Abstract.
Histoplasma capsulatum is the causative agent of histoplasmosis and this fungus inhabits soils rich in phosphorus and nitrogen that are enriched with bird and bat manure. The replacement of organic matter in agroecosystems is necessary in the tropics, and the use of organic fertilizers has increased. Cases and outbreaks due to the presence of the fungus in these components have been reported. The Instituto Colombiano Agropecuario resolution 150 of 2003 contains the parameters set by the Colombian Technical Standard (NTC 5167) on the physicochemical and microbiological features of fertilizers, but it does not regulate the search for H. capsulatum. The aim of this study was to demonstrate H. capsulatum presence in organic fertilizers by nested polymerase chain reaction (PCR). A total of 239 samples were collected: 201 (84.1%) corresponded to organic fertilizers, 30 (12.5%) to bird excrement, and 8 (3.4%) to cave soils. The Hc100 nested PCR had a detection limit of 0.1 pg/µL and a specificity of 100%. A total of 25 (10.5%) samples were positive and validated by sequencing. Seven of the positive samples represented locations where H. capsulatum was previously detected, suggesting the persistence of the fungus. No significant correlations were detected between the physicochemical and microbiological parameters with the presence of H. capsulatum by nested PCR, indicating the fungus existence in organic fertilizers that complied with the NTC 5167. The Hc100 nested PCR targeting H. capsulatum standardized in this work will improve the evaluation of organic fertilizers and ensure the prevention of outbreaks and cases due to manufacturing, marketing, and use of fertilizers contaminated with H. capsulatum.
INTRODUCTION
Histoplasma capsulatum is a thermal and nutritionally dimorphic fungus. At temperature of 28°C, H. capsulatum grows as a mold composed of septate hyaline hyphae, tuberous macroconidia (7 to 15 µm), and thin-walled microconidia (2 to 6 µm). Microconidia in conjunction with hyphal fragments constitute the infective particles. The fungus transforms into the yeast form in the host or in culture at 37°C, and this yeast form comprises blastoconidia measuring 2 to 4 μm.1,2
Histoplasma capsulatum infection is an accidental event that occurs when a contaminated source is disturbed and the infective particles become aerosolized. These particles reach the alveoli upon inhalation, where they convert into the yeast form.1,3 The development of histoplasmosis depends on both host and fungal conditions. In the host, disease development depends on the status of the host immune system and lung structural defects, whereas in the fungus, it depends on the amount of inhaled fungus and the virulence.1,4 As a result, clinical manifestations of the disease can range from asymptomatic infection to a progressive and fatal disseminated form.1,2,5 Furthermore, a latent focus has been found in the host that can be reactivated at the onset of risk factors that reduce the immune response, such as transplants, hematologic malignancies, therapy with corticosteroids, or the advanced stages of human immunodeficiency virus/acquired immunodeficiency syndrome.1,6,7
Since the first isolation by Emmons,8–10 numerous studies have confirmed the presence of H. capsulatum in soils rich in nitrogen and phosphorus, such as soils containing bird and bat excrement.10–14 In 1968, Lockwood and Garrison15 demonstrated the ability of H. capsulatum to grow with different concentrations of nitrogen (N), which was shown to promote fungal growth. The same year, Brandsberg16 described fungi isolated together from soils harboring H. capsulatum, but the author noted that a more complete physicochemical and microbiological characterization of the soils where H. capsulatum was isolated was needed to identify areas with a greater risk of infection. Based on studies of cases and outbreaks and areas where this fungus has been recovered, the populations at increased risk of becoming infected include workers who perform construction and demolition activities; miners, archaeologists and speleologists who visit caves inhabited by bats during their work; individuals working in aviculture, poultry manure, or bat guano collection and the production of organic fertilizers with these excrements; and farmers, housewives, and anyone who handles these fertilizers. Therefore, histoplasmosis has been designated as an occupational and recreational disease from rural and urban areas. However, the people who practice these activities are often unaware of the risk to which they are exposed.6,14,17–19
Although the prevalence of histoplasmosis in Colombia is unknown because its reporting is not mandatory, histoplasmosis is thought to be the most common endemic systemic mycosis. The most comprehensive studies of H. capsulatum infection were conducted by Restrepo et al.20 and Carmona Fonseca,21 which showed 10% to 32% reactivity to intradermoreaction in the national territory. More than 420 cases have been reported through a national survey in recent years.22 In addition, 18 outbreaks were registered in the Colombian Andean region, three of which were associated with poultry manure and composted fertilizers.23,24 These findings demonstrate the need for methods to detect the presence of this fungus in this type of material and to characterize the sources of infection.
Recently, the Colombian poultry industry experienced sustained growth. For example, in 2010, there was an increase of 5.5% in the production of broilers and table eggs25 that created the need to seek alternatives for the disposal of excrement. This increase combined with the need to restore organic matter in agroecosystems26 generated an increase in the production of organic fertilizers made from poultry manure, which must be stabilized and sanitized. Composting is the most commonly used stabilization procedure.25
Because of the boom in the production and use of composted organic fertilizers in Colombia, the Ministry of Agriculture and the Instituto Colombiano Agropecuario (ICA) generated policies for the handling of organic fertilizer that were contained in resolution 150 of 2003.27 This policy outlines the parameters of the technical regulation of fertilizers and soil conditioners and defines organic fertilizers in terms of the physical, chemical, and microbiological variables contained in the Colombian Technical Standard (NTC 5167).28 The goal of microbiological studies is to find Salmonella spp., facultative mesophiles, and total enterobacteria and fungi; culture media such as Sabouraud or potato dextrose agar are used for the latter purpose. It is very difficult to recover H. capsulatum from these media because this fungus grows slowly (3 to 6 weeks); thus, fast growing environmental bacteria and mold overcome H. capsulatum in most cases.
Traditionally, the search for H. capsulatum in samples highly contaminated by saprophytic microorganisms (i.e., soil samples) was performed using methods such as inoculation in mice or conventional culture. However, the difficulty in isolating H. capsulatum has led to the use of molecular tools based on polymerase chain reaction (PCR), which allows the rapid detection of the fungus in samples. In 1999, Reid and Schafer29 standardized a nested PCR targeting the internal transcribed spacer of the 5.8S ribosomal ribonucleic acid that would allow the detection of H. capsulatum in soil samples. However, their assay was only useful when H. capsulatum was the sole microorganism present in the sample. Other assays have been designed to detect the fungus primarily in clinical samples. For example, in 2002, Bialek et al.30 published a nested PCR assay to detect H. capsulatum DNA in samples of clinical origin quickly and with high sensitivity and specificity. This PCR targeted a 100 kDa protein (Hc100-PCR) that was constitutive in the H. capsulatum genome.31 In 2003, two studies with conventional PCRs were published; these PCRs were designed to amplify a portion of the sequence encoding two fungal antigens (the H32 antigen and M33 antigen). Both studies showed less sensitivity and specificity than that reported by Bialek for the Hc100-PCR. Therefore, our goal is to standardize and optimize the Hc100 nested PCR for the study of organic fertilizers because conventional techniques are expensive, have low sensitivity, and require a lot of time and skills to be performed.6,32,34,35
The availability of a protocol to detect H. capsulatum in organic fertilizers, soils, and bird excrement based on a molecular biology technique will provide an agile, sensitive, and specific diagnosis to facilitate the design of preventive strategies and environmental interventions to identify sources of outbreaks and cases caused by this fungus. In addition, the inclusion of the protocol designed in our study within the parameters required by the NTC 5167 for the physicochemical and microbiological characterization of organic fertilizers and amendments will contribute to the knowledge of the soil and environmental conditions that promote the development and permanence of the fungus, thereby enabling the prevention of the emergence of cases and outbreaks during the production and handling of materials contaminated with H. capsulatum.
MATERIALS AND METHODS
Sample features.
Samples were collected from organic fertilizers, which were the compost products from a source of organic matter (i.e., food scraps, pruning material, straw, or sawdust) and a nitrogen source (i.e., excrement from animals [primarily poultry or other birds], cave floors, and bird droppings).
The collection was casual and was performed from 2010 to 2014. A total of 239 samples with the following characteristics were collected and analyzed: 175 fertilizers and organic amendments analyzed according to NTC 5167; a total of 26 composted organic fertilizers that were not analyzed according to NTC 5167; a total of eight soils samples from caves in the Rio Claro region, Antioquia, Colombia; and a total of 30 bird depositions collected in the northern municipality of Medellin.
Determination of the physicochemical and microbiological characteristics of the organic fertilizers.
Organic manure samples collected by the Interdisciplinary Group of Molecular Studies (GIEM) were analyzed using the methodology and parameters described in NTC 5167 (updated in 2011).
The assessed variables were as follows:
Essential nutrients: nitrogen (N), phosphorus (P2O5), potassium (K2O), sodium (Na), calcium (CaO), magnesium (MgO), zinc (Zn), aluminum (Al), and carbon (C);
Heavy metals: chromium, cadmium, lead, and nickel;
Microorganisms: mesophiles, thermophiles, mold and yeast, nematodes and/or protozoa, Enterobacteriaceae, and Salmonella spp.;
Physicochemical characteristics: ash, C/N ratio, capacity to retain water, cation exchange capacity (CEC), moisture, pH, density, particle size, germination, and respirometric index.
Histoplasma capsulatum culture.
Isolate 13475 H. capsulatum was isolated from a patient who visited the Corporation for Biological Research (Corporación para Investigaciones Biológicas, CIB). This isolate was transformed into yeast by incubation at 37°C with 5% CO2 in 5% brain heart infusion (BHI) agar medium (Ref. 211065; Brain Hearth Infusion, BBL™, Franklin Lakes, NJ) prepared as described by the manufacturer’s instructions and supplemented with 1% glucose (Ref. G5400-250G; Sigma, St. Louis, MO), 0.01% L-cysteine (Ref. C-7755; Sigma), 0.1% antibiotic solution (gentamicin and penicillin) and sheep blood until abundant yeast growth was observed.
DNA extraction from H. capsulatum isolates in yeast phase.
The phenol–chloroform–isoamyl alcohol method was used for DNA extraction from the 13475 isolate in the yeast phase.36 The DNA concentration and quality were evaluated with a NanoDrop ND1000 spectrometer (Thermo Scientific, Wilmington, DE) and agarose gel electrophoresis, respectively.
DNA extraction from organic fertilizer, soil, and bird dropping samples using the “FastDNA SPIN Kit for Soil.”
The FastDNA SPIN Kit For Soil® (MP Biomedicals, Santa Ana, CA) was used to extract DNA from composted fertilizers using the manufacturer’s instructions with some modifications. