Abstract
With the genomes available for many animal clades, including the early-branching metazoans, one can readily study the functional conservation of genes across a diversity of animal lineages. Ectopic expression of an animal protein in, for instance, a mammalian cell line is a generally used strategy in structure–function analysis. However, this might turn out to be problematic in case of distantly related species. Here we analyzed the GC content of the coding sequences of basal animals and show its impact on gene expression levels in human cell lines, and, importantly, how this expression efficiency can be improved. Optimization of the GC3 content in the coding sequences of cadherin, alpha-catenin, and paracaspase of Trichoplax adhaerens dramatically increased the expression of these basal animal genes in human cell lines.
Keywords: Trichoplax adhaerens, Placozoa, genome evolution, GC content, ectopic expression
Introduction
Genomic sequences for several early diverging animals have become available during the last decade. These include the placozoan Trichoplax adhaerens (Placozoa), the demosponge Amphimedon queenslandica (Porifera) and the sea anemone Nematostella vectensis (Cnidaria) (Putnam et al. 2007; Srivastava et al. 2008; Srivastava et al. 2010). They help us to gain insight into the evolution of animal multicellularity, in development and in the mechanisms for division, adhesion, death, and specialization of cells. The transition from simple unicellular eukaryotes to multicellular organisms necessitates proteins that are able to mediate connections between individual cells. Members of the cadherin and armadillo superfamilies are essential key players in this important process. For instance, our previous research on the evolution of the cadherin and armadillo superfamilies revealed that only a few representative members of these superfamilies can be found in those early branching metazoans, including T. adhaerens and N. vectensis (Hulpiau and van Roy 2011; Gul et al. 2017).
Placozoans are small (1–3 mm) disk-shaped marine invertebrates that were initially discovered in 1883 on the walls of a seawater aquarium (Schulze 1883; for review see Schierwater 2005). Since then the enigmatic placozoans have drawn much attention from biologists because of their morphological simplicity and their basal position in metazoan evolution (Schierwater et al. 2016). In contrast to other metazoans, placozoans have half a dozen somatic cell types and lack an extracellular matrix (ECM) and any obvious basal lamina (Schierwater et al. 2009; Smith et al. 2014). This simple body plan also includes the lack of any organs, a nervous system, muscle cells and of any kind of symmetry. Although the genome of the placozoan T. adhaerens was sequenced as early as 2008 (Srivastava et al. 2008), there is unfortunately no method described to specifically introduce DNA constructs in Placozoa. Alternatively, a most commonly preferred strategy to study the function of ancestral genes is to use human or mouse cell lines as an ectopic expression host (Luthringer et al. 2011; Selvan et al. 2015; Siau et al. 2016). However, it is not straightforward to perform (co) immunoprecipitation experiments, subcellular localization or any other type of functional conservation study in a heterologous animal host expression system whose protein synthesis machinery is separated by hundreds of millions of years of evolution. In ongoing work in our laboratories, where we study T. adhaerens and N. vectensis cadherins and catenins, we encountered problems in expressing the recombinant T. adhaerens proteins in human cell lines. Hence, we analyzed the GC content of coding sequences from basal metazoan animals and investigated its impact on gene expression levels in human cell lines, and how these can be improved.
In this study, we have expressed variants of a human, N. vectensis and T. adhaerens cadherin (CDH1) and catenin (CTNNA2) in HEK293T and HeLa cell lines to show that altering the GC3 to the level of the host expression system dramatically improves the expression of the GC poor genes.
Materials and Methods
Expression Plasmids
Plasmids expressing the full-length (FL) human αN-catenin (encoded by CTNNA2 in man) and FL human E-cadherin (encoded by CDH1 in man) were described previously (Goossens et al. 2007; Pieters et al. 2016). cDNA fragments encoding an N-terminal fragment of human αN-catenin (amino acids [AA] 24–206) or the cytoplasmic tail of human E-cadherin (AA 729–882 = C-terminal end) were amplified for Gateway cloning (Invitrogen) with gene-specific primers containing the AttB sites (supplementary table S1, Supplementary Material online). The amplified fragments were precipitated with polyethylene glycol and inserted in pDONR207 (Invitrogen) by the BP recombination reaction, yielding pDONR207-Hs-Ctnna2-Nterm and pDONR207-Hs-Cdh1-Cyto. These entry clones were transferred to pdcDNAMyc by Gateway LR cloning (Invitrogen), yielding pdcDNAMyc-Hs-Ctnna2-Nterm and pdcDNAMyc-Hs-Cdh1-Cyto.
