin RNA constructs or a scrambled sequence that were produced in Human Embryonic Kidney 293 cells (HEK293T) as detailed in [1], [10], [11]. Silencing efficiency was checked by Western Blot in each experiment.
2.3. Detection of reactive oxygen species
ROS production was determined by using the fluorescent probes DHE (Sigma-Aldrich; D7008) and MitoSOXTM Red (Thermo Fisher Scientific; M36008). DHE has been commonly used to detect cytosolic superoxide anions (O2.−) and MitoSOXTM Red, as a cationic derivative of DHE, is rapidly and selectively targeted to the mitochondria where it is oxidized mainly by superoxide anions (O2.−). Detection of ROS abundance with MitoSOX and DHE by in situ imaging is discouraged due to detection of potential artifacts [16]. Therefore we used a flow cytometry procedure as described below and accompanied it with a test using mitochondria-specific and cytosolic inducers of ROS, as described in the supplementary information. Rat neonatal cardiomyocytes transduced with Endog-shRNA constructs and mouse neonatal cardiomyocytes from Endog+/+ and Endog-/- hearts, both of them treated with N-acetyl-L-cysteine (NAC) 0.2 mM for the indicated times, were washed twice in PBS, tripsinized and counted using Neubauer cell chamber. Equal amount of cells from each population were incubated in PBS containing 5 μM MitoSOXTM Red for 10 min or 10 μM DHE for 30 min at 37 °C in the dark prior to ROS measurement. The fluorescence of the cell population is proportional to the levels of intracellular ROS generated and measured with a BD FACSCanto II cytometer (Becton Dickinson, Mountain View, CA, USA) using 488 nm laser excitation and detection with BP 585/42 filter. Fluorescence values varied between days. Therefore, before statistical comparisons, the full set of data for each experiment was normalized to the mean value obtained the same day for Endog+/+ mouse cardiomyocyte cultures or scrambled-transduced neonatal rat cardiomyocyte cultures.
2.4. Subcellular fractionation, protein extraction and expression analysis by Western Blot
Subcellular fractionation of primary neonatal cardiomyocytes from Endog+/+ and Endog-/- hearts was performed with the Subcellular Protein Fractionation kit for cultured cells (Thermo Scientific-Pierce; 78840) following manufacturer's guideline. In addition, Nuclear fraction was obtained with the Nuclei EZ Prep Nuclei Isolation kit (Sigma-Aldrich; NUC101) which includes a gentle and nonionic detergent, Igepal CA-630 (NP-40), following supplier's instructions. Protein expression was analyzed in total protein extracts from tissues, cell cultures and subcellular fractions diluted in Tris-buffered 2% SDS solution at pH 6.8 and SDS-PAGE was performed as described [10], [11], [12]. Western blots were performed as reported in [10], [11], [12]. Antibody specifications are described in Supplementary S1 table.
2.5. Mitochondrial DNA sequencing
Whole mtDNA was amplified from 8 Endog+/+ and 8 Endog-/- mice in one amplicon by long range PCR (Takara LA PCR kit) using 50–100 ng of DNA and specific mtDNA primers (F:5’-GGTTCGTTTGTTCAACGATTAAAGTCCTACGTG-3’, R: 5’-GAGGTGATGTTTTTGGTAAACAGGCGGGGT-3’). Quantification of amplified mtDNA was performed using Qubit 2.0 Fluorimeter (Qubit dsDNA BR Assay kit; Invitrogen). Sample libraries were prepared with Nextera XT DNA Sample Preparation kit (Illumina) according to the manufacturer's protocol for fragments to 150pb and pair-end runs. One ng of amplified mtDNA was used to prepare each library. After PCR clean up with Ampure beads XT (Beackman Coulter), libraries were normalized and pooled. Pooled libraries were carried out into the MiSeq Reagent kit V2 (300 cycles and 2×150 chemistries) (Illumina) and sequenced in the MiSeq platform and analyzed using MiSEq Control Software and MiSeq Reporter Software. Libraries were multiplexed to obtain 3000× medium coverage. The reference sequence was from Mus musculus C57BL/6 J strain (GenBank Ref. NC_005089.1).
2.6. Quantification of mtDNA replication in organello
Heart mitochondria from adult Endog-/- and Endog+/+ mice (n=7/genotype) were isolated by differential centrifugation as described [17], [18]. Briefly, fresh heart tissue was homogenized in cold MIB mitochondria isolation buffer (MIB, 320 mM sucrose, 1 mM EDTA, 10 mM Tris-HCI, pH 7.4) with a Dounce homogenizer and centrifuged at 1,000 × g at 4 °C for 5 min. The supernatant was centrifuged at 9,000 × g at 4 °C for 10 min to obtain the mitochondrial pellet. For the in organello labeling of de novo synthesized DNA we followed the protocol described in [1]. Briefly, the mitochondrial pellets were washed 3 times in MIB and once in incubation buffer (25 mM sucrose, 75 mM sorbitol, 100 mM KCl, 10 mM K2HPO4, 0.05 mM EDTA, 5 mM MgCl2, and 10 mM Tris-HCl, pH 7.4), 250 µg of protein of the mitochondrial pellet (Coomassie Plus Assay Kit, Thermo Scientific, Rockford IL) were resuspended at 1 mg/ml in incubation buffer supplemented with 1 mM ADP, 10 mM glutamate, 2.5 mM malate and 1 mg/mL fatty acid-free bovine serum albumin, as well as 1 µM of dATP, dCTP and dGTP and 1 µM of radiolabeled dTTP ([methyl-3H]dTTP) and incubated at 37 °C in a rotary shaker for 30 min. As a control of even radionucleotide incorporation between samples, total mitochondrial label was measured by scintillation counting of 25 µl of homogenate (Beckman Coulter LS 6500, Brea, CA). Mitochondria were then pelleted (13,000 × g for 1 min) and washed twice with 10% glycerol, 0.15 mM MgCl2, and 10 mM Tris-HCl (pH 6.8). For DNA extraction, the pellet was lysed with 500 µL of 20 mM Hepes-NaOH, (pH 7.4) 75 mM NaCl, 50 mM EDTA, 20 mg/mL proteinase K, and incubated at 4 °C for 45 min. Then, 17 µL of 30% N-Lauroylsarcosine were added and the mixture was incubated at 50 °C for 45 additional minutes. After classical phenol:chloroform:IAA extraction, DNA was resuspended in TE buffer for 12 h at 4 °C and quantified (Quant-iT PicoGreen dsDNA Reagent, Invitrogen). The DNA-incorporated radiolabel was measured by scintillation counting. De novo mtDNA replication was quantified as the apparent fmoles of 3H-labeled nucleotide per ng DNA, calculated from the specific radioactivity of the radiolabeled nucleotide used. Data are represented as the values of Endog-/- vs. Endog+/+ mitochondria obtained, processed and quantified in parallel. Statistical significance of the differences was calculated using the sign test for paired data (SPSS software, IBM Analytics).
