Abstract
Chemoreception is a widespread biological function that is essential for the survival, reproduction, and social communication of animals. Though the molecular mechanisms underlying chemoreception are relatively well known in insects, they are poorly studied in the other major arthropod lineages. Current availability of a number of chelicerate genomes constitutes a great opportunity to better characterize gene families involved in this important function in a lineage that emerged and colonized land independently of insects. At the same time, that offers new opportunities and challenges for the study of this interesting animal branch in many translational research areas. Here, we have performed a comprehensive comparative genomics study that explicitly considers the high fragmentation of available draft genomes and that for the first time included complete genome data that cover most of the chelicerate diversity. Our exhaustive searches exposed thousands of previously uncharacterized chemosensory sequences, most of them encoding members of the gustatory and ionotropic receptor families. The phylogenetic and gene turnover analyses of these sequences indicated that the whole-genome duplication events proposed for this subphylum would not explain the differences in the number of chemoreceptors observed across species. A constant and prolonged gene birth and death process, altered by episodic bursts of gene duplication yielding lineage-specific expansions, has contributed significantly to the extant chemosensory diversity in this group of animals. This study also provides valuable insights into the origin and functional diversification of other relevant chemosensory gene families different from receptors, such as odorant-binding proteins and other related molecules.
Keywords: chemosensory gene family, gustatory receptors, ionotropic receptors, acari, spiders, scorpions
Introduction
The i5k initiative (Robinson et al. 2011) has greatly boosted the complete genome sequencing and functional annotation of a number of arthropod species. The currently available genome data were obtained from species chosen for their significance as model organisms in diverse areas, such as agriculture, medicine, food safety or biodiversity, or for their strategic phylogenetic position in evolutionary studies on the diversification of the major arthropod lineages (Adams et al. 2000; Colbourne et al. 2011; Cao et al. 2013; Chipman et al. 2014; Sanggaard et al. 2014; Gulia-Nuss et al. 2016). As expected, the first sequencing initiatives focused on insects, although the number of sequenced noninsect genomes has increased considerably over time, especially in chelicerates. The recent genome sequence data from chelicerate species (Cao et al. 2013; Sanggaard et al. 2014; Gulia-Nuss et al. 2016) are disrupting the strongly biased taxonomic distribution of arthropod genomes hitherto available. More importantly, these new data have greatly facilitated studies on the origin and evolutionary divergence of this highly diverse animal subphylum (Kenny et al. 2016; Schwager et al. 2017), which has important impacts on translational research such as silk production in spiders, biomedical applications of spider and scorpion venom toxins, or plague control in acari (Mille et al. 2015; Hoy et al. 2016; Babb et al. 2017; Gendreau et al. 2017; Pennisi 2017).
Chemoreception is a paradigmatic example of a relatively well-known biological system in insects, but it is not as well characterized in other arthropods despite numerous practical applications as pest control strategies, biosensors or electronic nose sensors (Berna et al. 2009; Wei et al. 2017). In chelicerates, as in other animals, the chemosensory system (CS) is critical for the survival, reproduction, and social communication of individuals. The detection and integration of environmental chemical signals, including smell and taste, allow organisms to detect food, hosts, and predators and frequently play a crucial role in social communication (Joseph and Carlson 2015). In Drosophila, peripheral events occur in specialized hair-like cuticular structures (sesilla) that are distributed throughout the body surface, with a prominent concentration in antennae and maxillary palps (olfactory sensilla) or on the distal tarsal segments of the legs (gustatory sensilla) (Pelosi 1996; Shanbhag et al. 2001). In this species, chemoreceptor proteins, which are located in the membranes of sensory neurons innervating the sensillum lymph, convert the external chemical signal into an electrical one, which is, in turn, processed in higher brain regions (de Bruyne and Baker 2008; Sánchez-Gracia et al. 2009; Sato and Touhara 2009). The sensillum lymph contains a set of highly abundant small globular proteins (hereafter termed “binding proteins”) that are thought to bind to, solubilize and transport chemical cues to the space surrounding chemoreceptors (Vogt and Riddiford 1981; Pelosi et al. 2006). The genome of the fruit fly encodes two different kinds of membrane chemoreceptors that are phylogenetically unrelated. The first group comprises the superfamily of insect olfactory (Or) and gustatory (Gr) receptors, which encode seven-transmembrane receptors with an atypical membrane topology and heteromeric function, and share a common origin (Missbach et al. 2015). Interestingly, and despite performing analogous functions, these receptors are structurally and genetically unrelated to their vertebrate counterparts, where G protein-coupled receptors are involved in chemoreception (Kaupp 2010). The second group of chemoreceptors encodes the ionotropic receptor (Ir) gene family, a highly divergent lineage that is related to the ionotropic glutamate receptors superfamily (iGluR) associated with both olfaction and taste functions (Robertson and Wanner 2006; Benton et al. 2009; He et al. 2013; Missbach et al. 2014). The extracellular binding proteins of Drosophila include the odorant binding protein (Obp), chemosensory protein (Csp), chemosensory proteins A and B (CheA and CheB) and Niemann–Pick Type C2 (Npc2) families (Li et al. 2008; Dani et al. 2011; Iovinella et al. 2011). Moreover, sensory neuron membrane proteins (SNMPs), which are related to the CD36 receptor family and expressed in specific Drosophila pheromone-responding sensory neurons, also play a key role in sensory perception by facilitating the contact between ligand and receptor (Gomez-Diaz et al. 2016). It is worth noting that there is a lack of evidence that all CS family members actually possess a true chemosensory function, and they are usually classified as chemosensory-related genes based on their sequence similarity with previously examined members (Kitabayashi et al. 1998; Wanner et al. 2005; Ishida et al. 2013; Joseph and Carlson 2015).
There are few comprehensive studies of the characterization and classification of CS gene families in noninsect genomes, with only six noninsect arthropod species investigated to date: The crustacean Daphnia pulex, the myriapods Strigamia maritima and Trigoniulus corallinus, and the chelicerates Ixodes scapularis, Metaseiulus occidentalis and Tetranychus urticae (Colbourne et al. 2011; Chipman et al. 2014; Kenny et al. 2015; Gulia-Nuss et al. 2016; Hoy et al. 2016; Ngoc et al. 2016). Moreover, we and others have also reported transcriptome data for various chelicerate species (Frías-López et al. 2015; Qu et al. 2016; Eliash et al. 2017; Vizueta et al. 2017). These works confirm that chelicerates contain members of all insect CS gene families, with the single exception of the Or family (Benton et al. 2007; et al. 2011; Missbach et al. 2014), which likely emerged from a Gr ancestor during the diversification of flying insects (Missbach et al. 2015). The recent identification of two novel candidate CS families in chelicerates, the Obp-like and the candidate carrier protein (Cpp) families, is also remarkable (Vizueta et al. 2017). The Obp-like family, which encodes proteins with some sequence and structural similarity to canonical insect OBPs, has also been identified in centipedes (Vizueta et al. 2017), and this finding makes unclear the evolution of these gene families in arthropods. The Ccp family, which was first discovered in the transcriptome of D. silvatica, contains members that are differentially expressed in the putative chemosensory appendages of this spider. Although OBP-like and CCPs share some common structural features with other CS proteins, their potential functional roles as chemosensory proteins and the extent to which these proteins are present in arthropods remain to be elucidated (Renthal et al. 2017; Vizueta et al. 2017).
The ancestor of all extant chelicerates can be traced back to the Cambrian period (∼530 Ma); therefore, this group colonized land independently of the other arthropod lineages (Hexapoda, Crustacea, and Myriapoda; Rota-Stabelli et al. 2013). As there are no OR-encoding genes, other proteins likely perform OR’s function. Current experimental data from noninsect arthropods, such as the specific gene expression and electrophysiological recording data for some IR members in the olfactory structures of lobsters and hermit crabs (Corey et al. 2013; Groh-Lunow et al. 2015) and RNA-seq of the palps and first pair of legs of spiders (Vizueta et al. 2017) and centipede antennas (C. Frias-López, F.C. Almeida, S. Guirao-Rico, R. Jenner, A. Sánchez-Gracia and J. Rozas, unpublished results), indicate that this receptor family contains the best candidates for actual olfactory receptors. The specific organs and molecules responsible for gustatory function are less well understood; nevertheless, as some Gr and Ir family members are differentially expressed across some body parts in these species, contact chemoreceptors appear to be the best candidates. Given this difference in functional roles of the various CS families, it is highly relevant to gain further comprehensive insights into their evolution in arthropods other than insects/hexapods.
Here, we carried out an enhanced comparative genomic analysis of the CS families across 11 chelicerate genomes. We applied powerful sequence similarity-based searches using state-of-the-art methodologies and expressly considered the fragmented nature of the surveyed genomes. We conducted a comprehensive phylogenetic analysis of chemosensory genes from different gene families and characterized the turnover rates of chemoreceptor families across chelicerates after accurate estimation of the number of gene duplications and gene losses in each lineage. We also contribute new knowledge about some interesting questions that are not yet fully resolved, such as the evolutionary relationship between OBP and OBP-like proteins or the extent in which CCP and CSP are present in chelicerates.
Materials and Methods
Genomic Data
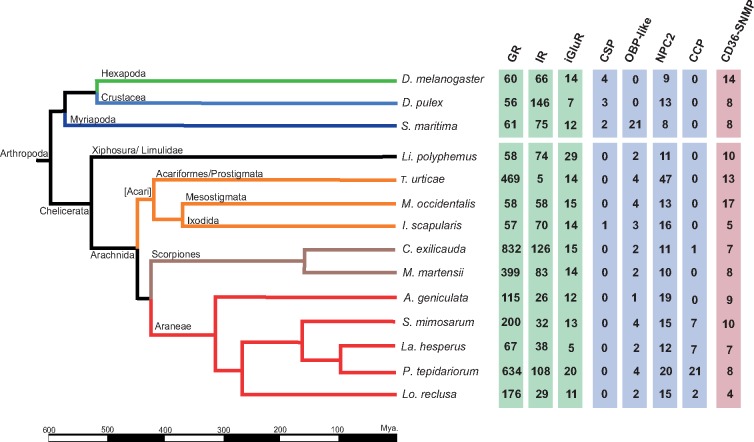
We retrieved all genomic sequences, annotations, and predicted peptides of 14 arthropods, including 11 chelicerates, from public databases (fig. 1). Specifically, we used the genome information of the fruit fly Drosophila melanogaster (r6.05, FlyBase) (Adams et al. 2000), the crustacean Daphnia pulex (r1.26, Ensembl Genomes) (Colbourne et al. 2011), and the centipede Strigamia maritima (r1.26, Ensembl Genomes) (Chipman et al. 2014). The chelicerate genomes included the horseshoe crab Limulus polyphemus (v2.1.2, NCBI Genomes) (Nossa et al. 2014); the acari Tetranychus urticae (r1.26, Ensembl Genomes) (Grbić et al. 2011), Metaseiulus occidentalis (v1.0, NCBI Genomes) (Hoy et al. 2016), and Ixodes scapularis (r1.26, Ensembl Genomes) (Gulia-Nuss et al. 2016); the scorpions Centruroides exilicauda (bark scorpion, genome assembly version v1.0, annotation version v0.5.3; Human Genome Sequencing Center [HGSC]) and Mesobuthus martensii (v1.0, Scientific Data Sharing Platform Bioinformation [SDSPB]; Cao et al. 2013); and the spiders Acanthoscurria geniculata (tarantula, v1, NCBI Assembly, BGI; Sanggaard et al. 2014), Stegodyphus mimosarum (African social velvet spider, v1, NCBI Assembly, BGI; Sanggaard et al. 2014), Latrodectus hesperus (western black widow, v1.0, HGSC), Parasteatoda tepidariorum (common house spider, v1.0 Augustus 3, SpiderWeb and HGSC; Schwager et al. 2017), and Loxosceles reclusa (brown recluse, v1.0, HGSC).
Fig. 1.—
Phylogenetic relationships among the 14 surveyed species. Divergence times are given in millions of years. Some branches representative of major lineages are shaded in different colors. Green, insects; light blue, crustaceans; dark blue, myriapods; black, horseshoe crabs; orange, acari; brown, scorpions; red, spiders. Numbers in the right part of the figure indicate the number of CS encoding sequences separated per each family (SMIN values).
Query Data Sets and Protein Search Protocol
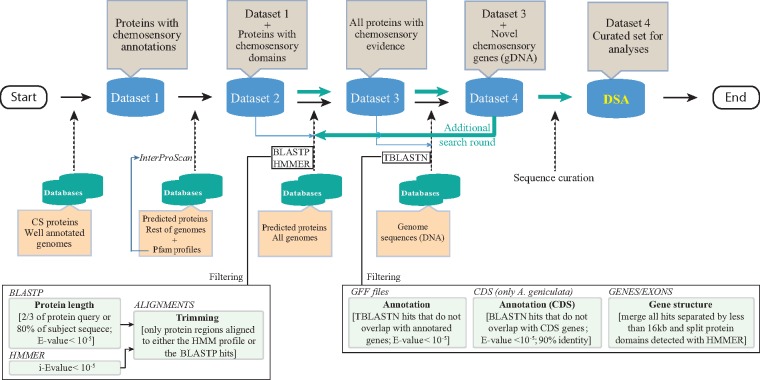
Our comprehensive CS search protocol included the creation of three data sets, which were iteratively used as queries in successive hierarchical rounds of sequence similarity- and profile-based searches (fig. 2).
Fig. 2.—
Workflow showing the steps used for the identification and annotation of the chemosensory gene families.
Data Set 1
The starting data set contained the CS proteins from publicly available, well-annotated genomes. This data set included the protein sequences of the Gr, Ir/iGluR, Or, Csp, Obp, Npc2, and Snmp-Cd36 families from 1) the hexapods D. melanogaster (Benton et al. 2009; Vogt et al. 2009; Vieira and Rozas 2011; Pelosi et al. 2014), T. castaneum (Sánchez-Gracia et al. 2009; Croset et al. 2010; Dippel et al. 2014), A. pisum (Zhou et al. 2010), and A. mellifera (Robertson and Wanner 2006; Forêt et al. 2007; Nichols and Vogt 2008); 2) the crustacean D. pulex (Peñalva-Arana et al. 2009); 3) the myriapod S. maritima (Chipman et al. 2014); and 4) the ticks I. scapularis (Gulia-Nuss et al. 2016), M. occidentalis (Hoy et al. 2016), and T. urticae (Ngoc et al. 2016).
Data Set 2
This data set included the sequences of data set 1 (DS1) plus the new identified CS protein sequences with specific CS protein domains (see Table S1 in Vizueta et al. [2017] for details). We applied InterProScan (5.4.47; Jones et al. 2014) against genome-wide predicted peptides without a functional chemosensory annotation (i.e., in chelicerate genomes that were not used in the step to build DS1). Furthermore, we also included in data set 2 (DS2) the members of the Cpp family identified in Vizueta et al. (2017), as well as those found in current chelicerate genomes, after conducting several rounds of BlastP searches (version 2.2.30; Altschul 1997).
Data Set 3
This data set resulted from incorporating some additional highly curated sequences (a second search round against all surveyed genomes) into DS2. For that, we built for each CS family a multiple sequence alignment (MSA) of all DS2 proteins and the corresponding Pfam profile as a guide (using the HMMER software; Eddy 2011). We used these MSAs to build new (more specific) HMM profiles, with one per gene family (generically named CS-F-HMM). For the second search round of predicted peptides from all genomes, we used as queries both the CS-F-HMM profiles (in HMMER searches; i-E-value < 10−5) and the sequences of DS2 (in BlastP searches; E-value < 10−5). Moreover, we only retained the BlastP-positive hits for which the alignment between the query and the subject either covered at least two-thirds of the query length or included at least 80% of the subject peptide. Finally, we trimmed all the fragments not aligned between queries and the subject sequences and added the alignment region to DS2 to build data set 3 (DS3).
Data Set 4 and Data Set for the Analyses
Data set 4 (DS4) is the most curated and inclusive data set used for searches. The new information in DS4 was obtained after conducting exhaustive searches for CS-encoding regions directly on the DNA genome sequences using DS3 peptides as queries in a TBlastN search (E-value < 10−5). Positive blast hits on regions that were not annotated in the GFF files were considered putative novel CS family members. For the genome of A. geniculata, where there is no GFF information, we checked for the presence of any protein-coding region in the available transcriptomic data.
The TBlastN search allowed essentially the identification of exonic regions. To expand these regions to cover complete genes (as much as possible), we concatenated all sequences with hits located in the same scaffold and separated by <16 kb. We chose a 16-kb cut-off value because it corresponds to the 95th percentile of the intron length distribution in the studied genomes (i.e., fragments separated by higher distances are unlikely to be exons of the same gene). Next, we translated the nucleotide sequences according to the TBlastN reading frame. To avoid generating chimeric proteins from physically close but different genes, we used the specific CS-F-HMM profile to determine whether the number of different domains of each new protein after concatenation was compatible with a single gene (HMMER search; i-E-value < 10−5). In addition to the “16-kb cut-off approach,” and to try to extend a putative incomplete gene because the putative exons might be located in different scaffolds, we also applied the ESPRIT algorithm (Dessimoz et al. 2011) to join these partial fragments using DS1 as a guide. Finally, all the newly discovered CS-encoding sequences were added to DS3 to generate DS4. These protein data in DS4 were then used as a query to conduct an additional search round (in the same way as in the DS3 and DS4 steps). Finally, we conducted a semiautomatic step to curate the newly identified sequences from putative errors introduced in the search process (deletion of putative artefactual stop codons generated by TBlastN searches, splitting different genes erroneously fused in the same sequence, removing very small fragments). With the curated data, we established the final chelicerate CS protein data set, named DSA (data set for the analyses), which was used in further comparative genomic and evolutionary analyses (supplementary table S1A, Supplementary Material online). All new CS-proteins (including incomplete fragments) identified in this study are provided in the supplementary material, Supplementary Material online.