RNA from various developmental stages of N. vectensis were isolated with Trizol reagent (Life Technologies) and pooled. RNA from T. adhaerens animals was prepared likewise. 1.5 µg of total RNA was subjected to 5′-RACE with a GeneRacer kit (Invitrogen) according to the manufacturer’s protocol. The human cDNA sequence of αN-catenin was searched against N. vectensis and T. adhaerens genomic scaffolds and expressed sequence tags (ESTs) using tBLASTn (https://genome.jgi.doe.gov/pages/blast-query.jsf?db=Nemve1; last accessed March 15, 2018) or (https://genome.jgi.doe.gov/portal/Triad1/Triad1.download.html; last accessed March 15, 2018), respectively. N. vectensis and T. adhaerens E-cadherin sequences were previously described (Hulpiau and van Roy 2011). Primers were designed from the EST sequences and used to amplify cDNA fragments from the N. vectensis and T. adhaerens cDNA libraries generated as described above. N. vectensis and T. adhaerens cDNAs for αN-catenin E-cadherin were PCR amplified with gene-specific primers (supplementary table S1, Supplementary Material online), containing the attB sites and stop codons for subsequent Gateway cloning (Invitrogen). Amplified fragments were precipitated and inserted into pDONR207 (Invitrogen) by the BP recombination reaction, yielding pDONR207 Gateway Entry clones. LR reactions between these Gateway Entry clones and the destination vector pdcDNAMyc produced the expression plasmids: pdcDNAMyc-Nv-Ctnna2-Nterm (encoding AA 33–267 of N. vectensis Ctnna2), pdcDNAMyc-Nv-Cdh1-Cyto (AA 4175–4357 of N. vectensis Cdh1), pdcDNAMyc-Ta-Ctnna2-Nterm (AA 9–262 of T. adhaerens Ctnna2), pdcDNAMyc-Ta-Cdh1-Cyto (AA 4082–4270 of T. adhaerens Cdh1). The protein and DNA sequences encoded by the cloned N. vectensis and T. adhaerens cDNAs are provided in the supplementary table S2, Supplementary Material online. A plasmid expressing the (FL) T. adhaerens paracaspase (Pcasp) was described previously (Hulpiau et al. 2016). AttB-site-containing Ta-Cdh1-Cyto-GC45 (guanine and cytosine [GC] content: 45), Ta-Cdh1-Cyto-GC50, Ta-Cdh1-Cyto-GC60, Ta-Ctnna2-Nterm-GC60, Ta-Pcasp-GC40, Ta-Pcasp-GC45, Ta-Pcasp-GC50 and Ta-Pcasp-GC55 oligonucleotides with increased GC contents were ordered from Integrated DNA Technologies as gBlocks. Synthetic DNAs were amplified using the attB elongation primers (supplementary table S1, Supplementary Material online) and inserted into pDONR207 (cadherin and catenin constructs) (Invitrogen) or pDONR221 (paracaspase constructs) (Invitrogen) by the BP recombination. LR reactions between these Gateway Entry clones and the destination vectors pdcDNAMyc and pdcDNA-FLAGMyc produced the expression plasmids; pdcDNAMyc-Ta-Cdh1-Cyto-GC45 (AA 4082–4270), pdcDNAMyc-Ta-Cdh1-Cyto-GC50 (AA 4082–4270), pdcDNAMyc-Ta-Cdh1-Cyto-GC60 (AA 4082–4270), pdcDNAMyc-Ta-Ctnna2-Nterm-GC60 (AA 9–262), pdcDNA-FLAGMyc-Ta-Pcasp-GC40 (AA 1–480), pdcDNA-FLAGMyc-Ta-Pcasp-GC45 (AA 1–480), pdcDNA-FLAGMyc-Ta-Pcasp-GC50 (AA 1–480), and pdcDNA-FLAGMyc-Ta-Pcasp-GC55 (AA 1–480).
Cell Lines and Transfections
HeLa and HEK293tsA1609neo (in short HEK293T) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 2 mM glutamine, and 0.4 mM sodium pyruvate. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. For transient transfection of HeLa and HEK293T cells, 5 × 106 cells were seeded the day before in a 75 cm2 cell culture flask. Cells were allowed to reach about 70% confluency and then transfected with the calcium phosphate method using 5 µg of Myc-tagged expression vector (see above). At 6–8 h post transfection, the medium was replaced with 15 ml of fresh medium.
SDS-PAGE and Western Blotting
Forty-eight hours posttransfection cells were washed once with ice-cold phosphate buffered saline (PBS) and lysed with ice-cold Laemmli lysis buffer. For subsequent SDS-PAGE analysis, we used an 8% polyacrylamide gel and loaded a total of 60 µg protein per lane. After blotting to PVDF membranes (Millipore), a subsequent blocking step was performed in the presence of 0.1% (vol/vol) Tween-20 (Sigma) and 5% (w/vol) nonfat dry milk. Membranes were then immunostained overnight with primary antivinculin (monoclonal mouse antivinculin, Sigma, dilution 1/400) or anti-Myc (monoclonal mouse anti-Myc-HRP, Sigma, dilution 1/10,000) or anti-Flag (monoclonal mouse anti-Flag-HRP, Sigma, dilution 1/1,000) antibodies in the same buffer at 4°C. After several washing steps, the membranes were incubated with secondary horseradish peroxidase (HRP)-conjugated antibody and detection was by enhanced chemiluminescence (ECL) (Amersham GE Healthcare).
