summary
Glia adopt remarkable shapes that are tightly coordinated with the morphologies of their neuronal partners. Glia and neurons exhibit coordinated morphological changes on the time scale of minutes and on size scales ranging from nanometers to hundreds of microns. Here, we review recent studies that reveal the highly dynamic, localized morphological changes of mammalian neuron-glia contacts. We then explore the power of Drosophila and C. elegans models to study coordinated changes at defined neuron-glia contacts, highlighting the use of innovative genetic and imaging tools to uncover the molecular mechanisms responsible for coordinated morphogenesis of neurons and glia.
Introduction
Glia have been renowned for their exquisite morphology since long before their functions were known. Now, the functions of glia are increasingly well understood (see reviews [1,2]), yet glial morphogenesis remains as mysterious as ever. Astrocytes, once called “spider cells,” extend innumerable radiating processes that wrap synapses with puzzle-piece-like precision[3-6]. How do glia attain their remarkable shapes, and how are they coordinated with neuronal morphologies? Here, we review recent striking examples of coordinated neuron-glia morphogenesis in mammals. We then highlight the power of simple model organisms such as Drosophila and C. elegans to address these questions.
The scope of the problem: Highly dynamic and localized morphological changes
The intimate associations between glia and synapses exhibit highly dynamic morphological changes on the order of minutes[7-11]. Landmark studies using time-lapse confocal imaging of rodent brain slices revealed that post-synaptic dendritic spines and astrocytic processes do not change shape in perfect register, yet generally grow or shrink together over time[7,9].Remarkably, this coordinated growth is achieved even though glia-spine interactions undergo rapid structural changes, the extent of glial coverage of spines varies, and astrocytic processes tend to exhibit even greater motility than their dendritic spine counterparts[7].
Two recent, complementary studies provided evidence for a mechanism that may help to explain the coordinated growth of astrocytic processes and dendritic spines. Perez-Alvarez et al. and Bernardinelli et al. examined coordinated morphological changes of astrocytic processes and dendritic spines in response to patterns of neuronal activity that increase the size of dendritic spines[12-14]. By observing single synapses, these groups found that stimulation of the presynaptic neuron also triggers a rapid, transient increase in the motility of the astrocytic processes associated with the dendritic spine (Fig. 1A, top). This effect is mediated by metabotropic glutamate receptors on astrocytes, which are bound by glutamate released by the presynaptic neuron upon stimulation. Activated receptors initiate intracellular calcium transients in astrocytes that are both necessary and sufficient for increased process motility. This spike in motility tends to expand astrocytic coverage of spines, such that subsequent stabilization of astrocytic processes (within ∼30 min) produces a sustained increase in the extent of astrocytic wrapping of spines. These coordinated morphological changes in astrocytes and spines were shown to affect neural function[12,13]. Interestingly, similar changes were also seen following sensory stimulation in vivo, consistent with previous EM studies[13,15].
Figure 1. Glia and neurons undergo dynamic, localized morphological changes in mammals and invertebrates.
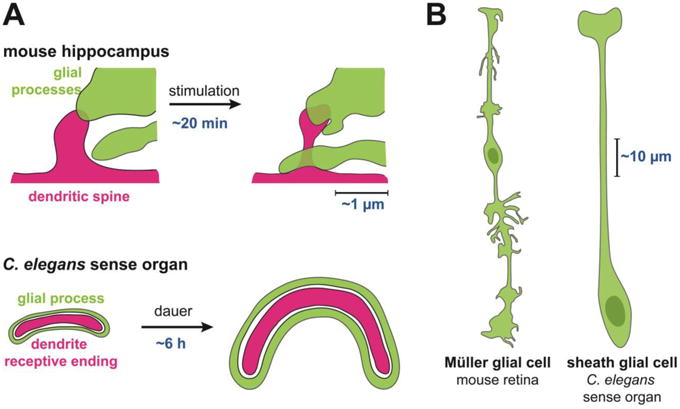
(A) Glial processes (green) exhibit coordinated changes with dendritic spines or receptive endings (red). These occur over minutes to hours on the scale of microns in response to experimental stimulation or environmental conditions. (B) A single glial cell can exhibit highly localized morphological patterning that is coordinated with its local neuronal environment, on the scale of tens to hundreds of microns.
Not only are these morphological changes highly dynamic over time, but they are also exquisitely restricted in space - even to the level of specific synapses. Photoactivation of astrocytic calcium signaling at a single synapse causes changes in astrocytic process motility at that synapse, but does not affect nearby glia-spine contacts on the same dendrite[12]. This suggests that astrocytes can respond to the unique demands of each synapse, which is especially remarkable given that a single astrocyte may contact nearly 100,000 synapses in rodents and up to two million synapses in humans[16].
An impressive anatomical study recently underscored the idea that glia exhibit highly localized morphological patterning that is coordinated with their local neuronal environment. Wang et al. demonstrated that a single Müller glial cell that spans the entire thickness of the retina (∼200 μm in adult mice) adopts strikingly different morphologies at each retinal layer (Fig. 1B, left) [17]. Its glial process courses in thin sheets through the outer nuclear layer but becomes wildly branched in the synapse-rich environment of the inner plexiform layer (∼50 μm each layer).In fact, the specificity of glial shape is so tailored to the local environment that Müller glia even elaborate distinct structures at different sublaminar positions within the inner plexiform layer. Such localized morphological differences were also noted in a recent characterization of radial glia-like stem cells in the dentate gyrus[18]. These cells extend thin, twisting processes through the granule cell layer (∼30 μm) and then arborize abruptly upon encountering the molecular layer, where they ensheathe synapses.
While these studies demonstrate that glial shapes are precisely patterned at the level of tens to hundreds of microns, another recent study suggests that astrocytic processes are also patterned at the submicron scale. Pannasch et al. found that glial wrapping is actively restricted to regions of the spine adjacent to the synapse[19]. Loss of the gap junction protein Connexin 30 (Cx30) causes inappropriate invasion of astrocytic processes into the synaptic cleft, promoting increased astrocytic uptake of glutamate and thus reducing neurotransmission. An elegant combination of pharmacological and genetic manipulations showed that this morphological intrusion was regulated not by the channel-forming properties of Cx30, but rather by signaling activity mediated through its intracellular region. These results raise the intriguing possibility that such fine-scale morphological changes might be actively regulated to modulate synaptic transmission.
These examples highlight the scope of the challenge in understanding coordinated neuron-glia morphogenesis: structural changes occur on the time scale of minutes, with highly specialized patterning that spans from the ultrastructural level to the tissue level. Additionally, advances in single-cell transcriptomics have revealed previously unrecognized heterogeneity among glia[20-22], raising the possibility of another dimension of complexity: specific subtypes of glia may preferentially coordinate with synapses from different types of neurons. Ideally, single glia-synapse contacts between defined neuronal and glial partners would be directly observed in intact, living animals, but these approaches are technically challenging amid the complexity of the mammalian nervous system. In contrast, such approaches have proven more feasible in invertebrate model organisms. The remainder of this review will focus on the use of these powerful models to understand coordinated neuron-glia morphogenesis.
The power of invertebrate systems
Simple anatomy, sophisticated genetics
The highly simplified nervous systems of invertebrates make these organisms tremendously powerful models for studying cooperative changes in cell shape. In addition to their simple anatomy and rapid developmental timeline, Drosophila melanogaster and C. elegans offer an impressive array of tools for facile genetic manipulation and unbiased forward genetic screens.
