Abstract
Colitis-associated colorectal cancer (CACC) is one of the most serious complications of inflammatory bowel disease (IBD), particularly in ulcerative colitis (UC); it accounts for approximately 15% of all-causes mortality among IBD patients. Because CACC shows a worse prognosis and higher mortality than sporadic colorectal cancer, early detection is critical. Colonoscopy is primarily recommended for surveillance and several advanced endoscopic imaging techniques are emerging. In addition, recent studies have reported on attempts to develop clinically relevant biomarkers for surveillance using various biosamples, which may become high-performance screening tools in the future, so the best approach and technique for cancer surveillance in long-standing UC patients remain under debate. This review gives a comprehensive description and summary about what progress has been made in terms of early CACC detection.
Keywords: Ulcerative colitis, colitis-associated colorectal cancer, endoscopy, biomarker, surveillance
Introduction
Inflammatory bowel disease (IBD) is a group of nonspecific chronic inflammatory diseases of the gut, which includes Crohn’s disease (CD), ulcerative colitis (UC) and indeterminate colitis. The pathogenesis of IBD remains unclear, and it is characterized by long-lasting and relapsing intestinal inflammation. Colitis-associated colorectal cancer (CACC) is one of the most serious complications of long-duration IBD, particularly UC. Although CACC contributes to only 1–2% of colorectal cancers (CRC) in the overall population, it accounts for approximately 15% of all-causes mortality among IBD patients [1] and its risk is 1.5–2.4-fold higher in IBD patients than in the normal population [2, 3]. In China, according to a large population-based study from Hong Kong, CACC incidence in UC patients is 0.81% [4], which is lower than in Western countries (3.7%) [5]. However, IBD incidence is increasing (up to 30 times in last decade) and CACC incidence is expected to increase rapidly. CACC risk is 0.5–1% per year following UC diagnosis and it increases over time, i.e. 1.6% at 10 years, 8.3% at 20 years and 18.4% at 30 years after UC onset [5], and CACC is frequently diagnosed at an advanced stage [6]. Although several recent reports have indicated a slight decline in CACC incidence in IBD patients [7, 8], which might be attributed to the widespread use of 5-aminosalicylic acid and immunosuppressive drugs [9, 10], as well as to widespread endoscopic screening and early coloproctectomy, current guidelines continue to recommend a regular surveillance for early detection [11]. Moreover, because of worse prognosis and higher mortality in CACC than in sporadic CRC [12], early CACC detection is crucial. Colonoscopy is primarily recommended for surveillance; however, newer technologies and approaches, such as molecular biomarkers in different biosamples, have emerged and the best approach and technique for surveillance remain under debate. Here we provide an overview of the methods for surveillance, focusing on the present and future, to optimize early CACC detection in UC patients.
Colonoscopic surveillance
CACC derives from the repeated inflammation–dysplasia–carcinoma model [13], which is different from the classical model of sporadic CRC (accumulated mutations–adenoma–cancer); thus, CACC emphasizes on the importance of the early detection of dysplastic alterations through endoscopy, which has been the primary and conventional method for CACC screening in UC patients over the past few decades. Indeed, evidence has suggested that surveillance colonoscopy decreases CACC incidence and mortality in UC [14–16]. It is generally recommended that endoscopic screening for CACC should be initiated between 6 and 10 years after the diagnosis or onset of symptoms of UC, 1 year after the diagnosis of primary sclerosing cholangitis (PSC) [17–20]; however, recommended surveillance intervals vary. Knowledge of risk factors (such as the age at diagnosis, disease duration, extent of inflammation, family history and combination of PSC) is also important to classify UC patients requiring frequent surveillance or intense treatment. Both the British Society of Gastroenterology (BSG) [21] and the American Gastroenterological Association (AGA) [22] agree that regular endoscopic surveillance is required in long-standing UC patients, and they support the use of a risk-stratified approach to determine surveillance intervals. Since 2014, the American Society for Gastrointestinal Endoscopy has recommended that these intervals can be appropriately extended for patients with no apparent endoscopic and histological abnormalities found in two successive endoscopies [19], which would improve the allocation of limited medical resources.
Standard white-light endoscopy with randomized biopsies
Traditionally, primary dysplasia lesions in long-standing UC have been considered to be flat and multifocal. Together with the complex background of mucosal changes due to chronic inflammation, it is difficult to pinpoint dysplastic alterations through the naked eye under general surveillance gastroenteroscopy because these lesions can be either invisible or very difficult to identify. Consequently, the previous AGA and BSG guidelines recommend white-light endoscopy (WLE) with random biopsies (four random biopsies should be collected at every 10 cm of the entire colorectum and the total number of biopsies can be ≥33) [23, 24]. Navaneethan et al. [25] showed an increased dysplasia detection with WLE (random biopsies) in patients with UC and PSC. Another study has shown that at least 34 and 64 biopsies should be ‘blindly’ taken in order to achieve 90% and 95% confidence, respectively [26]. However, even when an adequate number of biopsies were obtained, evidence showed that this approach only sampled approximately 0.3% of the colonic mucosa [27], and it is time-consuming and less cost-effective.
High-definition endoscopy or chromoendoscopy with targeted biopsies
With dramatic advances in endoscopic devices [such as high-definition (HD) endoscopy], most UC dysplasia can be endoscopically visualized [28, 29] and the approach focusing on targeted biopsies of any mucosal abnormalities has been recommended [19]. According to a recent randomized controlled trial (n = 246) by Watanabe, targeted and random biopsies demonstrated similar abilities to identify neoplasia under surveillance endoscopy; however, the targeted biopsy approach appeared to be superior to random biopsy when cost-effectiveness analysis was considered [30]. In addition, the detection and further delineation of any mucosal abnormalities are thought to be improved by the application of chromoendoscopy (CE) as well as HD endoscopy. HD endoscopy has a high-definition monitor and a high-resolution charge-coupled device. HD endoscopy can provide images of substantially higher resolution than standard video endoscopy, but it requires no additional time or skills in comparison to standard WLE. Therefore, HD endoscopy may represent an alternative to CE with similarly improved dysplasia detection and minimal drawbacks. In a recent retrospective study in IBD patients who underwent surveillance colonoscopies, HD endoscopy showed a higher dysplasia detection rate compared with standard WLE [31]. CE can improve the imaging of subtle mucosal changes that are suggestive of neoplasia in the presence of staining with methylene blue or indigo carmine in the intestine; indigo carmine contrast dye highlights irregularities in the mucosal architecture. A recent meta-analysis has found an 8.9-fold greater likelihood of detecting dysplasia, a 5.2-fold greater likelihood of detecting nonpolypoid dysplasia and a 14-fold lower likelihood of missing dysplasia with CE (targeted biopsies) than with WLE (random biopsies) [32]. Furthermore, it has been suggested that high-definition CE leads to a better detection of dysplastic lesions as compared to HD endoscopy alone. Notably, the latest guidelines from AGA and European Crohn's and Colitis Organisation have also indicated that targeted biopsies with the help of CE are more appropriate for endoscopic surveillance than random biopsies under WLE if the operator is well trained or experienced [12, 17], and the SCENIC consensus statement also suggests that endoscopists consider using CE to enhance the detection of dysplasia in surveillance [33]. However, criticisms such as difficulty in standardization of techniques, requirement for specialized disposable equipment, the need for additional training and longer procedure time remain important concerns, and more research over the best method of endoscopic surveillance for long-term UC patients is in need.