Functional and Structural Classification of CS Sequences
We classified the novel sequences in different categories based on structural and functional criteria. First, we examined the presence of premature stop codons; these features could represent real nonfunctional copies (pseudogenes), errors in sequencing or genome assembly steps or inaccuracies in our automatic annotation step based on TBlastN hits. All sequences encoding complete proteins (CPs) that were free of stop codons were included in the first category (CP set). Operationally, we considered a CP when its length was >80% of the corresponding average protein domain length. In addition, and only for the GR family, we also required that the CP members contained a minimum of 5 of the 7 transmembrane domains (defined by the software TMHMM version 2.0c; Krogh et al. 2001; Phobius version 1.01; Käll et al. 2004). For the CP Ir/iGluR members, we required the presence of the two ligand-binding domains, namely, PF00060 (ligand-gated ion channel) and PF10613 (ligand ion channel L-glutamate- and glycine-binding site), which are present in all Ir/iGluR subfamilies, i.e., kainate, AMPA, NMDA, conserved IRs (Ir25a/Ir8a), and divergent IRs (Croset et al. 2010). The third domain exhibited by some members of the family, PF01094 (ANF receptor), was not used in this step. The remaining sequences that were free of stop codons and did not pass the length filter criteria were classified as incomplete proteins (IP set). Finally, the CP and IP sequences exhibiting some in-frame stop codons (that could represent pseudogenes, among other features; Ψ) were incorporated into two extra data sets (CPΨ and IPΨ sets, respectively).
We used three different estimators of the number of copies of a particular CS family (family size). In addition to the straightforward number of CPs in a particular genome (SCP), we also determined the minimum number of sequences that could be unequivocally attributed to different functional genes (SMIN) and the maximum number of members in cases where all the incomplete protein fragments were actually different functional genes (SMAX). We estimated these numbers by aligning all protein sequences (both CP and IP) within a family using the CS-F-HMM profile as a guide and examining the matching distribution of all fragments aligned along the protein. The SMIN was obtained by adding to the total number of sequences present in the CP set, the minimum number of sequences of the IP set that could be unequivocally attributed to different family members. This minimum amount was determined by counting the number of partial sequences aligned in the most covered protein region of the CS-F-HMM profile-guided MSA. The SMAX is the total number of both CP and IP copies identified (supplementary table S1B and C, Supplementary Material online).
Phylogenetic Analyses
As the divergence between some members of the same CS family is huge (i.e., their most recent common ancestor traces back far before the split of the major arthropod lineages, ∼600 Ma; Hedges et al. 2006), building a reliable MSA to estimate the phylogenetic relationships is not straightforward. To address this long-standing problem, we applied the MSA-free HMM distance-based method (Bogusz and Whelan 2017) implemented in the PaHMM-Tree software, which outperforms MSA-based methods when dealing with the high alignment uncertainty that is usually associated with large divergences. All the phylogenies except those of the IR family (see Results for more details about this family) were based on complete sequences. We used the iTOL web server (Letunic and Bork 2007) to format and display the trees.
Gene Turnover Rates
We estimated the gene family turnover rates using a gene tree–species tree reconciliation approach. The ultrametric species tree required for the analysis was inferred by fitting the amino acid variation of all 88 putative single-copy orthologs to the most accepted topology for the 11 species. For the analysis, we used OrthoMCL (v2.0.9; Li et al. 2003) to identify 1:1 orthologs by clustering the sequences by similarity and then generated an MSA (for each ortholog group) with T-Coffee v11.00 (mcoffe mode; Notredame et al. 2000). After filtering the MSAs with trimAl v1.4 (-automated1 option; Capella-Gutiérrez et al. 2009), we estimated the best-fit amino acid substitution model for each MSA with the program jModelTest based on the Akaike information criteria for model selection (Guindon and Gascuel 2003; Darriba et al. 2012) and concatenated all MSA, keeping the individual coordinate information to be used as a partition for the phylogenetic analysis. We used RAxML software (option –f e) to obtain ML estimates of branch lengths and r8s software v 1.80 (Sanderson 2003) to linearize the unrooted ML using the penalized likelihood algorithm. For the last step, we constrained the ages of two internal nodes according to the fossil calibrations: 1) the root (on the range 528–445 Myr; Dunlop and Selden 2009) and 2) the split between scorpions and spiders (at a minimum of 428 Myr; Jeyaprakash and Hoy 2009).
We analyzed the family turnover rates for the two largest gene families in Arachnida, Gr and Ir/iGluR, using a gene tree–species tree reconciliation approach. For each family and lineage, we estimated separately the birth (β) and death (δ) rates, which measure the number of sequence gains and losses per sequence per million years, respectively. For the global analysis, we estimated the average values across all branches, excluding Li. polyphemus, which was used to root the tree. We used the software OrthoFinder (Emms and Kelly 2015) to obtain orthogroups (i.e., all groups of N: N orthologs) and gene trees to calculate the number of gene gain and loss events in each lineage with the program Notung (Chen et al. 2000). Finally, we estimated the global turnover rates (β and δ) from these events using formulas 1 and 2 in Almeida et al. (2014), whereas the net turnover rates (Δ) were directly estimated as Δ = β − δ.
Results
The Chemosensory Subgenome of Chelicerates
Our comprehensive search protocol revealed 6,026 CS protein-coding sequences across the 11 surveyed chelicerate genomes (supplementary table S1A, Supplementary Material online). Surprisingly, nearly 85% of them (5,086) had previously inaccurate genome annotations, including 4,131 nonannotated sequences (without a GFF record) and another 955 that, despite having structural annotation data in the GFF file, lacked functional information (as putative CS proteins) in the GFF field. Nevertheless, only 2,646 of the 6,026 sequences (supplementary table S1B, Supplementary Material online) encoded complete (or nearly complete) CS proteins free of stop codons (CP set). Among the remaining sequences, 1,895 were incomplete (but without stop codons in frame) (IP set) and 1,485 showed one or more premature stop codons (including both CP and IP sequences). Globally, the actual number of putative functional CS genes ranged from 4,255 (SMIN) to 4,541 (SMAX), although only 2,646 of them were complete (SCP) (supplementary table S1C, Supplementary Material online). Remarkably, although canonical insect Obp and Or genes were absent in chelicerate genomes, we found a huge and unexpected number of novel Gr-coding (108 uncharacterized peptides plus 3,331 novel genomic sequences) and Ir/iGluR-coding (525 plus 694) sequences. Furthermore, it is noteworthy that Csp members were absent in all genomes, except in the tick I. scapularis, and Ccp family members were identified only in spiders and scorpions (fig. 1).
Chemoreceptors
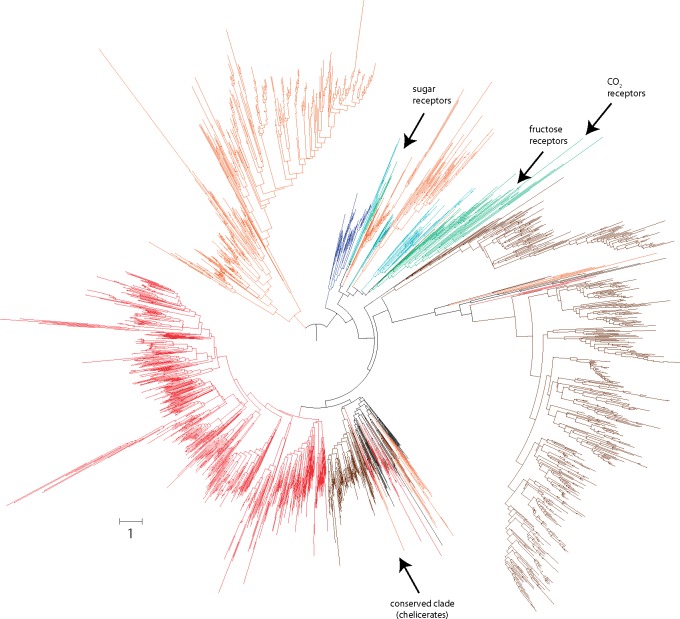
We found that the Gr family is the largest CS gene family in chelicerates (SMIN = 3,074, SMAX = 3,157, and SCP = 2,032, considering only putative functional sequences; fig. 1; supplementary table S1B, Supplementary Material online). Moreover, we also identified 1,097 putative Gr pseudogenes (see Discussion). Remarkably, there are extraordinary differences in the family size across chelicerates; although some species exhibit >400 copies, such as the scorpion C. exilicauda (SMIN = 832), the tick T. urticae (SMIN = 469) or the spider P. tepidariorum (SMIN = 643), others have <60, such as I. scapularis (SMIN = 57) and Li. polyphemus (SMIN = 58) (supplementary table S1C, Supplementary Material online). These results cannot be explained by putative differences in the assembly quality across genomes because the same trend was observed with SMAX and SCP values. In fact, there is no relationship between the values of our three estimates of the real number of Gr genes across genomes and the N50, the number of scaffolds or the number of predicted peptides in these genomes (supplementary table S1C, Supplementary Material online). Strikingly, even the most closely related species, the spiders La. hesperus and P. tepidariorum, greatly differ in their repertory size (fig. 1), revealing a highly dynamic evolution. These differences are clearly shown in the phylogenetic tree as large monophyletic groups (mostly species-specific clades). Despite these findings, the tree also reveals a distinctive monophyletic group of apparently less dynamic sequences with representatives from all chelicerates (fig. 3; supplementary fig. S1, Supplementary Material online). However, we did not detect any GR protein closely related to the functionally characterized carbon dioxide, sweet taste, and fructose insect receptors in chelicerates (Jones et al. 2007; Miyamoto et al. 2012; Fujii et al. 2015).
Fig. 3.—
Phylogenetic tree of the Gr family members across arthropods. The different species are depicted in colors as in figure 1. The scale bar represents one amino acid substitutions per site.
The Ir/iGluR is the second largest CS family (SCP = 323, SMIN = 825, and SMAX = 979). Again, but less pronounced than in the Gr family, we also detected a highly uneven distribution of copies across lineages. Interestingly, the repertory sizes of these two families do not correlate across chelicerates (Pearson correlation, P-value > 0.05); for instance, T. urticae encodes very few Ir/iGluR copies (SMIN = 19) but a large number of Gr genes (SMIN = 469). Similar to the Gr family, the relationship of the Ir/iGluR family size across species is very similar regardless of the use of SCP, SMIN, or SMAX values, suggesting that the assembly quality has no influence.
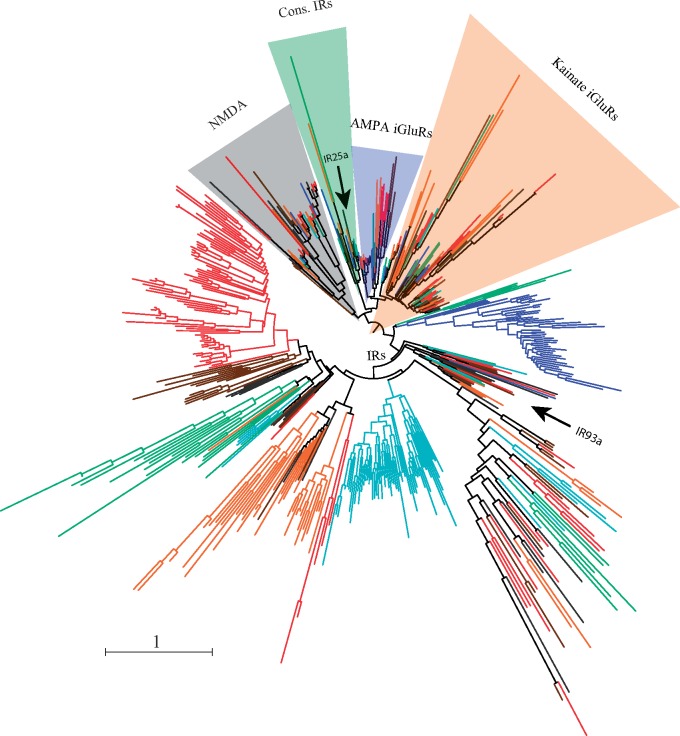
The phylogenetic analysis using sequences with the complete ligand channel domain reproduced the established relationships of the five major arthropod Ir/iGluR subfamilies (fig. 4; supplementary fig. S2, Supplementary Material online; Croset et al. 2010; Vizueta et al. 2017). The gene topology allowed us to identify 249 IR proteins (or truthful IR set, t-IR) (200 with the two ligand-binding domains plus another 49 with only the ligand channel domain; supplementary table S1C, Supplementary Material online), which would represent the minimum number of functional IR copy candidates to perform a chemosensory function. The phylogenetic analysis also revealed the absence of members of the Ir25a/Ir8a-conserved IR subfamily in M. martensii, S. mimosarum, A. geniculata, and La. hesperus. However, a more comprehensive analysis of the IP set revealed that, in fact, all these species encode one IR25a receptor (supplementary table S2 and fig. S3, Supplementary Material online). Interestingly, we failed to detect any putative homologs of IR8a in all chelicerates, except in the horseshoe crab Li. polyphemus (LpolIR11 sequence). Still, we could detect putative homologs of two Drosophila antennal IRs, IR93a and IR76b. The first member was identified in all species, excluding A. geniculata and S. mimosarum, whereas IR76b was present in Daphnia, the horseshoe crab, the two scorpions and the spiders P. tepidariorum and La. hesperus (supplementary table S2 and fig. S3, Supplementary Material online). Nonetheless, we did not find putative homologs of the other Drosophila antennal IRs with orthologous copies in insects, such as IR21a and IR40a (Croset et al. 2010; Eyun et al. 2017).
Fig. 4.—
Phylogenetic tree of the Ir/iGluR family members across arthropods. The tree is based on LCD domain sequences (PF00060). Different lineages are colored as in figure 1. The three main subfamilies of iGluRs and the conserved IR clade are shaded in different colors. The scale bar represents one amino acid substitution per site.
Other Chemosensory Families
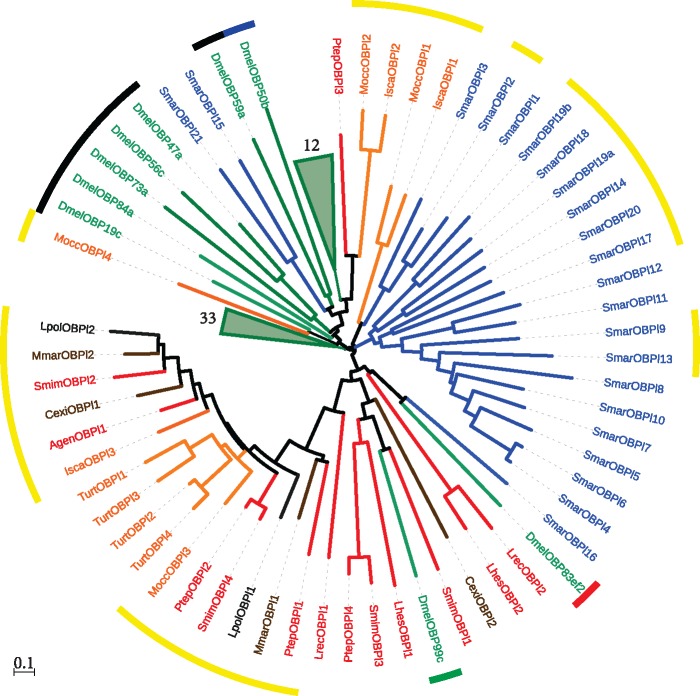
We identified several novel and complete OBP-like encoding sequences in chelicerates (fig. 1; supplementary table S1A, Supplementary Material online). In addition to the described members in I. scapularis, M. occidentalis, S. mimosarum, and S. maritima (Renthal et al. 2017; Vizueta et al. 2017), we identified a total of 26 new (out of 30) OBP-like proteins in chelicerates. All the chelicerates encode at least one member of this family, with repertory sizes ranging from 1 to 4 copies. Additionally, and very surprisingly, we detected 19 novel (out a total of 21) Obp-like genes in the centipede S. maritima. Our phylogenetic analysis of canonical OBP (from insects) and OBP-like proteins (fig. 5, supplementary fig. S4, Supplementary Material online) does not support the reciprocal monophyly of these two gene families. Although some OBP-like sequences (such as MoccOPBl2, IscaOBPl2 and PtepOBPl3) are phylogenetically close to the OBP Plus-C subfamily, others, for example, DmelOBP99c (a member of the insect minus-C subfamily), are more related to the chelicerate OBP-like sequences than to the insect OBP sequences. Moreover, the phylogenetic analysis revealed three major clades, each almost exclusively containing sequences of the given arthropod subphylum (i.e., D. melanogaster, S. maritima, and chelicerates).
Fig. 5.—
Phylogenetic relationships of the Obp-like and insect (D. melanogaster) Obp family members. Lineages and species names are colored as in figure 1. For clarity, two D. melanogaster nodes with 12 and 33 descent sequences are collapsed. The color of the inner circle indicates the Obp subfamily: Classic (black), Minus-C (green), Plus-C (blue) and Dimer (red). The outer circle in yellow indicates the members from noninsect species with PBP/GOBP domain (IPR006170). The scale bar represents 0.1 amino acid substitutions per site.
The size of the Npc2 family has remained relatively constant during the diversification of the major chelicerate lineages, ranging from 10 to 20 (SMIN values, supplementary table S1C, Supplementary Material online), with the outstanding exception of T. urticae, which encodes 47 genes. Nevertheless, nearly half of the Npc2 members of some species are incomplete fragments or show premature stop codons, resulting in much greater difficulty in drawing a firm conclusion about the real sizes of this family compared with the other families. In this case, we found a strong positive correlation between N50 and SCP, SMIN, and SMAX values (Pearson correlation coefficient, r > 0.80; P < 0.05; supplementary table S1C, Supplementary Material online), indicating that the observed variation in the number of Npc2 genes across species is clearly associated with genome assembly continuity. This result is probably due to the fact that the length of the genomic region that includes the target sequences of the similarity searches is the longest (jointly with the Cd36-Snmp family, see below) among the families surveyed in this work. Unlike chemoreceptors and Obp-like members, NPC2 proteins are not arranged in large species-specific phylogenetic clades (supplementary fig. S5, Supplementary Material online), suggesting a less dynamic evolution of this family compared with chemoreceptors and OBP-like proteins.
Our searches for members of the recently discovered Ccp gene family (Vizueta et al. 2017) only provided positive results in spiders and in Centruroides exilicauda (the Bark scorpion), although the sequence identity of the copy detected in this last species is low. We found important differences in family size across species, from 2 in Lo. reclusa to 21 in P. tepidariourum (SMIN). Like in D. silvatica, most CCPs exhibited an identifiable signal peptide sequence and a conserved cysteine pattern, supporting their putative role in the extracellular binding and transport of chemical cues (Vizueta et al. 2017). The phylogenetic analysis of this family revealed relatively short branches and clades likely representing orthologous genes (supplementary fig. S6, Supplementary Material online). Even so, the 21 copies of P. tepidariorum (11 of them forming a species-specific clade) is a remarkable exception and could be associated with an adaptive event linked to this family in this lineage. The high-quality assembly and annotation of the P. tepidariorum genome may be good enough to have a closer look at the genomic location of Cpp genes and to search in this family for signatures of the lineage-specific bursts of tandem duplications stated by Schwager et al. (2017).
The Cd36-Snmp Family
The Cd36-Snmp family size has also remained relatively constant during the diversification of chelicerates, especially with respect to the SMAX values (ranging from 8 to 19). Nevertheless, as in the Npc2 family, nearly half of the positive hits encode incomplete proteins, most of which are in spiders and scorpions (supplementary table S1B, Supplementary Material online). Consistent with the large size of the target genomic regions of this family, we also found a positive correlation between N50 and SCP and SMIN (but not SMAX) values for this family (Pearson correlation coefficient, r > 0.56; P < 0.05; supplementary table S1C, Supplementary Material online), although weaker than in the case of NPC2. The phylogenetic analysis (supplementary fig. S7, Supplementary Material online) showed that only one of three phylogenetic clades described by Nichols and Vogt (2008) has remained monophyletic across all arthropods (i.e., the group including the SNMP protein of D. melanogaster). However, many sequences do not form monophyletic groups and, therefore, cannot be unambiguously assigned to a given subfamily group, suggesting a more complex grouping than those observed in insects (Nichols and Vogt 2008).
Gene Turnover Rates of Chemoreceptors
We estimated gene family turnover rates for the two largest Chelicerata gene families, Gr and Ir/iGluR, using Li. polyphemus to root the tree (fig. 6, supplementary fig. S8, Supplementary Material online). As the analysis could have been compromised by the use of three different estimates of family size (per CS family), we first evaluated the behavior of these size estimates with respect to the turnover rates. We found that the number of gene duplications and losses calculated using SCP (only for the Gr family), SMIN, and SMAX values strongly correlated across lineages (r > 0.94; P-values < 10−5); therefore, we did not expect important relative rate differences among the three estimates. Thus, we calculated birth and death rates only with SMIN because this estimate likely represented the true number of copies in most genomes.
Fig. 6.—
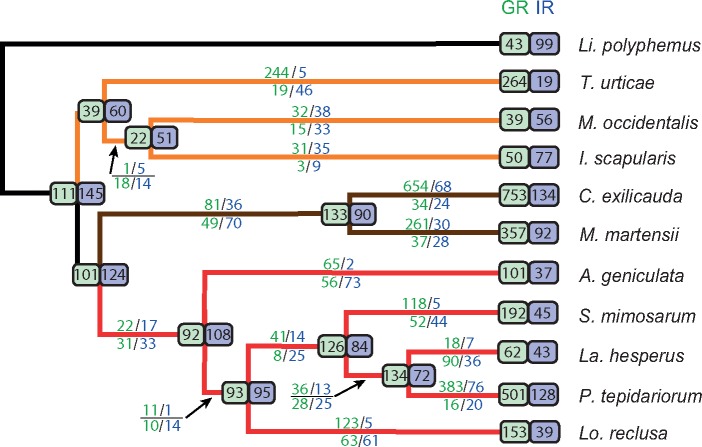
Gene turnover of chemoreceptors across chelicerates. Estimates obtained from the data set used to estimate SMIN. Numbers above and below each branch indicate lineage-specific gene duplications and losses, respectively. Green, GR family; blue, IR/iGluR family. Estimates in very short and outgroup branches have large uncertainty and are not showed. Numbers in the ancestral nodes show the estimated family sizes. Numbers at the tips indicate the number of sequences used for the analysis; such values can differ from SMIN because only sequences that clustered in an orthogroup (with three or more sequences) were included in the analysis.
We found that the global (across all phylogenetic tree) gene turnover rates of Gr and Ir/iGluR showed important differences (supplementary fig. S8, Supplementary Material online). In Gr, the net turnover rates were positive (Δ = 0.003), indicating an overall expansion of gustatory receptor repertory during arachnid diversification. In contrast, the Ir/iGluR family showed an overall contraction (Δ = −0.002). These results should be considered with caution because global turnover rates are strongly affected by the presence of specific phylogenetic branches with extreme values. In the Gr family, for instance, the external lineages leading to T. urticae (β = 0.015), C. exilicauda (β = 0.030), and P. tepidariorum (β = 0.030) have β values that are much higher than the global rates (β = 0.007); in contrast, other branches, such as the internal lineage leading to acari (δ = 0.008) and the external lineage leading to La. hesperus (δ = 0.007), show death rates that clearly exceed global estimates (δ = 0.004).
The Ir/iGluR family exhibits smaller turnover rate differences among the lineages than those observed for Gr. Even so, the external branches of C. exilicauda (β = 0.005), and especially of P. tepidariorum (β = 0.011), are clear outliers and the only ones that show a clear expansion of the Ir/iGluR repertory during the diversification of arachnids. It should be noted that the Ir/iGluR data set includes the sequences of five subfamilies of this highly functional, diverse family of receptors, which show very dissimilar turnover rates in insects. In fact, the Ir subfamily, which is the only subfamily encoding putative chemosensory receptors, is the most dynamic family of insects. Therefore, to disentangle subfamily-specific effects, we estimated the gene turnover rates using only the IR copies from SMIN and the t-IR set (fig. 4). As expected, birth and death rates estimated from the SMIN and t-IR sets did not show big differences (results not shown), suggesting a major effect of the Ir subfamily on gene turnover estimates in the Ir/iGluR family. Indeed, the t-IR estimates were even more variable across lineages than those obtained for the whole family, especially for birth rates, with slightly higher average rates. Especially noteworthy is the case of the P. tepidariorum lineage, which not only confirmed the findings of the SMIN set analysis but also showed that the gene number expansion (supplementary fig. S2, Supplementary Material online) was definitively caused by the birth of new Ir genes (t-Ir set based estimates, β = 0.020, δ = 4 x 10−4).
Discussion
The early diversification of arthropods predated the colonization of land by animals (Rota-Stabelli et al. 2013). Chemical communication strategies associated with this terrestrialization, therefore, should have been invented several times independently in their major lineages (Hexapoda, Crustacea, Myriapoda, and Chelicerata). It is likely that proteins involved in the first peripheral chemosensory perception steps, which are commonly associated with medium-size gene families, played a central role. Hence, these gene families represent an important fraction of arthropod genomes and contribute significantly to gene turnover dynamics in insects (Sánchez-Gracia et al. 2009, 2011). The recent availability of the complete genome sequences from various chelicerates has provided insights into their CS family members. Nevertheless, the quality of the genome assembly and functional annotation is far from satisfactory. Some genomes are highly fragmented, with an absence of functional annotations or annotations obtained using only nonexhaustive automated protocols. Here, we report the first comparative analysis of the actual copy number and gene turnover evolution of CS families in 11 nonhexapod genomes. This study is in fact the first comprehensive comparative genomics study that, although enriched in Arachnida species, covers most of the chelicerate diversity (see Eyun et al. [2017], Palmer and Jiggins [2015], and Sanggaard et al. [2014] for examples of previous studies based on many fewer genomes).
The Outstanding Chemoreceptor Repertory of Chelicerata Genomes
The most important challenge for understanding gene family evolution is having well-characterized copies and accurate functional annotations of their members. This is particularly relevant when using highly fragmented genome assemblies generated from short-read sequencing data. To circumvent this problem, we applied a very comprehensive identification and characterization protocol that combined both protein and DNA sequence data, including HMM profiles and protein domain signatures, in a series of sequential searches with accurate filters based on our biological knowledge of the CS system. Our study revealed a surprisingly large number of novel Gr- and Ir-encoding sequences. This feature can be mostly explained by the poor functional annotation status of some genomes. In fact, in those genomes in which CS families had been explicitly characterized (the three acari species, D. melanogaster, D. pulex, and S. marítima), our search protocol largely matched with previously annotations. This characteristic, therefore, indicated that the novel CS-encoding sequences were not false positives caused by a misleading search protocol.
We also found that some of the newly identified CS genes were highly fragmented, which is also a consequence of the low quality of assemblies and, therefore, of the poor annotation of gene structures in most surveyed genomes. Most genes are distributed across many different scaffolds, preventing the calculation of the exact number of functional copies in a particular genome. This feature led us to define three repertory size statistics, which not only provided an approximate idea of true values but also allowed for harmonized comparisons across genomes and lineages. As expected, the largest discrepancy occurred between size estimates based on complete genes (SCP) and those including information of incomplete gene fragments (SMIN and SMAX). Despite this difference, however, all three data sets yielded very similar estimates of gene turnover rates; therefore, all of them are good approximations of true CS family sizes and are appropriated to study gene family dynamics across chelicerates. Although SMIN and SMAX values were generally similar, two families showed very important discrepancies: Ir/iGluR and Cd36-Snmp. These discrepancies could be explained by the fact that these genes (and the encoding region including introns) are larger than in the other families, and therefore, it is more likely that the encoding region was fragmented in different scaffolds. In fact, this effect was not observed in genomes with more contiguity (based on the N50 values of the genome assemblies), as observed in T. urticae, M. occidentalis, S. mimosarum, and P. tepidariorum. Finally, we also found numerous sequences with in-frame stop codons, which we have preliminarily classified as putative pseudogenes. It should be taken into account that not all sequences with evidence of stop codons must be nonfunctional copies; indeed, some of these stop codons may be introduced during gene assembly from dispersed TBlastN hits (which has been done in a semiautomatic way). Only with the use of additional, high-quality assembled genomes will it be possible to obtain accurate information concerning the nature and number of these putative pseudogenes.
CS Gene Turnover in Chelicerates: Complex Evolutionary Dynamics
We have shown that although chelicerate have larger Gr gene repertories than nonchelicerates, the estimated birth and death rates for the Gr family are almost the same as those in insects (Almeida et al. 2014). The disparate family sizes might be explained by former differences in the ancestors of each of these two lineages. In fact, at least two ancient and independent whole-genome duplications (WGD) have been proposed for chelicerates, one in the ancestor of spiders and scorpions (∼450 Ma; Schwager et al. 2017), and the other likely occurred in the lineage of horseshoe crabs (Kenny et al. 2016; Schwager et al. 2017). Thus, it is tempting to hypothesize that evolutionary forces and genomic mechanisms underlying the long-term birth and death dynamics of chemosensory families were essentially the same in all arthropods, although eventually promoted by lineage-specific genome-scale events such as WGD. Nevertheless, not all of our results are compatible with such an evolutionary scenario. For instance, the results obtained for the Ir subfamily do not agree with those observed for Gr. The birth and death rates of these putative chemoreceptors differ between chelicerates and nonchelicerates, and they do not show the footprint of the WGD preceding the diversification of spiders and scorpions. In fact, net turnover rate of this family has the opposite pattern as GRs, suggesting an important contraction of ionotropic receptors in chelicerates.
Furthermore, the occurrence of WGD events could not satisfactorily explain the full evolutionary history of most of the surveyed families, not even for the Gr family. For instance, T. urticae shows very high GR repertoires (SMIN = 469) and a very low IR (SMIN = 6) compared with the other acari, and this pattern is unequivocally not explained by the use of a particular family size SMIN statistic (the three estimators point to the same feature). Although we cannot completely rule out the possibility of a WGD in this lineage, there is no compiled evidence in support of this phenomenon (Grbić et al. 2011; Kenny et al. 2016). Second, the closest phylogenetic lineages in our study (La. hesperus and P. tepidariorum, with the most recent common ancestor tracing back approximately 100 Ma) show enormous differences in Gr and Ccp family sizes. Finally, estimation of the turnover rates in a pair of phylogenetically close species (C. exilicauda and M. martensii; La. hesperus and P. tepidariorum) is difficult to reconcile with a constant birth and death process. Therefore, the evolutionary process was rather complex and cannot be entirely explained by WGD. Here, we have demonstrated that other processes affecting specifically chemosensory families, such as long-term birth-and-death evolution associated with high turnover rates occurred in parallel to these whole genomic changes. In addition, more episodic, and probably lineage-specific, expansions and/or contractions also contributed to determine current sizes, as suggested in other studies (Chipman et al. 2014; Schwager et al. 2017). In order to know the relative role of these different processes in shaping actual CS family sizes and their functional meaning, it is imperative to improve the quality of existing genomes and include in the analysis new, more closely related genomes (i.e., increase the phylogenetic coverage).
Phylogenetic Analysis of CS Genes in Arthropods
Despite the above-mentioned limitations, our phylogenetic analysis can shed light on the diversification pattern of CS families. As arthropod CS families are very old and many of their members, especially chemoreceptors, are distantly related, the use of the standard MSA alignment method could be inappropriate for building robust phylogenies. A common method to circumvent this problem is filtering poorly aligned positions and, therefore, considering only highly conserved sites for phylogenetic analyses (Croset et al. 2010; Wu et al. 2016). This approach nevertheless results in a significant loss of relevant amino acid positions that likely contain valuable information on functional and structural features related to the molecular specificity and diversification. Here, we used, for the first time in highly divergent CS families, a method to estimate gene trees using an MSA-free approach, which takes into account alignment uncertainty. For the sake of comparison, we reconstructed the same phylogenetic trees using RAxML based on HMM profile-guided MSAs (Stamatakis 2014: Supplementary file 4). Major differences between PaHMM-Tree and RAxML were found at internal nodes and nodes with low bootstrap support in ML trees (< 70% from 500 replicates). Although bootstrap values increased when filtering poorly aligned positions (Capella-Gutiérrez et al. 2009), the number of informative sites retained after removing these unreliable positions was very low, causing the ML trees to be based on a very small number of positions. These trees may not be reflecting the real evolutionary history of the chemosensory proteins. Besides, for very large families, such as the Gr, the bootstrap analysis was unfeasible in the practice due to excessive computation times. Given that PaHMM-Tree is an alignment-free approach, which allow us to utilize all the amino acid positions to reconstruct the trees, and that the results obtained by Bogusz and Whelan (2017) point to a better performance of this approach for highly divergent sequences without the need for a previous filtering step, here, we decided to report the results based on this method. However, a more exhaustive study comparing these and other tree reconstruction methods, using both real and simulated data and under different degrees of divergence, would be necessary to know whether this method actually improves the phylogenetic analysis. Our phylogenetic analysis correctly recovered all previously known (and accepted) relationships among subfamilies and revealed new aspects of the diversification of CS genes.