Drosophila has been an exceptionally strong model for probing glial biology, as the fly nervous system contains ramified glia with clear parallels to mammalian astrocytes. Detailed characterization of astrocyte morphologies and synaptic structures during Drosophila metamorphosis revealed that astrocytes invade the neuropil concomitant with synaptogenesis[23]. Thus, fine-scale changes in neuronal shape occur coordinately with large-scale morphogenetic changes in glia. Moreover, electron microscopy and fluorescence imaging of synapses and astrocytic processes in the larval ventral nerve cord have established that, as in mammals, astrocytes associate closely with synaptic structures and respond to local synaptic cues, although they do not ensheathe individual synapses[24]. These close associations are driven by astrocytic invasion of the neuropil. Analysis of candidate mutants and cell-specific rescue experiments demonstrated that this invasion requires activation of the Drosophila FGF receptor on astrocytes, most likely in response to neuronally-derived FGFs[24]. This suggests that glial morphologies can be specified by the neurons they contact.
Although these studies clearly highlight the power of Drosophila, this system has its drawbacks: with an estimated 250,000 neurons and 25,000 glia, using Drosophila to visualize asingle, defined cell-cell contact at high resolution is still technically difficult. Limitations on live imaging across development and the time-consuming nature of unbiased forward genetic screens further restrict studies of cooperative glia-synapse morphogenesis in Drosophila[25].
C. elegans offers a complementary model system to overcome these challenges. Each cell in the nematode is uniquely identifiable and derived from a known lineage, such that the nervous system of the adult hermaphrodite consists of exactly 302 neurons, 50 neural-derived glia, and 6 mesoderm-derived “glial-related” cells. Remarkably, each of these cells adopts a stereotyped shape and reproducible set of cell-cell contacts. All of these neuron-glia contacts have been catalogued and, as described below, can be grouped into three main classes. Most importantly, this means that a single, defined neuron-glia contact can be revisited across many individuals in wild-type or mutant backgrounds, and can be visualized and manipulated in live animals using a well-developed toolbox of cell-specific promoters to target the expression of transgenes to individual cells. This capacity for labeling single contacts is especially powerful given the transparency and small size of the nematode, which facilitate time-lapse imaging across development and super-resolution imaging of live, anesthetized animals (see Box). Together with the ease of unbiased forward genetic screens, these tools make C. elegans a tremendously potent model system for studying questions of coordinated neuron-glia morphogenesis. Yet this system has its own drawbacks: in contrast to Drosophila, glia in C. elegans are not clearly analogous to specific types of mammalian glia, such as astrocytes or oligodendrocytes. Nevertheless, parallels are emerging between the functions and molecular determinants of C. elegans and mammalian glia, as illustrated by the studies described below.
Overview of neuron-glia contacts in C. elegans
Prodigious efforts to fully reconstruct the architecture of the nervous system by electron microscopy have led to an unprecedented map of C. elegans neuron-glia contacts at the single-cell level[26-28]. All glia in this organism associate with sensory neurons to create sense organs[29,30]. Most of these are located in the animal's head and contain two glial cells of different types, called the sheath and the socket. Each glial cell extends an unbranched process that fasciculates with the dendrites of its neuronal partners and terminates at the tip of the nose.
Three classes of neuron-glia contacts have been described in C. elegans. The first is comprised of contacts between the distal-most region of the sheath glial process and the ciliated receptive endings (REs) of the sensory dendrites of its neuronal partners (Fig. 2A,i). The glial cell ensheathes these dendrites and forms epithelial-like junctions around them; the sheath also intimately wraps a subset of ciliated REs that are embedded within it. These ciliated REs are functionally analogous to dendritic spines; both are highly specialized compartments that contain machinery required to sense cues in the extracellular environment – be that the animal's surroundings or the synaptic cleft[31]. In each case, the tightly interlocking shapes of the two cells allude to precise coordination of neuronal and glial morphologies.
Figure 2. C. elegans as a model for coordinated neuron-glia morphogenesis.