Image-enhanced endoscopy
So far, novel digital image-enhanced endoscopy techniques, including Fujinon intelligent colour enhancement (FICE) and i-Scan digital contrast (i-Scan), which emphasize subtle changes of the mucosal surface through the reconstruction of a certain wavelength (computed spectral estimation technology), have already been developed to improve the positive percentage of primary neoplasia, although relevant research and data are currently limited and their potential for surveillance colonoscopy is currently under investigation [34, 35]. One recent piece of research found HD colonoscopy combination with i-Scan is superior in the detection of adenomas and advanced lesions compared to HD colonoscopy alone. Other advanced endoscopic imaging techniques, such as narrow-band imaging (NBI) and magnifying endoscopy, can also contribute to the early detection of CACC. NBI allows a better assessment of the vascular pattern and mucosal structures. Efthymiou et al. [36] have reported that NBI is not superior to CE in terms of sensitivity to pinpoint dysplasia, but it is more convenient to enhance the sensitivity of targeted biopsies because it only requires one press on a particular button to reveal a suspicious lesion. Magnifying endoscopy may assist us to further visualize delicate surface patterns, although chronic inflammation and its sequela in UC patients make pit pattern analysis less useful [37]. Overall, these novel virtual CE techniques have the advantages of being available on demand, easy to use and relatively inexpensive. Nevertheless, no published study has reported on the utility of these digital endoscopy for CACC surveillance in UC.
Confocal laser endomicroscopy
Confocal laser endomicroscopy (CLE) integrates a confocal laser microscope into the distal tip of a conventional endoscope; it makes real-time microscopy available in vivo during examination. CLE has demonstrated high agreement with true histopathology for the diagnosis of dysplasia in patients with UC [38]. Given this advantage, if CLE is combined with CE or HD for further histological evaluation, then a quicker real-time diagnosis can be obtained and a faster judgment regarding whether a resection is required can be rendered; several studies also have shown that CLE could increase the diagnostic yield of intraepithelial neoplasia in UC patients [39, 40]. However, in consideration of the limitations of this approach, such as increased procedure time, the need for extra equipment and training, and the requirement for expertise to interpret the images, it still could not be carried out as a routine clinical practice.
Full-spectrum endoscopy
In addition to the above techniques, full-spectrum endoscopy (FUSE) is emerging as a novel HD endoscopic technique that incorporates camera lenses to the right and left sides of the colonoscopy tip along with the forward-viewing lens, thereby allowing endoscopists to observe behind folds and in blind spots. In a recent prospective crossover study, Leong et al. [41] found that FUSE increases the number of detected dysplastic lesions compared with that detected by conventional forward-viewing colonoscopy in IBD patients. FUSE has the advantages of feasibility, usability and safety, although several studies have argued about the longer withdrawal times compared with conventional colonoscopy; further data on FUSE in dysplasia surveillance are needed.
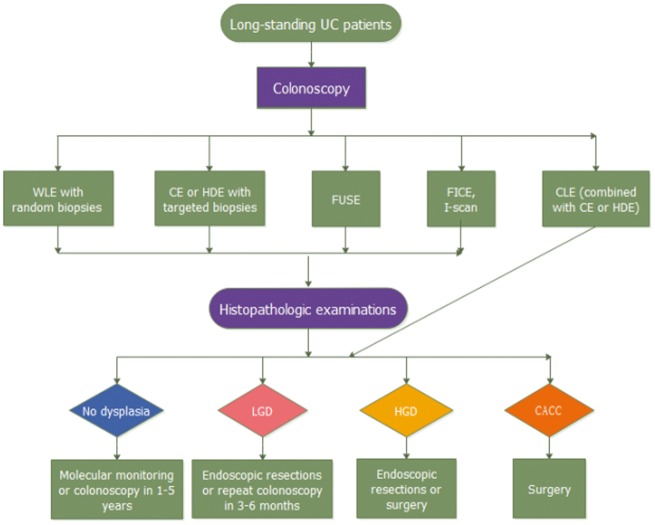
As discussed above, quite a few endoscopic procedures have been developed and applied for CACC surveillance in UC. These endoscopic procedures are summarized in Table 1. Sufficient attention should be drawn on optimally choosing and integrating these techniques so that they can completely benefit high-risk patients (Figure 1).
Table 1.
A summary of current available endoscopic procedures for colitis-associated colorectal cancer surveillance
| Type of endoscopy | Clinical application | Limitations | References |
|---|---|---|---|
| Standard white-light endoscopy (with ramdom biopsies) | Increases dysplasia detection rate | Longer procedure times and cost | [24–26] |
| High-definition endoscopy (with targeted biopsies) | Provides images of substantially higher resolution for dysplasia detection | Cost | [31] |
| Chromoendoscopy (with targeted biopsies) | Contrast dye highlights irregularities in the mucosal architecture | Requirement for specialized equipment, additional training and longer procedure time | [30, 32, 33] |
| Fujinon intelligent colour enhancement and i-Scan digital contrast | Enhances subtle changes of the mucosal surface | Limited relevant data | [34, 35] |
| Narrow-band imaging | Enhances mucosal surface contrast | Lower sensitivity to detect dysplasia | [36] |
| Confocal laser endomicroscopy | Makes real-time microscopy available in vivo during examination | Longer procedure time, the need for extra equipment and training, and requirement for interpreting the images | [38–40] |
| Full-spectrum endoscopy | Increases visual field to increase mucosal visualization | Longer withdrawal and total procedure time | [41] |
Figure 1.
Flow chart of colonoscopy surveillance in patients with ulcerative colitis (UC). It illustrates how these colonoscopy surveillance methods can affect disease management, leading to different treatment strategies or further monitoring. WLE, white-light endoscopy; CE, chromoendoscopy; HDE, high-definition endoscopes; FUSE, full-spectrum endoscopy; CLE, confocal laser endomicroscopy; CACC, Colitis-associated colorectal cancer; LGD, low-grade dysplasia; HGD, high-grade dysplasia.
Molecular biomarkers for surveillance
Although conventional surveillance colonoscopy may contribute to a decline in CACC incidence, given that it can detect and remove some dysplasia lesions, it still has certain disadvantages that may impair an accurate evaluation. For example, both endoscopic and histopathological examinations are dependent on expert knowledge and experience; thus, these examinations are slightly subjective, and an inexperienced or a careless expert may overlook the presence of dysplasia, making endoscopy incompetent for such surveillance. In addition, some studies have highlighted endoscopy surveillance failures [42]; in 50–80% of cases with colitis-associated neoplasms, lesions remain undetected on endoscopy [43]. In contrast, molecular monitoring has the advantage of high compliance and objectivity and may have a potential clinical value as a complement tool for screening dysplasia/cancer among UC patients.
