Abstract
Cetaceans (whales, dolphins, and porpoises) are a group of specialized mammals that evolved from terrestrial ancestors and are fully adapted to aquatic habitats. Taking advantage of the recently sequenced finless porpoise genome, we conducted comparative analyses of the genomes of seven cetaceans and related terrestrial species to provide insight into the molecular bases of adaptation of these aquatic mammals. Changes in gene sequences were identified in main lineages of cetaceans, offering an evolutionary picture of cetacean genomes that reveal new pathways that could be associated with adaptation to aquatic lifestyle. We profiled bone microanatomical structures across 28 mammals, including representatives of cetaceans, pinnipeds, and sirenians. Subsequent phylogenetic comparative analyses revealed genes (including leptin, insulin-like growth factor 1, and collagen type I alpha 2 chain) with the root-to-tip substitution rate significantly correlated with bone compactness, implicating these genes could be involved in bone mass control. Overall, this study described adjustments of the genomes of cetaceans according to lifestyle, phylogeny, and bone mass.
Keywords: cetacean, comparative genomics, aquatic adaptation, bone microanatomical structure
Introduction
Whales, dolphins, and porpoises are collectively termed cetaceans and have evolved from terrestrial ancestors to occupy aquatic niches and fully aquatic life histories (Thewissen et al. 2007). These mammals are a fascinating example of evolution because the return to a fully aquatic mode of life required wholesale anatomical rearrangements. For example, they lost hindlimbs, their forelimbs transformed into fins, and some species can dive and tolerate low levels of oxygen (Gatesy et al. 2013). In addition, the specialization to aquatic life occurred concurrently with the further divergence of modern whales to two suborders, Odontoceti (toothed whales) and Mysticeti (baleen whales), around 34 Ma (Gingerich et al. 1983; Zhou et al. 2011). Whereas the toothed whales have evolved echolocation (Oelschläger 1992), baleen whales have evolved a novel filter-feeding apparatus consisting of keratinized baleen plates (Deméré et al. 2008). Other features reported in cetaceans include extreme longevity (>200 years in the case of the bowhead whale, Balaena mysticetus; George et al. 1999; George and Bockstoce 2008), reduced cancer incidence (e.g., in the blue whale, Balaenoptera musculus; Caulin and Maley 2011), and resistance to insulin (bottlenose dolphin, Tursiops truncatus; Venn-Watson et al. 2013). Recent sequencing efforts of several whales and dolphins have provided many insights into the potential evolutionary adaptations to aquatic lifestyle and physiology (Lindblad-Toh et al. 2011; Zhou et al. 2013, 2015; Yim et al. 2014; Foote et al. 2015; Keane et al. 2015; Tsagkogeorga et al. 2015). However, most of the genome-scale studies of molecular adaptation in cetaceans to date have been restricted to single or few cetacean species and have not included the porpoises (family Phocoenidae), which include six recognized species, representing a distinct group of smallest cetaceans characterized by spade-shaped teeth, a triangular dorsal fin and a short, blunt snout (Barnes 1984).
Although the most obvious morphological changes observed in cetaceans relate to their skeletons, dramatic modifications also occurred in the structural properties of bone tissue (Ricqles and Buffrenil 2001). The structural modifications of bone can be due to ecological, biomechanical, and behavioral characteristics of particular species. Bone microanatomy was documented to resolve locomotor mode of extant and extinct amniotes and tetrapods, including early cetaceans (e.g., Ricqles and Buffrenil 2001; Amson et al. 2014; Houssaye et al. 2015; Canoville et al. 2016). It has been suggested that osteological adaptations to aquatic life in cetaceans are a strategy for managing buoyancy (Taylor 2000). Nevertheless, recent discoveries have documented molecular variations that are potential responses to phenotypes (such as sensory adaptations, hairless, and echolocation) in cetaceans (Chen et al. 2013; Xu et al. 2013; Zhou et al. 2013; Zhu et al. 2014). The genetic footprints of the structural properties of the cetacean bone have not been examined.
Here, to identify genes putatively associated with the major morphological and physiological changes required for an obligate aquatic niche, we contrasted whales, dolphins, and porpoises with their terrestrial relatives. We first focused on gene family changes in the ancestral branch leading to cetaceans (toothed and baleen whales), the ancestral branch to toothed whales, and the ancestral branches to baleen whales and porpoises (represented by the Yangtze River finless porpoise, Neophocaena asiaeorientalis). In addition, the availability of a large number of cetacean genomes allowed us to test if the bone microanatomical structure underwent adaptive evolution in the cetaceans, and we further identified candidate genes associated with the major skeletal changes facilitating an aquatic lifestyle.