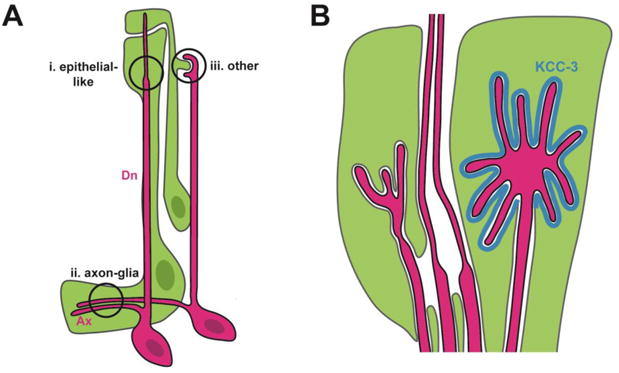
(A) Three main classes of neuron-glia contacts in C. elegans. Neurons, red; glia, green. Ax, axons; Dn, dendrite. (B) The glial cell can create specialized subdomains tailored for specific neuronal partners, as evidenced by the localization of the KCC-3 ion co-transporter (blue) in the glial cell membrane exclusively adjacent to a single dendrite receptive ending.
The second class of neuron-glia contacts consists of axon-glia interactions in which a sheath glial cell envelops axons in the central neuropil and promotes synaptogenesis via secreted cues (Fig. 2A,ii) [32-34].Along with the previously described glia-RE contacts, these have been the focus of all existing studies on glia in the nematode. However, a third class of neuron-glia contact has been observed by electron microscopy. These contacts also involve glia and dendritic REs, but they are more reminiscent of adhesion between mammalian glia and dendritic spines because they lack the epithelial-like junctions described above [26,28]. In one unique example, the “BAG” neuron RE precisely and reproducibly wraps a protrusion extending from its glial partner (Fig. 2A,iii). This raises several questions: what determines the exquisite morphology of a neuron-glia contact? How do the cells communicate to coordinate changes in cell shape? Is coordinated morphogenesis driven by the neuron, the glial cell, or both?
Coordinated morphogenesis of single neuron-glia contacts in C. elegans
Several studies have demonstrated that C. elegans neurons and glia undergo coordinated changes in morphology at sites of neuron-glia contact. First, in studies of glia-axon interactions, unbiased forward genetic screens have shown that synapses in the central neuropil are initially positioned by glia[32]. These synaptic positions are then maintained by epidermal signals that coordinate glial morphogenesis with larval growth [32,34]. Second, during embryonic development, morphogenesis of the sheath glial process and neuronal dendrites are coordinated, such that mutants with failures in dendrite extension show equivalent defects in glial process morphology[35]. Similarly, mutants that disrupt glial development also lead to defects in dendrite extension[33]. These effects are seen at size scales on the order of 100 μm (Fig. 1B, right).
Glia also play a role in the fine sculpting of REs at the scale of a few microns, comparable to the size of a mammalian synapse. When animals enter into an alternative life stage called dauer, some neurons undergo extensive structural remodeling over a period of about 6h[36].The sheath glial cell and the RE of one of its neuronal partners remodel in concert, expanding to encompass more than half the circumference of the head (Fig. 1A, bottom) [37-39]. In this case, the glial cell physically constrains the expansion of the neuronal RE, though glial remodeling occurs independently of neuronal changes. Further, in studies using standard (i.e. non-dauer) growth conditions, laser ablation of single glial cells and cell-specific inhibition of secretion in glia revealed that glia are also required to maintain the shape of neuronal REs embedded within them[40,41]. For example, RNAi-mediated knockdown of glial-enriched genes demonstrated that the transcription factor PROS-1/Prospero functions in glia to regulate the shape of associated dendritic REs[42].
Recent work by Singhvi and colleagues demonstrates that interactions at RE-glia contacts can be uniquely tailored to the identity of the neuron[41]. Candidate screens for morphology mutants identified a potassium-chloride co-transporter, KCC-3, that is expressed by the sheath glial cell. Remarkably, KCC-3 localizes specifically to the region of glial membrane adjacent to just one of the 12 REs ensheathed by the glial cell (Fig. 2B). Mutations in this co-transporter could be rescued by supplying high exogenous levels of potassium and chloride ions, implying that the co-transporter modulates the morphology of this neuronal RE by regulating ion concentrations in the interstitial space. This suggests the sheath glial cell can generate specialized subdomains tailored for particular neuronal partners. Future work will need to address how this specificity is achieved: how do neurons and glia recognize the identity of their partners? Do mammalian astrocytes similarly discriminate among dendritic spines from different neuron types?
In each of the examples discussed above, glia exert control over neuronal shape. Preliminary evidence suggests that neuronal cues can also regulate the fine-scale structure of glia in C. elegans[43,44]. For example, electron micrographs of mutants whose sensory neurons lack dendritic spine-like ciliary endings reveal changes in the fine-scale morphology of the sheath glial cell[44,45]. While the identities of neuronal signals remain enigmatic, the power and ease of unbiased genetic screens in C. elegans promise to uncover the molecular mechanisms by which neurons drive cell shape changes in glia.
Conclusion and Perspectives
Studies in vertebrate systems have described tightly interlocking neuronal and glial morphologies and have revealed striking examples of the coordinated morphogenesis of these cells. However, the detailed structures of glia-synapse contacts are highly heterogeneous and difficult to study at high temporal and spatial resolution in the mammalian brain. In contrast, an expanding toolbox in Drosophila and C. elegans will allow single defined neuron-glia contacts to be observed during development, throughout life, and in response to environmental and genetic perturbations. This experimental framework provides a powerful means to uncover the cellular and molecular mechanisms that govern coordinated neuron-glia morphogenesis.
Box 1. Technological innovations for visualizing coordinated neuron-glia morphogenesis in C. elegans.
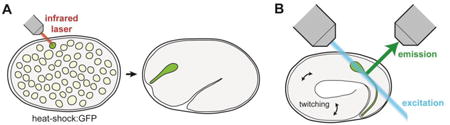
While single, defined neuron-glia contacts can be readily visualized in the mature C. elegans nervous system, a major remaining challenge for studying coordinated neuron-glia morphogenesis is to see these structures in the developing embryo. This challenge stems from two technical impediments. First, cell-specific markers from the mature adult are often not active in the early embryo, making it difficult to label single neuron-glia attachments. Second, rapid embryonic twitching movements – combined with the drug-impermeable eggshell that prevents the use of paralytics – necessitate high temporal resolution when imaging live embryos.
Recently, major inroads have been made to address each of these hurdles. First, single-cell heat-shock induction, schematized in part A of this figure, as well as photoconversion of fluorescent proteins, have been used to achieve cell-specific labeling in the embryo[35,46]. Second, advances in light microscopy – especially the invention of Dual-View Selective Plane Illumination Microscopy (diSPIM), schematized in part B of this figure – have made it possible to image cells in the developing embryo at high spatial and temporal resolution over the entirety of embryogenesis[47,48]. This allows acquisition of clear images through the complete volume of the embryo despite embryonic twitching movements. In addition to these advances in image acquisition, novel post-processing techniques have made it feasible to track single cell movements within the moving embryo[49,50]. Armed with these advances, efforts are now underway to generate large-scale atlases of all cellular positions throughout embryogenesis[49]. In conjunction with recent advances in EM of defined embryonic stages[51], these techniques will facilitate additional studies of neuron-glia contact formation by revealing how neurons and glia move relative to one another in the developing embryo.
Highlights.
Glial and neuronal morphogenesis is coordinated at size scales from <1μm to 100 μm.
Presynaptic signals help to coordinate changes in dendritic spines and glial processes.
C. elegans has highly stereotyped contacts between defined neurons and glia.
C. elegans glia can actively shape their neuronal partners.
Acknowledgments
We thank Dr. Keith Murai, Dr. Aakanksha Singhvi, and Dr. Shai Shaham for thoughtful comments on the manuscript. ERL received support from an NSF Graduate Research Fellowship and NIH F31NS103371. MGH received support from NIH R01GM108754.