Inflammation is an important initial factor in CACC, which is quite distinct from sporadic CRC. Repeated inflammation and intestinal mucosa repair can stimulate oxidative stress reactions and produce more reactive oxygen and nitrogen species in a microenvironment, which then impair DNA and induce mutations and consequently trigger tumor initiation. Meanwhile, the inflammatory environment activates some pro-survival and antiapoptotic pathways in the intestinal epithelial cells, contributing to tumor promotion [44]. Carcinogenesis in UC is facilitated by several molecular alterations, such as chromosomal and microsatellite instability, aneuploidy and p53 mutations, some of which are detectable before dysplasia [45–47]. Using advanced molecular techniques, several studies have identified tumor-associated biomarkers in CACC, making it possible to supervise UC carcinogenesis via the detection of key biomarkers in different biosamples, such as colorectal biopsies, which are the most commonly used samples, and biomarkers from blood, stool and urine samples.
Biomarkers from colorectal biopsies
Aneuploidy
Aneuploidy is the presence of an abnormal number of chromosomes in a cell. It generally develops during cell division when chromosomes are not properly divided into two cells and a missing or extra chromosome in a fetus is a common cause of genetic disorders. An increased incidence of aneuploidy has been observed in correlation with dysplastic mucosal changes [48] and Rubin et al. [26] have reported that DNA aneuploidy precedes dysplasia by 1–2.5 years in colonic biopsies, which makes it act as a possible predictive factor for the development of dysplasia in UC. Moreover, Meyer et al. [49] have reported aneuploidy in non-malignant mucosa adjacent to CACC, which appears to be irrespective of dysplasia. A recent meta-analysis has indicated that aneuploidy is as competent as dysplasia in assessing CACC risk and a combinational analysis of the two parameters is superior to that of a single parameter [50].
Genetic susceptibility
Inflammatory stresses, such as reactive oxygen species and some free radicals, may cause genetic changes and are considered factors in CACC development. Although the duration of inflammation is considered the most important risk factor, recent studies have found a significant difference in CACC progression between individuals with similar disease durations [51], suggesting that other carcinogenic factors, in addition to inflammation, affect CACC development. Genome-wide association studies have identified >200 susceptibility loci for IBD [52], whereas very few loci have been identified for CACC. However, Watanabe et al. [53] identified 20 genes showing differential expression phenotypes in nontumor tissues of UC carcinoma and noncarcinoma patients, including cancer-relevant genes such as GBP4, SAMSN1, NOD27, NOL3, CYP27B1, RUNX3 and CYP27B1.
P53 mutation
Chronic inflammation is assumed to lead to genomic changes that increase the risk for carcinogenesis, and differences in genetic alterations between CACC and sporadic CRC lie in the differential gene expression on time sequence and frequency. APC deletion is a common early event in sporadic CRC, which is quite rare and occurs late in CACC. In contrast, p53 mutation occurs late in sporadic CRC but early in CACC and is often detected in nondysplastic or dysplastic areas in UC patients [47]. A recent study by Cooks et al. [54] has revealed that mice with mutant p53 exposed to dextran sodium sulfate not only develop a more frequent inflammation-associated colon cancer but also develop carcinoma much earlier than those with a knockout of one p53 allele, which suggests that the early presence of p53 mutant in the inflamed colon of IBD patients drives the subsequent progression to carcinoma. Shigaki et al. [55] have suggested that a combinational analysis of p53 expression and grain protein reveals a sensitivity of 66.7% and specificity of 80%, respectively, for the evaluation of high-grade dysplasia in UC patients and that the coexpression of p53 and other related genes is useful in predicting CACC risk in UC patients.
DNA methylation
DNA methylation refers to the addition of methyl groups to DNA strands without changing its sequence. An abnormal DNA methylation plays a role in CACC carcinogenesis because, when DNA methylation occurs in a gene promoter, it can silence the corresponding critical transcription. Attention should be paid to the methylation of key genes as biomarkers to predict cancer risk in UC patients. miR-124 has an anti-tumor effect, and chronic inflammation results in methylation and miR-124 silencing. Studies found that miR-124 methylation occurs in almost all CACCs, and the methylation state is 7.4 times higher in long-duration and pancolitis patients with UC than in those without any risk factor [56]. Gerecke et al. [57] have reported the methylation of ITGA4 and TFPI2 to be dense in malignant tissues of CACC and adenoma and absent in normal controls. Papadia et al. [58] have found a greater hypermethylation of FOXE1 and SYNE1 in tumor tissues of CACC than in healthy controls. Futhermore, a study by Scarpa et al. [59] has revealed a much higher methylation state of APC, CDH13, MGMT, MLH1 and RUNX3 in non-neoplastic mucosa of CACC than in normal controls and UC patients.
Protein molecule
Protein associated with UC neoplastic progression included proteins related to mitochondrial function, oxidative activity and calcium-binding. Studies have suggested that changes in protein expression occur very early in the neoplastic process before the histologic changes [60]. Immunohistochemical (IHC) analysis has shown that DNA methyltransferase 3 b (DNMT3b) expression is significantly higher in tumor tissues of CACC than in nontumor tissues of UC patients [61], which implies that IHC staining of DNMT3b facilitates early CACC detection. In addition, the RNA-binding motif protein 3 (RBM3) could induce multiple stress reactions involved in the refractory clinical process of IBD and CACC development. A recent study has revealed a significantly increased RBM3 expression in a CACC population and significantly enhanced cell apoptosis in a RBM3-deficient mouse CACC model [62]. Given that IHC staining is an easy and convenient method to assess the expression of a particular protein, such diagnostic protein molecules may be of great value in discriminating UC with and without dysplasia/cancer in the future.
MicroRNAs
MicroRNAs (miRs) are small noncoding RNA molecules generally containing 20–25 nucleotides. Although the first miRNA was reported in 1993 [63], great achievements have been made since then. Inflammation can regulate miR expression and can further influence the expression of target functional genes via the inhibition or degradation of tissue-specific miRNA translation after transcription. These processes are involved in cell metabolism, proliferation, differentiation, apoptosis, inflammation, and immune function and tumor responses [64]. Several studies have demonstrated that expression phenotypes of miRNAs are different among tumors, and miRs may function as either proto-oncogenes or tumor suppressors. In 2008, Wu et al. [65] first reported that miRNA expression varies in colonic tissues from UC patients; since then, more and more IBD-associated miRNAs have been reported. In 2011, Olaru et al. [66] reported that miR-31 expression in colonic tissue increases with disease progression from normal to IBD to IBD-associated neoplasia (IBDN). The progressive increase from normal to chronically inflamed mucosae to dysplasia and cancer mirrors the UC-related carcinoma sequence. Of note, no difference in miR-31 expression was found between IBD-dysplasia and IBD-carcinoma, which implys that miR-31 upregulation is an early event in the neoplastic transformation, and its expression level has a potential to predict early carcinogenesis. Two years later, Olaru et al. [67] reported that miR-224 level also increased successively with the progression of carcinogenesis in colonic tissues of IBD patients and differentiated cancers from normal or chronically inflamed IBD tissues, suggesting the involvement of miR-224 in both chronic inflammation and carcinogenesis. Furthermore, this study indicated that miR-224 participates in cell-cycle regulation and induces G1-S release in colon-cancer cells, which substantiate the etiologic role of miR-224 in the development of IBD-related carcinoma. In addition, Benderska et al. [68] revealed an increased miR-26 b expression in tissues of CACC compared with UC and a down-regulation of miR-26 b in SCC in 2015, suggesting miR-26 b could serve as an early biomarker for inflammation-associated processes in gastrointestinal diseases. Moreover, in silico functional pathway analysis revealed that the common cellular pathways affected by miR-26 b are highly related to colonic cancerogenesis. And, because of down-regulation of miR-26 b in sporadic CRC, it could also discriminate between CACC and sporadic CRC. Furthermore, a number of other miRNAs have also been reported to play certain roles in the pathogenesis of CACC recently [69, 70] and a better understanding of miRNA expression alterations in CACC may contribute to improved diagnosis and management, ultimately leading to improved survival and quality of life for patients at high risk of CACC.
Signaling pathways
Previous studies have suggested that the substance P-neurokinin-1 receptor-epidermal growth factor receptor (SP-NK-1 R-EGFR) pathway is involved in colitis and colitis-associated cancer [71, 72]; binding of SP and NK-1 R triggers transactivation of the EGFR, with subsequent activation of other pathways, which can lead to increased inflammation and proliferation, along with decreased apoptosis and differentiation [73, 74]. NK-1r and EGFR expressions are significantly more up-regulated in sporadic CRC and CACC than in normal controls [75]. Metastasis associated in colon cancer-1 (MACC1) is a newly identified regulator of the HGF-MET signaling pathway, and a recent study by Harpaz et al. [76] has indicated that MACC1 expression is remarkably higher in IBD-associated dysplasia than in the corresponding inflammatory or normal colonic tissues and that its level is further elevated during the progression from dysplasia to CACC.
As discussed above, a series of biomarker candidates for cancer surveillance have been identified from colonic biopsies (Table 2) but, because it is not easy to take colonic biopsies during colonoscopy and it carries risks such as bleeding, perforation and cardiorespiratory complications, researchers have turned to other non-invasive examinations. Biomarkers identified from blood, stool and urine samples have emerged in view of their painlessness, easy accessibility and low cost, although not enough data and convincing findings regarding these biomarkers are currently available.
Table 2.
A summary of clinically relevant biomarkers for colitis-associated colorectal cancer surveillance in patients with ulcerative colitis
| Type of specimen | Analyte | Biomarkers | References |
|---|---|---|---|
| Colon tissue | DNA | Aneuploidy | [48–50] |
| Cancer genes: GBP4, SAMSN1, NOD27, NOL3, CYP27B1, RUNX3 and CYP27B1 | [53] | ||
| DNA methylation | ITGA4 and TFPI2; FOXE1 and SYNE1 | [57, 58] | |
| APC, CDH13, MGMT, MLH1 and RUNX3 | [59] | ||
| Protein | P53 mutation | [47, 54, 55] | |
| DNMT3b and RBM3 | [61, 62] | ||
| microRNA | miR-31, miR-224, miR-26b, miR-214, miR-375 | [64, 66–70] | |
| Stool | DNA methylation | EYA4, BMP3 and NDRG4; SLIT2 | [77, 78] |
| RNA | miR-17-92 and miR-135; miR-21 | [79, 80] | |
| Upregulation of 7 miRNAs (miR-21, miR-106a, miR-96, miR-203, miR-20a, miR-326 and miR-92) | [81] | ||
| Decrease of 7 miNRAs (miR-320, miR-126, miR-484-5p, miR-143, miR-145, miR-16 and miR-125b) | |||
| Protein | Calprotectin and M2-PK; haptoglobin | [82, 83] | |
| Gut microbiome | Bifidobacterium, Fusobacterium nucleatum, Bacteroides clarus, Clostridium, Porphyromonas, Lachnospiraceae and Enterobacteriaceae | [84–87] | |
| Blood | DNA | mSEPT9, NEUROG1, THBD and C9orf50 | [88–90] |
| RNA | miR-375 and miR-26b | [69, 70] | |
| Protein | CA19-9, CRP and ESR | [91, 92] | |
| Urine | Metabolites | Oxidatively modified DNA bases/nucleosides | [94, 95] |
Stool biomarkers
The fundamental principle of stool-based biomarkers for CACC surveillance is based on mechanisms resulting in the presence of these tumor markers in stool, which can be attributed to leakage, exfoliation or secretion, making this a rich source of biomarkers. Although considerable efforts are being made to identify the presence of novel biomarkers in stool, stool-based studies for surveillance are currently limited.
DNA-based biomarkers
As previously mentioned, changes in methylation occur early in carcinogenesis and can be detected in both stool and blood, making these markers ideal candidates for a non-invasive test for cancer surveillance. In a prospective study by Kisiel et al. [77], the methylation states of EYA4, BMP3 and NDRG4 were assessed in stool, which revealed that the expression greatly differs between colorectal neoplasia associated with IBD (IBD–CRN) and IBD controls. A recent study by Azuara et al. [78] demonstrated that SLIT2 gene methylation in a stool sample is more frequently methylated in high-risk IBD patients for dysplasia or cancer (4/16) than in low-risk patients (0/37) (p = 0.006).
RNA-based biomarkers
The detection of RNA markers in stool has not been as extensively studied as DNA markers, partly because mRNA is less stable than DNA in stool. miRNAs are involved in tumorigenesis, including angiogenesis and metastasis, and are more stable than mRNAs in stool. In 2010, Koga et al. [79] found that miR-17–92 cluster and miR-135 in colonocytes isolated from stool are overexpressed in CRC patients compared with healthy individuals. In addition, a study conducted by Wu et al. [80] revealed that, compared with healthy controls, CRC patients have significantly higher miR-21 (p < 0.01) and miR-92a (p < 0.0001) levels in stool. Data from the study by Ahmed et al. [81] revealed an increased expression of seven miRNAs (miR-21, miR-106a, miR-96, miR-203, miR-20a, miR-326 and miR-92) and a decreased expression of seven miRNAs (miR-320, miR-126, miR-484–5p, miR-143, miR-145, miR-16 and miR-125 b) in the stool of CRC patients compared with that of UC patients.