Materials and Methods
Gene Family and Positively Selected Genes
All mammalian genomes with significant sequence coverage from the current Entrez Genome Project at NCBI were used in this study (a total of 24 organisms). The genome sequence of the bowhead whale was retrieved from The Bowhead Whale Genome Resource (http://www.bowhead-whale.org/). Gene families were constructed using TreeFam (Li et al. 2006). The phylogenetic tree was estimated using 3,911 single-copy genes shared by the finless porpoise (107× coverage), bowhead whale (150× coverage), minke whale Balaenoptera acutorostrata (128× coverage), sperm whale Physeter macrocephalus (72× coverage), Yangtze river dolphin Lipotes vexillifer (115× coverage), killer whale Orcinus orca (200× coverage) and bottlenose dolphin (2.59× coverage), and 10 other mammals (cow Bos taurus, sheep Ovis aries, pig Sus scrofa, Alpaca Vicugna pacos, horse Equus caballus, dog Canis lupus familiaris, cat Felis catus, large flying fox Pteropus vampyrus, hedgehog Erinaceus europaeus, shrew Sorex araneus) using the maximum-likelihood algorithm as implemented in RAxML software (Stamatakis 2006). The divergence time for the analyzed taxa was estimated with Reltime (Tamura et al. 2012) on the basis of 4-fold-degenerate codon sites. The branch-site model (Zhang et al. 2005) was used to detect positive selection along a target branch. P value of each gene was computed using the likelihood ratio tests (LRTs) and corrected for multiple testing by the false discovery rate (FDR) method.
Technical Processing of the Ribs and Compactness Profile Parameters
Ribs from eight mammals (minke whale, Yangtze river dolphin, finless porpoise, bottlenose dolphin, cow, sheep, pig, and dog) were sampled and were further photographed. The sample preparation was following the standard procedure suggested by Canoville et al. (2016) and de Buffrénil et al. (2010). Using binary images of thin-section, quantification analysis of the distribution of bone density were performed by software Bone Profiler (Girondot and Laurin 2003) to obtain parameters S (relative width of the transition zone between the medullary and the cortical regions) and P (proportional to the size of the medullary cavity) for each section. Compactness of center/periphery/whole of the bone sections was calculated by the image analysis software Image-Pro Plus (Version 6.2, Media Cybernetics). Bone section pictures of other mammalian species analyzed in this study, including platypus (Ornithorhynchus anatinus), tammar wallaby (Macropus eugenii), African elephant (Loxodonta africana), manatee (Trichechus manatus), rat (Rattus norvegicus), marmoset (Callithrix jacchus), western gorilla (Gorilla gorilla), hedgehog, David's myotis (Myotis davidii), big brown bat (Eptesicus fuscus), large flying fox, black flying fox (Pteropus alecto), Weddell seal (Leptonychotes weddellii), walrus (Odobenus rosmarus), polar bear (Ursus maritimus), cat, horse, white rhinoceros (Ceratotherium simum), camel (Camelus dromedarius), and killer whale (O. orca) were retrieved from Canoville et al. (2016) and measured using the method described above.
Analysis of Associations between dN/dS and Bone Global Compactness
Genes with significant associations between dN/dS and bone microstructure were identified across the mammalian phylogeny. The “root-to-tip” dN/dS, which was defined as accumulated dN/dS extending from the last common ancestor of mammals to terminal branch, was estimated for each of the 28 species scored for ribs data. This measurement was recognized as an index of selection that takes the entire evolutionary history of a lineage from a common ancestor into account and negates the issue of temporal effects on dN/dS (Montgomery et al. 2011). Using Orthomcl Software-v2.0.9 (Li et al. 2003), we generated a base set of 3,293 single copy orthologues shared by 28 mammals. We also searched for genes that underlie processes such as skeletal system development, ossification, bone remodeling, osteoblast proliferation, osteoclast differentiation, and osteoclast proliferation in Gene Ontology and pathway such as osteoclast differentiation in KEGG database (Kanehisa et al. 2016). We identified 350 genes associated with possible roles in bone development and formation. The coding sequences were aligned and selected by PRANK (Loytynoja and Goldman 2008) and Gblocks (Castresana 2000). Root-to-tip dN/dS values were estimated with PAML 4.4 (Yang 2007) using the free ratio model. Bone compactness was measured and analyzed using Bone Profiler (Girondot and Laurin 2003). Phylogenetic and statistical analyses were performed using R (R Core Team 2013) and R packages “phytools” (Revell 2012) and “phylolm” (Ho and Ané 2014). Regression by generalized least-squares method was used to identify a correlation between bone traits and root-to-tip dN/dS of genes. Robustness of regression was evaluated using a two-step verification procedure. First, regression was repeated by excluding a point with the largest residue error (“P value.robust”), so the potential effect from outliers on the overall relationship could be reduced. Next, we repeated regression by removing one species at a time to calculate the maximal (i.e., least significant) P value (“P value.max”) for each gene, ensuring that the correlation was not dependent on a particular species (Ma et al. 2015).
Results and Discussion
Evolution of Genes in the Ancestor Branch of Cetaceans
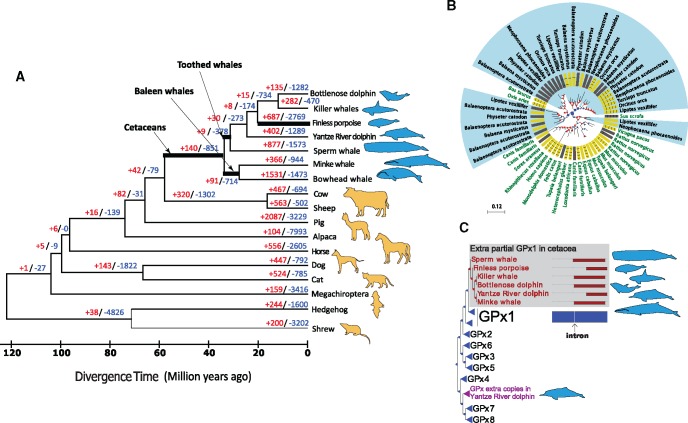
We compiled 3,911 single-copy orthologs that shared by seven cetaceans and 10 other mammals (fig. 1a). In the ancestral branch leading to all cetaceans, we identified 140 expanded and 851 contracted gene families. The expanded gene families were involved in “cellular response to oxidative stress” (3 genes, P = 4.4 × 10−3), “oxidation reduction” (10 genes, P = 2.5 × 10−2), and “response to hydrogen peroxide” (3 genes, P = 9.1 × 10−3; supplementary fig. S1 and table S1, Supplementary Material online). Under hypoxic conditions, reactive oxygen species (ROS) are generated, that may damage cellular molecules, such as DNA, proteins, and lipids. We also detected an expansion of the peroxiredoxin (PRDX) family, which has an important role in eliminating peroxides and in redox signaling. This expansion has been previously reported for the minke whale and dolphin genomes (Yim et al. 2014). Our analysis showed that all cetaceans harbored several copies of PRDX1, though most of these appear to be pseudogenes (ranged one copy in bottlenose dolphin to four copies in killer whale). A similar situation was observed in the case of the glutathione peroxidase (GPx) family (fig. 1b and c), which also degrades hydroperoxides and protects against oxidative damage. For example, a partial extra copy of GPx1 was identified in most of the cetacean genomes. The overall conservation of these genes with GPx1 was quite low, and their ratio of nonsynonymous to synonymous substitutions, dN/dS, was 0.87, approaching the value expected under neutral evolution (dN/dS = 1.0). Together with a lack of supporting evidence from transcriptome data, these observations suggest that the extra copies are unlikely to be pseudogenes that are not functional.
Fig. 1.
—Gene families and aquatic adaptations in cetacean genomes. (a) Phylogenetic tree and divergence times estimated for the cetaceans and their relatives. Numbers associated with each branch designate the number of gene families that have expanded (red) and contracted (blue) since the split from the common ancestor. (b) Phylogenetic tree of PRDX1 family in mammals showing expanded PRDX1 in cetaceans (highlighted by blue background). Positions of introns along the protein sequence are shown with black lines. (c) Phylogenetic tree of the GPx family in mammals showing expanded GPX1 sequences in cetaceans.
Contracted gene families in the ancestral cetacean lineage were mainly associated with sensory perception, including “sense of smell” (106 genes, P = 5.2 × 10−99) and “taste” (4 genes, P = 6.5 × 10−3). These findings are consistent with the recent studies proposing that cetaceans have a greatly reduced sense of smell, and cannot detect basic taste modalities (sweet, sour, and umami; the umami, also known as savory taste, described as meaty) except for salt, and have a reduced ability to sense bitter taste (Zhou et al. 2013; Zhu et al. 2014). Genes associated with “immune response” (25 genes, P = 4.8 × 10−5) were also contracted in cetaceans. These were further categorized into different groups according to their biological function: “defense response to bacterium” (10 genes), “cellular defense response” (4 genes), “acute inflammatory response” (5 genes), and “innate immune response” (6 genes; supplementary table S2, Supplementary Material online). Similarly, a recent study demonstrated that toothed whales (i.e., dolphins and porpoises) have lost two genes associated with antiviral activity (MX1 and MX2; Braun et al. 2015). We speculate that the immune system of cetaceans is modified to fit the aquatic environment. Recent studies support this hypothesis. For example, cetacean Toll-like receptor 4 has undergone adaptive evolution, against a background of purifying selection in a lipopolysaccharide (LPS) interaction domain (Shen et al. 2012), and LPS treatment rapidly increases the expression of immune genes in leukocytes from the bottlenose dolphin (Ohishi et al. 2011).
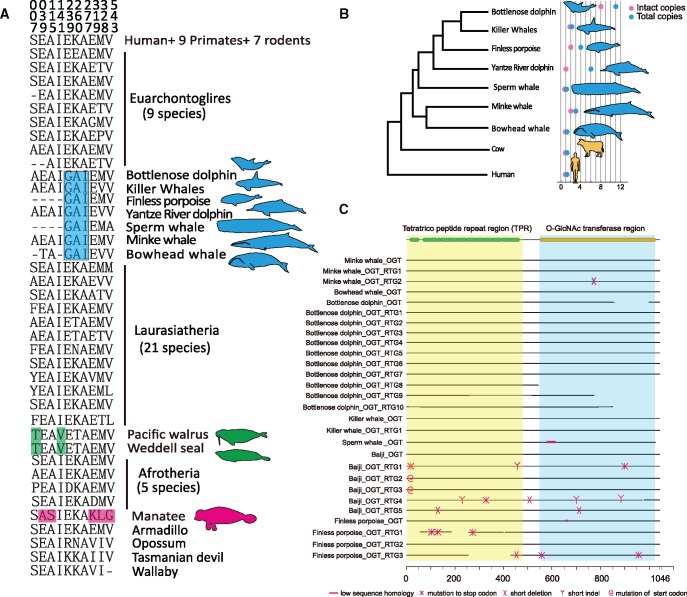
Adaptive evolution can also be manifested by rapid evolution of genes. Thirty-nine PSGs (positively selected genes; supplementary table S3, Supplementary Material online) were identified in the common branch leading to cetaceans using the branch-site model (Zhang et al. 2005). Three reproduction-related categories, “development of primary female sexual characteristics” (P = 5.2 × 10−4), “ovarian follicle development” (P = 3.9 × 10−3), and “multicellular organism reproduction” (P = 2.4 × 10−2) were significantly enriched by these PSGs (supplementary table S4, Supplementary Material online). In particular, PSGs included follicle-stimulating hormone receptor (FSHR) and INHBA genes that encode subunits of the FSH inhibitor, and FSHR is essential for reproduction and mutations in this gene were found to be associated with human pathologies manifested by altered ovarian function (Beau et al. 1998; De Leener et al. 2008; Kuechler et al. 2010). Intriguingly, compared with 60 terrestrial mammals, we found several nonsynonymous substitutions in FSHR that were unique to cetaceans and other marine mammals (fig. 2a). For example, an isoleucine present at position 277 in cetacean FSHR is located at a repeat domain critical for activation of the receptor by FSH (Jiang et al. 2012). Whereas the reproductive organs of marine mammals are clearly mammalian in appearance, some specialization to accommodate body streamlining and marine environment is evident (Berta and Sumich 1999). For example, the duration of a recognizable corpus albicans, which can increase FSH secretion, varies between species and individuals across marine mammals; and remnants may persist for several years in some species (Laws and Sinha 1993).
Fig. 2.
—Distinct substitutions of FSHR in marine mammals and copy number variation of OGT gene in cetaceans. (a) Alignment of FSHR (Follicle Stimulating Hormone Receptor) residues under positive selection in cetaceans. Unique substitutions identified in marine mammals are shown in blue (cetaceans), green (walrus and seal), and pink (manatee). (b) Mammalian phylogeny was used to represent the number of OGT (O-Linked N-Acetylglucosamine (GlcNAc) Transferase) gene uncovered in the cetacean genomes. (c) A schematic map showing the gene structure and sequence variation of OGT gene in cetacean genomes.
In the common branch leading to toothed whales, genes associated with “intermediate filament organization” (two genes, KRT9 and KRT20, P = 1.4 × 10−3) were significantly contracted (supplementary table S5, Supplementary Material online). Both genes play significant role in maintaining keratin filament organization (Szeverenyi et al. 2008). Toothed whales are nearly hairless, whereas baleen is formed from keratin. Our findings are consistent with the recent studies attributing the loss of keratin genes in cetacean genomes to their hairless phenotype (Nery et al. 2014; Strasser et al. 2015). Contraction of genes associated with “neuron differentiation” (eight genes, APC, CNTNAP2, CTNNA2, DMD, HES5, HOXA2, RELN, and STMN3; P = 1.3 × 10−2) was observed in baleen whales (supplementary table S6, Supplementary Material online). Cetaceans are believed to be highly intelligent, possessing large brains and complex behavioral patterns, with toothed whale brains being larger and having significantly more neuronal cells than baleen whales (Marino et al. 2007; Mortensen et al. 2014). We speculate that the loss of genes associated with neuronal differentiation in baleen whales reflects their comparatively solitary life histories compared with their toothed whale counterparts. Consistently, gene families expanded in toothed whales showed enrichment of “learning” (two genes, P = 6.8 × 10−4) and “cognition” (three genes, P = 1.9 × 10−2; supplementary tables S7 and S8, Supplementary Material online) GO term categories.
A previous study attributed expansion of OGT (O-GlcNActransferase; O-GlcNAc) in a baleen whale (minke whale) and several toothed whales to resistance against cellular stressors such as hypoxia, oxidative stress, and osmotic stress (Yim et al. 2014). By examining all publicly available cetacean genomes, we found that the sperm whale (which is known as deep diver) has only one intact copy of OGT (fig. 2b and c), implying that the function of this gene in hypoxic adaptation is limited. Indeed, OGT encodes an enzyme that glycosylates proteins involved in diverse processes including memory, metabolism and immunity (Bond and Hanover 2015). Because overexpression of OGT has been associated with insulin resistance and dyslipidemia in mice (Yang et al. 2008), expansion of OGT (11 copies) in the bottlenose dolphin genome may be underlying the unusual response of this species to insulin (Venn-Watson et al. 2013). Taken together, these findings provide insights into how evolution shaped genes associated with aquatic adaptation of cetaceans.