References
- 1.Stogsdill JA, Eroglu C. The interplay between neurons and glia in synapse development and plasticity. Current Opinion in Neurobiology. 2017;42:1–8. doi: 10.1016/j.conb.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields RD, Woo DH, Basser PJ. Glial regulation of the neuronal connectome through local and long-distant communication. Neuron. 2015;86:374–386. doi: 10.1016/j.neuron.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spacek J. Three-dimensional analysis of dendritic spines. III. Glial sheath. Anatomy and embryology. 1984;171:245–252. doi: 10.1007/BF00341419. [DOI] [PubMed] [Google Scholar]
- 4.Ventura R, Harris KM. Three-dimensional relationships between hippocampal synapses and astrocytes. The Journal of Neuroscience. 1999;19:6897–6906. doi: 10.1523/JNEUROSCI.19-16-06897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grosche J, Matyash V, Möller T, Verkhratsky A, Reichenbach A, Kettenmann H. Microdomains for neuron–glia interaction: parallel fiber signaling to Bergmann glial cells. Nature neuroscience. 1999;2:139–143. doi: 10.1038/5692. [DOI] [PubMed] [Google Scholar]
- 6.Grosche J, Kettenmann H, Reichenbach A. Bergmann glial cells form distinct morphological structures to interact with cerebellar neurons. Journal of neuroscience research. 2002;68:138–149. doi: 10.1002/jnr.10197. [DOI] [PubMed] [Google Scholar]
- 7.Haber M, Zhou L, Murai KK. Cooperative astrocyte and dendritic spine dynamics at hippocampal excitatory synapses. Journal of Neuroscience. 2006;26:8881–8891. doi: 10.1523/JNEUROSCI.1302-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lippman JJ, Lordkipanidze T, Buell ME, Yoon SO, Dunaevsky A. Morphogenesis and regulation of Bergmann glial processes during Purkinje cell dendritic spine ensheathment and synaptogenesis. Glia. 2008;56:1463–1477. doi: 10.1002/glia.20712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirrlinger J, Hülsmann S, Kirchhoff F. Astroglial processes show spontaneous motility at active synaptic terminals in situ. European Journal of Neuroscience. 2004;20:2235–2239. doi: 10.1111/j.1460-9568.2004.03689.x. [DOI] [PubMed] [Google Scholar]
- 10.Sala C, Segal M. Dendritic spines: the locus of structural and functional plasticity. Physiological reviews. 2014;94:141–188. doi: 10.1152/physrev.00012.2013. [DOI] [PubMed] [Google Scholar]
- 11.Nishida H, Okabe S. Direct astrocytic contacts regulate local maturation of dendritic spines. Journal of Neuroscience. 2007;27:331–340. doi: 10.1523/JNEUROSCI.4466-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Bernardinelli Y, Randall J, Janett E, Nikonenko I, König S, Jones EV, Flores CE, Murai KK, Bochet CG, Holtmaat A, Muller D. Activity-dependent structural plasticity of perisynaptic astrocytic domains promotes excitatory synapse stability. Current Biology. 2014;24:1679–1688. doi: 10.1016/j.cub.2014.06.025. By examining single synapses in mouse hippocampal slices, the authors demonstrate that neuronal activity alters the motility of astrocytic processes, resulting in increased ensheathment of stimulated synapses. Thus, bidirectional communication between these cells controls coordinated changes in cell shape. [DOI] [PubMed] [Google Scholar]
- 13••.Perez-Alvarez A, Navarrete M, Covelo A, Martin ED, Araque A. Structural and functional plasticity of astrocyte processes and dendritic spine interactions. Journal of Neuroscience. 2014;34:12738–12744. doi: 10.1523/JNEUROSCI.2401-14.2014. Here, the authors examine mouse hippocampal slices and show that patterns of neuronal activity induce changes not only in the structure of dendritic spines, but also in the astrocytic processes that contact them. The two cells change shape coordinately, and this is important for normal neural transduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genoud C, Quairiaux C, Steiner P, Hirling H, Welker E, Knott GW. Plasticity of astrocytic coverage and glutamate transporter expression in adult mouse cortex. PLoS Biol. 2006;4:e343. doi: 10.1371/journal.pbio.0040343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberheim NA, Wang X, Goldman S, Nedergaard M. Astrocytic complexity distinguishes the human brain. Trends in neurosciences. 2006;29:547–553. doi: 10.1016/j.tins.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 17••.Wang J, O'Sullivan ML, Mukherjee D, Puñal VM, Farsiu S, Kay JN. Anatomy and spatial organization of Müller glia in mouse retina. Journal of Comparative Neurology. 2017;525:1759–1777. doi: 10.1002/cne.24153. This impressive anatomical study demonstrates that a single Müller glial cell adopts strikingly different morphologies at each layer of the mammalian retina. The precision of these shapes suggests active sculpting of the glial cell by neuronal neighbors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss J, Gebara E, Bushong EA, Sánchez-Pascual I, O'Laoi R, El M'Ghari I, Kocher-Braissant J, Ellisman MH, Toni N. Fine processes of Nestin-GFP–positive radial glia-like stem cells in the adult dentate gyrus ensheathe local synapses and vasculature. Proceedings of the National Academy of Sciences. 2016;113:E2536–E2545. doi: 10.1073/pnas.1514652113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Pannasch U, Freche D, Dallérac G, Ghézali G, Escartin C, Ezan P, Cohen-Salmon M, Benchenane K, Abudara V, Dufour A. Connexin 30 sets synaptic strength by controlling astroglial synapse invasion. Nature neuroscience. 2014;17:549–558. doi: 10.1038/nn.3662. This study identified a surprising role for connexin 30 in actively regulating astrocyte contacts with the synaptic cleft. Such contacts are important for regulating synaptic glutamate levels. [DOI] [PubMed] [Google Scholar]
- 20.Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nature neuroscience. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell stem cell. 2016;18:591–596. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haim LB, Rowitch DH. Functional diversity of astrocytes in neural circuit regulation. Nature Reviews Neuroscience. 2017;18:31–41. doi: 10.1038/nrn.2016.159. [DOI] [PubMed] [Google Scholar]
- 23.Muthukumar AK, Stork T, Freeman MR. Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nature neuroscience. 2014;17:1340–1350. doi: 10.1038/nn.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stork T, Sheehan A, Tasdemir-Yilmaz OE, Freeman MR. Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron. 2014;83:388–403. doi: 10.1016/j.neuron.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman MR. Drosophila central nervous system glia. Cold Spring Harbor perspectives in biology. 2015;7:a020552. doi: 10.1101/cshperspect.a020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. Journal of Comparative Neurology. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- 27.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans: the mind of a worm. Phil Trans R Soc Lond. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 28.Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. eLife. 2014. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans; p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizeracka K, Heiman MG. The many glia of a tiny nematode: studying glial diversity using Caenorhabditis elegans. Wiley Interdisciplinary Reviews: Developmental Biology. 2015;4:151–160. doi: 10.1002/wdev.171. [DOI] [PubMed] [Google Scholar]
- 30.Shaham S. Glial development and function in the nervous system of Caenorhabditis elegans. Cold Spring Harbor perspectives in biology. 2015;7:a020578. doi: 10.1101/cshperspect.a020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaham S. Chemosensory organs as models of neuronal synapses. Nature Reviews Neuroscience. 2010;11:212–217. doi: 10.1038/nrn2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colón-Ramos DA, Margeta MA, Shen K. Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science. 2007;318:103–106. doi: 10.1126/science.1143762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimura S, Murray JI, Lu Y, Waterston RH, Shaham S. mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development. 2008;135:2263–2275. doi: 10.1242/dev.019547. [DOI] [PubMed] [Google Scholar]