mRNA Quantification
Cells were washed with PBS after which Trizol reagent (Life Technologies) was added. Following a short incubation time, cells were scraped from the tissue culture flasks and harvested. Total RNA was isolated using the Bio-Rad RNA extraction kit. cDNA synthesis was done with the iScript cDNA synthesis kit according to the manufacturer’s instructions (Bio-Rad). RT-qPCR reactions were performed using the SensiFAST SYBR No-ROX Kit (GC Biotech) and detection was performed using a LightCycler 480 Instrument (Roche Diagnostics). For normalization of gene expression levels the geometric mean of the most stable reference genes was used (Vandesompele et al. 2002). Two primer pairs were designed for the Myc tag (forward #1Myc 5′-TCTTTTTGCAGGATCCCA-3′, forward #2Myc 5′-TGCAGGATCCCATCGATT-3′, reverse #1Myc 5′-GCTTTTGCTCCATTTCATTC-3′, reverse #2Myc 5′-GCTCCATTTCATTCAAGTCCTC-3′, and one primer pair for the Flag tag (forward #1Flag 5′-AGAACCCACTGCTTACTGGCT-3′ and reverse #1Flag 5′-CATCGTCATCCTTAGTGTCCA-3′). The primers for the reference genes were designed as follows: RPL13A primers (forward 5′-CCTGGAGGAGAAGAGGAAAGAGA-3′, reverse 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′), UBC primers (forward 5′-ATTTGGGTCGCGGTTCTTG-3′, reverse 5′-TGCCTTGACATTCTCGATGGT-3′) and YWHAZ primers (forward 5′-ACTTTTGGTACATTGTGGCTTCAA-3′, reverse 5′-CCGCCAGGACAAACCAGTAT-3′).
Data Acquisition and Sequence Analyses
Trichoplax adhaerens proteomics data covering the expression abundance of 6,500 T. adhaerens proteins were taken from Ringrose et al. (2013). Uniprot accession IDs were converted to the NCBI RefSeq Nucleotide IDs using the Retrieve/ID mapping tool of the UniProt database (http://www.uniprot.org/; last accessed March 15, 2018). The nucleotide sequences were imported into the CLC Main workbench (http://www.clcbio.com/products/clc-main-workbench/; last accessed March 15, 2018) and coding sequences were extracted. To calculate the counts of nucleotides, frequency of codons and nucleotide frequency in codon positions, sequence statistics for each entry were created using the CLC Main workbench. This data was then merged and compared with the Log 10 abundance factors of the proteomics data. Codon usage data of Homo sapiens were obtained from http://gtrnadb.ucsc.edu/; last accessed March 15, 2018. To calculate the frequency of codons and nucleotide frequency in codon positions of representative bilaterians, coding sequences for protein-coding genes of H. sapiens, Mus musculus, Gallus gallus, Danio rerio, Ciona intestinalis, Lingula anatine, Crassostrea gigas, Octopus bimaculoides, Daphnia pulex, Tribolium castaneum, Drosophila melanogaster, and Caenorhabditis elegans were downloaded from Ensembl, (http://www.ensembl.org/info/data/ftp/index.html; last accessed March 15, 2018) or (http://metazoa.ensembl.org/info/website/ftp/index.html; last accessed March 15, 2018). The coding sequence data for N. vectensis and Monosiga brevicollis were obtained from http://genome.jgi.doe.gov/; last accessed March 15, 2018, for Mnemiopsis leidyi and Pleurobrachia bachei from https://neurobase.rc.ufl.edu/; last accessed March 15, 2018, for Oscarella carmela and Haliclona amboinensis from http://www.compagen.org/;last accessed March 15, 2018, for Capsaspora owczarzaki and Dictyostelium discoideum from http://protists.ensembl.org/info/website/ftp/index.html, last accessed March 15, 2018, and for Amphimedon queenslandica from http://amphimedon.qcloud.qcif.edu.au/downloads.html; last accessed March 15, 2018. Detailed information on these data sets and calculations are provided in supplementary table S3, Supplementary Material online. To calculate the nucleotide frequency in codon positions the Biopython SeqUtils module was used. The GC and GC3 plots were created in R using the ggplot2 package.
The nucleotide sequences of the expression plasmids (pdcDNAMyc backbone plus insert) were subjected to a compositional sequence analysis using the CpGPlot and Isochore tools from EMBOSS (European Molecular Biology Open Software Suite, (http://www.ebi.ac.uk/Tools/seqstats/; last accessed March 15, 2018). All profiles were obtained using an overlapping window of 500 base pairs.