Association of Root-to-Tip dN/dS and Bone Compactness
It has been suggested that bone microanatomy is critical for habitat preferences, facilitating transition of terrestrial mammals to the aquatic environment (see Introduction, Gray et al. 2007; Canoville et al. 2016). While most studies use appendicular skeleton, such as the humerus, femur, or vertebral column to measure their effects of lifestyle, the ribs are an important part of the axial skeleton are rarely analyzed (Canoville et al. 2016). In addition to the environmental pressures and respiratory function, biomechanical constraints linked to locomotion and body support would strongly affect the ribs and rib cage, particular in limbless tetrapods (e.g., Bramble and Carrier 1983; Carrier 1996; Houssaye et al. 2013, 2014; Fujiwara et al. 2009). The buoyant force of water relaxes constraints on skeletal mass; however, cetaceans have adapted to diverse habitats. Therefore, investigating microstructural features of bone over the main lineages of cetaceans as well as other marine mammals can help revealing the adaptive changes of bone microanatomy in aquatic environment.
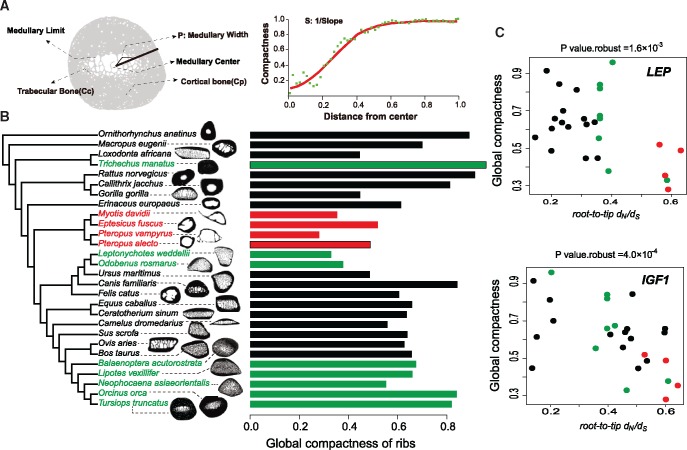
We compiled data on rib compactness from eight marine mammals (finless porpoise, minke whale, Yangtze river dolphin, killer whale, bottlenose dolphin, Weddell seal, walrus, and manatee) and another 23 mammals (fig. 3a and b), for which we obtained full genome data. We employed the root-to-tip dN/dS (the ratio of nonsynonymous to synonymous substitutions) for each species to test the association between selection pressure and phenotype. Such “accumulated” dN/dS values from root to each tip branch allow taking a common ancestor of each node into account. Of 3,293 single gene copy orthologues shared by these 28 mammals, 134 were found with a significant correlation (with P value, P value.robust [from regression by excluding the point with largest residue error] and P value.maximum [defined as the maximal P value from regression with species excluded separately] < 0.05) between rib globe compactness (Cg) and root-to-tip dN/dS rations.
Fig. 3.
—Root-to-tip dN/dS correlates with global compactness of ribs across 28 mammals. (a) Diagrammatic sketch showing bone microanatomy of the rib and parameters measured in this study. (b) Variation in global compactness of ribs across 28 mammals in a phylogenetic context. Representatives from marine, terrestrial, and flying mammals are in green, black, and red, respectively. (c) Root-to-tip dN/dS of genes LEP (top panel) and IGF1 (bottom panel) negatively correlates with global compactness. Points are colored according to taxonomical orders (same color scheme as in b).
The identified genes were enriched for functional categories “DNA metabolic process” (P = 3.7 × 10−3), “cell proliferation” (P = 4.0 × 10−3), and “cholesterol transport” (three genes, APOA1, LEP, and LIPG, P = 6.0 × 10−4; supplementary tables S9 and S10, Supplementary Material online). Pathways associated with transport of cholesterol are interesting because high-density lipoprotein cholesterol and bone mineral density are genetically linked, and high-density lipoprotein can interact directly with both osteoblasts and osteoclasts (Ackert-Bicknell 2012). It has been demonstrated that levels of serum leptin (encoded by LEP) may play a role in regulating bone mass in humans, and leptin is central neuronal regulator for bone formation (Pasco et al. 2001; Takeda et al. 2002). Conversely, root-to-tip dN/dS of several genes (IGF1, P value.robust= 4.0 × 10−4; CTSK, P value.robust= 3.9 × 10−3; PLA2G4A, P value.robust= 8.3 × 10−3) with known function in bone mineralization and resorption showed significant correlation with global rib compactness (fig. 3c). For example, IGF1 is likely to stimulate bone formation by a direct effect on osteoblasts and higher plasma IGF1 levels were found positively correlated with bone mineral density and bone mineral content in humans (Bianda et al. 1998; Yanovski et al. 2000).
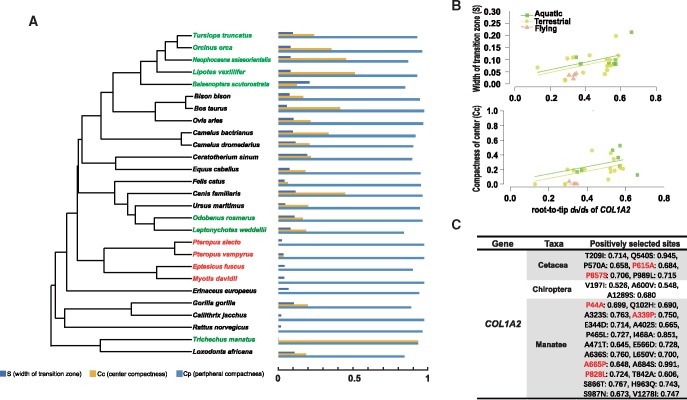
In addition to global compactness (Cg), three other indices of bone microstructure, that is, bone compactness of center (Cc), width of transition zone (S), and compactness of periphery (Cp) have been influenced by body size or lifestyle (supplementary table S11, Supplementary Material online). Regression analyses were performed between these three parameters for a subset of 27 mammals and root-to-tip dN/dS values of 350 genes involved in bone development or formation (supplementary table S12, Supplementary Material online). Significant correlation (defined by P value <0.05) was identified between root-to-tip dN/dS of 96 genes and Cc, 12 genes and Cp as well as 38 genes and S, respectively (supplementary table S13, Supplementary Material online). In addition, 18 genes showed significant correlation between root-to-tip dN/dS and both Cc and S (AHSG, BMP6, COL1A2, GAB2, GLI3, HDAC7, IFNGR1, LTF, MMP14, NCF2, NCF4, PDGFRA, PDGFRB, PIK3CB, TGFB1, TGFBI, TNFRSF11B, WWTR1), 2 genes between Cp and S (SOX2 and FSHR), and 2 genes between Cp and Cc (PIK3R1 and OCSTAMP). Genes with positive selection in the branch leading to the cetacean’s ancestor were identified based on this 350-gene data set (fig. 4a and supplementary table S14, Supplementary Material online).
Fig. 4.
—Root-to-tip dN/dS from 350 bone-associated genes that correlate with the bone microstructure of ribs in 27 mammals. (a) Phylogenetic tree for 27 species with bone section bone microstructure and histogram of five bone parameters. (b) Scatter plot of significant relationships between S, Cc, and root-to-tip values. (c) Positively selected sites for Cetaceans, bats, and manatees.
Bone microstructure is extremely complex and variable with a full hierarchical structure, with mineralized collagen fibril composed of collagen protein with the mineral hydroxyapatite (Fratzl 2008). Two genes (COL1A1 and COL1A2, encoding the α1(I) and α2(I) tropocollagen chain) were suggested to be related to abnormal changes in the collagen triple-helical structure (Fratzl et al. 2004). In the present study, root-to-tip dN/dS values of COL1A2 were significantly correlated with Cc (P value.robust < 0.001, P value.max < 0.01) and S (P value.robust < 0.01, P value.max < 0.05; fig. 4b). In addition, COL1A2 appeared to be a positively selected gene in cetaceans (P < 0.01, ω = 197.77), manatee (T. manatus; P < 0.05, ω = 4.785), and the ancestor of Chiroptera (P < 0.05, ω = 131.667; fig. 4c). Manatees are fully aquatic herbivores that feed on plants in shallow waters and are characterized by compact bones (de Buffrénil et al. 2010). The bats, as the only group of flying mammals, exhibit simple bone microanatomy, with thin cortices, and few (or none) trabeculae bones in the medullary region. It is reasonable to hypothesize that these substitutions in fully aquatic cetaceans may have modified the collagen triple helix related to bone mass. The overall evolutionary pattern suggests that gene level changes potentially drive phenotypic alterations in the mammalian bone structure. Future studies aiming to disentangling the observed changes by studying the expression and function of these genes in diverse taxa, could be rewarding.
Conclusions
Comparative genomic analyses of cetaceans and their terrestrial relatives provided several novel insights into the distinct evolutionary scenarios of adaptation to a fully aquatic lifestyle. Genes associated with oxidation–reduction and immune process were found to be accompanied by pseudogene copies. Genes under positive selection in the cetaceans were related to reproduction, keratin protein, learning, and energy turnover. This was interesting given their special lifestyle compared with other mammals. Our study also documented the bone microstructure across mammals and marine mammals, and for the first time, revealed the benefit of using a phylogenetic comparative approach to study the evolution of bone compactness. Our findings offer valuable information on genes critical for adaptation to aquatic life of mammals in diverse environments.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Key Project of the National Natural Science Foundation of China (NSFC) (grant no. 31630071 to G.Y.), the National Science Fund for Distinguished Young Scholars (grant no. 31325025 to G.Y.), Ministry of Science and Technology of China (grant no. 2016YFC0503200), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and NIH grants GM065204 and AG047200.
Literature Cited
- Ackert-Bicknell CL. 2012. HDL cholesterol and bone mineral density: is there a genetic link? Bone 502:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amson E, de Muizon C, Laurin M, Argot C, de Buffrénil V.. 2014. Gradual adaptation of bone structure to aquatic lifestyle in extinct sloths from Peru. Proc Biol Sci. 2811782:20140192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes LG. 1984. Fossil odontocetes (Mammalia: cetacea) from the Almejas Formation, Isla Cedros, Mexico. PaleoBios 42:1–46. [Google Scholar]
- Beau I, et al. , 1998. A novel phenotype related to partial loss of function mutations of the follicle stimulating hormone receptor. J Clin Invest. 1027:1352–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta A, Sumich JL.. 1999. Marine mammals Evolutionary biology. San Diego (CA: ): Academic Press. [Google Scholar]
- Bianda T, Glatz Y, Bouillon R, Froesch ER, Schmid C.. 1998. Effects of short-term insulin-like growth factor-I (IGF-I) or growth hormone (GH) treatment on bone metabolism and on production of 1, 25-dihydroxycholecalciferol in GH-deficient adults 1. J Clin Endocrinol Metab. 831:81–87. [DOI] [PubMed] [Google Scholar]
- Bond MR, Hanover JA.. 2015. A little sugar goes a long way: the cell biology ofO-GlcNAc. J Cell Biol. 2087:869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramble DM, Carrier DR.. 1983. Running and breathing in mammals. Science 2194582:251–256. [DOI] [PubMed] [Google Scholar]
- Braun BA, Marcovitz A, Camp JG, Jia R, Bejerano G.. 2015. Mx1 and Mx2 key antiviral proteins are surprisingly lost in toothed whales. Proc Natl Acad Sci USA. 11226:8036–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoville A, de Buffrénil V, Laurin M.. 2016. Microanatomical diversity of amniote ribs: an exploratory quantitative study. Biol J Linn Soc Lond. 1184:706–733. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Bio Evol. 174:540–552. [DOI] [PubMed] [Google Scholar]
- Caulin AF, Maley CC.. 2011. Peto's Paradox: evolution's prescription for cancer prevention. Trends Ecol Evol. 264:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier DR. 1996. Ontogenetic limits on locomotor performance. Physiol Zool. 693:467–488. [Google Scholar]
- Chen Z, Wang Z, Xu S, Zhou K, Yang G.. 2013. Characterization of hairless (Hr) and FGF5 genes provides insights into the molecular basis of hair loss in cetaceans. BMC Evol Biol. 13:34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leener A, et al. , 2008. Identification of the first germline mutation in the extracellular domain of the follitropin receptor responsible for spontaneous ovarian hyper stimulation syndrome. Hum Mutat. 291:91–98. [DOI] [PubMed] [Google Scholar]
- de Buffrénil V, Canoville A, D’Anastasio R, Domning DP.. 2010. Evolution of sirenian pachyosteosclerosis, a model-case for the study of bone structure in aquatic tetrapods. J Mamm Evol. 172:101–120. [Google Scholar]
- Deméré TA, McGowen MR, Berta A, Gatesy J.. 2008. Morphological and molecular evidence for a stepwise evolutionary transition from teeth to baleen in mysticete whales. Syst Biol. 571:15–37. [DOI] [PubMed] [Google Scholar]
- Foote AD, et al. , 2015. Convergent evolution of the genomes of marine mammals. Nat Genet. 473:272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratzl P, Gupta HS, Paschalis EP, Roschger P.. 2004. Structure and mechanical quality of the collagen–mineral nano-composite in bone. J Mater Chem. 1414:2115–2123. [Google Scholar]
- Fratzl P. 2008. Collagen: structure and mechanics, an introduction. New York: Springer. [Google Scholar]
- Fujiwara SI, Kuwazuru O, Inuzuka N, Yoshikawa N.. 2009. Relationship between scapular position and structural strength of rib cage in quadruped animals. J Morphol. 2709:1084–1094. [DOI] [PubMed] [Google Scholar]
- Gatesy J, et al. , 2013. A phylogenetic blueprint for a modern whale. Mol Phylogenet Evol. 662:479–506. [DOI] [PubMed] [Google Scholar]
- George JC, et al. , 1999. Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can J Zool. 774:571–580. [Google Scholar]
- George JC, Bockstoce JR.. 2008. Two historical weapon fragments as an aid to estimating the longevity and movements of bowhead whales. Polar Biol. 316:751–754. [Google Scholar]
- Gingerich PD, Wells NA, Russell DE, Shah SI.. 1983. Origin of whales in epicontinental remnant seas: new evidence from the early Eocene of Pakistan. Science 2204595:403–406. [DOI] [PubMed] [Google Scholar]
- Girondot M, Laurin M.. 2003. Bone Profiler: a tool to quantify, model, and statistically compare bone-section compactness profiles. J Vert Paleontol. 23:458–461. [Google Scholar]
- Gray NM, Kainec K, Madar S, Tomko L, Wolfe S.. 2007. Sink or swim? Bone density as a mechanism for buoyancy control in early cetaceans. Anat Rec (Hoboken). 2906:638–653. [DOI] [PubMed] [Google Scholar]
- Tung Ho LS, Ané C.. 2014. A linear-time algorithm for Gaussian and nonGaussian trait evolution models. Syst Biol. 633:397–408. [DOI] [PubMed] [Google Scholar]
- Houssaye A, Boistel R, Böhme W, Herrel A.. 2013. Jack-of-all-trades master of all? Snake vertebrae have a generalist inner organization. Naturwissenschaften 10011:997–1006. [DOI] [PubMed] [Google Scholar]
- Houssaye A, Tafforeau P, Herrel A.. 2014. Amniote vertebral microanatomy – what are the major trends? Biol J Linnean Soc. 1124:735–746. [Google Scholar]
- Houssaye A, Tafforeau P, De Muizon C, Gingerich PD.. 2015. Transition of Eocene whales from land to sea: evidence from bone microstructure. PLOS ONE. 102:e0118409.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, et al. , 2012. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc Natl Acad Sci USA. 10931:12491–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M.. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44(D1):D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, et al. , 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 101:112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler A, et al. , 2010. An unbalanced translocation unmasks a recessive mutation in the follicle-stimulating hormone receptor (FSHR) gene and causes FSH resistance. Eur J Hum Genet. 186:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws RM, Sinha AA.. 1993. Reproduction In: Laws RM, editor. Antarctic seals. Cambridge: Cambridge University Press; p. 228–267. [Google Scholar]