Stool protein biomarkers
Although protein markers have mostly been detected in blood samples to date, the presence of proteins such as calprotectin and lactoferrin in stool has already been applied to evaluate therapeutic response and relapse in IBD patients. Fecal calprotectin and M2 pyruvate kinase (M2-PK) are the two most studied fecal proteins for colon-cancer screening, although their potential roles in the development of colon cancer are not well understood and the results have been inconsistent. A recent meta-analysis of eight studies of M2-PK has shown that this marker has a pooled CRC sensitivity, specificity and accuracy of 79%, 80% and 85%, respectively [82]. A study conducted in Australia has shown that the use of haptoglobin, which is an acute-phase response protein, as a stool biomarker can identify CRC patients with 92% sensitivity and 98% specificity, with an increasing sensitivity of 100% when combined with occult blood testing [83].
Gut microbiome
Recent studies have suggested that dynamic changes in the gut microbiome structure occur prior to the first signs of macroscopic tumor formation and a community-wide effect involving the gain and loss of bacterial populations and general metabolic functions likely plays a critical role in cancer development [84]. Building upon these studies, when focused on the composition of microbiota in stool samples from the gut, Yusuf et al. [85] have reported a disappearance of the dominant band in the Bifidobacterium group in all CRC, whereas colitis and internal hemorrhoid samples did not show such a disappearance. Another study conducted in Asia revealed significantly more abundance of Fusobacterium nucleatum (Fn) in CRC than in controls (p < 0.0001) and an improved diagnostic ability using the combination of four bacterial markers [Fn, Bacteroides clarus (Bc), Clostridium hathewayi (Ch) and one undefined species (m7)] [86]. Similarly, data from a study conducted in the USA have suggested that CRC patients show an increase in the relative abundance of Fusobacterium, Porphyromonas, Lachnospiraceae and Enterobacteriaceae and a decrease in the relative abundance of Bacteroides, Lachnospiraceae and Clostridiales than healthy individuals [87].
Blood biomarkers
Blood also represents an ideal diagnostic biosample for screening tests owing to its low cost and easy accessibility.
DNA-based biomarkers
An increasing number of studies have reported the presence of methylated DNA in the plasma of colon-cancer patients. A recent multicenter prospective trial has shown that CRC sensitivity and specificity of circulating aberrant methylated SEPT9 DNA (mSEPT9) are 48% and 91.5%, respectively [88]. In 2011, Herbst et al. [89] revealed 91% specificity and 61% sensitivity of NEUROG1 DNA methylation for the detection of CRC. Recently, Lange et al. [90] have identified two novel DNA-methylation markers—THBD and C9orf50—for early CRC detection through a systematic genome-scale marker discovery and verification study and revealed that these markers outperform carcinoembryonic antigen (CEA) for CRC screening.
RNA-based biomarkers
A recent study has suggested that blood-based miRNA-375 is significantly up-regulated in the CACC cohort compared with the UC cohort, and plasma miRNA has the potential to act as a novel biomarker to monitor cancer in UC patients [70]. In a chemically induced murine model of CACC, Benderska et al. [68] have indicated an increased miR-26 b expression in the serum of colon-cancer mice compared with that of untreated controls, and its expression is downregulated in SCC, which helped to discriminate between CACC and SCC.
Protein markers
CEA was first considered specific for CRC, but it lacks sufficient sensitivity for early CRC detection, and elevated CEA levels were later detected in other diseases such as IBD and other malignancies, and its potential role in the development of CRC is unclear; therefore, CEA is not recommended for CRC screening. Several studies have suggested that the sensitivity of cancer antigen 19–9 (CA 19–9) is much inferior to that of CEA, and elevated CA19–9 levels are indicative of poor prognosis [91]. Persistent inflammation is an essential factor for carcinogenesis in UC. Ananthakrishnan et al. [92] have found that elevated levels of serum inflammatory markers, C-reactive protein and erythrocyte sedimentation rate, are associated with a high CACC risk. In addition, a recent study has shown that CRC is associated with median albumin levels (p = 0.03) and increased CRP–albumin scores (p = 0.0002) [93].
Urinary biomarkers
Thus far, there have been limited urine-based studies on CACC surveillance. Oxidative stress linked with chronic inflammation is associated with CACC etiology and a study conducted in Poland assessed the diagnostic utility of the urinary excretion of oxidatively modified DNA bases/nucleosides in CRC patients and healthy individuals and found that urinary DNA modifications may reflect the oxidative stress/chronic inflammation in CRC, but its diagnostic performance for early detection is insufficient [94]. Given that field asymmetric ion mobility spectrometer (FAIMS) identifies IBD patients by analysing shifts in volatile organic compound patterns in both urine and stool, Arasaradnam et al. [95] have demonstrated that CRC patients can be distinguished from healthy individuals through FAIMS analysis of urine with 88% sensitivity and 60% specificity.
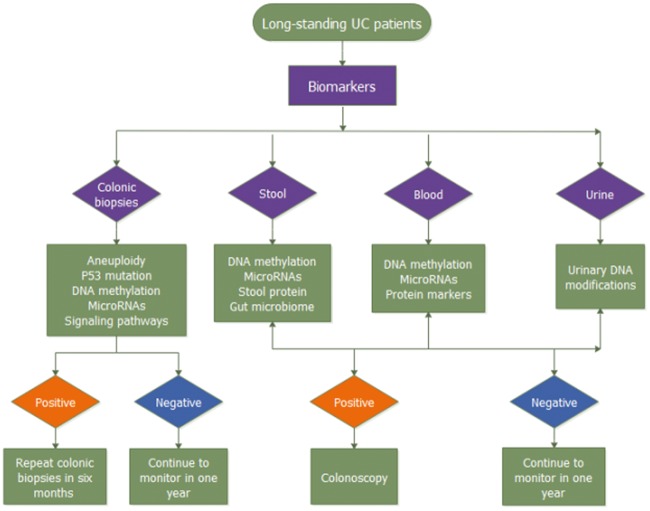
The non-invasive methods mentioned above (Table 2) can be used to decrease unnecessary colonoscopies and biopsies, thereby decreasing patients’ burden as well as lowering medical costs (Figure 2).
Figure 2.
Flow chart of molecular monitoring in patients with ulcerative colitis. It illustrates how these molecular monitoring surveillance methods can affect disease management strategies or further monitoring.
Conclusion
With increasing incidence of CACC [8, 96, 97], early CACC detection has remained a clinical challenge. Although traditional endoscopic screening has been performed for years, it may contribute to a decline in CACC incidence; however, its efficiency is mainly based on operator experience and knowledge and improvements in the endoscopic facility. Using a risk-stratified approach to determine surveillance intervals would improve the allocation of limited medical resources. Furthermore, molecular monitoring can improve the clinical management of neoplastic risk in UC patients, since the process could be based on the earliest pathways and molecular markers of UC carcinogenesis, and research on molecular detection is expanding and will likely lead to other high-performance diagnostic or screening tools in the future, but practical values for these markers remain to be confirmed because of limited findings and small sample sizes in most studies. Hence, it is necessary to find more accurate, convenient, economical and effective screening methods to manage patients in a personalized way to achieve the ultimate goal of decreasing patient morbidity and mortality from CACC.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81570502) and Research Fund for the Doctoral Program of Higher Education of China (No. 20130181120041). Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (No. 201416851110)
Conflict of interest statement: none declared.
References
- 1. Breynaert C, Vermeire S, Rutgeerts P. et al. Dysplasia and colorectal cancer in inflammatory bowel disease: a result of inflammation or an intrinsic risk? Acta Gastroenterol Belg 2008;71:367–72. [PubMed] [Google Scholar]
- 2. Herrinton LJ, Liu L, Levin TR. et al. Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology 2012;143:382–9. [DOI] [PubMed] [Google Scholar]
- 3. Jess T, Rungoe C, Peyrin-Biroulet L.. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol 2012;10:639–45. [DOI] [PubMed] [Google Scholar]
- 4. So J, Tang W, Leung WK. et al. Cancer risk in 2621 Chinese patients with inflammatory bowel disease: a population-based cohort study. Inflamm Bowel Dis 2017;23:2061–8. [DOI] [PubMed] [Google Scholar]
- 5. Eaden JA, Abrams KR, Mayberry JF.. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 2001;48:526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kavanagh DO, Carter MC, Keegan D. et al. Management of colorectal cancer in patients with inflammatory bowel disease. Tech Coloproctol 2014;18:23–8. [DOI] [PubMed] [Google Scholar]
- 7. Jess T, Simonsen J, Jorgensen KT. et al. Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology 2012;143:375–81.e1. [DOI] [PubMed] [Google Scholar]
- 8. Beaugerie L, Svrcek M, Seksik P. et al. Risk of colorectal high-grade dysplasia and cancer in a prospective observational cohort of patients with inflammatory bowel disease. Gastroenterology 2013;145:166–75. [DOI] [PubMed] [Google Scholar]
- 9. Baars JE, Looman CW, Steyerberg EW. et al. The risk of inflammatory bowel disease-related colorectal carcinoma is limited: results from a nationwide nested case-control study. Am J Gastroenterol 2011;106:319–28. [DOI] [PubMed] [Google Scholar]
- 10. van Schaik FD, van Oijen MG, Smeets HM. et al. Thiopurines prevent advanced colorectal neoplasia in patients with inflammatory bowel disease. Gut 2012;61:235–40. [DOI] [PubMed] [Google Scholar]
- 11. Farraye FA, Odze RD, Eaden J. et al. AGA technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology 2010;138:746–74. [DOI] [PubMed] [Google Scholar]
- 12. Dugum M, Lin J, Lopez R. et al. Recurrence and survival rates of inlfammatory bowel disease-associated colorectal cancer following postoperative chemotherapy: a comparative study. Gastroenterol Rep (Oxf) 2017;5:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Itzkowitz SH, Yio X.. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol 2004;287:G7–17. [DOI] [PubMed] [Google Scholar]
- 14. Ananthakrishnan AN, Cagan A, Cai T. et al. Colonoscopy is associated with a reduced risk for colon cancer and mortality in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol 2015;13:322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi PM, Nugent FW, Schoetz DJ. et al. Colonoscopic surveillance reduces mortality from colorectal cancer in ulcerative colitis. Gastroenterology 1993;105:418–24. [DOI] [PubMed] [Google Scholar]
- 16. Suzuki K, Muto T, Shinozaki M. et al. Results of cancer surveillance in ulcerative colitis. J Gastroenterol 1995;30 (Suppl 8):40–2. [PubMed] [Google Scholar]
- 17. Van Assche G, Dignass A, Bokemeyer B. et al. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 3: special situations. J Crohns Colitis 2013;7:1–33. [DOI] [PubMed] [Google Scholar]
- 18. Mowat C, Cole A, Windsor A. et al. Guidelines for the management of inflammatory bowel disease in adults. Gut 2011;60:571–607. [DOI] [PubMed] [Google Scholar]
- 19. Shergill AK, Lightdale JR, Bruining DH. et al. The role of endoscopy in inflammatory bowel disease. Gastrointest Endosc 2015;81:1101–21. [DOI] [PubMed] [Google Scholar]
- 20. Gu J, Remzi FH, Lian L. et al. Practice pattern of ileal pouch surveillance in academic medical centers in the United States. Gastroenterol Rep 2016;4:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cairns SR, Scholefield JH, Steele RJ. et al. Guidelines for colorectal cancer screening and surveillance in moderate and high risk groups (update from 2002). Gut 2010;59:666–89. [DOI] [PubMed] [Google Scholar]
- 22. Farraye FA, Odze RD, Eaden J. et al. AGA institute medical position panel on diagnosis and management of colorectal neoplasia in inflammatory bowel disease: AGA medical position statement on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology 2010;138:738–45. [DOI] [PubMed] [Google Scholar]
- 23. Eaden JA, Mayberry JF.. Guidelines for screening and surveillance of asymptomatic colorectal cancer in patients with inflammatory bowel disease. Gut 2002;51:V10–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winawer S, Fletcher R, Rex D. et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology 2003;124:544–60. [DOI] [PubMed] [Google Scholar]
- 25. Navaneethan U, Kochhar G, Venkatesh PG. et al. Random biopsies during surveillance colonoscopy increase dysplasia detection in patients with primary sclerosing cholangitis and ulcerative colitis. J Crohns Colitis 2013;7:974–81. [DOI] [PubMed] [Google Scholar]
- 26. Rubin CE, Haggitt RC, Burmer GC. et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology 1992;103:1611–20. [DOI] [PubMed] [Google Scholar]
- 27. East JE. Colonoscopic cancer surveillance in inflammatory bowel disease: what’s new beyond random biopsy? Clin Endosc 2012;45:274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rutter MD, Saunders BP, Wilkinson KH. et al. Most dysplasia in ulcerative colitis is visible at colonoscopy. Gastrointest Endosc 2004;60:334–9. [DOI] [PubMed] [Google Scholar]
- 29. Rubin DT, Rothe JA, Hetzel JT. et al. Are dysplasia and colorectal cancer endoscopically visible in patients with ulcerative colitis? Gastrointest Endosc 2007;65:998–1004. [DOI] [PubMed] [Google Scholar]
- 30. Watanabe T, Ajioka Y, Mitsuyama K. et al. Comparison of targeted vs random biopsies for surveillance of ulcerative colitis-associated colorectal cancer. Gastroenterology 2016;151:1122–30. [DOI] [PubMed] [Google Scholar]
- 31. Subramanian V, Ramappa V, Telakis E. et al. Comparison of high definition with standard white light endoscopy for detection of dysplastic lesions during surveillance colonoscopy in patients with colonic inflammatory bowel disease. Inflamm Bowel Dis 2013;19:350–5. [DOI] [PubMed] [Google Scholar]
- 32. Soetikno R, Subramanian V, Kaltenbach T. et al. The detection of nonpolypoid (flat and depressed) colorectal neoplasms in patients with inflammatory bowel disease. Gastroenterology 2013;144:1349–52. [DOI] [PubMed] [Google Scholar]