Results
Low GC(3) Content of T. adhaerens Genes in Comparison with Other Organisms
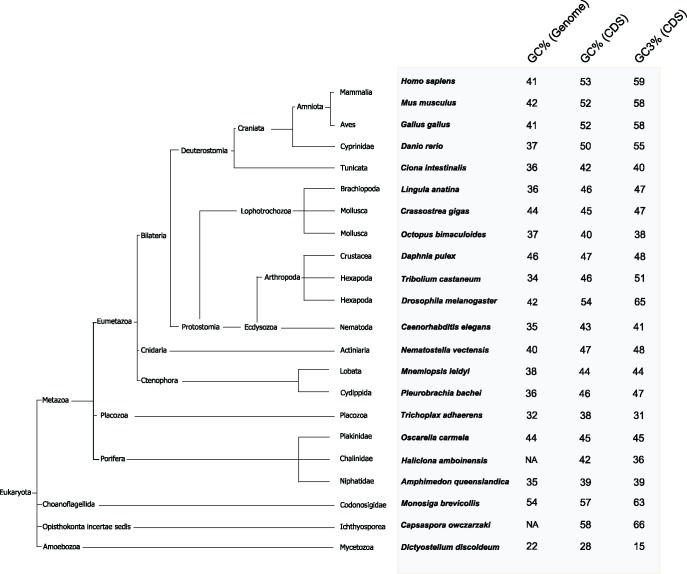
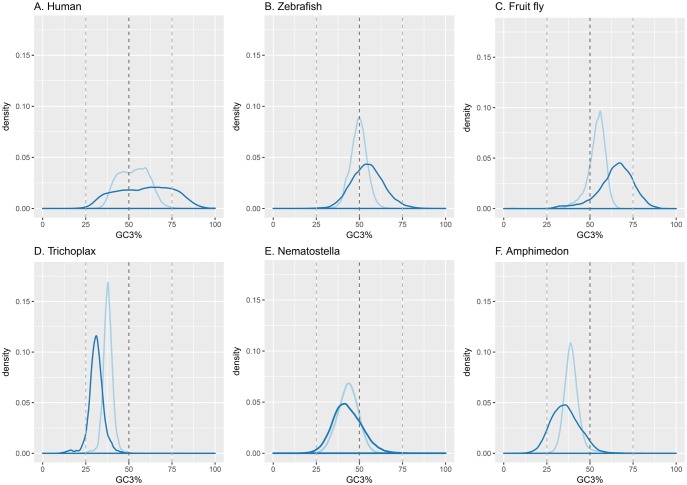
At the onset of this project we investigated the GC (or Guanine-Cytosine content) and GC3 (proportion of G and C in the third position of the codons) content of the protein coding sequences (CDS) of 22 metazoan and nonmetazoan species using the available data from the genome databases (supplementary table S3, Supplementary Material online). Figure 1 shows that the mean GC3 content of the four vertebrate CDS is above 55%. In contrast, none of the GC3 contents of the investigated early diverging metazoans (or nonbilaterian animals) exceeds 50%. In figure 2, we have compared the distribution of the GC and GC3 contents of the CDS of the vertebrate species and nonbilaterian animals. The mean of the GC content of the nonbilaterian T. adhaerens CDS is 38%, and thus the lowest seen in any metazoan genome so far investigated (fig. 1). Figure 2D shows the distribution of the GC content of the 11,520 predicted T. adhaerens CDS. Remarkably, 80% of the T. adhaerens CDS have GC3 contents between 25% and 35% (fig. 2D), with a mean of 31%. In sharp contrast, the GC3 content of the mammalian CDS ranges from 10% to almost 100% (Alvarez-Valin et al. 2002), with a mean of 59% (figs. 1 and2A). Figure 2D confirms the narrow range of the GC3 content of T. adhaerens CDS and emphasizes the differences to other bilaterian and nonbilaterian metazoan species.
Fig. 1.
—The GC% and GC(3) in genomes and coding sequences (CDS) across metazoan and nonmetazoan species. (Left) Cladogram showing the evolutionary relationships among the organisms used in this study. (Right) The percentages of the GC content of the whole genome, of the CDS, and the third-codon position GC-content (GC3) of the CDS.
Fig. 2.
—The distribution of GC and GC3 content of the protein coding sequences in vertebrate, insect and nonbilaterian species. GC (light blue) and GC3 (dark blue) density plots of the CDS of vertebrates (A) Homo sapiens (human) and (B) Danio rerio (zebrafish), of (C) Drosophila melanogaster (fruit fly), and of non-bilaterians (D) Trichoplax adhaerens (placozoan), (E) Nematostella vectensis (cnidarian), and (F) Amphimedon queenslandica (porphyrian).
Expression of T. adhaerens cDNAs in Human Cell Lines
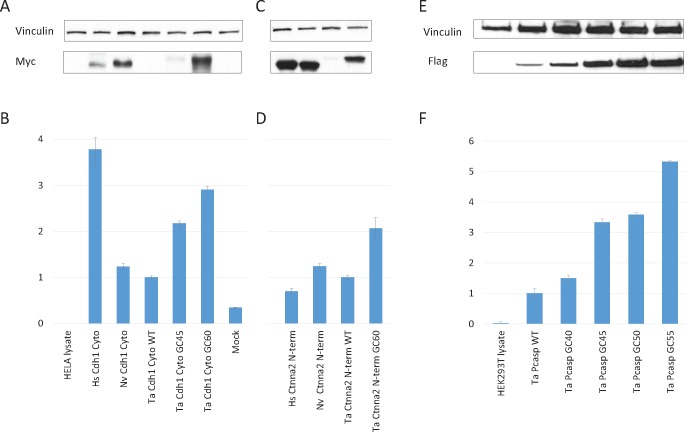
In a first set of experiments, we compared the protein and mRNA expression levels in HeLa cells for constructs encoding Myc-tagged cytoplasmic tails of cadherins from human (Hs-Cdh1-Cyto), the starlet sea anemone N. vectensis (Nv-Cdh1-Cyto), and the placozoan T. adhaerens (Ta-Cdh1-Cyto). These coding sequences were cloned downstream the CMV promoter in the pdcDNAMyc vector. Except for these insert regions, the plasmid constructs used were the same for all three expression vectors (supplementary fig. S1, Supplementary Material online) and equal amounts of pdcDNAMyc-(Hs/Nv/Ta)-Cdh1-Cyto vectors were transfected. After 2 days of incubation, the cells were lysed and the amount of the pdcDNAMyc-(Hs/Nv/Ta)-Cdh1-Cyto proteins expressed was quantified by Western-blotting using an anti-Myc antibody.
The wild-type (WT) Hs-Cdh1-Cyto and Nv-Cdh1-Cyto protein fragments, encoded by GC3-rich sequences (57% and 63%, respectively), were readily expressed in transfected HeLa cells (fig. 3A). However, expression of the GC3-poor Ta-Cdh1-Cyto (GC3: 21%; WT sequence) was undetectable. Next, we quantified the mRNA expression levels of (Hs/Nv/Ta)-Cdh1-Cyto using real-time RT-PCR. The amount of Hs-Cdh1-Cyto mRNA was over 3-fold higher than the amount of Nv-Cdh1-Cyto and Ta-Cdh1-CytoWT mRNAs. Despite the high Hs-Cdh1-Cyto mRNA levels, the expression of the Hs-Cdh1-Cyto protein was not higher than the expression of the Nv-Cdh1-Cyto protein (fig. 3A and B). Similarly, while the Nv-Cdh1-Cyto and Ta-Cdh1-CytoWT mRNA levels were comparable, the amount of expressed Nv-Cdh1-Cyto and Ta-Cdh1-CytoWT proteins were remarkably different (fig. 3A and B). We have extended this observation by an expression experiment in HeLa cells for the N-terminal region of αN-catenin (Ctnna2) of, respectively, H. sapiens, N. vectensis, and T. adhaerens. Quite similar to the previous experiment, the levels of Hs-Ctnna2-Nterm and Nv-Ctnna2-Nterm proteins were readily detectable; however no Ta-Ctnna2-NtermWT was detectable (fig. 3C), despite high expression levels of the encoding mRNA (fig. 3D). Similar results were obtained when the expression efficiencies of (Hs/Nv/TaWT)-Cdh1-Cyto or of (Hs/Nv/TaWT)-Ctnna2-Nterm constructs were compared in HEK293T cells (supplementary fig. S2, Supplementary Material online).
Fig. 3.
— Expression of human (Hs), N. vectensis (Nv), and T. adhaerens (Ta) cDNAs in HeLa and HEK293T cells. Equal amounts of pdcDNAMyc-(Hs/Nv/Ta)-Cdh1-Cyto, pdcDNAMyc-(Hs/Nv/Ta)-Ctnna2-Nterm and pdcDNAFlag-Ta-Pcasp plasmids were used to transfect HeLa or HEK293T cells. (A, C, E) Protein levels were analyzed by Western blotting using an anti-Myc antibody or an anti-Flag antibody as indicated; an anti-vinculin antibody was used as a control. (B, D, F) Expression of (Hs/Nv/Ta)-Cdh1-Cyto, (Hs/Nv/Ta)-Ctnna2-Nterm and Ta-Pcasp mRNA. After 24 h, total cellular RNA was isolated and analyzed by qRT-PCR. As negative controls, untransfected HeLa or HEK293T cells were used as well as transfection with empty pdcDNAMyc plasmid (mock). Values have been normalized such that the values of the different WT constructs of T. adhaerens are equal to 1. The results are representative of three experiments.
To test whether the expression of the T. adhaerens cDNAs could be modulated by changing their GC content, we designed two synthetic variants of Ta-Cdh1-Cyto. The GC3 content of the first variant has been enriched from 21% (WT, with GC: 35%) to 51% (with GC: 45%), and the second enhanced variant had a GC3 content as high as 99% (with GC: 60%). All of these codon modifications were synonymous changes and therefore did not change the resulting AA sequence of the Ta-Cdh1-Cyto protein (supplementary fig. S3, Supplementary Material online). Remarkably, Ta-Cdh1-Cyto protein synthesis increased significantly with increasing GC3/GC content of the encoding cDNA. Although the WT Ta-Cdh1-Cyto protein was not detectable at all, the Ta-Cdh1-Cyto-GC45 encoded protein was expressed at weak but detectable levels (fig. 3A). When the GC3 content of the Ta-Cdh1-Cyto mRNA was further increased to 99% (GC: 60%), the encoded protein was expressed 11-fold stronger than in case of the protein encoded by Ta-Cdh1-Cyto-GC45. qPCR experiments demonstrated a similar tendency between GC content and mRNA levels in HeLa cells. Ta-Cdh1-Cyto-GC45 and Ta-Cdh1-Cyto-GC60 mRNAs were expressed, respectively, 2- and 3-fold stronger than the WT Ta-Cdh1-Cyto mRNA (fig. 3B). We have repeated the ectopic expression of Ta-Cdh1-Cyto constructs in HEK293T cells, and in addition to the two previously used GC enriched versions of Ta-Cdh1-cyto (GC45 and GC60), we expressed a version of Ta-Cdh1-cyto with a GC content of 50%. Supplementary figure S4, Supplementary Material online shows the gradual increase in mRNA and protein levels when the GC3 content of Ta-Cdh1-Cyto was increased. We obtained similar results when we tested the GC enhanced version of Ta-Ctnna2-Nterm. When the GC3 content of Ta-Ctnna2-Nterm was increased from 36% to 94%, there was an 18- and 2-fold increase at protein and mRNA levels, respectively (fig. 3C and D). These results were confirmed upon transfection of HEK293T cells (supplementary fig. S2, Supplementary Material online). The expression experiments discussed above were performed on partial gene fragments of cadherins and catenins. To demonstrate the wider applicability of our findings, we have performed an additional expression experiment on a full-length T. adhaerens paracaspase (Ta-Pcasp) construct (Hulpiau et al. 2016). In addition to the WT Ta-Pcasp (with GC: 37% and GC3: 32%), we have designed and expressed four different GC enriched Ta-Pcasp constructs in HEK293T cells: Ta-Pcasp-GC40 (with GC3: 40), Ta-Pcasp-GC45 (with GC3: 55%), Ta-Pcasp-GC50 (with GC3: 70%) and Ta-Pcasp-GC55 (with GC3: 86%). The resulting Western blot and qPCR data confirmed the gradual increase in mRNA and protein levels when the GC content of this additional T. adhaerens cDNA was enriched (fig. 3E and F). Altogether, these experiments indicate that the GC content in CDS strongly affects the expression efficiency of the T. adhaerens genes in human cells. In view of the data presented here (fig. 3 and supplementary figs. S2 and S4, Supplementary Material online), we cannot exclude that both transcription and translation efficiencies are enhanced in function of increasing GC3 content. Hence, the majority of the T. adhaerens genes may require a major nucleotide optimization to increase their ectopic expression in mammalian cell lines.
Correlation between T. adhaerens Gene Expression and GC Content
It has been shown that the human genome is highly compositionally compartmentalized (Bernardi 1995), and abundantly expressed genes are localized in the GC-richest isochores (Bernardi 2007). As we showed above, the T. adhaerens CDs have a low GC and GC3 content compared with those of vertebrates. Figure 4 shows the relationship between the GC content and the respective abundance factor of T. adhaerens proteins using the published expression data (Ringrose et al. 2013). Remarkably, these results suggest that, similar to the human genes, abundantly expressed T. adhaerens genes have a higher GC content (fig. 4).
Fig. 4.
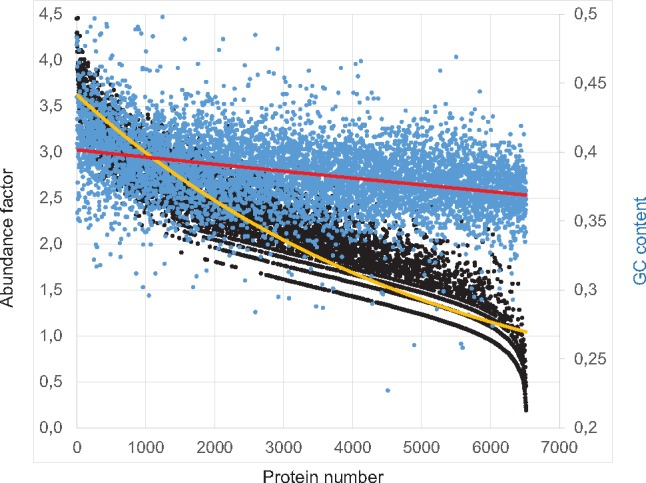
—Correlation between GC content and gene expression efficiency in T. adhaerens. Protein abundance factors per animal for 6,500 proteins (Ringrose et al. 2013) are plotted against the GC content of the corresponding genes. The yellow line represents the trend line for the abundance factors and the red line represents the trend line for the GC content.
Next, based on the available expression data we have generated a codon usage table for T. adhaerens and compared this with the codon usage table for the human genome. Supplementary figure S5, Supplementary Material online, shows that, for both data sets, 14 (out of 61) sense codons have a usage above 2%. However, while for T. adhaerens six out of seven pure A/T codon combinations are present in this window, we found only one for H. sapiens (AAA = K codon, 2.44% usage). Moreover, similar to the Drosophila melanogaster genome (Vicario et al. 2007), the codon usage in the human genome is biased toward G/C-ending codons (or GC3), as 11 out of 14 GC3 codons have ≥ 2% usage (supplementary fig. S5B, Supplementary Material online). In contrast, in the T. adhaerens genome, the codon usage is biased toward A/T-ending codons (12 out of 14 AT3 codons have ≥ 2% usage) (supplementary fig. S5A, Supplementary Material online).
Discussion
Over the last two decades, GC(3) content dynamics in the context of vertebrate evolution have been extensively studied (Costantini et al. 2009; Romiguier et al. 2010). Most of the mammalian genomes are composed of GC-poor (L1 and L2 families, GC% ranging between 36% and 39%) and GC-rich isochores (H1-H3 families, GC% ranging between 43% and 55%) (Costantini et al. 2006). Previous studies indicate that GC3 can be used to detect isochore existence and structure (Bernardi [2001]; Romiguier et al. [2010]; but see Elhaik et al. [2009] for opposing arguments), and GC3-rich human genes are generally located in H3 isochores (Alvarez-Valin et al. 2002). Similar to other vertebrate genomes, the mean GC3 content of the human coding sequences is above 55% (fig. 1). In nonvertebrate metazoans the mean GC3 content of coding sequences is often significantly lower than 50% (supplementary table S3, Supplementary Material online), and can drop to as low as 31% in T. adhaerens. In contrast, the fruit fly (D. melanogaster) has a much higher GC3% of 65% (figs. 1 and2). In this study, we analyzed the GC(3) content of the genes in three nonbilaterian species N. vectensis, T. adhaerens, and A. queenslandica and analyzed the ectopic expression in human cell lines of two proteins from both N. vectensis and T. adhaerens.
The data presented in figure 3 and supplementary figures S2 and S4, Supplementary Material online, clearly show that “optimizing” the GC3 content of the T. adhaerens genes causes an increase in both protein and mRNA expression levels in mammalian cell lines. It should be noted that in order to obtain such increased expression for T. adhaerens genes, we did not specifically optimize either the codon usage in view of the host (human), like in the previously reported “preferred human codon-optimized method” (Kudla et al. 2006; Inouye et al. 2015), or the Kozak sequences. Instead, we changed the GC content in the third position of each codon without change on the encoded AA (supplementary fig. S3 and table S2, Supplementary Material online). This suggests that synonymous substitutions in CDS towards G/C codon combinations have a positive effect on the expression levels in mammalian cell lines. At least in part, this was due to increased steady-state mRNA levels, as observed before for human proteins and green fluorescent protein (GFP) (Kudla et al. 2006; Inouye et al. 2015), but we do not exclude an additional and significant enhancing effect at the translational level.
The positive effect of GC3 content on gene expression also raises important questions regarding the neutral theory of evolution (Kimura 1968, 1983), which assumes that most mutations are either “silent” or have a negligible effect on fitness. Especially synonymous substitutions, which do not alter the AA sequence, are considered neutral and thought to be propagated by chance alone. This assumption is also the basis for dN/dS calculations (Hurst 2002), which are commonly used to detect positively selected genes. The ratio of synonymous (“neutral”) substitutions per synonymous site is considered the background mutation rate and compared with the ratio of nonsynonymous (protein altering) substitutions per nonsynonymous site. However, more recently it has been suggested that mutations at synonymous sites are not necessarily neutral (for reviews, see Chamary et al. 2006; Shabalina et al. 2013). Just as one example, in human it has been found that the GC3 content and synonymous substitution rates differ between constitutive and alternatively expressed exons of the same gene (Iida and Akashi 2000), suggesting varying constraint between different synonymous sites. There is also strong evidence that a great portion of 4-fold degenerate sites (all changes are synonymous) in mammalian and avian genes are under selective constraint (Eory et al. 2010; Kunstner et al. 2011). Similarly, the effect of GC3 content on gene expression reported here suggests that synonymous substitutions are not always neutral and may have an effect on an organism’s fitness. It thus further corroborates the view that dN/dS ratios need to be cautiously interpreted for estimating natural selection.
Optimizing heterologous gene expression is a key step to produce and study proteins of interest. It has been shown that high GC content leads to increased mRNA levels in mammalian cells (Kudla et al. 2006). This phenomenon might cause a major problem for scientists who perform experimental studies to gain insight into the functional conservation of certain genes and gene families. More than 5,000 T. adhaerens CDSs, including the orthologs of E-cadherin, α- and β-catenin, p53, and DNA Topoisomerase II, have low GC values (<39%). These GC-poor coding sequences may behave as GC-poor families in the mammalian context and probably cannot be expressed in mammalian cell lines as we clearly demonstrated for T. adhaerens E-cadherin and α-catenin in HeLa (fig. 3) and HEK293T (supplementary figs. S2 and S4, Supplementary Material online) cells. Our work, together with previous studies of expression of human proteins, GFP and luminescence proteins in mammalian cell lines (Kudla et al. 2006; Inouye et al. 2015), suggest that GC optimization is essential for the efficient production of proteins of interest in mammalian cell lines. Our work demonstrates that this is especially true for proteins of phylogenetically distant animal groups, such as those of lower metazoans like T. adhaerens, which are important for studying protein evolution in particular and animal evolution in general.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank our colleagues and Prof. Dr Klaas Vandepoele (Ghent University) for helpful discussions. This work was supported by the Research Foundation—Flanders (FWO-Vlaanderen, Award G.0320.11 N), the Belgian Science Policy (Interuniversity Attraction Poles—Award IAP7/07), and the Special Research Fund of Ghent University (Award BOF 01J14211). B.S. acknowledges support from the German Science Foundation (DFG Schi-277/27-1).