- Li H, et al. , 2006. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34:D572–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 139:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. , 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 4787370:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N.. 2008. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 3205883:1632–1635. [DOI] [PubMed] [Google Scholar]
- Ma S, et al. , 2015. Organization of the mammalian metabolome according to organ function, lineage specialization, and longevity. Cell Metab. 222:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino L, et al. , 2007. Cetaceans have complex brains for complex cognition. PLoS Biol. 55:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Venditti C, Barton RA, Mundy NI.. 2011. Adaptive evolution of four microcephaly genes and the evolution of brain size in anthropoid primates. Mol Biol Evol. 281:625–638. [DOI] [PubMed] [Google Scholar]
- Mortensen HS, et al. , 2014. Quantitative relationships in delphinid neocortex. Front Neuroanat. 8:132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery MF, Arroyo JI, Opazo JC.. 2014. Increased rate of hair keratin gene loss in the cetacean lineage. BMC Genomics. 15:869.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschläger HA. 1992. Development of the olfactory and terminalis systems in whales and dolphins In: Doty RL, Müller-Schwarze D. (Eds.), Chemical Signals in Verterbrates VI. New York: Plenum Press, pp. 141–147. [Google Scholar]
- Ohishi K, et al. , 2011. Lipopolysaccharide-induced innate immune factors in the bottlenose dolphin (Tursiops truncates) detected in expression sequence tag analysis. Microbiol Immunol. 5511:790–797. [DOI] [PubMed] [Google Scholar]
- Pasco JA, et al. , 2001. Serum leptin levels are associated with bone mass in nonobese women 1. J Clin Endocrinol Metab. 865:1884–1887. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 32:217–223. [Google Scholar]
- Ricqlès AD, Buffrénil VD.. 2001. Bone histology, heterochronies and the return of tetrapods to life in water: where are we In: Mazin JM, Buffrénil V, editors. Secondary adaptation of tetrapods to life in water. München (Germany: ): Verlag; p. 289–310. [Google Scholar]
- Shen T, et al. , 2012. Adaptive evolution and functional constraint at TLR4 during the secondary aquatic adaptation and diversification of cetaceans. BMC Evol Biol. 121:39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RaxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2221:2688–2690. [DOI] [PubMed] [Google Scholar]
- Strasser B, Mlitz V, Fischer H, Tschachler E, Eckhart L.. 2015. Comparative genomics reveals conservation of filaggrin and loss of caspase-14 in dolphins. Exp Dermatol. 245:365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeverenyi I, et al. , 2008. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum Mutat. 293:351–360. [DOI] [PubMed] [Google Scholar]
- Takeda S, et al. , 2002. Leptin regulates bone formation via the sympathetic nervous system. Cell 1113:305–317. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. , 2012. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci USA. 10947:19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MA. 2000. Functional significance of bone ballastin in the evolution of buoyancy control strategies by aquatic tetrapods. Histor Biol. 14(1–2):15–31. [Google Scholar]
- Thewissen JGM, Cooper LN, Clementz MT, Bajpai S, Tiwari BN.. 2007. Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 4507173:1190–1194. [DOI] [PubMed] [Google Scholar]
- Tsagkogeorga G, et al. , 2015. A phylogenomic analysis of the role and timing of molecular adaptation in the aquatic transition of cetartiodactyl mammals. R Soc Open Sci. 29:150156.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venn-Watson S, et al. , 2013. Blood-based indicators of insulin resistance and metabolic syndrome in bottlenose dolphins (Tursiops truncatus). Front Endocrinol. 4:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. , 2013. Adaptive evolution of the osmoregulation-related genes in cetaceans during secondary aquatic adaptation. BMC Evol Biol. 13:189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, et al. , 2008. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 4517181:964–969. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yanovski JA, Sovik KN, Nguyen TT, Sebring NG.. 2000. Insulin-like growth factors and bone mineral density in African American and White girls. J Pediatr. 1376:826–832. [DOI] [PubMed] [Google Scholar]
- Yim HS, et al. , 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 461:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2212:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhu K, et al. , 2014. The loss of taste genes in cetaceans. BMC Evol Biol. 14:218.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Seim I, Gladyshev VN.. 2015. Convergent evolution of marine mammals is associated with distinct substitutions in common genes. Sci Rep. 5:16550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. , 2013. Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat Commun. 4:2708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xu S, Yang Y, Zhou K, Yang G.. 2011. Phylogenomic analyses and improved resolution of Cetartiodactyla. Mol Phylogenet Evol. 612:255–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.