2.7. MtDNA recovery in MEF mtDNA-depleted cells
We analysed mtDNA copy number recovery after Ethidium bromide-forced depletion essentially as previously reported [19]. MEF from 5 Endog +/+ and 5 Endog -/- mice were cultured with Ethidium bromide solution (EtBr) 25 ng/ml (Sigma-Aldrich; E1510) during 1 week until the cells approached a state containing 10–20% of the normal amount of mtDNA. After the treatment period, a subsequent complete medium replacement without BrEt allowed recovery of mtDNA. At the times required according to the experimental designs, cells were harvested by trypsinization, washed with PBS, pelleted and stored at −20 °C until DNA isolation. Total DNA was isolated from cell pellets (QuiAmpDNA Mini kit, Qiagen), dissolved in 10 mM Tris–HCl (pH 8.0) and quantified by spectrophotometry (NanoDrop Spectrophotometer, Thermo Scientific). Duplicates were assessed for each experimental condition. mtDNA copy number was calculated as mtDNA/nDNA ratio and detection of mtDNA and nuclear DNA was performed as a multiplex PCR reaction. Relative quantitation of mtDNA (16 S rRNA and ND4 genes) versus nuclear DNA (ANG1 single copy gene) was performed using an ABI PRISM® 7500 real-time PCR system (Applied Biosystems). Custom designed primers and probes used for assessing mtDNA copy number are listed in S2 Table, as previously described [20]. Changes in the mtDNA amount were calculated using the 2-ΔΔCt method [21] and represented as fold changes relative to the indicated control. Trajectories over time for mtDNA were assessed using natural splines that allow fitting nonlinear trends [22]. To assess the effect of BrEt (comparing Endog +/+ and Endog -/- cultures) mixed effect models [23] were used, including interaction term of cells type (Endog +/+ vs. Endog -/-) with time. The additional effect of NAC was assessed including the interaction with NAC. These analyses were performed with the R software (R Core Team, https://www.R-project.org/).
2.8. Mitochondrial proteomics
Mouse cardiac mitochondria were isolated from Endog +/+ and Endog -/- hearts following a differential centrifugation method as previously [24]. Two independent mitochondria enriched samples per genotype were pooled and protein was quantified by Bradford assay. Briefly, proteins were digested using the filter aided sample preparation (FASP) protocol and the resulting peptides labeled with Isobaric tagging for relative and absolute quantitation (iTRAQ), fractionated by cation exchange and analyzed by LC-MS/MS. Quantitative data were analyzed using statistical models developed in our laboratory. Detailed description of the procedures and references can be found in our previous work [12].
2.9. Analysis of respiratory chain complex activity
Hearts were obtained from all mice and snap-frozen to be stored subsequently at −80 °C until further use. Tissue homogenization was always performed at 0–5 °C in mannitol buffer, pH 7.2 (mannitol 225 mM, sucrose 75 mM, Tris–HCl 10 mM and EDTA 0.1 mM) and Complex I, complex II, complex II+III, complex IV and citrate synthase (CS) activities were determined as previously described [25]. Protein content was determined using Coomassie Plus protein assay reagent (Pierce, Rockford, IL, USA). Enzyme activities were expressed as nmol/min.mg protein and results were expressed as relative activity (specific respiratory chain complex activity / citrate synthase activity).
2.10. Quantification of cardiac ATP content
ATP was quantified using 1H NMR spectra of tissue extracts using Chenomx software (Chenomx Inc, Canada) as described previously [26].
2.11. Statistical analysis
Statistics were performed with GraphPad Prism (GraphPad Software, San Diego, CA, USA), unless indicated otherwise. The effect of Endog expression on the experimental values was assessed by the Mann-Whitney U test. The Kruskal-Wallis test was used when comparing more than two groups, followed by the Dunn's test for selected post hoc comparisons, and the Dunnett test when comparing to a control. Moreover, two-way analysis of variance (ANOVA) was performed to determine the impact of Endog expression and drug addition on the experimental values and for possible interaction. In order to assess the different effect of HN concentration by Endog expression, a linear regression model with interaction was fitted for cardiomyocyte cross-sectional area (in logarithm scale for model validation purposes) and adjusted means with 95% confidence interval were obtained in the original scale with the R software (R Core Team). The same approach was used to assess the differential effect of NAC, GSH and the superoxide dismutase (SOD) mimetic MitoTEMPO to block hypertrophy in cardiomyocytes with low ENDOG levels. SPSS Software was used to run the sign test to analyze differences between pairs of observations in the mtDNA replication in organelle experiment, and the R software was used to analyze the mtDNA recovery experiments, as described in the corresponding sections. Statistical analysis of quantitative proteomics data was performed using the WSPP model developed by Dr. Jesús Vázquez (CNIC, Madrid, Spain) as described previously [12]. Experiments were performed at least three times, the exact number is specified in each figure legend. All statistical tests were two-sided at a significance level of 0.05.
3. Results
3.1. Endog deficiency triggers cardiomyocyte hypertrophy dependent on ROS accumulation
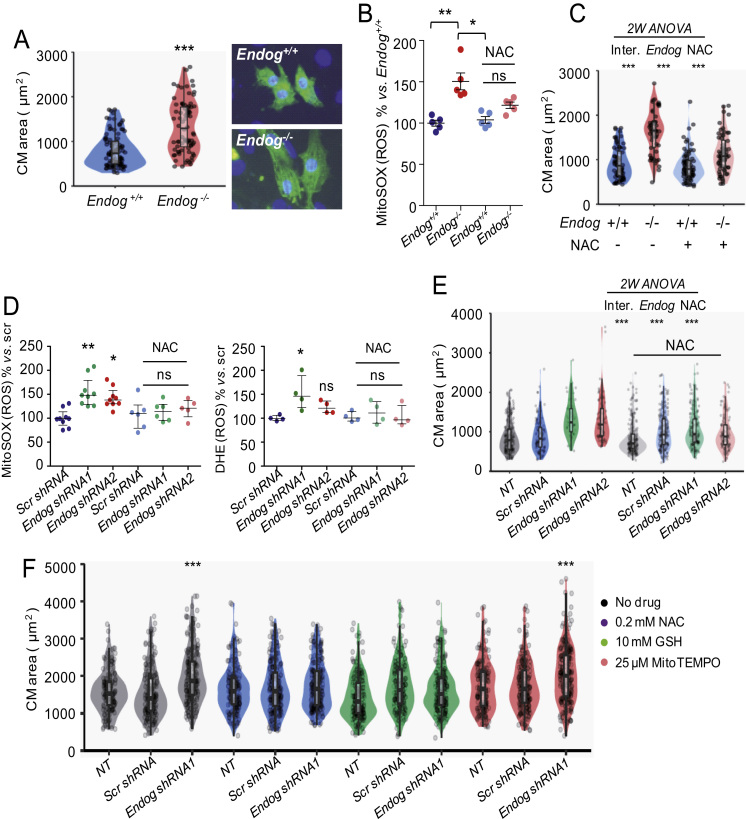
We previously showed that cardiomyocytes of adult Endog-/- mice were bigger than those of Endog+/+ mice, by using a histological procedure [1]. Here we dissected and digested neonatal hearts, seeded the cells in culture dishes and stained them with α-actinin to identify cardiomyocytes. The mean cross-sectional area of neonatal cardiomyocytes from cultures of both genotypes was quantified. Cardiomyocytes of Endog-/- neonatal mice were bigger than those of Endog-/- mice (Fig. 1A) and the increase in size was accompanied by an increase in ROS accumulation as measured using the O2•- indicator MitoSOX by flow cytometry (Fig. 1B), following the recommendations published elsewhere [16], [27], which are commented in the discussion and material and methods sections. Mitochondria were the most probably source of ROS in Endog-deficient cells given the published specificity of MitoSOX to react with ROS in these organelle [27]. Nevertheless, we checked the specificity of MitoSOX for mitochondrial ROS in our model by quantifying MitoSOX signal in myocytes treated with the mitochondrial superoxide-inducer Antimycine A or with phorbol myristate acetate (PMA), an activator of cytosolic NADPH oxidase [27], confirming that MitoSOX signal increased only in myocytes with enhanced mitochondrial ROS production (Fig.S1). Increased ROS accumulation was hampered by the presence of 0.2 mM N-acetyl-L-cysteine (NAC) in the culture medium (Fig. 1B) and NAC addition was associated with a reduction in the cross-sectional area of Endog-/- cardiomyocytes (Fig. 1C). These results were complemented with the analysis of cardiomyocyte size, ROS accumulation and effects of NAC in rat neonatal cardiomyocytes transduced with control (scrambled, Scr) or two different Endog-specific shRNA constructs to silence Endog expression (shRNA1, 2). Endog knockdown was sufficient to induce ROS accumulation in rat cardiomyocytes (Fig. 1D), and hypertrophy (Fig. 1E) and NAC addition suppressed both effects (Fig. 1D,E). This suggested that, whatever the ROS generated, they were transformed in H2O2, as NAC has been described to neutralize ROS mostly through enhancing glutathione (GSH) synthesis and GSH-dependent detoxification of H2O2 by glutathione peroxidase. In our model, NAC addition induced increased ThiolTrackerTM signal (an indicator of GSH abundance, according to the manufacturer) in cells with low expression of Endog (Fig.S2). Based on these results, we assessed the efficiency of GSH and the superoxide dismutase (SOD) mimetic MitoTEMPO to block hypertrophy in cardiomyocytes with low ENDOG levels. Only NAC and GSH, which act mainly by neutralizing H2O2, but not MitoTEMPO, which transforms O2.- in H2O2, prevented hypertrophy (Fig. 1F), indicating the potential role of H2O2 in Endog-deficient cardiomyocyte hypertrophy. These results showed that Endog deficiency induces the increase of cardiomyocyte size at the neonatal stage and that blocking the concomitant mitochondrial production and accumulation of ROS is sufficient to hamper hypertrophy.