Literature Cited
- Alvarez-Valin F, Lamolle G, Bernardi G.. 2002. Isochores, GC3 and mutation biases in the human genome. Gene 300(1–2):161–168. [DOI] [PubMed] [Google Scholar]
- Bernardi G. 1995. The human genome: organization and evolutionary history. Annu Rev Genet. 29:445–476. [DOI] [PubMed] [Google Scholar]
- Bernardi G. 2001. Misunderstandings about isochores. Part 1. Gene 276(1–2):3–13. [DOI] [PubMed] [Google Scholar]
- Bernardi G. 2007. The neoselectionist theory of genome evolution. Proc Natl Acad Sci U S A. 104(20):8385–8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD.. 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 7(2):98–108. [DOI] [PubMed] [Google Scholar]
- Costantini M, Cammarano R, Bernardi G.. 2009. The evolution of isochore patterns in vertebrate genomes. BMC Genomics 10:146.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini M, Clay O, Auletta F, Bernardi G.. 2006. An isochore map of human chromosomes. Genome Res. 16(4):536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaik E, Landan G, Graur D.. 2009. Can GC content at third-codon positions be used as a proxy for isochore composition?. Mol Biol Evol. 26(8):1829–1833. [DOI] [PubMed] [Google Scholar]
- Eory L, Halligan DL, Keightley PD.. 2010. Distributions of selectively constrained sites and deleterious mutation rates in the hominid and murid genomes. Mol Biol Evol. 27(1):177–192. [DOI] [PubMed] [Google Scholar]
- Goossens S, et al. , 2007. A unique and specific interaction between alphaT-catenin and plakophilin-2 in the area composita, the mixed-type junctional structure of cardiac intercalated discs. J Cell Sci. 120(Pt 12):2126–2136. [DOI] [PubMed] [Google Scholar]
- Gul IS, Hulpiau P, Saeys Y, van Roy F.. 2017. Evolution and diversity of cadherins and catenins. Exp Cell Res. 358(1):3–9. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, Driege Y, Staal J, Beyaert R.. 2016. MALT1 is not alone after all: identification of novel paracaspases. Cell Mol Life Sci. 73(5):1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F.. 2011. New insights into the evolution of metazoan cadherins. Mol Biol Evol 28(1):647–657. [DOI] [PubMed] [Google Scholar]
- Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18(9):486.. [DOI] [PubMed] [Google Scholar]
- Iida K, Akashi H.. 2000. A test of translational selection at ‘silent’ sites in the human genome: base composition comparisons in alternatively spliced genes. Gene 261(1):93–105. [DOI] [PubMed] [Google Scholar]
- Inouye S, Sahara-Miura Y, Sato J, Suzuki T.. 2015. Codon optimization of genes for efficient protein expression in mammalian cells by selection of only preferred human codons. Protein Expr Purif. 109:47–54. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217(5129):624–626. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M.. 2006. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 4(6):e180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunstner A, Nabholz B, Ellegren H.. 2011. Significant selective constraint at 4-fold degenerate sites in the avian genome and its consequence for detection of positive selection. Genome Biol Evol. 3:1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthringer B, et al. , 2011. Poriferan survivin exhibits a conserved regulatory role in the interconnected pathways of cell cycle and apoptosis. Cell Death Differ. 18(2):201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters T, et al. , 2016. p120 Catenin-mediated stabilization of E-cadherin is essential for primitive endoderm specification. PLoS Genet. 12(8):e1006243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. , 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317(5834):86–94. [DOI] [PubMed] [Google Scholar]
- Ringrose JH, et al. , 2013. Deep proteome profiling of Trichoplax adhaerens reveals remarkable features at the origin of metazoan multicellularity. Nat Commun. 4:1408. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJ, Galtier N.. 2010. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 20(8):1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B. 2005. My favorite animal, Trichoplax adhaerens. Bioessays 27(12):1294–1302. [DOI] [PubMed] [Google Scholar]
- Schierwater B, de Jong D, Desalle R.. 2009. Placozoa and the evolution of Metazoa and intrasomatic cell differentiation. Int J Biochem Cell Biol. 41(2):370–379. [DOI] [PubMed] [Google Scholar]
- Schierwater B, et al. , 2016. Never ending analysis of a century old evolutionary debate: “Unringing” the urmetazoon bell. Front Ecol Evol. 4:5. [Google Scholar]
- Schulze FE. 1883. Trichoplax adhaerens, nov. gen., nov. spec. Zool Anz. 6:92–97. [Google Scholar]
- Selvan N, et al. , 2015. The early metazoan Trichoplax adhaerens possesses a functional O-GlcNAc system. J Biol Chem. 290(19):11969–11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Spiridonov NA, Kashina A.. 2013. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 41(4):2073–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siau JW, et al. , 2016. Functional characterization of p53 pathway components in the ancient metazoan Trichoplax adhaerens. Sci Rep. 6:33972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, et al. , 2014. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr Biol. 24(14):1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, et al. , 2008. The Trichoplax genome and the nature of placozoans. Nature 454(7207):955–960. [DOI] [PubMed] [Google Scholar]
- Srivastava M, et al. , 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466(7307):720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, et al. , 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3(7):RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR.. 2007. Codon usage in twelve species of Drosophila. BMC Evol Biol. 7:226.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.