- 33. Laine L, Kaltenbach T, Barkun A. et al. SCENIC Guideline Development Panel. SCENIC international consensus statement on surveillance and management of dysplasia in inflammatory bowel disease. Gastroenterology 2015;148:639–51. [DOI] [PubMed] [Google Scholar]
- 34. Basford PJ, Longcroft-Wheaton G, Higgins B. et al. High-definition endoscopy with i-Scan for evaluation of small colon polyps: the HiSCOPE study. Gastrointest Endosc 2014;79:111–8. [DOI] [PubMed] [Google Scholar]
- 35. Koo JS. Equipment-based image-enhanced endoscopy for differentiating colorectal polyps. Clin Endosc 2014;47:330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Efthymiou M, Allen PB, Taylor AC. et al. Chromoendoscopy versus narrow band imaging for colonic surveillance in inflammatory bowel disease. Inflamm Bowel Dis 2013;19:2132–8. [DOI] [PubMed] [Google Scholar]
- 37. Oka S, Tanaka S, Chayama K.. Detection of nonpolypoid colorectal neoplasia using magnifying endoscopy in colonic inflammatory bowel disease. Gastrointest Endosc Clin N Am 2014;24:405–17. [DOI] [PubMed] [Google Scholar]
- 38. Su P, Liu Y, Lin S. et al. Efficacy of confocal laser endomicroscopy for discriminating colorectal neoplasms from non-neoplasms: a systematic review and meta-analysis. Colorectal Dis 2013;15:e1–12. [DOI] [PubMed] [Google Scholar]
- 39. Kiesslich R, Goetz M, Lammersdorf K. et al. Chromoscopy-guided endomicroscopy increases the diagnostic yield of intraepithelial neoplasia in ulcerative colitis. Gastroenterology 2007;132:874–82. [DOI] [PubMed] [Google Scholar]
- 40. Hurlstone DP, Kiesslich R, Thomson M. et al. Confocal chromoscopic endomicroscopy is superior to chromoscopy alone for the detection and characterisation of intraepithelial neoplasia in chronic ulcerative colitis. Gut 2008;57:196–204. [DOI] [PubMed] [Google Scholar]
- 41. Leong RW, Ooi M, Corte C. et al. Full-spectrum endoscopy improves surveillance for dysplasia in patients with inflammatory bowel diseases. Gastroenterology 2017;152:1337–44. [DOI] [PubMed] [Google Scholar]
- 42. Wang YR, Cangemi JR, Loftus EJ. et al. Rate of early/missed colorectal cancers after colonoscopy in older patients with or without inflammatory bowel disease in the United States. Am J Gastroenterol 2013;108:444–9. [DOI] [PubMed] [Google Scholar]
- 43. Ekbom A, Helmick C, Zack M. et al. Ulcerative colitis and colorectal cancer: a population-based study. N Engl J Med 1990;323:1228–33. [DOI] [PubMed] [Google Scholar]
- 44. O’Connor PM, Lapointe TK, Beck PL. et al. Mechanisms by which inflammation may increase intestinal cancer risk in inflammatory bowel disease. Inflamm Bowel Dis 2010;16:1411–20. [DOI] [PubMed] [Google Scholar]
- 45. Brentnall TA, Crispin DA, Bronner MP. et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res 1996;56:1237–40. [PubMed] [Google Scholar]
- 46. O’Sullivan JN, Bronner MP, Brentnall TA. et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet 2002;32:280–4. [DOI] [PubMed] [Google Scholar]
- 47. Brentnall TA, Crispin DA, Rabinovitch PS. et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology 1994;107:369–78. [DOI] [PubMed] [Google Scholar]
- 48. Sjoqvist U, Hertervig E, Nilsson A. et al. Chronic colitis is associated with a reduction of mucosal alkaline sphingomyelinase activity. Inflamm Bowel Dis 2002;8:258–63. [DOI] [PubMed] [Google Scholar]
- 49. Meyer KF, Nause SL, Freitag-Wolf S. et al. Aneuploidy characterizes adjacent non-malignant mucosa of ulcerative colitis-associated but not sporadic colorectal carcinomas: a matched-pair analysis. Scand J Gastroenterol 2013;48:679–87. [DOI] [PubMed] [Google Scholar]
- 50. Meyer R, Freitag-Wolf S, Blindow S. et al. Combining aneuploidy and dysplasia for colitis’ cancer risk assessment outperforms current surveillance efficiency: a meta-analysis. Int J Colorectal Dis 2017;32:171–82. [DOI] [PubMed] [Google Scholar]
- 51. Connelly TM, Berg AS, Harris LR. et al. Ulcerative colitis neoplasia is not associated with common inflammatory bowel disease single-nucleotide polymorphisms. Surgery 2014;156:253–62. [DOI] [PubMed] [Google Scholar]
- 52. Khor B, Gardet A, Xavier RJ.. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Watanabe T, Kobunai T, Yamamoto Y. et al. Predicting ulcerative colitis-associated colorectal cancer using reverse-transcription polymerase chain reaction analysis. Clin Colorectal Cancer 2011;10:134–41. [DOI] [PubMed] [Google Scholar]
- 54. Cooks T, Pateras IS, Tarcic O. et al. Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 2013;23:634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shigaki K, Mitomi H, Fujimori T. et al. Immunohistochemical analysis of chromogranin A and p53 expressions in ulcerative colitis-associated neoplasia: neuroendocrine differentiation as an early event in the colitis-neoplasia sequence. Hum Pathol 2013;44:2393–9. [DOI] [PubMed] [Google Scholar]
- 56. Ueda Y, Ando T, Nanjo S. et al. DNA methylation of microRNA-124a is a potential risk marker of colitis-associated cancer in patients with ulcerative colitis. Dig Dis Sci 2014;59:2444–51. [DOI] [PubMed] [Google Scholar]
- 57. Gerecke C, Scholtka B, Lowenstein Y. et al. Hypermethylation of ITGA4, TFPI2 and VIMENTIN promoters is increased in inflamed colon tissue: putative risk markers for colitis-associated cancer. J Cancer Res Clin Oncol 2015;141:2097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Papadia C, Louwagie J, Del RP. et al. FOXE1 and SYNE1 genes hypermethylation panel as promising biomarker in colitis-associated colorectal neoplasia. Inflamm Bowel Dis 2014;20:271–7. [DOI] [PubMed] [Google Scholar]
- 59. Scarpa M, Scarpa M, Castagliuolo I. et al. Aberrant gene methylation in non-neoplastic mucosa as a predictive marker of ulcerative colitis-associated CRC. Oncotarget 2016;7:10322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brentnall TA, Pan S, Bronner MP. et al. Proteins that underlie neoplastic progression of ulcerative colitis. Prot Clin Appl 2009;3:1326.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ueda H, Tanaka H, Ichikawa K. et al. Immunohistochemical analysis of the DNA methyltransferase 3b expression is associated with significant improvements in the discrimination of ulcerative colitis-associated neoplastic lesions. Surg Today 2013;43:1275–80. [DOI] [PubMed] [Google Scholar]
- 62. Sakurai T, Kashida H, Komeda Y. et al. Stress response protein RBM3 promotes the development of colitis-associated cancer. Inflamm Bowel Dis 2017;23:66–74. [DOI] [PubMed] [Google Scholar]
- 63. Lee RC, Feinbaum RL, Ambros V.. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–54. [DOI] [PubMed] [Google Scholar]
