Abstract
Improved insight into cancer cell populations responsible for relapsed disease will lead to better outcomes for patients. Here, we report a single-cell study of B-cell precursor acute lymphoblastic leukemia at diagnosis that revealed hidden developmentally dependent cell signaling states uniquely associated with relapse. With mass cytometry, we simultaneously quantified 35 B-cell developmental proteins in 60 primary diagnostic samples. Each leukemia cell was then matched to it’s nearest healthy B-cell population by a developmental classifier that operated at the single-cell level. Machine learning identified 6 features of expanded leukemic populations sufficient to predict patient relapse at diagnosis. These features implicated pro-BII cells with activated mTOR signaling, and pre-BI cells with activated and unresponsive pre-B-cell receptor signaling, to be associated with relapse. This model, termed Developmentally Dependent Predictor of Relapse (DDPR), significantly improves currently established risk stratification methods. DDPR features exist at diagnosis and persist at relapse. Leveraging a data-driven approach, we demonstrate the predictive value of single-cell ‘omics’ for patient stratification in a translational setting and provide a framework for application in human cancers.
Introduction
Despite high rates of initial response to frontline treatment, cancer mortality largely results from relapse or metastasis. Although there is debate as to whether resistant cancer cells are present at the time of initial diagnosis or whether they emerge under the pressure of therapy, many studies have suggested that it is the former1–4. Such cells can be rare and are not accurately represented in animal models or patient-derived xenografts5,6. Hence, the identification and study of the cellular species underlying cancer persistence will require high-throughput single-cell analyses of primary human tissues and new analytical tools to align these rare populations with clinical outcomes.
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is a common childhood malignancy. Despite dramatic improvements in survival using current treatment regimens, relapse is the most frequent cause of cancer-related death among children with BCP-ALL7. BCP-ALL is characterized by the clonal proliferation of blast cells in the bone marrow and/or peripheral blood that bear the hallmarks of immature B cells. Known molecular alterations stall the development of B lymphocytes (B lymphopoiesis) in BCP-ALL8–12.
Healthy B lymphopoiesis occurs through sequential developmental stages marked by losses and appearances of surface proteins, intracellular mediators of DNA rearrangement, and activation of signaling pathways that regulate decisions of cell fate13,14. We previously applied single-cell cytometry by time-of-flight (CyTOF; mass cytometry) to align developing B cells into a unified trajectory, which enabled us to better define human pre-pro-B, pro-B, and pre-B cells and their regulatory signaling during early developmental checkpoints14.
Currently, for children with BCP-ALL, risk prediction strategies integrate clinical, genetic, and treatment response features gathered during the first months of treatment15. As in most risk-prediction scenarios, prediction is imperfect. We reasoned that performing deep phenotypic single-cell studies of diagnostic leukemic samples could identify cell populations predictive of relapse and discover novel aspects of resistance to treatment in this disease.
Building on our study of normal early B lymphopoiesis, we performed a mass cytometry analysis of primary diagnostic BCP-ALL samples. Aligning individual BCP-ALL cells with developmental states along the normal B-cell trajectory demonstrated expansion across the pre-pro-B to pre-BI transition. Applying machine learning to proteomic features extracted from these expanded cell populations, we constructed a predictive model of relapse that was validated in an independent patient cohort. This model revealed six cellular features that implicated a developmental phenotype and behavioral identity of two cell populations in portending relapse. Analysis of matched diagnosis-relapse pairs confirmed the persistence of these predictive features at relapse. Thus, BCP-ALL samples viewed through a lens of high-resolution developmental maturity indicated that a unique and reproduced cellular behavior across patients is a main driver of relapse.
Results
Deep phenotyping reveals developmental heterogeneity in BCP-ALL
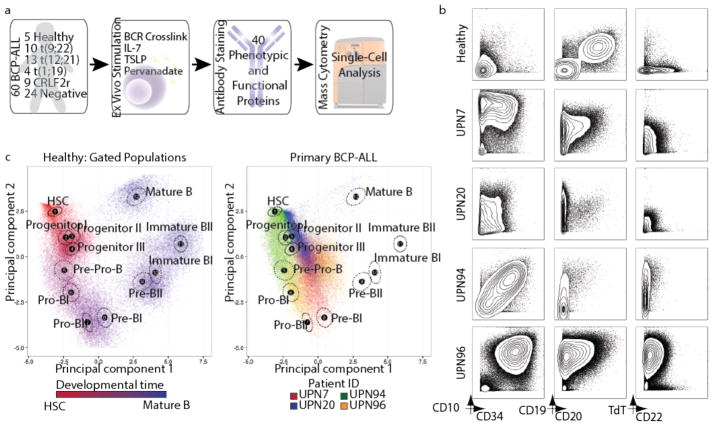
To understand the extent to which childhood BCP-ALL mimics the differentiation of its tissue of origin, we profiled 60 primary diagnostic bone marrow aspirates with diverse clinical genetics by single-cell mass cytometry in comparison to normal bone marrow from five healthy donors (Fig. 1a and Supplementary Tables 1–3). Examining expression of proteins routinely used in diagnostic flow cytometry on leukemic blasts revealed expected patterns of expression, with overexpression of CD10 and CD34 as compared to healthy bone marrow (Fig. 1b). To visualize similarity to normal developing B cells, we compared BCP-ALL cells to their healthy bone marrow counterparts using principal component analysis (PCA) (Fig. 1c and Supplementary Fig. 1). Healthy developing B cells occupied a remarkably clear path in this representation space (Fig. 1c, left). Once projected into the same space, BCP-ALL cells from individual patients fell into areas with similarity to multiple healthy populations, with a heavy skewing towards early stages of B lymphopoiesis (Fig. 1c, right), as expected8. We thus reasoned that aligning individual leukemic cells to their closest developmental state would enable us to view each BCP-ALL sample as a set of aberrant developing B-cell populations, potentially uncovering novel aspects of BCP-ALL biology.
Figure 1. Mass cytometry analysis of BCP-ALL reveals phenotypic heterogeneity of leukemic cells.
(a) Summary of primary BCP-ALL sample processing for mass cytometry analysis (see Supplementary Tables 1–3 for patient information, antibody panel, and perturbation conditions, respectively). 60 primary BCP-ALL samples and 5 healthy control bone marrow aspirates were included. Prognostic cytogenetic translocations identified at diagnosis, as well as relevant ex vivo perturbations used to uncover cell state, are indicated. ‘Negative’ patients were negative for any of the prognostic cytogenetic translocations analyzed. (b) Mass cytometry analysis of commonly used diagnostic antigens expressed by lineage-negative bone marrow cells (see Supplementary Fig. 1a for gating) from 4 representative BCP-ALL patients and 1 healthy donor. (c) Left panel: 5,000 cells from 12 manually gated stages of B-cell development in healthy bone marrow demonstrate phenotypic progression during normal B lymphopoiesis (1,000 cells sampled from each of n = 5 donors). The first 2 principal components were constructed using 11 markers defining B-cell developmental populations (see Supplementary Figs. 1b–d for gating, marker weights, and variance captured by each principal component). The developmental time color scale was defined by setting hematopoietic stem cells as red and mature B cells as blue. Intermediate populations were placed on this red-to-blue color gradient at equal intervals. For each stage, a black dot indicates the population centroid, and the surrounding circle indicates standard error based on 5 healthy donors. Right panel: Data from 4 patients in (b) shown projected onto healthy B-cell progression. Each sample uniquely occupies the PCA space, while overlapping with multiple healthy populations and other patient samples. BCR, B-cell receptor; TSLP, thymic stromal lymphopoietin; PCA, principle component analysis.