Fig. 1.
ENDOG deficiency induces hypertrophy and ROS accumulation in neonatal cardiomyocytes that can be prevented by ROS scavengers. (A) Violin plots showing areas of mouse cardiomyocytes (CM) isolated from 8–10 P2–3 neonatal Endog+/+ and Endog-/- pups seeded, fixed and stained with anti-α-actinin (sarcomere, green) and Hoescht 33342 stain (nucleus, blue). Cell area was quantified with the ImageJ software as described in the Materials and Methods section in at least 100 cardiomyocytes per genotype obtained in three independent experiments; *** P<0.001 (Mann-Whitney U test). Points are individual data. Standard boxplot is also included. Representative images are shown on the right. (B) ROS abundance was detected with the MitoSOXTM mitochondrial superoxide indicator in five independent CM cultures of P2–3 neonatal Endog+/+ and Endog-/- mice cultured 48 hours in the presence or absence of 0.2 mM NAC and calculated as indicated in the M&M section. Medians ± interquartile ranges are shown. The Kruskall-Willis test (P<0.01) followed by the Dunn's test were performed. **, P<0.01 vs. Endog+/+; *, P<0.05 vs. NAC, ns = not significant differences vs. Endog+/+. (C) CM areas were measured as in (A) for each of the treatments described in (B); Two-way ANOVA indicated ***, P<0.001 for interaction, Endog expression and NAC treatment. Points are individual data. (D) MitoSOXTM and DHE signals in neonatal rat cardiomyocytes transduced with two different Endog shRNA or scrambled (Scr shRNA) lentiviral vectors and cultured 48 hours in the absence or presence of 0.2 mM NAC. Medians ± interquartile ranges are shown. The Kruskall-Wallis test (P<0.001) followed by the Dunn's test were performed. *, P<0.05; **, P<0.01, ns = not significant differences vs. Scr. N=5–8 (MitoSOX) and N=5 (DHE) independent experiments. (E) Rat neonatal CM areas were measured as in (A) from 3 independent experiments (at least 100 CM/condition) in non-transduced cultures (NT) or cultures transduced with two different Endog shRNA or scrambled (Scr shRNA) lentiviral vectors and cultured 48 hours in the absence or presence of 0.2 mM NAC; Two-way ANOVA indicated ***, P<0.001 for interaction, Endog expression and NAC treatment. Points are individual data. (F) Cross-sectional cardiomyocyte areas were measured as in (A) after treatment of non-transduced (NT), Scr or Endog shRNA-transduced cultures in absence or presence of the ROS scavengers NAC, GSH or MitoTEMPO for 48 hours. Between 150 and 200 areas were recorded per condition and shown in the violin plot, which includes a standard boxplot. The Dunnett's post hoc test comparing all conditions against NT indicated significant differences of Endog shRNA1 without ROS scavenger and with MitoTEMPO. ***, P<0.001. Additional two-way ANOVA showed same results, with significant interaction (P<0.001).
3.2. Lack of Endog expression induces changes in the AKT/PKB and Class-II HDAC transduction signaling pathways in cardiomyocytes
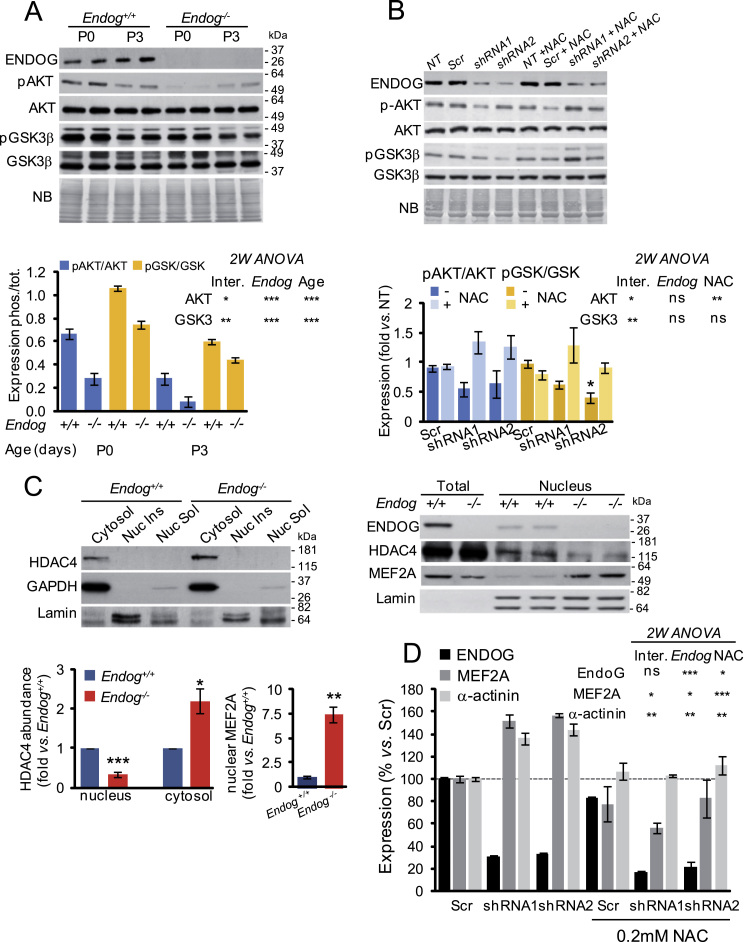
The above results showed that cardiomyocyte hypertrophy induced by Endog deficiency was dependent on ROS accumulation. The cellular redox status has been shown to modify signaling transduction involved in cell growth, including the AKT/PKB pathway [3], [4], [5] and the nucleo-cytoplasmatic shuttling of Class-IIa HDACs, in particular HDAC4 [6]. Therefore, we characterized both pathways comparing control and Endog-deficient cardiomyocytes. Analysis of AKT phosphorylation at Ser473 (pAKT), which is an indicator of its kinase activity, showed that the Endog-/- myocardium had a marked reduction of the pAKT/AKT ratio both at birth (P0) and at three days of extra-uterine life (P3). In agreement with this finding, phosphorylation of its direct target GSK3B was reduced in the Endog-/- myocardium (Fig. 2A). Replication of the experiment in vitro using neonatal rat cardiomyocytes allowed checking the effects of NAC addition. Endog silencing by two independent shRNA constructs caused a decrease in the phosphorylation of AKT and GSK3β and, thus, in the pAKT/AKT and pGSK3B/GSK3β ratios (Fig. 2B). However, the effects were not as marked as in Endog-/- myocytes probably because of the residual Endog expression after gene knockdown (Fig. 2B). The presence of the antioxidant compound NAC was able to block the changes in the phosphorylation status of both kinases (Fig. 2B, graph). Because hypertrophy has been associated with higher pAKT/AKT ratio [3], [4], [5] and our