- 64. Raisch J, Darfeuille-Michaud A, Nguyen HT.. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol 2013;19:2985–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu F, Zikusoka M, Trindade A. et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008;135:1624–35. [DOI] [PubMed] [Google Scholar]
- 66. Olaru AV, Selaru FM, Mori Y. et al. Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm Bowel Dis 2011;17:221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Olaru AV, Yamanaka S, Vazquez C. et al. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis 2013;19:471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Benderska N, Dittrich AL, Knaup S. et al. miRNA-26b overexpression in ulcerative colitis-associated carcinogenesis. Inflamm Bowel Dis 2015;21:2039–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Polytarchou C, Hommes DW, Palumbo T. et al. MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology 2015;149:981–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Patel M, Verma A, Aslam I. et al. Novel plasma microRNA biomarkers for the identification of colitis-associated carcinoma. Lancet 2015;385:S78.. [DOI] [PubMed] [Google Scholar]
- 71. Pagan B, Isidro AA, Coppola D. et al. Effect of a neurokinin-1 receptor antagonist in a rat model of colitis-associated colon cancer. Anticancer Res 2010;30:3345–53. [PMC free article] [PubMed] [Google Scholar]
- 72. Koon HW, Zhao D, Na X. et al. Metalloproteinases and transforming growth factor-alpha mediate substance P-induced mitogen-activated protein kinase activation and proliferation in human colonocytes. J Biol Chem 2004;279:45519–27. [DOI] [PubMed] [Google Scholar]
- 73. Castagliuolo I, Valenick L, Liu J. et al. Epidermal growth factor receptor transactivation mediates substance P-induced mitogenic responses in U-373 MG cells. J Biol Chem 2000;275:26545–50. [DOI] [PubMed] [Google Scholar]
- 74. Witsch E, Sela M, Yarden Y.. Roles for growth factors in cancer progression. Physiology (Bethesda) 2010;25:85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Isidro RA, Cruz ML, Isidro AA. et al. Immunohistochemical expression of SP-NK-1R-EGFR pathway and VDR in colonic inflammation and neoplasia. World J Gastroenterol 2015;21:1749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Harpaz N, Taboada S, Ko HM. et al. Expression of MACC1 and MET in inflammatory bowel disease-associated colonic neoplasia. Inflamm Bowel Dis 2014;20:703–11. [DOI] [PubMed] [Google Scholar]
- 77. Kisiel JB, Yab TC, Nazer HF. et al. Stool DNA testing for the detection of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther 2013;37:546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Azuara D, Rodriguez-Moranta F, de Oca J. et al. Novel methylation panel for the early detection of neoplasia in high-risk ulcerative colitis and Crohn’s colitis patients. Inflamm Bowel Dis 2013;19:165–73. [DOI] [PubMed] [Google Scholar]
- 79. Koga Y, Yasunaga M, Takahashi A. et al. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila) 2010;3:1435–42. [DOI] [PubMed] [Google Scholar]
- 80. Wu CW, Ng SS, Dong YJ. et al. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 2012;61:739–45. [DOI] [PubMed] [Google Scholar]
- 81. Ahmed FE, Jeffries CD, Vos PW. et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics 2009;6:281–95. [PubMed] [Google Scholar]
- 82. Uppara M, Adaba F, Askari A. et al. A systematic review and meta-analysis of the diagnostic accuracy of pyruvate kinase M2 isoenzymatic assay in diagnosing colorectal cancer. World J Surg Oncol 2015;13:48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xing PX, Young GP, Ho D. et al. A new approach to fecal occult blood testing based on the detection of haptoglobin. Cancer 1996;78:48–56. [DOI] [PubMed] [Google Scholar]
- 84. Zackular JP, Baxter NT, Iverson KD. et al. The gut microbiome modulates colon tumorigenesis. MBio 2013;4:e00692-13–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yusuf F, Ilyas S, Damanik HA. et al. Microbiota composition, HSP70 and caspase-3 expression as marker for colorectal cancer patients in Aceh, Indonesia. Acta Med Indones 2016;48:289–99. [PubMed] [Google Scholar]
- 86. Liang Q, Chiu J, Chen Y. et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 2017;23:2061–70. [DOI] [PubMed] [Google Scholar]
- 87. Zackular JP, Rogers MA, Ruffin MT. et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014;7:1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Church TR, Wandell M, Lofton-Day C. et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Herbst A, Rahmig K, Stieber P. et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol 2011;106:1110–8. [DOI] [PubMed] [Google Scholar]
- 90. Lange CP, Campan M, Hinoue T. et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One 2012;7:e50266.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lumachi F, Marino F, Orlando R. et al. Simultaneous multianalyte immunoassay measurement of five serum tumor markers in the detection of colorectal cancer. Anticancer Res 2012;32:985–8. [PubMed] [Google Scholar]
- 92. Ananthakrishnan AN, Cheng SC, Cai T. et al. Serum inflammatory markers and risk of colorectal cancer in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol 2014;12:1342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Koutroubakis IE, Regueiro M, Schoen RE. et al. Multiyear patterns of serum inflammatory biomarkers and risk of colorectal neoplasia in patients with ulcerative colitis. Inflamm Bowel Dis 2016;22:100–5. [DOI] [PubMed] [Google Scholar]
- 94. Rozalski R, Gackowski D, Siomek-Gorecka A. et al. Urinary 5-hydroxymethyluracil and 8-oxo-7, 8-dihydroguanine as potential biomarkers in patients with colorectal cancer. Biomarkers 2015;20:287–91. [DOI] [PubMed] [Google Scholar]
- 95. Arasaradnam RP, McFarlane MJ, Ryan-Fisher C. et al. Detection of colorectal cancer (CRC) by urinary volatile organic compound analysis. PLoS One 2014;9:e108750.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Watanabe T, Konishi T, Kishimoto J. et al. Ulcerative colitis-associated colorectal cancer shows a poorer survival than sporadic colorectal cancer: a nationwide Japanese study. Inflamm Bowel Dis 2011;17:802–8. [DOI] [PubMed] [Google Scholar]
- 97. Choi CH, Rutter MD, Askari A. et al. Forty-year analysis of colonoscopic surveillance program for neoplasia in ulcerative colitis: an updated overview. Am J Gastroenterol 2015;110:1022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]