Organizing BCP-ALL with a single-cell developmental classifier
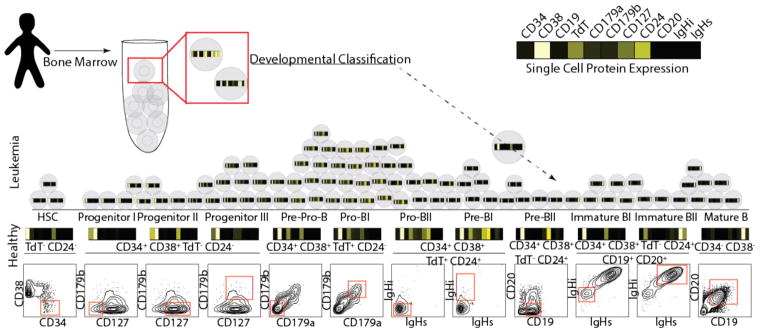
To approximate the developmental state of each leukemic cell, we constructed a single-cell developmental classifier. Building on our recent study14, we manually partitioned healthy bone marrow cells into twelve developing B-cell populations and three populations of mixed progenitors or mature non-B cells (Supplementary Fig. 1b). These fifteen populations were defined by expression of eleven B-cell developmental proteins, providing a phenotypic maturity ‘barcode’ (Fig. 2). To assign each cell to its closest developmental population, we used Mahalanobis distance (Supplementary Fig. 2a). We used manually gated healthy populations to test the reliability of this approach. In a 10-fold cross-validation, this classifier had an excellent predictive performance for each population and a 92% overall accuracy (Supplementary Figs. 2b–d). If a cell was misclassified, it was most likely to fall into a neighboring population, suggesting that the biological effect of misclassification is likely small (Supplementary Fig. 2c). We found that Mahalanobis distance was superior to other distance metrics (cosine14,16, Euclidian17, and Manhattan18) in assigning cells to the correct developmental population (Supplementary Fig. 2d). Moreover, additional phenotypic markers did not improve classification relative to the original eleven proteins (Supplementary Fig. 2e).
Figure 2. Single-cell developmental classifier for BCP-ALL.
Healthy bone marrow aspirates from 5 donors were manually gated into 12 consecutive developmental stages of B lymphopoiesis (final gate is shown as a red box on a contour plot in the bottom, while the text above indicates prior gate(s) on lineage-negative cells; see Supplementary Figs. 1a and 1b for complete gating strategy). The mean arsinh-transformed expression of 11 proteins with relevance to normal B lymphopoiesis, shown in the heat-bar, was determined for each normal cell population (shown above the contour plots, where black indicates low expression and white – high expression). Single cells from each BCP-ALL sample were then assigned to their most similar normal population based on the shortest Mahalanobis distance calculated from expression of the same 11 proteins. Cells with distance above the classification threshold to all developmental populations remained unclassified (<1% for each patient). IgHi, intracellular immunoglobulin heavy chain; IgHs, surface immunoglobulin heavy chain.
We used this single-cell classifier to assign each leukemic cell into the most phenotypically similar developmental stage. We emphasize that in this case we select the closest developmental stage and do not imply that a leukemic cell is equivalent to a normal developing B cell (e.g. a leukemic cell classified as pre-BI is not suggested to be a pre-BI cell, but is rather considered to be a ‘pre-BI-like’ cell). Leveraging prior knowledge of normal cellular differentiation to subset cancer cells into biologically meaningful populations enabled direct comparisons of these populations among healthy donors and patients.
BCP-ALL expands across the pre-pro-B to pre-BI developmental transition
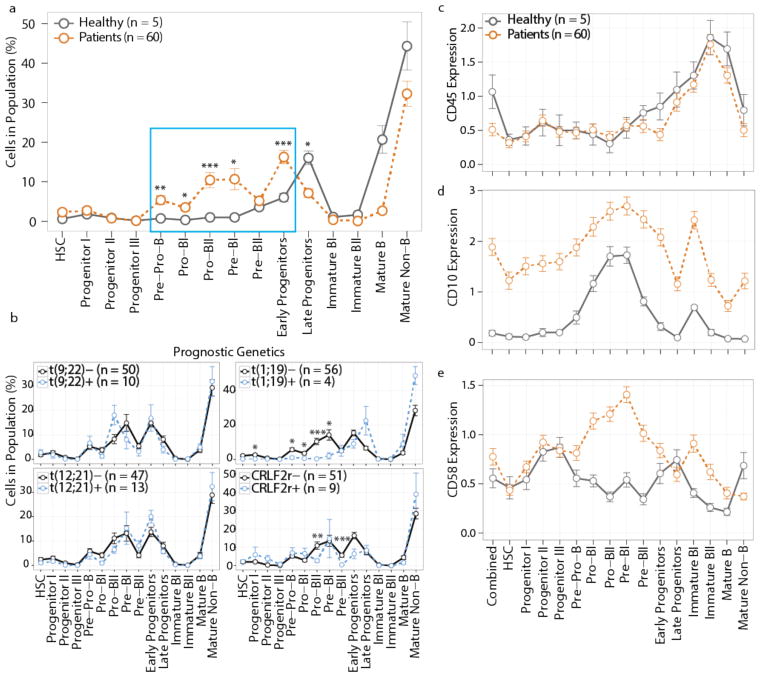
Once classified, the frequency of cells in each developmental compartment was examined across all samples. Compared to healthy controls, we found a significant expansion across the pre-pro-B to pre-BI transitional populations in leukemic samples (Fig. 3a). Despite the clonal nature of leukemia, phenotypic heterogeneity was observed within individual ALL samples, such that in 100% of cases there was an expansion of cells in more than one developmental population (mean ± s.d. = 5.3 ± 1.9; Supplementary Fig. 3a).
Figure 3. Developmental classification reveals that BCP-ALL expands across the pre-proB to pre-BI transition.
(a) Percentage of cells from healthy donor (n = 5, gray line) or diagnostic BCP-ALL patient (n = 60, orange line) bone marrow classified into each developmental population. Cell populations significantly expanded in leukemic samples are shown in the blue box (pre-pro-B p = 0.0012, pro-BI p = 0.011, pro-BII p = 0.00013, pre-BI p = 0.011, early progenitors p = 0.00013); late progenitors contracted (p = 0.036), and the remaining populations did not change significantly (p ≥ 0.05). (b) Percentage of cells in each developmental patient population grouped by diagnostic cytogenetics: i) translocation t(9;22)(q34;q11) BCR/ABL1: t(9;22)- (n = 50) vs. t(9;22)+ (n = 10); ii) translocation t(1;19)(q23;p13) TCF3/PBX1: t(1;19)- (n = 56) vs. t(1;19)+ (n = 4): progenitor I p = 0.015, pre-pro-B p = 0.037, pro-BI = 0.026, pro-BII p = 6.2×10−6, pre-BI p = 0.022; iii) translocation t(12;21)(p13;q22) ETV6/RUNX1: t(12;21)- (n = 47) vs. t(12;21)+ (n = 13), and iv) CRLF2-rearranged: CRLF2r- (n = 51) vs. CRLF2r+ (n = 9): pro-BII p = 0.0033, pre-BII p = 0.00017. (c–e) Antigen expression on bone marrow developmental populations from healthy donors (n = 5, gray line) or patients (n = 60, orange line): CD45 (c), CD10 (d), and CD58 (e). Mean ± s.e.m.; p-values in (a) and (b) are from unpaired two-tailed Welch’s t test accounting for multiple comparisons using Bonferroni correction. “Combined” in (c–e) denotes expression in all cells without developmental classification. *p < 0.05, **p < 0.01, ***p < 0.001.
Since prognosis and risk stratification in BCP-ALL are based partially upon recurrent chromosomal rearrangements, we determined whether there was any association between these translocations and leukemic developmental classification. All known genetic drivers showed little correlation to overall developmental classification, although in some cases expansion of particular populations did reach statistical significance (Fig. 3b). Specifically, patients with translocation 1;19 (TCF3/PBX1) had a contraction in immature B-cell populations when compared to patients without this translocation, corroborating prior findings19,20. In CRLF2-rearranged ALL, pro-BII and pre-BII compartments were contracted. In some cases, we had access to matched bone marrow and peripheral blood samples at diagnosis. In general, the overall classification did not substantially change based on the use of bone marrow vs. peripheral blood samples, but some pro-B to pre-B transitional populations were not as frequently seen in the peripheral blood as in the bone marrow (Supplementary Fig. 3b). Weak association of single-cell developmental classification with recurrent chromosomal rearrangements emphasizes the general applicability of our approach across BCP-ALL samples with diverse clinical genetics.
BCP-ALL maintains the developmental structure of early B cells
Comparing classified leukemia cell subsets to their normal B-cell counterparts, three major patterns emerged: i) an expression pattern similar to healthy B cells (CD45, IKAROS), ii) overexpression in all cell populations (CD10, PAX5), and iii) a developmentally inappropriate expression pattern (CD58, CD123, CD43) (Figs. 3c–e and Supplementary Figs. 3c–e). Surprisingly, despite aberrant expression of three (of 24 examined) developmental proteins, BCP-ALL cells generally maintained the expected developmental progression of protein expression in leukemic populations compared to healthy bone marrow. Interestingly, PAX5, a master B-cell transcription factor frequently mutated in BCP-ALL21, was highly overexpressed on all leukemic cell populations, even in the case of two patients who harbored heterozygous PAX5 deletions based on genomic analysis (Supplementary Fig. 3f and Supplementary Table 4). In contrast to the general maintenance of phenotypic structure in leukemic cells, there was a higher frequency of BCP-ALL cells with activated basal signaling than in controls. In particular, the frequency of cells with active ribosomal protein S6 (rpS6), activated during protein translation and downstream of mTOR, or with activated cAMP-response element-binding protein (CREB) expression, was markedly elevated in leukemic populations as compared to normal B-cells (Supplementary Fig. 3g). Thus, using healthy B-cell progenitor populations to organize BCP-ALL provides a refined view of aberrations in phenotypic and regulatory molecule expression in leukemic cell populations.
Developmentally Dependent Predictor of Relapse, a data-driven model based on BCP-ALL cellular features at diagnosis
Long-term survival without recurrence of disease is the primary clinical indicator of BCP-ALL therapeutic success. For patients for whom ≥3 years of follow-up data were available (n = 54), 31% relapsed (n = 17), a slightly higher rate than the 15–20% expected relapse rate for childhood BCP-ALL15. We first determined whether developmental classification alone could stratify patients who would go on to relapse from those who would remain in remission. Cells from patients who relapsed were not enriched in a particular developmental state relative to cells from patients who did not relapse (Supplementary Fig. 4a), suggesting that relapse is not strictly connected to a particular phenotypic developmental state.
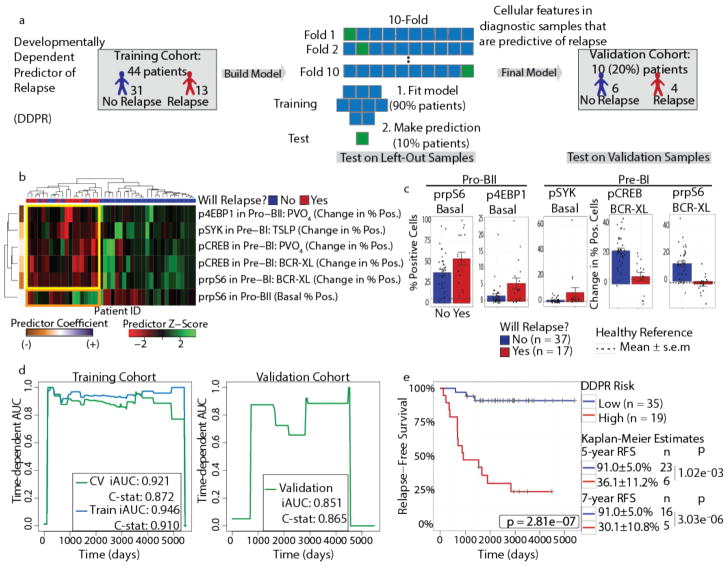
Utilizing the classified cellular features at diagnosis for each patient, we constructed a model to: i) predict clinical outcome (relapse vs. continued remission) and ii) identify a short list of leukemic cell features at diagnosis that are sufficient to predict relapse (Fig. 4a). To do this, we employed a machine learning approach, termed elastic net, designed to identify a small set of predictive features while preserving predictive power22. Since we cannot exclude the possibility that some patients may relapse after the last follow-up date, we modeled relapse as a time-to-event outcome in an elastic net-regularized Cox model23. We applied this method to a set of cell features from the expanded leukemic cell populations, including frequency of cells in each population, expression of surface and intracellular proteins, and frequency of cells with activated signaling molecules in the unperturbed state and in response to each perturbation (Supplementary Table 5). We also included patient age and white blood cell count at diagnosis, as these features are considered important clinically (Supplementary Table 1). This resulted in 352 features per patient across 54 patients, for a total of 19,008 data points.
Figure 4. DDPR predicts which patients will go on to relapse based on features of expanded BCP-ALL populations at diagnosis.
(a) Construction of the Developmentally Dependent Predictor of Relapse (DDPR) model that predicts relapse in BCP-ALL. Data from 54 patients with ≥3 years of follow-up were divided into training (n = 44) and validation (n = 10) cohorts. Cellular features available to DDPR included signaling in the basal state, changes in signaling state following perturbations, mean arsinh-transformed expression of surface and intracellular proteins, and frequency of cells in the expanded developmental populations. DDPR performance was estimated using 10-fold cross-validation (CV) within the training cohort to yield pre-validated relative risk for each patient. The final DDPR model (elastic net-regularized Cox model) was then built using all training cohort samples. Once constructed, DDPR was applied to predict relative risk for samples in the validation cohort. (b) Hierarchical clustering of 6 predictive features of relapse identified by DDPR within the training cohort. The last documented relapse status is shown above the heatmap as relapse (red) or continuous complete remission (blue). Coefficients of predictors are shown on the left of the heatmap. Yellow box indicates 5 features with negative correlation to relapse. Orange box indicates 1 feature with positive correlation to relapse. (c) Bar plots show mean ± s.e.m of key DDPR cellular features in pro-BII and pre-BI cells in all patients (n = 54); p-values are not shown, because these features were selected to be different and non-redundant between classes (unpaired two-tailed Welch’s t test from left to right would yield: p = 0.055, p = 0.044, p = 0.13, p = 5.7×10−6, p = 1.6×10−7). Dashed lines indicate mean levels in the corresponding developmental populations within healthy bone marrow aspirates of 5 healthy donors; dotted lines indicate standard error. (d) Time-dependent AUC curves showing performance for relapse prediction in the training (left) and validation (right) cohorts. Integrated dynamic/cumulative AUC (iAUC) and C-statistic (C-stat) summary measures are shown for each curve built using pre-validated (green, left), overall model fit (blue, left), and predicted (green, right) relative risk of relapse with reference to the sample average. (e) Kaplan-Meier analysis of relapse-free survival (RFS) of all patients with ≥3 years of follow-up (n = 54) stratified by DDPR risk group. An estimate for relative risk of relapse was used to assign a risk group to each patient (pre-validated in the training cohort; predicted in the validation cohort; see Methods). P-values were calculated using the log-rank test. Log-rank tests for the training cohort alone: p = 5.6×10−6; validation cohort alone: p = 0.040. RFS estimates, standard error, number of patients at risk, and p-values for both groups at 5 and 7 years are shown on the right (5 years p = 1.02×10−3, 7 years p = 3.03×10−6). BCR-XL, B-cell receptor crosslink; TSLP, thymic stromal lymphopoietin; PVO4, pervanadate.
Using random sampling, we divided the patient cohort into a training cohort (80% of patients, n = 44) and a validation cohort (20% of patients, n = 10). We applied pre-validation24,25 to estimate performance of the relapse prediction model within the training cohort, and then validated the final model on the independent validation cohort. We therefore assessed the predictive performance of our model twice: within the training cohort and then within the validation cohort. We termed the resulting model Developmentally Dependent Predictor of Relapse (DDPR; pronounced “deeper”).
Activated signaling in pro-BII and pre-BI cells at diagnosis predicts relapse
Of the 352 features, hierarchical clustering of six cellular features identified by DDPR almost perfectly separated patients according to their last documented relapse status (Fig. 4b). Examining individual features revealed two developmentally dependent patterns: i) the ability of cells to respond to ex vivo stimulation was associated with continuous remission (Fig. 4b, yellow box); and ii) an increased frequency of cells with basally active rpS6 signaling was associated with relapse (Fig. 4b, orange box). All six features were confined to pro-BII and pre-BI cell populations. In pro-BII cells of patients who ultimately relapsed, we observed high basal activity of rpS6 and a lack of response to pervanadate, as assessed by phosphorylated eukaryotic translation initiation factor 4E-binding protein 1 (p4EBP1), due to high basal activation of 4EBP1 (Fig. 4c, left; Supplementary Fig. 4b). Similarly, in pre-BI cells, the response to pre-BCR crosslinking or pervanadate, as assessed by prpS6 and pCREB levels, and the response to TSLP stimulation, as assessed by phosphorylated spleen tyrosine kinase (pSYK), were significantly blunted in patients who would go on to relapse (Fig. 4c, right; Supplementary Fig. 4b, right; Supplementary Fig. 4c). In essence, the signaling features predictive of relapse relate to high basal activation of the mTOR pathway in pro-BII cells, as well as high basal activation and a lack of response to stimulation of the pre-BCR pathway in pre-BI cells. Differences in these features were apparent even when applied to a single common genetic subgroup of patients with translocations ETV6/RUNX1 (t(12;21)(p13;q22); Supplementary Fig. 4d), who generally have a favorable prognosis.
To assess DDPR performance, we calculated an integrated cumulative/dynamic area under the curve (iAUC)26 and a C-statistic27, the measures appropriate for censored time-to-event data. In the training cohort, DDPR had a predicted (cross-validated) iAUC of 0.92 and a C-statistic of 0.87 (Fig. 4d, left). Applying the model to the independent validation cohort resulted in an iAUC of 0.85 and a C-statistic of 0.87 (Fig. 4d, right), indicating a strong model performance. To determine whether predictive cellular features could be detected in bulk leukemia cells without developmental classification, we repeated this analysis using features from all cells in the blast gate. Using unclassified bulk data, the model performed inferiorly to DDPR (training cohort iAUCDDPR = 0.92 vs. iAUCbulk = 0.71; validation cohort iAUCDDPR = 0.85 vs. iAUCbulk = 0.66; Supplementary Fig. 4e). DDPR performed well as a risk stratification method at diagnosis in predicting relapse-free survival (RFS) in a retrospective analysis of both cohorts (p = 2.8×10−7; Fig. 4e and Supplementary Fig. 4f). Thus, organizing data from primary diagnostic leukemia samples using single-cell developmental classification was critical for predicting future clinical outcomes.
DDPR synergizes with current risk stratification methods
Current risk prediction integrates a combination of clinical (NCI/Rome criteria28) and genetic (cytogenetic) features at diagnosis, as well as early response to therapy (prednisone response and/or MRD29,30). Final risk, which guides clinical decisions, integrates all of these features and is generally determined three months following initiation of treatment, although for some patients risk may be known earlier.
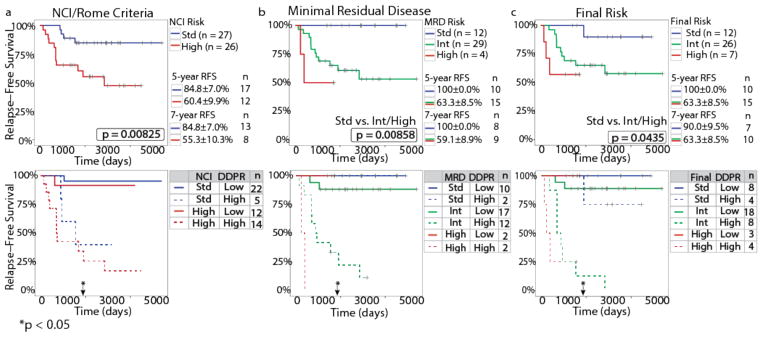
As expected, NCI/Rome criteria successfully stratified 53 patients with available risk data according to RFS (log-rank p = 0.0083; Fig. 5a, top). However, integrating DDPR with NCI/Rome criteria resulted in a significant risk stratification improvement, as assessed by the integrated discrimination improvement index (IDI), continuous net reclassification improvement (NRI), and median improvement (MI) for time-to-event data31,32 at five years following diagnosis (Fig. 5a, bottom; Supplementary Fig. 5a). Within the 45 patients for whom MRD and final risk were available (per AIEOP-BFM 2000 protocol definitions), either MRD risk (p = 0.0086; Fig. 5b, top) or final risk (p = 0.044; Fig. 5c, top) alone performed well in stratifying patients according to RFS. Combining DDPR prediction with either MRD risk (Fig. 5b, bottom) or final risk (Fig. 5c, bottom) significantly improved patient stratification at five years following diagnosis (Supplementary Fig. 5a).
Figure 5. DDPR synergizes with existing risk stratification methods to improve relapse-free survival prediction for patients with BCP-ALL.
(a) A Kaplan-Meier analysis showing RFS for patients with ≥3 years of follow-up data and known NCI/Rome criteria (n = 53), stratified by NCI/Rome criteria alone (top) or NCI/Rome criteria and DDPR (bottom). (b–c) RFS as in (a) for patients with known MRD risk (b) or final risk (c), as determined by protocol definitions (n = 45), stratified by the clinical risk group alone (top) or risk group and DDPR (bottom). An estimate for relative risk of relapse was used to assign a DDPR risk group to each patient (pre-validated in the training cohort; predicted in the validation cohort; see Methods). Kaplan-Meier estimates with standard error and the number of people at risk are shown for 5-year and 7-year RFS in the top plots (5-year and 7-year p-values: NCI/Rome criteria: p = 0.033 and p = 0.040, MRD risk: p = 0.157 and p = 0.084, final risk: p = 0.157 and p = 0.169). The p-values were calculated using the log-rank test; p-values in (b) and (c) are between standard risk and intermediate/high risk groups due to low number of patients in the high risk group. Black arrows with asterisks indicate a significant improvement in patient risk stratification at 5 years following diagnosis achieved by adding DDPR to each established criteria: continuous net reclassification improvement (NRI) for NCI/Rome criteria: p = 0.027, MRD risk: p = 0.033, final risk: p = 0.013; see Supplementary Fig. 5a for NRI estimates and 95% confidence intervals. MRD, minimal residual disease; RFS, relapse-free survival.
In clinical practice, relapse risk determines treatment decisions for patients. Key DDPR features were clearly different in patients who went on to relapse from those who remained in continuous remission, regardless of their NCI/Rome or MRD status (Supplementary Figs. 5b and 5c), suggesting that cellular phenotypes identified by DDPR may provide rational drug targets for patients at risk of relapse.
Cells characterized by activated mTOR and deficits in pre-BCR signaling persist from diagnosis to relapse
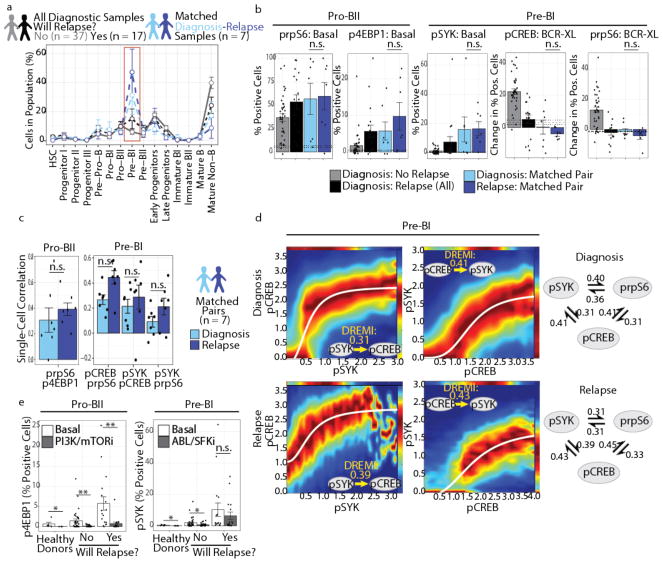
The analyses described thus far do not establish whether features identified by DDPR are present at relapse. To understand how BCP-ALL remodels under the pressure of treatment, seven matched diagnosis-relapse sample pairs were analyzed following developmental classification (Fig. 6a). These samples comprised diverse underlying prognostic genetics, including good risk (ETV6/RUNX1; n = 1), poor risk (BCR/ABL1; n = 2), and without known prognostic genetic aberrations (n = 4). In contrast to the initial diagnostic samples, which expanded to span the pre-pro-B to pre-BI transition, at relapse the expansion narrowed almost exclusively to the pre-BI population, but was also present in the diagnosis specimen (Fig. 6a, red box). In these paired samples, DDPR predictive features were present at diagnosis and were either maintained or exacerbated at relapse (Fig. 6b). Taken together, these results indicate that the cellular populations and features associated with poor outcome exist at diagnosis and persist at relapse.
Figure 6. Cells with DDPR features pre-exist at diagnosis and persist at relapse.
(a) Percentage of cells in each developmental population (mean ± s.e.m.) of all diagnostic BCP-ALL samples from patients who stayed in continuous remission for ≥3 years (gray, n = 37) or went on to relapse (black, n = 17), compared to matched diagnosis-relapse pairs (diagnosis: blue, relapse: purple, n = 7). Red box highlights a significant (p = 0.0030) expansion of pre-BI population at relapse compared to diagnostic samples of patients who did not relapse. P-values were calculated using a two-sided Tukey’s honest significance test and were corrected for multiple comparisons using Bonferroni correction. (b) Bar plots (mean ± s.e.m.) showing key DDPR features in all diagnostic samples compared to matched diagnosis-relapse pairs, as in (a): percent of pro-BII cells with phosphorylated (p) rpS6 or 4EBP1 in non-stimulated (basal) state, percent of pre-BI cells with pSYK in basal state, and change from basal state in percent of pre-BI cells with pCREB or prpS6 signaling following pre-BCR cross-linking (BCR-XL). (c) Bar plots (mean ± s.e.m.) showing Spearman's rank correlation coefficient for key DDPR features listed in (b) was calculated for matched diagnosis-relapse pairs (n = 7): single-cell correlation of arsinh-transformed values between prpS6 and p4EBP1 in pro-BII cells (left), or between pCREB and prpS6, pSYK and pCREB, or pSYK and prpS6 in pre-BI cells (right). None of the DDPR features changed significantly from diagnosis to relapse in (b-c) (paired two-tailed Welch’s t test applied to matched diagnosis-relapse pairs only). (d) DREMI analysis and DREVI visualization for DDPR features in pre-BI cells. Up to 5,000 pre-BI cells from matched diagnosis-relapse pairs (n = 7) were sampled and pooled prior to analysis. Left: Estimated conditional density functions for pSYK-to-pCREB signaling response (pSYK→pCREB) and pCREB→pSYK at diagnosis and relapse; sigmoidal response functions were fitted to each plot. Right: Quantification for strengths of pairwise signaling relationships within the network formed by pSYK, pCREB, and prpS6 at diagnosis and relapse. (e) Bar plots (mean ± s.e.m.) showing response of DDPR features (basal p4EBP1 in pro-BII cells and basal pSYK in pre-BI cells) to short-term ex vivo treatment (see Supplementary Table 3) in healthy donors (n = 5) or diagnostic samples (no relapse: n = 37, relapse: n = 17). Shown are the effects of BEZ235 (a dual PI3K and mTOR inhibitor, PI3K/mTORi) in pro-BII cells (healthy p = 0.023, no relapse p = 0.0032, relapse p = 0.0092) and of dasatinib (a dual BCR-ABL and SRC family kinase inhibitor, ABL/SFKi) in pre-BI cells (healthy p = 0.031, no relapse p = 0.048, relapse p = 0.22). Effects were assessed using two- tailed Welch’s t test. n.s, not significant (p ≥ 0.05), *p < 0.05, **p < 0.01.
Since we are able to examine the concomitant expression of proteins in the same cell, we examined single-cell pairwise correlation between prpS6 and p4EBP1 in pro-BII cells from the matched specimens. We found that the levels of these proteins were correlated at both diagnosis and relapse (Spearman’s rho±s.e.m. for prpS6-p4EBP1: diagnosis 0.31±0.10, relapse 0.39±0.05 ; Fig. 6c, left; Supplementary Fig. 6a). Similarly, pre-BI cells displayed a moderate correlation between their implicated molecules pSYK, pCREB, and prpS6 (e.g. pCREB-prpS6: diagnosis 0.27±0.04, relapse 0.45±0.05; Fig. 6c, right; Supplementary Fig. 6b). To quantify the strengths of signaling relationships between these proteins in the pro-BII and pre-BI populations, we utilized conditional-Density Resampled Estimate of Mutual Information (DREMI) to estimate the dependency between each pair of proteins33. In leukemic pre-BI cells, pSYK, pCREB, and prpS6 were indeed likely to belong to cross-correlated signaling networks that persisted from diagnosis to relapse (Fig. 6d). These pre-BI cells cannot respond to pre-BCR crosslinking by further increasing the levels of pCREB and prpS6 (Supplementary Fig. 6b). Similarly, in the pro-BII population, DREMI confirmed an expected dependency between p4EBP1 and prpS6 (Supplementary Fig. 6d). Interestingly, for both p4EBP-prpS6 and pSYK-pCREB dependencies, the strength of the signaling relationship was enhanced at relapse, indicating that there was either a modest strengthening of the relationship between these molecules, or an increased homogeneity of cell populations. In either case, the evidence points towards the importance of these signaling relationships during progression towards an aggressive tumor state. By comparison, in patients who did not relapse, co-activation of CREB, rpS6, and SYK at baseline was diminished in pre-BI cells, and these cells had the ability to co-activate CREB and rpS6 in response to pre-BCR crosslinking (Supplementary Fig. 6c).
To determine if the predicted cellular signaling phenotypes can be therapeutically targeted, we treated primary diagnostic samples ex vivo with chemical inhibitors relevant to the activated pathways. Treatment of healthy or leukemic cells with BEZ235, a dual PI3K/mTOR inhibitor (PI3K/mTORi) reduced the frequency of pro-BII cells with activated p4EBP1 (Fig. 6e, left). However, prpS6 was not as strongly inhibited by BEZ235 in leukemic pro-BII and pre-BI cells, suggesting multiple routes to rpS6 activation (Supplementary Fig. 6e). By contrast, treatment with dasatinib, a dual ABL/SRC family kinase inhibitor (ABL/SFKi), reduced the frequency of pre-BI cells with activated SYK in patients who remained in remission to levels approaching that in normal bone marrow; yet, its effect was minor in patients who would go on to relapse (Fig. 6e, right). Dasatinib was also able to slightly reduce the frequencies of pro-BII cells with activated rpS6 and 4EBP1, as well as of pre-BI cells with activated rpS6 and CREB (Supplementary Fig. 6f). Together, these data indicate that some DDPR features may be therapeutically targetable, but knowledge of underlying developmental signaling must guide the design of therapeutic approaches for improving patient outcomes.
Discussion
Deep phenotyping of primary cancer samples provides a unique opportunity to examine intratumoral heterogeneity and link patient outcomes to specific cellular populations. Yet, it remains challenging to organize these massive datasets into meaningful models. Data from single-cell analyses of primary tumors must be stratified to understand how cellular diversity of tumors impacts disease progression or treatment response.
This study leveraged the phenotypic profiling depth enabled by mass cytometry to perform high-parameter single-cell analyses of primary patient samples, without extended ex vivo culture or passage through an immunodeficient mouse, to preserve biology as it exists in the patient. We addressed the intrinsic heterogeneity of the single-cell data by showing that BCP-ALL cells could be reliably aligned to a developmental continuum of normal cell populations. In doing so, we organized diverse data from multiple patients into a ‘universal’ physiologic standard allowing relevant comparisons. This approach not only allowed for the identification of key cell populations and their behaviors in relation to clinical outcome, but also provided mechanistic insight into the persistent disease state.
Direct comparison of BCP-ALL cells to human B-cell progenitors enabled identification of developmental states most vulnerable to malignant transformation: the pre-pro-B to pre-BI transition. During this transition, normal B cells rearrange their immunoglobulin heavy chain locus with coordinated cell fate decisions. We have previously demonstrated changes in network structure surrounding the differentiation of pro-B cells into pre-B cells, particularly regarding the transition between IL-7 receptor and pre-BCR signaling pathways14. Signaling through the IL-7 receptor in normal pro-B cells activates JAK/STAT and PI3K pathways34,35. Subsequently normal developing B cells proceed to further expansion and differentiation after receiving a strong pre-BCR signal informing them of a successful heavy chain rearrangement34. DDPR identified activation of signaling molecules in the pre-BCR (pSYK, pCREB) and PI3K/mTOR pathways (prpS6, p4EBP1) around this developmental transition in portending relapse. Aberrant BCR-related signaling is known to be pathogenic in other B-cell malignancies, such as diffuse large B-cell lymphoma36. Dysregulation of pre-BCR signaling has been described in Ph+ BCP-ALL, likely mediated by upregulation or mutation of inhibitory regulators37,38,39. Our results suggest that leukemia may exploit this normal developmental process to maintain pre-BCR signaling at a ‘just right’ level. ABL/SFKi was effective in targeting pre-BCR signaling in pre-BI cells, as assessed by pSYK, but only in patients who would not go on to relapse. Thus, an understanding of normal developmental signaling states, and how leukemic cells maintain or diverge from these states in their developmental context, can guide therapeutic efforts to impact patient outcomes.
As we have demonstrated previously, cellular behavior is a key determinant of resistance to therapy40–44. It is not just the outward identity of leukemia cells, as measured by surface molecules, that determines therapy resistance, but instead how those cells behave, as measured by intracellular signaling states. This model cannot currently determine the origin of leukemic transformation, but our data suggest that the drive of leukemic cells to differentiate remains strong as they attempt to continue their developmental program. Given the limited size of the patient cohort studied, DDPR must be applied to larger cohorts of patients. It will be of interest to extend this type of analysis to adult BCP-ALL, as it is well understood that older age at diagnosis carries a higher risk of relapse. Moreover, this developmental model of BCP-ALL would benefit from integration with genomic, epigenetic, and transcriptomic investigations of both healthy and leukemic populations. Defining the mechanisms that control survival and proliferation decisions at the pro-BII to pre-BI developmental transition will inform how BCP-ALL exploits these pathways and how genetic lesions associated with leukemia cooperate with the underlying developmental program.
Altogether, this study shows that aligning transformed cells to their normal developmental path can improve the risk stratification system and identify, with improved precision, the most relevant cell populations for further study and therapeutic targeting. More broadly, this study highlights the translational value of understanding cancer at the single-cell level and applying machine learning to guide treatment paradigms for patients with cancer.
Online Methods
Patient and healthy control bone marrow samples
Reagents and methods relevant to this work can also be found in the Life Sciences Reporting Summary. Fresh human bone marrow was obtained from healthy adult donors (n = 5; 3 females: 20, 27, 44 y.o., 2 males: 26, 28 y.o., average: 29 y.o.; AllCells, Alameda, CA, USA). De-identified pediatric BCP-ALL bone marrow specimens were obtained under informed consent from Lucile Packard Children’s Hospital at Stanford (Stanford, CA, USA; Ph+ samples, n = 9) and from Pediatric Clinic University of Milan Bicocca (Monza, Italy; n = 51) for a total of 60 primary diagnostic patient samples. Use of these samples was approved by the Institutional Review Boards at both institutions. All relevant ethical regulations were followed in this study. No a priori power calculation was performed, as this is the first study of its kind. Patients were included based on availability of sufficient cells for mass cytometry study and with at least 3 years of follow-up data from date of diagnosis. MLL-rearranged infants were excluded from this study. Clinical data were available for these samples, including minimal residual disease (MRD) risk group and final risk assignment as per AIEOP-BFM ALL 2000 protocol (ClinicalTrials.gov identifier: NCT00613457)45, diagnostic cytogenetics, age at diagnosis, gender, white blood cell count at diagnosis, date of diagnosis, date of relapse, and date of the last follow-up (Supplementary Table 1). Median follow-up time was 5.5 years. In agreement with the AIEOP-BFM ALL 2000 study45, median time-to-relapse was 2.0 years. Median follow-up time for patients in continuous complete remission (CCR) was 7.6 years. Gene copy number and mutational analysis for genes IKZF1, P2RY8-CRLF2, CDKN2A/B, PAX5, ETV6, BTG1, RB1, and ERG was carried out using multiplex ligation-dependent probe amplification (MLPA) for 20 patients; IGH@-CRLF2 rearrangement was tested by fluorescent in situ hybridization (FISH) for 20 patients; and JAK2 mutations were identified by high-resolution melt (HRM) for 16 patients (Supplementary Table 4).
Clinical protocol definitions
Treatment protocols for each patient are indicated in Supplementary Table 1. Therapy for all patients was based on the Berlin-Frankfurt-Munster (BFM) backbone46. For Italian patients (the majority in this study), MRD risk and final risk were assigned as per the AIEOP-BFM ALL 2000 clinical protocol45. Briefly, after 7-day monotherapy with prednisone and 1 dose of intrathecal methotrexate, treatment was complemented with corticosteroid, vincristine, daunorubicin, and L-asparaginase. Remission induction was followed by intravenous cyclophosphamide and cytarabine, intrathecal methotrexate, and oral mercaptopurine. Risk group assignment resulted from a combination of presenting features and response to therapy measured by peripheral blood morphology at day 8 and MRD at Time Point (TP) 1 (day 33) and TP2 (day 78) as assessed by polymerase chain reaction analysis of immunoglobulin gene rearrangements. Patients were determined to be standard risk (SR) if MRD was negative at both TP1 and TP2 and no adverse clinical features were observed; intermediate risk (IR) if MRD was positive at TP1 or TP2 but less than 10−3; MRD high risk (HR) if MRD was greater than 10−3 at TP2. The HR group included patients with any of the following criteria: Ph+ ALL, t(4;11) or MLL/AF4; “prednisone-poor-response” (PPR: >1,000 blasts/mL on day 8); failure to achieve CR on TP1; or high degree of MRD at TP2 (more than 1×103)45. Clinical remission is defined by the presence of less than 5% blasts in the bone marrow by morphologic inspection.
Mass cytometry
Samples were processed as previously described47. Briefly, viably preserved bone marrow cells were thawed and resuspended in 90% RPMI with 10% FCS supplemented with 20 U/mL sodium heparin (Sigma-Aldrich, St. Louis, MO, USA), 0.025 U/mL benzonase (Sigma-Aldrich), 1X L-glutamine, and 1X penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA). Cells were rested at 37°C for 30 minutes and stained for viability with cisplatin as described48. Following viability staining, cells were perturbed under the following conditions: pervanadate,
BCR crosslinking, IL-7, thymic stromal lymphopoietin (TSLP), dasatinib, BEZ-235, or tofacitinib; sources, concentrations, and timepoints are listed in Supplementary Table 3. Cells were then fixed with paraformaldehyde (PFA; Electron Microscopy Sciences, Hatfield, PA, USA) to a final concentration of 1.6% for 10 minutes at room temperature. Cells were barcoded using 20-plex palladium barcoding plates prepared in-house as described49. In order to control for batch effects, we included at least one healthy bone marrow reference sample within each barcoding plate. A total of 36 barcode plates were used in this study. Following barcoding, cells were pelleted and washed once with cell staining media (CSM; PBS with 0.5% BSA, 0.02% sodium azide) to remove residual PFA. Blocking was performed with Purified Human Fc Receptor Binding Inhibitor (eBioscience, San Diego, CA, USA) following the manufacturer’s instructions. Surface marker antibodies were added, yielding 50 or 100 μL final reaction volumes, and samples were incubated at room temperature for 30 minutes (Supplementary Table 2). Cells were pelleted and washed with CSM before permeabilization with 4°C methanol for 10 minutes at 4°C, then optionally stored at −80°C. Cells were washed with CSM and stained with intracellular marker and phospho-specific antibodies in 50 μL for 30 minutes at room temperature (Supplementary Table 2). Cells were washed once in CSM, then stained with 1:5000 191Ir/193Ir DNA intercalator (Fluidigm, South San Francisco, CA, USA) in PBS with 1.6% PFA for 20 minutes at room temperature. Cells were washed once with CSM, washed twice with double-distilled water, filtered to remove aggregates, and resuspended in 139La/142Pr/159Tb/169Tm/175Lu normalization beads50 immediately prior to analysis using a CyTOF1 mass cytometer (Fluidigm). Throughout analysis, cells were maintained at 4ºC with Sample Chiller and introduced at a constant rate of ~300 cells per second.
Lineage depletion of healthy samples
To enrich Ficoll-treated bone marrow from healthy donors for rare hematopoietic and B lymphocyte progenitors, cells were incubated with biotin-conjugated antibodies (Supplementary Table 6) for 30 minutes at a concentration of 5 million cells per 100 μL. Cells were washed with CSM twice then incubated with BD Streptavidin Particles Plus (BD Biosciences, Franklin Lakes, NJ, USA) at the manufacturer’s recommended concentration for 30 minutes at room temperature. Particle-labeled cells were resuspended in CSM to approximately 2–8×107 cells/mL and placed in a magnetic holder for 7 minutes. The supernatant was transferred to a new tube, and the beads/cells were washed and resuspended and placed back in the magnetic holder for an additional round of depletion and supernatant recovery. This washing procedure was repeated. Cells from the supernatant were then pelleted by centrifugation at 250 g for 5 minutes. Depleted healthy cells were then stimulated and fixed before being aliquoted for use as controls on each barcode plate. These samples were then stained and analyzed alongside the leukemia samples.
Processing of mass cytometry data
Data were normalized together using bead normalization50, and files were debarcoded as described49. After debarcoding, we obtained ≥600,000 single-cell events per patient. Single-cell protein expression data were extracted using Bioconductor software (www.bioconductor.org) and transformed using the inverse hyperbolic sine (arsinh) function with a cofactor of 5. To control for batch effects among barcoding plates, we performed percentile normalization using healthy reference bone marrow sample(s) included within each plate (normalization values across barcoding plates were in the range 0.97 – 1.18, indicating that batch effects prior to normalization were small). Expression of proteins in each population of interest was determined by calculating the mean level of expression after arsinh transformation. Percent positive cells for each phosphorylated protein was based on a mass cytometry cutoff of ≥10 counts. For single-cell correlation between two antigens, we calculated Spearman’s rank correlation coefficient using arsinh-transformed data for each patient separately. To perform DREMI (conditional-Density Resampled estimate of Mutual Information) analysis and DREVI (conditional-Density Rescaled Visualization) visualization, we sampled up to 5,000 cells from each patient and assessed strengths of pairwise interaction on pooled samples using a MATLAB-based software, simpledremi (www.c2b2.columbia.edu/danapeerlab/html/dremi.html)33.
Manual gating
Single cells were gated using Cytobank software (www.cytobank.org) based on event length and 191Ir/193Ir DNA content (in order to avoid debris and doublets) as described47. Following single-cell gating, live non-apoptotic cells were gated based on cleaved PARP (cPARP) and 195Pt content48. Platelets and erythrocytes were excluded by gating on CD61 and CD235a, respectively. The remaining fraction was gated to exclude T cells and myeloid cells based on CD3e and CD33/CD16, respectively. After further exclusion of CD38High plasma cells, the remaining fraction was defined as lineage-negative blasts (Lin− B+; see Supplementary Fig. 1a for gating). Further analysis was applied to this Lin− B+ fraction unless otherwise noted.
Single-cell developmental classification
Lin− B+ fraction from healthy human bone marrow was gated into 15 developmental populations of normal B lymphopoiesis, mixed progenitors, and mature non-B fractions as shown in Supplementary Fig. 1b. The distribution of each population was based on the expression of 11 B-cell developmental proteins that were used for manual gating: CD34, CD38, CD127, CD24, terminal deoxynucleotidyl transferase (TdT), CD179a, CD179b, intracellular immunoglobulin heavy chain (IgHi), surface IgH (IgHs), CD19, and CD20. Prior to classification, each leukemia sample was normalized to control for batch effects (see Processing of mass cytometry data). Lin− B+ cells from each leukemia sample were then assigned to the most similar healthy fraction based on the shortest Mahalanobis distance among distances to all healthy developmental populations in these 11 dimensions. For stability in calculating Mahalanobis distance, the covariance matrix eigenvalues were set to ≥0.2. A cell was designated “unclassified” if none of the distances were below the classification threshold (Mahalanobis distance = 11 based on the number of dimensions). These single-cell classification parameters were optimized via 10-fold cross-validation using cells with known population assignment from healthy bone marrow donors. Optimization parameters included additional protein markers as well as cosine, Manhattan, or Euclidian distance metrics. By using bone marrow from adult healthy donors as a reference, we assume that B lymphopoiesis proceeds through the same stages of development in both children and adults. The percentages of B-cell developmental fractions in healthy samples are not relevant to the performance of the single-cell developmental classifier. As such, the gradual decline in the B-cell output with age does not confound single-cell developmental classification under this assumption.
Relapse predictive modeling
To construct a predictive model of relapse, termed the Developmentally Dependent Predictor of Relapse (DDPR), we allocated 54 BCP-ALL patients with ≥3 years of follow-up data into training (80% of patients, n = 44) and validation (20% of patients, n = 10) cohorts using a random assignment function that preserves proportions of cases and controls. A complete set of mass cytometry features available to DDPR is listed in Supplementary Table 5. It includes frequency of cells in each of 5 expanded populations (pre-pro-B, pro-BI, pro-BII, pre-BI, early progenitors), average expression of 24 proteins in these populations (CD10, CD19, CD20, CD22, CD24, CD34, CD38, CD43, CD45, CD58, CD79b, CD123, CD127, CD179a, CD179b, HLA-DR, IgHi, IgHs, IKAROS, Ki-67, PAX5, RAG1, TdT, TSLPr), and frequency of cells with each of 9 activated signaling molecules (p4EBP1, pSTAT5, pPLCγ2, pAKT, pSYK, prpS6, pERK1/2, pCREB, pIKAROS) in the unperturbed state and in response to each of 4 perturbations (BCR crosslink, IL-7, TSLP, pervanadate; as per Supplementary Table 3). Two clinical features available to DDPR (age and white blood cell count at diagnosis) are in Supplementary Table 1. Missing values were imputed as a median of all values. All features were scaled using their mean and s.d. within the training cohort, and these scaling parameters were also applied to the validation cohort. DDPR was built using elastic net22, a regularized machine learning approach that utilizes both L1 and L2 penalty types to prevent overfitting. To take advantage of the follow-up data in our cohort, relapse was modeled as a time-to-event outcome in a regularized Cox proportional hazard model23. To estimate DDPR predictive performance, we used pre-validation24,25. In a 10-fold cross-validation, we sequentially constructed an elastic net-regularized Cox model using 90% of the training cohort samples and tested that model on the remaining 10% of the training cohort samples. We then used all training cohort samples to construct the final model. Feature scaling parameters and final DDPR coefficients are listed in Supplementary Table 7. To test DDPR predictive performance, we applied the final model to the validation cohort. To assess performance, we reported an integrated cumulative/dynamic area under the curve (iAUC)26 and a C-statistic27 for censored time-to event data calculated using predicted relative risk (with reference to the sample average; RR) values within each cohort. To stratify patients into low and high DDPR risk groups, we selected a RR threshold (0.9967) based on the optimal log-rank p-value calculated from the fitted RR values within the training cohort. We then applied this threshold to pre-validated (training cohort) and predicted (validation cohort) RR values, yielding the final DDPR risk group assignments for all patients (Supplementary Table 1).
Statistical analysis
Data analysis was performed using R statistical software (www.r-project.org). To test statistical significance between two groups, we applied two-tailed unpaired Student’s t test; when more than two groups were compared, we used Tukey’s honest significance difference test; Bonferroni correction was included for multiple comparisons; when equal variance assumption was not met based on an F test, Welch’s t test was used instead of Student’s t test; normality was assessed using the Shapiro-Wilk test. Paired Welch’s t test was used for paired samples. The Kaplan-Meier method was used to estimate relapse-free survival rates; differences between groups were assessed using the log-rank test. DDPR relapse-free survival curves were built using predicted risk group assignment for each patient (i.e. based on pre-validated RR for the training cohort and predicted RR for the validation cohort). To test for synergy between current risk stratification methods and DDPR, we calculated integrated discrimination improvement index (IDI), continuous net reclassification improvement (NRI), and median improvement (MI) for censored time-to-event data and their statistical significance at 5 years32,51.
Data availability statement
The mass cytometry data are available at https://github.com/kara-davis-lab/DDPR/releases.
Supplementary Material
Editorial summary.