results showed the opposite in the case of Endog-deficient myocytes, we also assessed the cytosolic and nuclear expression of HDAC4. In the nucleus, HDAC4 repress Myocyte Enhancer Factor-2 (MEF2)-dependent activation of hypertrophic genes [28]. Dissociation of HDAC4 from MEF2 and posterior nuclear export of HDAC4 allows MEF2-dependent gene transcription and hypertrophy [29], and this pathway has been shown to be also regulated by redox signaling [6]. We obtained the cytosolic fraction of neonatal Endog+/+ and Endog-/- mice's ventricles and assessed the expression of HDAC4. Cytosolic abundance of HDAC4 was higher in hearts of Endog-/- mice than in control hearts (Fig. 2C, upper panel). Using this technique, we were unable to detect HDAC4 in the nuclear fraction. However, when we used a specific protocol to obtain nuclear extracts, we observed lower HDAC4 presence in the nuclear fraction of Endog-/- mice's myocardium, coinciding with higher expression of MEF2A (Fig. 2C, right panel and bottom graph). Combined, these results were in agreement with lower repression of nuclear MEF2A by HDAC4 in the hearts of Endog-/- mice. Despite our attempts, we were unable of assessing the role of ROS in this event, due to the requirement of millions of mouse myocytes for the subcellular fractionation procedure and lack of rat HDAC4 specific antibodies. Nevertheless, MEF2A and α-actinin expression were higher in Endog-deficient rat cardiomyocytes in vitro compared to their controls (Fig. 2D) and addition of 0.2 mM NAC blunted this effect (Fig. 2D), supporting a role of ROS in the control of MEF2A-dependent transcription.
Fig. 2.
Lack of ENDOG induces downregulation of the AKT-GSK3β transduction signaling due to ROS accumulation and promotes the release MEF2 activity through nuclear export of HDAC4. (A) Expression of ENDOG, phospho-AKT Ser473 (pAKT), total AKT (AKT), phospho-GSK3β Ser21/9 (pGSK3β) and total GSK3β (GSK3β) in neonatal Endog+/+ and Endog-/- mice hearts by Western Blot. Representative Western blots with equal loading verified by naphtol blue (NB) membrane staining are shown in the upper panel. Quantitative analysis of densitometric data is shown in the graph. Bars depict means ± standard errors of the phosphorylated/total expression quotients from three independent experiments. Two-way ANOVA indicated that Endog expression and age influenced the ratios of pAKT/AKT and pGSK3/GSK3 *, P<0.05; **, P<0.01; ***, P<0.001.Inter: Interaction between both variables in the experimental value. (B) Analysis of the effect of NAC treatment (0.2 mM) on the phosphorylation of AKT and GSK3β in primary neonatal rat cardiomyocytes transduced with lentiviral vectors containing Endog shRNA or a scrambled sequence (Scr) or not transduced (NT). Expression was checked by Western Blot (upper panel) and equal loading was verified by membrane staining with naphtol blue (NB). Due to important inter-experiment signal differences, densitometric data were normalized dividing the values by the mean of the Scrambled signal and represented as means ± standard errors of three independent experiments (bar graph). Two-way ANOVA indicated whether Endog expression and NAC influenced the ratios of pAKT/AKT and pGSK3/GSK3. (C) Subcellular distribution of HDAC4 and MEF2A in primary mouse cardiomyocytes from neonatal Endog+/+ and Endog-/- hearts. Fractions were analyzed by Western Blot to detect HDAC4 and MEF2A in cytosolic (left panel) and nuclear fractions (right panel), using GAPDH as a cytosolic marker and Lamin as a nuclear marker to control fractionation. Quantification of HDAC4 and MEF2A Western Blot signals are shown in the graphs. Bars depict means ± standard errors of two independent experiments (8–10 hearts / condition / experiment). The Mann-Whitney U test indicated significant differences between groups *, P<0.05; **, P<0.01; ***P<0.001. Nuc Ins: nuclear insoluble fraction (membranes); Nuc Sol: nuclear soluble fraction. (D) Densitometric quantification of ENDOG, MEF2A and α-actinin expression in rat neonatal cardiomyocytes not transduced (NT), transduced with a scrambled construct (Scr) or transduced with Endog-specific shRNA1 and 2 viruses; in the absence or presence of 0.2 mM NAC. Bars are means ± standard errors of densitometric values related to their respective Scr signal. Two-way ANOVA indicated whether Endog silencing and NAC influenced the expression of ENDOG, MEF2A and α-actinin. *, P<0.05; **, P<0.01; ***, P<0.001; ns: not significant.
3.3. Endog loss-of-function compromises mitochondrial DNA replication independently of ROS accumulation
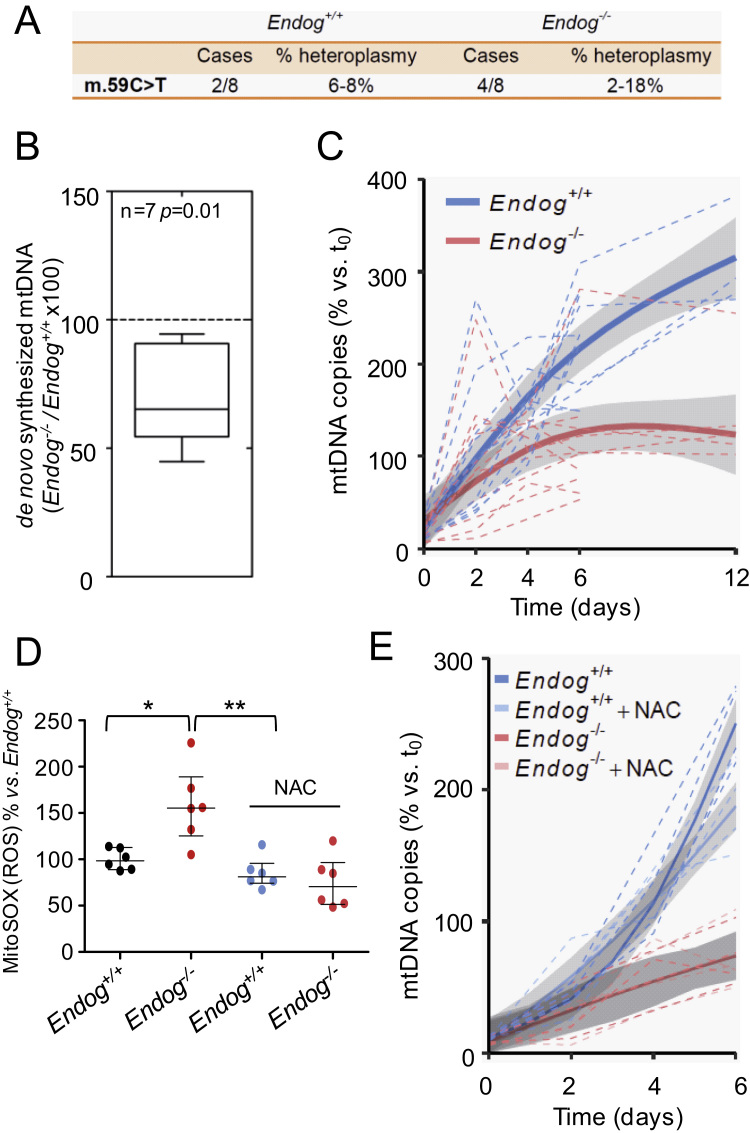
Once we found that ENDOG regulates cardiomyocyte hypertrophy through the control of ROS accumulation, we investigated the molecular events mediating this event. We reasoned that the influence of ENDOG on mtDNA biology could be upstream of ROS accumulation and ROS-dependent cardiomyocyte hypertrophy. We sequenced the cardiac mitochondrial genome of Endog+/+ and Endog-/- mice and found a single nucleotide polymorphism compared to the standard genome that was present at similar frequencies in both genotypes (Fig. 3A). These results suggested that ENDOG is not required to maintain mtDNA replication fidelity in the heart. Our previous results showed that lack of ENDOG associates with lower abundance of cardiac mtDNA [1]. We isolated cardiac mitochondrial preparations from Endog+/+ and Endog-/- mice and quantified the incorporation of labeled deoxynucleotides to mtDNA. Our results showed that incorporation of deoxynucleotide to the DNA of Endog-/- mitochondria was slower than in Endog+/+ mitochondria, suggesting the requirement of ENDOG to sustain normal mtDNA synthesis (Fig. 3B). To further investigate this event and the potential influence of ROS, we quantified the mtDNA content of Endog+/+ and Endog-/- mouse embryonic fibroblasts (MEF) in which we induced mtDNA depletion by ethidium bromide (EtBr) treatment (MEF-Depl) and then MEF-Depl were let to recover their mtDNA content in the absence of EtBr. Treatment with EtBr reduced MEF's mtDNA content at similar levels in both genotypes. Recovery of the mtDNA was faster in Endog+/+ MEF (P<0.001) and Endog-/- MEF-Depl were unable to sustain a net gain of mtDNA copies reaching a plateau by the end of the studied period (Fig. 3C). Then, we questioned whether ROS accumulation in Endog-/- MEF-Depl determined the lower capacity to mtDNA synthesis. The presence of NAC, which hampered ROS accumulation (Fig. 3D), did not improve mtDNA replication in Endog-/- MEF-Depl (P=0.9) (Fig. 3E). These results showed that Endog deficiency compromises mtDNA replication upstream, or at least independently, of ROS accumulation.
Fig. 3.
EndoG deficiency has no effect on mtDNA stability but hinders mtDNA synthesis independently of ROS accumulation. (A) Sequencing of mitochondrial DNA extracted from hearts of Endog+/+ and Endog-/- neonatal mice did not show any nucleotide variant associated to the Endog genotype. A single mutation (59 C>T) was detected in heteroplasmy (% frequency vs. reference sequence) within the Phe tRNA (MT-TF) gene in 2 Endog+/+ and 4 Endog-/- mice; N=8 per genotype, ns: not statistically significant differences. (B) Incorporation of dTTP-3H in mtDNA was quantified simultaneously in isolated Endog+/+ and Endog-/- cardiac mitochondria preparations; N=7. Non-parametric sign test for paired observations indicated significant differences due to genotype, P=0.01. (C) MtDNA of MEF from Endog+/+ and Endog-/- mice was depleted with EtBr (25 ng/ml) in order to reach minimum similar levels in both genotypes. Removal of the drug from the culture medium (day 0) allowed recovery of mtDNA content and the mtDNA copy number was determined in Endog+/+ and Endog-/- MEF at the indicated time intervals. The relative mtDNA copy number was quantitated and analyzed as described in the Materials and Methods section. N=5 per genotype. (D) Mitochondrial ROS abundance was quantified using the MitoSOXTM mitochondrial superoxide indicator in Endog+/+ and Endog-/- fibroblasts cultured in the absence or presence of ROS scavenger NAC (0.2 mM). Due to important inter-experiment basal signal oscillations, data were normalized divinding by the Endog+/+ mean before statistical treatment. The Kruskall-Wallis test (P=0.03) was followed by the Dunn's test for pair wise comparisons; *, P<0.05 vs. Endog+/+; **, P<0.01 vs. Endog+/++NAC. Medians ± interquartile ranges are shown. N=6. (E) MtDNA copy number recovery was analyzed in the presence of ROS scavenger NAC. After removal of BrEt (day 0), Endog+/+ and Endog-/- MEF were divided into two groups during one week recovery period. In both genotypes, one group was cultured in a medium with NAC (0.2 mM) and the other group served as control without NAC. At the indicated time intervals, the relative mtDNA copy number was determined in cells from each group as in Fig. 3.C. N=5 per genotype.
3.4. Electron Transport Chain subunit expression, respiratory chain complex activity and ATP concentration are normal in the Endog-/-myocardium
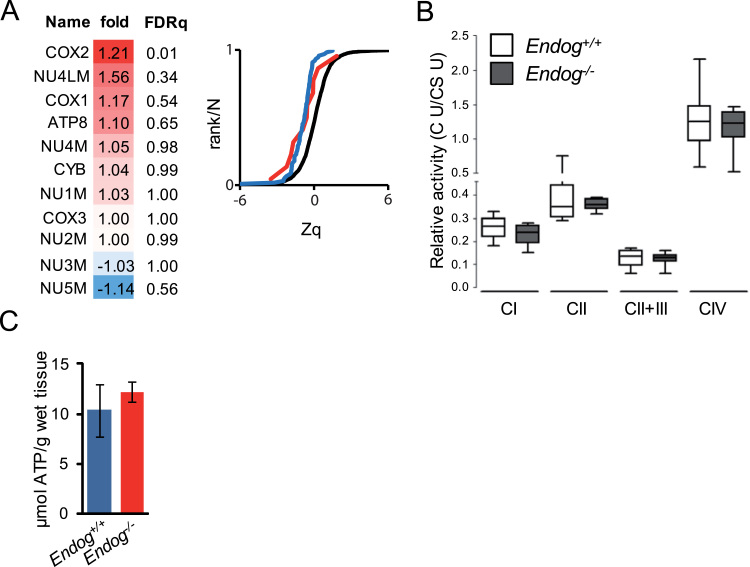
Mitochondrial Electron Transport Chain (ETC) activity is an important source of cellular ROS. Thirteen ETC subunits are coded by the mtDNA. We previously found lower mtDNA content in the heart of Endog-/- mice and here we report reduced mtDNA replication in Endog-deficient mitochondria. Thus, we assessed whether the expression of mtDNA-encoded ETC subunits was compromised in the heart of Endog-/- mice. An Isobaric tag for relative and absolute quantitation (iTRAQ)-based comparative proteomics of cardiac mitochondrial extracts from Endog+/+ and Endog-/- mice showed no overall significant differences by genotype between the abundance of eleven detected mtDNA-encoded ETC subunits with only the Cytochrome c Oxidase complex subunit-2 (COX2) showing very slight but significant increase in Endog-/- mitochondria (Fig. 4A, panel). No changes were observed in the abundance of the whole cardiac mitochondrial proteome (Fig. 4A, graph). This was in accordance with the lack of genotype-associated changes in the activity of the different ETC complexes, as quantified in vitro (Fig. 4B), and the cardiac ATP concentration (Fig. 4C). Together, these results show that reduced mtDNA abundance and impaired mtDNA replication due to Endog deficiency have no relevant impact on ETC expression and activity and, thus, the origin of mitochondrial ROS accumulation in Endog-deficient cells is probably due to external regulation of the ETC and/or independent of it.
Fig. 4.
Mitochondrial respiratory chain expression and activity is not affected by ENDOG deficiency.(A) Left, relative abundance profiles of mitochondria-encoded components of the Electron transport chain in both Endog+/+ and Endog-/- in cardiac mitochondria extracts (FDRq, false discovery rate quotient, significant difference FDRq<0.01). Right graph, sigmoid plot showing the cumulative distributions of log2-ratios in units of the standard deviation at each level (Zq, protein to grand mean variability) (Endog-/- / Endog+/+) of proteins belonging to all oxidative phosphorylation complexes in Endog-/- (red line) and in Endog+/+ (blue line) and the null hypothesis distribution (black line), where a trend to the left would denote increased abundance and a trend to the right would indicate lower abundance vs. the null hypothesis. (B) Complex I, complex II, complex II+III, complex IV and citrate synthase (CS) activities were determined in heart mitochondria extracts from Endog+/+ and Endog-/- mice and expressed as relative activity (specific respiratory chain complex activity / citrate synthase activity), see Materials and Methods section for procedure and statistics. N=5. (C) ATP content of Endog+/+ and Endog-/- mouse hearts. N=3. The Mann-Whitney U test showed no significant differences.
3.5. Humanin, a synthetic micropeptide based in an ORF within the mtDNA prevents ROS accumulation and cardiomyocyte growth induced by Endog deficiency
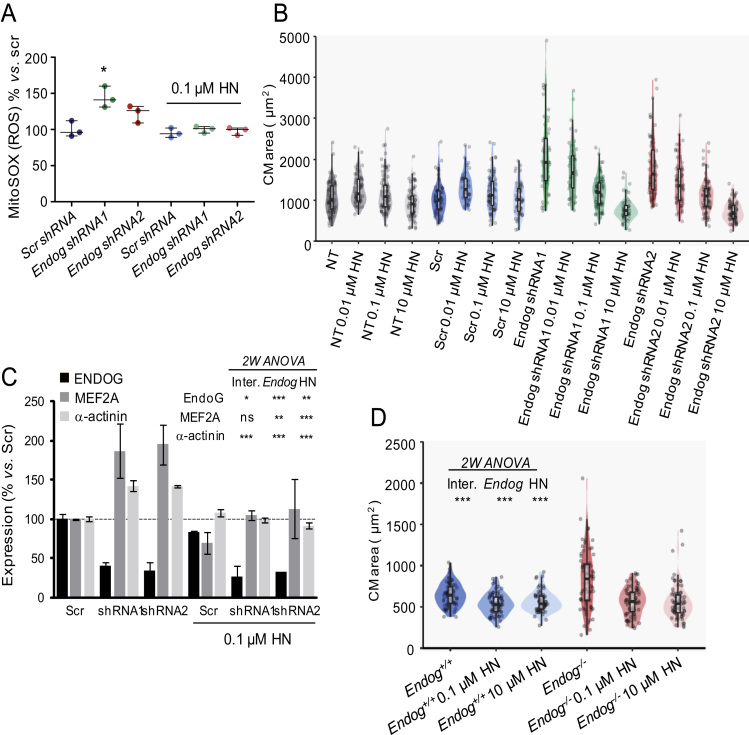
MtDNA has been suggested to contain small ORFs that could code for micropeptides [30]. Humanin was identified as an ORF within the sequence of the 16 S ribosomal RNA gene (MT-RNR2), potentially coding for a 2.6 kDa peptide and addition of synthetic HN to cell cultures has several metabolic effects [30], [31]. However, up to date, no reliable detection of the HN peptide or transcript has been published and attempts from our lab to detect HN in human and rodent samples suggest that commercial antibodies are not specific (data not shown). Nevertheless, we reasoned that reduced mtDNA content due to Endog deficiency, although insufficient to affect the abundance of the mtDNA-encoded ETC subunits, could hinder HN expression and, thus, addition of HN to Endog-deficient cardiomyocytes could affect ROS abundance and cell growth. Indeed, addition of 0.1 µM HN hampered mitochondrial ROS accumulation in Endog-deficient cardiomyocytes (Fig. 5A). The anti-hypertrophic effect of HN was markedly dose-dependent in cardiomyocytes with reduced Endog expression (Fig. 5B) and practically without effect in cardiomyocytes from control groups (Fig.S3). This pattern was accompanied by normalization of MEF2A and α-actinin expression (Fig. 5C). Humanin was also able of blocking abnormal growth of Endog-/--derived mouse cardiomyocytes cultured in vitro (Fig. 5D).
Fig. 5.
The micropeptide humanin reverses the effects on ROS abundance and hypertrophy induced by Endog deficiency. (A) ROS abundance was quantified in neonatal rat cardiomyocytes transduced with lentiviral vectors containing Endog shRNA1 or 2 to silence Endog expression, or a scrambled sequence (Scr) and cultured 48 h in the absence or presence of 0.01 µM humanin (HN). Due to important inter-experiment basal signal oscillations, data were normalized dividing by the Scr mean before statistical treatment. Medians ± interquartile ranges of 3 independent experiments are shown. The Kruskall-Wallis test (P=0.039) was followed by the Dunn's test for pair wise comparisons; *, P<0.05 vs. Scr.(B) Rat neonatal cardiomyocyte cross-sectional areas from non-transduced (NT), Scr or Endog shRNA-transduced cultures in the absence or the presence (48 hours) of HN (0.01, 0.1 or 10 µM), and immunostained with anti-α-actinin. Cell areas were quantified with the ImageJ software in at least 100 cardiomyocytes per genotype from 3 independent experiments (5–8 hearts/experiment). Statistical analysis was performed with a linear regression model with interaction. Adjusted means and 95% confidence intervals are shown in Fig.S3, which indicated significant effects of Endog expression, HN and their interaction on cell size (P<0.001). (C) Expression of muscle-specific factors MEF2A and α-actinin were analyzed by Western Blot in non-transduced (NT), scrambled-transduced (Scr) or Endog shRNA1 (1) or Endog shRNA2 (2) transduced cardiomyocytes, in the absence or presence of 0.1 µM humanin (HN). Bars are means ± standard errors of densitometric values related to their respective Scr signal. Two-way ANOVA indicated whether Endog silencing and HN influenced the expression of ENDOG, MEF2A and α-actinin. *, P<0.05; **, P<0.01; ***, P<0.001; ns: not significant. (D) Effects of 0.1 µM and 10 µM humanin (HN) treatment during 48 hours in the cross-sectional area of cultured cardiomyocytes of Endog+/+ and Endog-/- neonatal mice. Two-way ANOVA indicated ***, P<0.001 for interaction, Endog expression and HN treatment on cell size. Points are individual data.