- 34.Shao Z, Watanabe S, Christensen R, Jorgensen EM, Colón-Ramos DA. Synapse location during growth depends on glia location. Cell. 2013;154:337–350. doi: 10.1016/j.cell.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heiman MG, Shaham S. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell. 2009;137:344–355. doi: 10.1016/j.cell.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder NE, Androwski RJ, Rashid A, Lee H, Lee J, Barr MM. Dauer-specific dendrite arborization in C. elegans is regulated by KPC-1/Furin. Current Biology. 2013;23:1527–1535. doi: 10.1016/j.cub.2013.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Procko C, Lu Y, Shaham S. Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Development. 2011;138:1371–1381. doi: 10.1242/dev.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Procko C, Lu Y, Shaham S. Sensory organ remodeling in Caenorhabditis elegans requires the zinc-finger protein ZTF-16. Genetics. 2012;190:1405–1415. doi: 10.1534/genetics.111.137786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albert PS, Riddle DL. Developmental alterations in sensory neuroanatomy of the Caenorhabditis elegans dauer larva. Journal of Comparative Neurology. 1983;219:461–481. doi: 10.1002/cne.902190407. [DOI] [PubMed] [Google Scholar]
- 40.Bacaj T, Tevlin M, Lu Y, Shaham S. Glia are essential for sensory organ function in C. elegans. Science. 2008;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Singhvi A, Liu B, Friedman CJ, Fong J, Lu Y, Huang XY, Shaham S. A glial K/Cl transporter controls neuronal receptive ending shape by chloride inhibition of an rGC. Cell. 2016;165:936–948. doi: 10.1016/j.cell.2016.03.026. Here, Singhvi and colleagues demonstrate that a glial K/Cl co-transporter specifically localizes to the glial membrane surrounding just one of the twelve neuronal REs that the glial cell contacts. This remarkable localization of the glial K/Cl co-transporter controls neuronal RE shape. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace SW, Singhvi A, Liang Y, Lu Y, Shaham S. PROS-1/Prospero is a major regulator of the glia-specific secretome controlling sensory-neuron shape and function in C. elegans. Cell reports. 2016;15:550–562. doi: 10.1016/j.celrep.2016.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oikonomou G, Perens EA, Lu Y, Watanabe S, Jorgensen EM, Shaham S. Opposing activities of LIT-1/NLK and DAF-6/patched-related direct sensory compartment morphogenesis in C. elegans. PLoS Biol. 2011;9:e1001121. doi: 10.1371/journal.pbio.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perens EA, Shaham S. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Developmental cell. 2005;8:893–906. doi: 10.1016/j.devcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 45.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Developmental biology. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 46.Singhal A, Shaham S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nature communications. 2017;8:14100. doi: 10.1038/ncomms14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Wawrzusin P, Senseney J, Fischer RS, Christensen R, Santella A, York AG, Winter PW, Waterman CM, Bao Z. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nature biotechnology. 2013;31:1032–1038. doi: 10.1038/nbt.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar A, Wu Y, Christensen R, Chandris P, Gandler W, McCreedy E, Bokinsky A, Colón-Ramos DA, Bao Z, McAuliffe M. Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. nature protocols. 2014;9:2555–2573. doi: 10.1038/nprot.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santella A, Catena R, Kovacevic I, Shah P, Yu Z, Marquina-Solis J, Kumar A, Wu Y, Schaff J, Colón-Ramos D. WormGUIDES: an interactive single cell developmental atlas and tool for collaborative multidimensional data exploration. BMC bioinformatics. 2015;16:189. doi: 10.1186/s12859-015-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christensen RP, Bokinsky A, Santella A, Wu Y, Marquina-Solis J, Guo M, Kovacevic I, Kumar A, Winter PW, Tashakkori N. Untwisting the Caenorhabditis elegans embryo. Elife. 2015;4:e10070. doi: 10.7554/eLife.10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nechipurenko IV, Berciu C, Sengupta P, Nicastro D. Centriolar remodeling underlies basal body maturation during ciliogenesis in Caenorhabditis elegans. eLife. 2017;6:e25686. doi: 10.7554/eLife.25686. [DOI] [PMC free article] [PubMed] [Google Scholar]