Using mass cytometry data from diagnostic biopsies of children with B-cell precursor acute lymphoblastic leukemia (BCP-ALL), Good et al. match leukemic cells to their closest counterparts in normal B-cell development and use this information to identify a signaling state that predicts relapse.
Acknowledgments
We thank G. Fragiadakis, M.H. Spitzer, P.F. Gherardini, A. Tsai, R.M. Angelo, J. Levine, M. O’Brien, D. Pe’er, M. Shipp. and the ASH/EHA TRTH program for discussions. This work received funding from the Stanford Immunology NIH Training program (5T32AI007290-29, -30, -32; 2T32AI007290-31; Z.G.), Fondazione Italiana per la Ricerca sul Cancro (FIRC-AIRC, 19488; J.S.), M.Tettamanti Foundation and Benedetta è la vita ONLUS Foundation (J.S.), Damon Runyon Cancer Research Foundation Fellowship (DRG-2017-09; S.C.B.), NIH (K99GM104148-01; S.C.B.), and Associazione Italiana per la Ricerca sul Cancro (20564; A.B.). G.P.N. is supported by the NIH grants R01CA184968, 1R01GM10983601, 1R01NS08953301, 1R01CA19665701, R01HL120724, 1R21CA183660, R33CA0183692, 1R33CA183654-01, U19AI057229, 1U19AI100627, U54-UCA149145A, N01-HV-00242 HHSN26820100034C, and 5UH2AR067676; the NIH Northrop-Grumman Corporation Subcontract 7500108142; FDA grants HHSF223201210194C and BAA-15-00121; DOD grants OC110674 and W81XWH-14-1-0180; NWCRA Entertainment Industry Foundation; and Bill and Melinda Gates Foundation grant OPP1113682. K.L.D. is supported by the NetApp St. Baldrick’s Foundation Scholar award and a CureSearch Young Investigator award. Z.G. and G.P.N. are members of the Parker Institute for Cancer Immunotherapy, which supported the Stanford Cancer Immunotherapy Program.
Footnotes
Competing financial interests
S.C.B. and G.P.N. are paid consultants for Fluidigm, the manufacturer that produced some of the reagents and instrumentation used in this manuscript.
Author Contributions
Conceptualization: G.P.N. and K.L.D.; Methodology: Z.G., S.C.B., E.F.S., L.W., N.S., N.A., G.F., W.J.F., R.T., and K.L.D.; Software: Z.G., N.S., and R.T.; Validation: Z.G., J.S., A.J., and K.L.D.; Formal Analysis: Z.G. and R.T.; Investigation: J.S., A.J., and K.L.D.; Resources: N.J.L., G.G., A.B., K.L.D., and G.P.N.; Writing – Original Draft: Z.G. and K.L.D.; Writing – Review & Editing: Z.G., J.S., S.C.B., R.T., G.G., K.L.D., and G.P.N.; Supervision: G.G., K.L.D., and G.P.N.; Funding Acquisition: A.B., K.L.D., and G.P.N.
References
- 1.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2010;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 2.Walter MJ, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klco JM, et al. Functional Heterogeneity of Genetically Defined Subclones in Acute Myeloid Leukemia. Cancer Cell. 2014;25:379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duque-Afonso J, Cleary ML. The AML Salad Bowl. Cancer Cell. 2014;25:265–267. doi: 10.1016/j.ccr.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DDB, PCHP Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncology. 2013;14:e205–e217. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- 8.Greaves MF. Differentiation-linked leukemogenesis in lymphocytes. Science. 1986;234:697–704. doi: 10.1126/science.3535067. [DOI] [PubMed] [Google Scholar]
- 9.Bhojwani D. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan LE, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118:5218–5226. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullighan CG, et al. BCR–ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 13.LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teachey DT, Hunger SP. Predicting relapse risk in childhood acute lymphoblastic leukaemia. Br J Haematol. 2013;162:606–620. doi: 10.1111/bjh.12442. [DOI] [PubMed] [Google Scholar]
- 16.Spitzer MH, et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science. 2015;349:1259425. doi: 10.1126/science.1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samusik N, Good Z, Spitzer MH, Davis KL, Nolan GP. Automated mapping of phenotype space with single-cell data. Nat Methods. 2016 doi: 10.1038/nmeth.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP. A Continuous Molecular Roadmap to iPSC Reprogramming through Progression Analysis of Single-Cell Mass Cytometry. Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bicocca VT, et al. Crosstalk between ROR1 and the Pre-B Cell Receptor Promotes Survival of t(1;19) Acute Lymphoblastic Leukemia. Cancer Cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crist W, et al. Prognostic importance of the pre-B-cell immunophenotype and other presenting features in B-lineage childhood acute lymphoblastic leukemia: a Pediatric Oncology Group study. Blood. 1989;74:1252–1259. [PubMed] [Google Scholar]
- 21.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 22.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R. The lasso method for variable selection in the Cox model. Statist Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Tibshirani RJ, Efron B. Pre-validation and inference in microarrays. Statistical Applications in Genetics and Molecular Biology. 2002;1 doi: 10.2202/1544-6115.1000. Article 1. doi.org/10.2202/1544-6115.1000. [DOI] [PubMed] [Google Scholar]
- 25.Höfling H, Tibshirani R. A study of pre-validation. Ann Appl Stat. 2008;2:643–664. [Google Scholar]
- 26.Uno H, Cai T, Tian L, Wei LJ. Evaluating Prediction Rules for t-Year Survivors With Censored Regression Models. Journal of the American Statistical Association. 2007;102:527–537. [Google Scholar]
- 27.Uno H, Cai T, Pencina MJ, D'Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Statist Med. 2011;121 doi: 10.1002/sim.4154. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith M, et al. Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. J Clin Oncol. 1996;14:18–24. doi: 10.1200/JCO.1996.14.1.18. [DOI] [PubMed] [Google Scholar]
- 29.Basso G, et al. Risk of Relapse of Childhood Acute Lymphoblastic Leukemia Is Predicted By Flow Cytometric Measurement of Residual Disease on Day 15 Bone Marrow. Journal of Clinical Oncology. 2009;27:5168–5174. doi: 10.1200/JCO.2008.20.8934. [DOI] [PubMed] [Google Scholar]
- 30.Borowitz MJ, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children's Oncology Group study. Blood. 2008;111:5477–5485. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pencina MJ, D'Agostino RB, Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Statist Med. 2010;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uno H, Tian L, Cai T, Kohane IS, Wei LJ. A unified inference procedure for a class of measures to assess improvement in risk prediction systems with survival data. Statist Med. 2012;32:2430–2442. doi: 10.1002/sim.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnaswamy S, et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science. 2014;346:1250689. doi: 10.1126/science.1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesisthrough interplay of IL-7 receptorand pre-B cell receptor signalling. Nat Rev Immunol. 2013:1–12. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Reilly LA, et al. MEK/ERK-Mediated Phosphorylation of Bim Is Required to Ensure Survival of T and B Lymphocytes during Mitogenic Stimulation. The Journal of Immunology. 2009;183:261–269. doi: 10.4049/jimmunol.0803853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 37.Trageser D, et al. Pre–B cell receptor–mediated cell cycle arrest in Philadelphia chromosome–positive acute lymphoblastic leukemia requires IKAROSfunction. J Exp Med. 2009;206:1739–1753. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldhahn N, et al. Mimicry of a constitutively active pre-B cell receptor in acute lymphoblastic leukemia cells. J Exp Med. 2005;201:1837–1852. doi: 10.1084/jem.20042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shojaee S, et al. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat Med. 2016;22:379–387. doi: 10.1038/nm.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotecha N, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14:335–343. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibbs KD, Jr, et al. Decoupling of Tumor-Initiating Activity from Stable Immunophenotype in HoxA9-Meis1-Driven AML. Stem Cell. 2012;10:210–217. doi: 10.1016/j.stem.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irish JM, et al. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc Natl Acad Sci USA. 2010;107:12747–12754. doi: 10.1073/pnas.1002057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 44.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conter V, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115:3206–3214. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- 46.Al-Fannah Chemotherapy Regimens for Paediatric ALL. 2008:1–11. [Google Scholar]
- 47.Bendall SC, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pencina MJ, D'Agostino RB, Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Statist Med. 2010;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass cytometry data are available at https://github.com/kara-davis-lab/DDPR/releases.