4. Discussion
In the present study we aimed at identifying the mechanism by which Endog loss-of-function impacts on cardiomyocyte growth. This is relevant because the Endog locus was identified as a determinant of blood pressure-independent cardiac hypertrophy by integrative functional genomics and Endog deficiency was shown to induce cardiomyocyte hypertrophy [1] but the signaling involved remain to be characterized. The main novelty of our contribution is the identification of ROS as the major mediators of hypertrophy in Endog-deficient cardiomyocytes. To our knowledge, we also contribute for the first time biological data supporting that ENDOG is important to sustain mtDNA replication. Despite the lack of overt effects on the expression and activity of the mitochondrial ETC, reduced mtDNA content could hinder HN expression, a mtDNA-encoded micropeptide that has been suggested to be relevant for cell metabolism [30], [31]. Although we cannot yet provide evidence of possible changes in the expression of HN in Endog-deficient cardiomyocytes, our results show that addition of nanomolar concentrations of HN can revert and prevent ROS accumulation and abnormal cardiomyocyte growth induced by Endog deficiency, indicating that this micropeptide could hold therapeutic potential.
ENDOG activity on DNA was discovered three decades ago in cardiac mitochondria extracts [7]. Although its nuclease activity has been well characterized at the molecular level, its biological roles are not completely understood. Some reports show that ENDOG may have a role in DNA recombination in HeLa cells [32] and leukemia cells [33], but the interest about ENDOG rose for its role in cell death as the inducer of genomic DNA degradation [34] in certain cell types including cardiomyocytes [10], [11]. However, Endog was later identified as a determinant of blood pressure-independent cardiac hypertrophy [1] and cardiomyocytes with low ENDOG expression accumulated ROS [1]. Our results help to understand the events mediating Endog-dependent cardiomyocyte hypertrophy by showing the relevant contribution of ROS.
Cardiomyocyte hypertrophy proceeds through mechanical, endocrine- and neurohumoral-induced signal transduction pathways, which can imply the phosphorylation (activation) of the PI3K-AKT/MEK/ERK axes, phosphorylation (inhibition) of GSK3β, and mTOR complex activation, all promoting transcription and protein synthesis [35], and/or the nuclear export of HDAC4 that releases MEF2A to induce transcription of several genes including MEF2A itself [29]. These pathways can be stimulated by mitochondrial ROS [36] and, if sustained, lead to heart failure. Hydrogen peroxide has been shown to drive the redox-dependent signaling influencing cardiac hypertrophy [6], [36]. Our experimental approach combining MitoSOX and DHE fluorescence detection by flow cytometry, combined with the controls presented as supplementary information, suggest that O2.- is produced in the mitochondria with reduced abundance of ENDOG. However, electron paramagnetic resonance (EPR)-based or HPLC-based techniques are required to confirm this [16]. NAC and GSH but not the SOD mimetic MitoTEMPO, blunted Endog deficient cardiomyocyte growth suggesting that H2O2 is the main inductor of the signaling leading to cardiomyocyte hypertrophy. The results presented here suggest that the AKT pathway is not involved in Endog-deficient myocyte growth because Endog deficiency both in vivo and in vitro lead to a reduction, not induction, of AKT and GSK3β phosphorylation, despite ROS accumulation, and NAC restored ROS and pAKT to normal levels in parallel to alleviate cell growth. However, Endog-/- hearts had higher HDAC4 abundance in the cytosol and lower nuclear HDAC4 protein level than Endog+/+. These results together with the induced expression of MEF2A and α-actinin, support the role of the activation of the HDAC/MEF2 pathway. Unfortunately, technical limitations due to insufficient mouse cardiomyocyte yield to perform large cell culture experiments and lack of rat HDAC4-specific antibodies, prevented to settle an in vitro model to check the influence of ROS in HDAC4 export to the cytosol in Endog-deficient myocytes.
Radical oxygen species are important mediators of hypertrophy triggered by extracellular signals, either neurohumoral, endocrine or mechanical [36], [37]. However, Endog loss of function, which occurs in several widely studied rodent models of cardiac hypertrophy such as the spontaneously hypertensive rat (SHR) due to mutations in the Endog locus [1], could be an intrinsic mechanism driving ROS-dependent cardiomyocyte hypertrophy. These results suggest the interest of studying the impact of Endog expression and ROS abundance in human heart failure unrelated to adrenergic or endocrine cues.
Our findings also show that, concomitant to increased ROS accumulation but independently of it, lack of ENDOG hindered mtDNA replication and provoked a 35% drop of the mtDNA content. Decreased mtDNA content causes ROS accumulation and oxidative stress when mtDNA replication capacity is limited [38], [39]. Taken together, these facts suggest that the reduced mtDNA content due to Endog deficiency could be upstream of ROS accumulation and cardiomyocyte hypertrophy. However, we did not identify any overt change in mtDNA-encoded ETC subunit expression, mtDNA mutation rate nor in ETC activity that could justify ROS accumulation. A search for alternative links between the changes in mtDNA and ROS accumulation brought our attention to humanin, which has been identified as an ORF within the mitochondrial 16 S rRNA gene with metabolic and cytoprotective functions [31], [40]. We reasoned that a modest drop of mtDNA abundance could still sustain ETC subunit expression but compromise the production of the peptide HN. Despite reports claiming the detection of HN protein in muscle samples of humans [41] and rodents (Rattin) [40], our attempts to detect it led us to question the specificity of the commercial antibodies. In addition, a search in our iTRAQ-based cardiac proteomics database did not detect HN (nor Angiotensin-II), showing that the procedure is not presently prepared to detect such small peptides. Nevertheless, addition of HN to Endog-deficient cardiomyocytes was sufficient to block ROS accumulation and hamper cardiomyocyte growth in cardiomyocytes with reduced Endog expression. In conclusion, the results presented here together with the existing bibliography, suggest a potential link between ENDOG activity, mtDNA replication, HN expression, ROS production and cardiomyocyte hypertrophy that deserves further investigation due to its therapeutic potential.
Acknowledgements
We thank Prof. Montse Rué from the IRBLLEIDA's Biostatistics Unit (UBiostat) for her help in the design and statistical study of the MEF-Depl experiment.
Acknowledgments
Funding
This work was supported by Grant SAF2013–44942R from the Ministerio de Economía y Competitividad (MINECO) to DS, Grant 20153810 from Fundació La Marató de TV3 to DS, Program “Redes Temáticas de Investigación Cooperativa en Salud” (RETICS) Grants RD12/0042/0035, RD12/0042/0056 and RD12/0042/0021, Red de Investigación Cardiovascular (RIC) from the Instituto de Salud Carlos-III (ISCIII) to DS, DG-D and JV, Grant 2009SGR-346 from the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) from the Government of Catalonia to DS. AB is supported by Fundació La Marató de TV3 and GB is supported by a predoctoral contract from the Universitat de Lleida.
Conflict of interest
None.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.02.021.
Appendix A. Supplementary material
Supplementary material
References
- 1.McDermott-Roe C. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafstad A.D., Nabeebaccus A.A., Shah A.M. Novel aspects of ROS signalling in heart failure. Basic Res Cardiol. 2013;108:359. doi: 10.1007/s00395-013-0359-8. [DOI] [PubMed] [Google Scholar]
- 3.Rizzi E. β1-Adrenergic blockers exert antioxidant effects, reduce matrix metalloproteinase activity, and improve renovascular hypertension-induced cardiac hypertrophy. Free Radic Biol Med. 2014;73:308–317. doi: 10.1016/j.freeradbiomed.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 4.Kwon S.H., Pimentel D.R., Remondino A., Sawyer D.B., Colucci W.S. H(2)O(2) regulates cardiac myocyte phenotype via concentration-dependent activation of distinct kinase pathways. J Mol Cell Cardiol. 2003;35:615–621. doi: 10.1016/s0022-2828(03)00084-1. [DOI] [PubMed] [Google Scholar]
- 5.Wang H.X. NADPH oxidases mediate a cellular "memory" of angiotensin II stress in hypertensive cardiac hypertrophy. Free Radic Biol Med. 2013;65:897–907. doi: 10.1016/j.freeradbiomed.2013.08.179. [DOI] [PubMed] [Google Scholar]
- 6.Ago T. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 7.Cummings O.W., King T.C., Holden J.A., Low R.L. Purification and characterization of the potent endonuclease in extracts of bovine heart mitochondria. J Biol Chem. 1987;262:2005–2015. [PubMed] [Google Scholar]
- 8.Côté J., Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- 9.Li L.Y., Luo X., Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J. EndoG Links Bnip3-Induced Mitochondrial Damage and Caspase-Independent DNA Fragmentation in Ischemic Cardiomyocytes. Plos One. 2011;6 doi: 10.1371/journal.pone.0017998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahi N. Switch from caspase-dependent to caspase-independent death during heart development - Essential role of endonuclease G in ischemia-induced DNA processing of differentiated cardiomyocytes. Journal of Biological Chemistry. 2006;281:22943–22952. doi: 10.1074/jbc.M601025200. [DOI] [PubMed] [Google Scholar]
- 12.Cardona M. Executioner Caspase-3 and 7 Deficiency Reduces Myocyte Number in the Developing Mouse Heart. Plos One. 2015;10 doi: 10.1371/journal.pone.0131411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye J., Cardona M., Llovera M., Comella J.X., Sanchis D. Translation of Myocyte Enhancer Factor-2 is induced by hypertrophic stimuli in cardiomyocytes through a Calcineurin-dependent pathway. J Mol Cell Cardiol. 2012;53:578–587. doi: 10.1016/j.yjmcc.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Cornago M. Histone deacetylase inhibitors promote glioma cell death by G2 checkpoint abrogation leading to mitotic catastrophe. Cell death & disease. 2014;5:e1435. doi: 10.1038/cddis.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z. Epac2-Rap1 Signaling Regulates Reactive Oxygen Species Production and Susceptibility to Cardiac Arrhythmias. Antioxid Redox Signal. 2017;27:117–132. doi: 10.1089/ars.2015.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griendling K.K. Measurement of Reactive Oxygen Species, Reactive Nitrogen Species, and Redox-Dependent Signaling in the Cardiovascular System: A Scientific Statement From the American Heart Association. Circ Res. 2016;119:e39–e75. doi: 10.1161/RES.0000000000000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández-Vizarra E. Isolation of mitochondria for biogenetical studies: An update. Mitochondrion. 2010;10:253–262. doi: 10.1016/j.mito.2009.12.148. [DOI] [PubMed] [Google Scholar]
- 18.González-Vioque E., Torres-Torronteras J., Andreu A.L., Martí R. Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) PLoS Genet. 2011;7:e1002035. doi: 10.1371/journal.pgen.1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pontarin G., Ferraro P., Bee L., Reichard P., Bianchi V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc Natl Acad Sci U S A. 2012;109:13302–13307. doi: 10.1073/pnas.1211289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cámara Y. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Hastie T.J. Generalized additive models. In: Chambers J., Hastie T., editors. vol. 7. Pacific Grove; California: 1992. (Statistical Models in S). [Google Scholar]
- 23.Pinheiro J.C., Bates D. Statistics and Computing. Springer-Verlag; New York: 2000. Mixed-effects models in S and S-PLUS. [Google Scholar]
- 24.Ruiz-Meana M. Ischemic preconditioning protects cardiomyocyte mitochondria through mechanisms independent of cytosol. J Mol Cell Cardiol. 2014;68:79–88. doi: 10.1016/j.yjmcc.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Medja F. Development and implementation of standardized respiratory chain spectrophotometric assays for clinical diagnosis. Mitochondrion. 2009;9:331–339. doi: 10.1016/j.mito.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Barba I. High-fat diet induces metabolic changes and reduces oxidative stress in female mouse hearts. J Nutr Biochem. 2017;40:187–193. doi: 10.1016/j.jnutbio.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Dikalov S.I., Harrison D.G. Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid Redox Signal. 2014;20:372–382. doi: 10.1089/ars.2012.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miska E.A. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J., McKinsey T.A., Nicol R.L., Olson E.N. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S.J., Xiao J., Wan J., Cohen P., Yen K. Mitochondrially derived peptides as novel regulators of metabolism. J Physiol. 2017 doi: 10.1113/JP274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C., Yen K., Cohen P. Humanin: a harbinger of mitochondrial-derived peptides? Trends Endocrinol Metab. 2013;24:222–228. doi: 10.1016/j.tem.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson A.B., Robertson J., Fusser M., Klungland A. Endonuclease G preferentially cleaves 5-hydroxymethylcytosine-modified DNA creating a substrate for recombination. Nucleic Acids Res. 2014;42:13280–13293. doi: 10.1093/nar/gku1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gole B., Baumann C., Mian E., Ireno C.I., Wiesmüller L. Endonuclease G initiates DNA rearrangements at the MLL breakpoint cluster upon replication stress. Oncogene. 2015;34:3391–3401. doi: 10.1038/onc.2014.268. [DOI] [PubMed] [Google Scholar]
- 34.Samejima K., Earnshaw W.C. Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 35.Maillet M., van Berlo J.H., Molkentin J.D. Molecular basis of physiological heart growth: fundamental concepts and new players. Nat Rev Mol Cell Biol. 2013;14:38–48. doi: 10.1038/nrm3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai D.F. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Galphaq overexpression-induced heart failure. Circ Res. 2011;108:837–846. doi: 10.1161/CIRCRESAHA.110.232306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsushima S. Increased oxidative stress in the nucleus caused by Nox4 mediates oxidation of HDAC4 and cardiac hypertrophy. Circ Res. 2013;112:651–663. doi: 10.1161/CIRCRESAHA.112.279760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis W. Decreased mtDNA, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolesar J.E. Defects in mitochondrial DNA replication and oxidative damage in muscle of mtDNA mutator mice. Free Radic Biol Med. 2014;75:241–251. doi: 10.1016/j.freeradbiomed.2014.07.038. [DOI] [PubMed] [Google Scholar]
- 40.Muzumdar R.H. Acute humanin therapy attenuates myocardial ischemia and reperfusion injury in mice. Arterioscler Thromb Vasc Biol. 2010;30:1940–1948. doi: 10.1161/ATVBAHA.110.205997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gidlund E.K. Humanin skeletal muscle protein levels increase after resistance training in men with impaired glucose metabolism. Physiol Rep. 2016;4 doi: 10.14814/phy2.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material