We found that chelicerates virtually have their own GR repertoires with almost no phylogenetic clade containing members of insects, crustaceans, and myriapods. In fact, we did not find homologs of any of the GR functionally characterized in insects. Apparently, chelicerate genomes do not encode any protein sequence close to Drosophila sugar, fructose, or carbon dioxide receptors (Jones et al. 2007; Miyamoto et al. 2012; Fujii et al. 2015), questioning their ability to detect these substances. Nevertheless, chelicerates might be using other phylogenetically distant gustatory receptors to perform these tasks. Yet, the presence of a monophyletic clade with more conserved GR chelicerate sequences would suggest the existence of some other important biological function played by these receptors. The members of this clade could have a highly relevant function in chelicerates, evolving under lower evolutionary rates despite the tremendous diversification of this subphylum. Future functional studies combined with new evidence based on greater coverage phylogenetic analysis will definitely shed light on this interesting hypothesis.
Another remarkable result is the verification that most GR receptors found in species with very large repertories such as in P. tepidariorum or in C. exilicauda are monophyletic, pointing to important bursts of gene duplication events in relatively recent time periods. These events probably represent adaptive expansions of the gustatory repertory associated with chemosensory diversifications. In other cases, such as in T. urticae lineage, apparent species-specific family expansions might be just an artefact caused by the continued effect of the birth-and-death process in a very long terminal branch (i.e., reflecting the low phylogenetic coverage of this part of the tree).
Although the general phylogenetic pattern observed in the IR is very similar to that of the GR, we detected some Ir members with relatively conserved sequences across all arthropods. We can hypothesize that these receptors should have a very relevant and not easily replaceable function. For instance, IR25a, a receptor found in all arthropods surveyed to date, is a broadly expressed protein involved in trafficking to the membrane of other IRs in olfactory and taste organs that has been proposed to have also a coreceptor function in the membrane (Joseph and Carlson 2015). We also found a putative ortholog of IR8a in the horseshoe crab Li. polyphemus, which led us to reformulate the hypothesis of Eyun et al. (2017) suggesting that this member arose in the ancestor of myriapods and pancrustaceans, tracing back its origin, again, to at least the ancestor of arthropods.
Our analysis also supports the presence of a group of IR76b homologs outside the insect clade (Eyun et al. 2017) which was likely present in the arthropod ancestor. This receptor, proposed to play a coreceptor function for other IRs and associated with a gustatory function as a detector of low salt concentrations (Zhang et al. 2013), has been identified in all chelicerates except in the acari and some spider clades. Its absence in these arthropod groups suggests a secondary loss in the ancestor of these lineages. However, we could not fully refute the possibility that we were unable to detect this member in these genomes, especially in spiders, because of assembly fragmentation. Our current phylogenetic analysis failed to detect putative homologs of IR21a and IR40a in chelicerates. Though we found some week evidence for homologs of these receptors in the transcriptome of the spider D. silvatica (Vizueta et al. 2017), we rely more in the analysis applied herein, which is most comprehensive and uses an alignment-free method based on HMM profiles to generate the trees. These new evidences, together with previous genomic analyses, would indicate the presence of IR21a exclusively in panarthropods (Eyun et al. [2017] have recently found a putative homolog of the IR21a protein in copepods) and of IR40a exclusively in insects.
Notably, our study shows that all chelicerates and the centipede S. maritima carry members of the Obp-like family, a gene family that is closely related to insect OBPs (Renthal et al. 2017; Vizueta et al. 2017). This family, which is absent in crustaceans, might represent a remote homolog of canonical insect OBPs. The close relationship of a Drosophila minus-C OBP within an OBP-like chelicerates clade, in agreement with the results of Renthal et al. (2017) based on the disulfide bonding pattern, suggests that this subfamily represents an ancestral state of an OBP. Nonetheless, we cannot completely ignore the possibility that the similar sequence arose by structural convergence. As a canonical OBP, OBP-like has a signal peptide region, a predicted globular protein with the characteristic cysteine patterns of OBPs, and predicted folding similar to that of insect OBPs. Moreover, some experimental results have also confirmed the expression of some Obp-like members in specific chelicerates chemosensory appendages (Renthal et al. 2017). All compiled evidence, therefore, suggests that chelicerates and myriapod OBP-like may have a similar function to canonical OBPs, such as in solubilizing and transporting chemical cues. Regardless, the extraordinarily large repertory observed in S. maritima clearly merits further investigation. This is especially interesting because the genome paper of S. marticima reported a high number of tandem duplications (Chipman et al. 2014).
Intriguingly, we did not find CSP-encoding genes in the surveyed chelicerates, except the single copy found in the tick I. scapularis (Gulia-Nuss et al. 2016). Although Eyun et al. (2017) reported some sequences encoding CSP proteins in the bark scorpion C. exilicauda and the spider La. hesperus, our analysis of such sequences could not unequivocally establish that they encode real CSP proteins; indeed, these sequences are very short with multiple in frame stop codons and do not exhibit the characteristic cysteine CSP pattern, suggesting a false positive result. Our analysis also allowed us to identify members of the Ccp gene family in spiders, as well as a remote homolog in the bark scorpion C. exilicauda, suggesting that the origin of this rapidly evolving gene family traces back to the ancestor of these two groups. Remarkably, we observed a large expansion of some members (a lineage-specific expansion) in the house spider P. tepidariorum, a feature that reflects its greater number of chemoreceptors. We have established that the CCP-encoding genes have a signal peptide fragment and similar folding characteristics to the insect OBP and are differentially expressed in the putative chemosensory appendices of the spider D. silvatica (Vizueta et al. 2017). Therefore, although their actual function is unknown, it is tempting to assign a putative function to the transport and solubilization of chemical cues, a functional role equivalent to that of the canonical OBP, Nevertheless, given that the Ccp is a rapidly evolving gene family that emerged in some derived chelicerate lineages, it could provide new insights into the extracellular-binding protein functions and their roles in diversification and adaptation in arthropods.
Conclusions
Noninsect arthropods comprise a significant portion of earth’s biodiversity and include many species of economic and medical importance. Here, we conducted the first comprehensive comparative genomic analysis across 11 genomes of this old lineage and the first of this magnitude outside of insects. Despite that the high fragmentation of genome drafts prevented us from establishing the exact number of chemosensory genes in each species, our exhaustive search protocol exposed an unprecedented huge number of new family members. Remarkably, many of these new genes were not characterized or even not detected before and most of them encode chemoreceptors. Moreover, we found a remarkable disparity in chemoreceptor repertories across species that is difficult to explain without invoking lineage-specific adaptive expansions probably related with sensory diversification processes. Characterizing the intragenomic dynamics and the specific function of these recently expanded chemosensory genes is an exciting prospect that jointly with the improvement of existing genome assemblies and the reduction of the phylogenetic gap will allow researchers to move forward in the knowledge of chelicerate genomics and biology. This work aims to contribute to this advance and hopes to be the starting signal for many future comprehensive comparative genomic studies in a group of animals as fascinating as unknown.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Ministerio de Economía y Competitividad of Spain (CGL2013-45211 and CGL2016-75255) and the Comissió Interdepartamental de Recerca I Innovació Tecnològica of Catalonia, Spain (2014SGR-1055). J.V. was supported by an FPI grant (Ministerio de Economía y Competitividad of Spain, BES-2014-068437) and J.R was partially supported by ICREA Academia (Generalitat de Catalunya). The authors declare that they have no competing interests.
Author Contributions
A.S.-G. and J.R. conceived and designed the study. J.V. analyzed the data. J.V., J.R. and A.S.-G. wrote the manuscript.
Literature Cited
- Adams MD, et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Almeida FC, Sánchez-Gracia A, Campos JL, Rozas J.. 2014. Family size evolution in Drosophila chemosensory gene families: a comparative analysis with a critical appraisal of methods. Genome Biol Evol. 6(7):1669–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb PL, et al. , 2017. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat Genet. 49(6):895–903. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB.. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136(1):149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB.. 2007. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450(7167):289–293. [DOI] [PubMed] [Google Scholar]
- Berna AZ, Anderson AR, Trowell SC.. 2009. Bio-benchmarking of electronic nose sensors. PLoS One 4(7):e6406.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogusz M, Whelan S.. 2017. Phylogenetic tree estimation with and without alignment: new distance methods and benchmarking. Syst Biol. 66(2):218–231. [DOI] [PubMed] [Google Scholar]
- Cao Z, et al. , 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun. 4:2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M.. 2000. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 7(3–4):429–447. [DOI] [PubMed] [Google Scholar]
- Chipman AD, et al. , 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12(11):e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, et al. , 2011. The ecoresponsive genome of Daphnia pulex. Science 331(6017):555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey EA, Bobkov Y, Ukhanov K, Ache BW.. 2013. Ionotropic crustacean olfactory receptors. PLoS One 8(4):e60551.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, et al. , 2010. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6(8):e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani FR, et al. , 2011. Odorant-binding proteins and chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori. Chem Senses. 36(4):335–344. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Baker TC.. 2008. Odor detection in insects: volatile codes. J Chem Ecol. 34(7):882–897. [DOI] [PubMed] [Google Scholar]
- Dessimoz C, et al. , 2011. Comparative genomics approach to detecting split-coding regions in a low-coverage genome: lessons from the chimaera Callorhinchus milii (Holocephali, Chondrichthyes). Brief Bioinform. 12(5):474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippel S, et al. , 2014. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 15(1):1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop JA, Selden PA.. 2009. Calibrating the chelicerate clock: a paleontological reply to Jeyaprakash and Hoy. Exp Appl Acarol. 48(3):183–197. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliash N, et al. , 2017. Chemosensing of honeybee parasite, Varroa destructor: transcriptomic analysis. Sci Rep. 7(1):13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyun S, et al. , 2017. Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol Biol Evol. 34(8):1838–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forêt S, Wanner KW, Maleszka R.. 2007. Chemosensory proteins in the honey bee: insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol. 37(1):19–28. [DOI] [PubMed] [Google Scholar]
- Frías-López C, et al. , 2015. Comparative analysis of tissue-specific transcriptomes in the funnel-web spider Macrothele calpeiana (Araneae, Hexathelidae). PeerJ 3:e1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, et al. , 2015. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol. 25(5):621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau KL, et al. , 2017. House spider genome uncovers evolutionary shifts in the diversity and expression of black widow venom proteins associated with extreme toxicity. BMC Genomics 18(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Diaz C, et al. , 2016. A CD36 ectodomain mediates insect pheromone detection via a putative tunnelling mechanism. Nat Commun. 7:11866.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, et al. , 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479(7374):487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh-Lunow KC, Getahun MN, Grosse-Wilde E, Hansson BS.. 2015. Expression of ionotropic receptors in terrestrial hermit crab’s olfactory sensory neurons. Front Cell Neurosci. 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Gulia-Nuss M, et al. , 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 7:10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, et al. , 2013. The venom gland transcriptome of Latrodectus tredecimguttatus revealed by deep sequencing and cDNA library analysis. PLoS One 8(11):e81357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S.. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22(23):2971–2972. [DOI] [PubMed] [Google Scholar]
- Hoy MA, et al. , 2016. Genome sequencing of the phytoseiid predatory mite Metaseiulus occidentalis reveals completely atomized hox genes and superdynamic intron evolution. Genome Biol Evol. 8(6):1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovinella I, et al. , 2011. Differential expression of odorant-binding proteins in the mandibular glands of the honey bee according to caste and age. J Proteome Res. 10(8):3439–3449. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Ishibashi J, Leal WS.. 2013. Fatty acid solubilizer from the oral disk of the blowfly. PLoS One 8(1):e51779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA.. 2009. First divergence time estimate of spiders, scorpions, mites and ticks (subphylum: Chelicerata) inferred from mitochondrial phylogeny. Exp Appl Acarol. 47(1):1–18. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB.. 2007. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445(7123):86–90. [DOI] [PubMed] [Google Scholar]
- Joseph RM, Carlson JR.. 2015. Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet. 31(12):683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL.. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 338(5):1027–1036. [DOI] [PubMed] [Google Scholar]
- Kaupp UB. 2010. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 11(3):188.. [DOI] [PubMed] [Google Scholar]
- Kenny NJ, et al. , 2015. Genome of the rusty millipede, Trigoniulus corallinus, illuminates diplopod, myriapod and arthropod evolution. Genome Biol Evol. 7(5):1280–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, et al. , 2016. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity (Edinb) 116(2):190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi AN, Arai T, Kubo T, Natori S.. 1998. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem Mol Biol. 28:785–790. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, et al. , 2008. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem Biophys Res Commun. 372(3):464–468. [DOI] [PubMed] [Google Scholar]
- Mille BG, Peigneur S, Predel R, Tytgat J.. 2015. Trancriptomic approach reveals the molecular diversity of Hottentotta conspersus (Buthidae) venom. Toxicon 99:73–79. [DOI] [PubMed] [Google Scholar]
- Missbach C, et al. , 2014. Evolution of insect olfactory receptors. Elife 3:e02115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C, Vogel H, Hansson BS, Groβe-Wilde E.. 2015. Identification of odorant binding proteins and chemosensory proteins in antennal transcriptomes of the jumping bristletail Lepismachilis y-signata and the firebrat Thermobia domestica: evidence for an independent OBP-OR origin. Chem Senses. 40(9):615–626. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Slone J, Song X, Amrein H.. 2012. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 151(5):1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoc PCT, et al. , 2016. Complex evolutionary dynamics of massively expanded chemosensory receptor families in an extreme generalist chelicerate herbivore. Genome Biol Evol. 8(11):3323–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols Z, Vogt RG.. 2008. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem Mol Biol. 38(4):398–415. [DOI] [PubMed] [Google Scholar]
- Nossa CW, et al. , 2014. Joint assembly and genetic mapping of the Atlantic horseshoe crab genome reveals ancient whole genome duplication. Gigascience 3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J.. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 302(1):205–217. [DOI] [PubMed] [Google Scholar]
- Palmer WJ, Jiggins FM.. 2015. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol. 32(8):2111–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P. 1996. Perireceptor events in olfaction. J Neurobiol. 30(1):3–19. [DOI] [PubMed] [Google Scholar]
- Pelosi P, Iovinella I, Felicioli A, Dani FR.. 2014. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 5:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P, Zhou J-J, Ban LP, Calvello M.. 2006. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 63(14):1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva-Arana DC, Lynch M, Robertson HM.. 2009. The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol Biol. 9(1):79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. 2017. Spider genes put a new spin on arachnid's potent venoms, stunning silks, and surprising history. Posted in: Biology, Plants and Animals. ; doi:10.1126/science. aar2331. [Google Scholar]
- Qu S-X, Ma L, Li H-P, Song J-D, Hong X-Y.. 2016. Chemosensory proteins involved in host recognition in the stored-food mite Tyrophagus putrescentiae. Pest Manag Sci. 72(8):1508–1516. [DOI] [PubMed] [Google Scholar]
- Renthal R, et al. , 2017. The chemosensory appendage proteome of Amblyomma americanum (Acari: Ixodidae) reveals putative odorant-binding and other chemoreception-related proteins. Insect Sci. 24(5):730–742. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW.. 2006. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16(11):1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE, et al. , 2011. Creating a buzz about insect genomes. Science 331(6023):1386. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, Daley AC, Pisani D.. 2013. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol. 23(5):392–398. [DOI] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Almeida FC, Rozas J.. 2011. Comparative Genomics of the Major Chemosensory Gene Families in Arthropods. In:. Chichester: John Wiley & Sons, Ltd. [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Rozas J.. 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb) 103(3):208–216. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19(2):301–302. [DOI] [PubMed] [Google Scholar]
- Sanggaard KW, et al. , 2014. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 5:3765.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Touhara K.. 2009. Insect olfaction: receptors, signal transduction, and behavior. Results Probl Cell Differ. 47:121–138. [DOI] [PubMed] [Google Scholar]
- Schwager EE, et al. , 2017. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, et al. , 2001. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc Res Tech. 55(5):297–306. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FG, Rozas J.. 2011. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 3:476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizueta J, et al. , 2017. Evolution of chemosensory gene families in arthropods: insight from the first inclusive comparative transcriptome analysis across spider appendages. Genome Biol Evol. 9(1):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG, et al. , 2009. The insect SNMP gene family. Insect Biochem Mol Biol. 39(7):448–456. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM.. 1981. Pheromone binding and inactivation by moth antennae. Nature 293(5828):161–163. [DOI] [PubMed] [Google Scholar]
- Wanner KW, Isman MB, Feng Q, Plettner E, Theilmann DA.. 2005. Developmental expression patterns of four chemosensory protein genes from the Eastern spruce budworm, Chroistoneura fumiferana. Insect Mol Biol. 14(3):289–300. [DOI] [PubMed] [Google Scholar]
- Wei H-S, Li K-B, Zhang S, Cao Y-Z, Yin J.. 2017. Identification of candidate chemosensory genes by transcriptome analysis in Loxostege sticticalis Linnaeus. PLoS One 12(4):e0174036.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, et al. , 2016. De novo transcriptome analysis of the common New Zealand stick insect Clitarchus hookeri (Phasmatodea) reveals genes involved in olfaction, digestion and sexual reproduction. Hull, JJ, editor. PLoS One 11(6):e0157783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Ni J, Montell C.. 2013. The molecular basis for attractive salt-taste coding in Drosophila. Science 340(6138):1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-J, et al. , 2010. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol Biol. 19:113–122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.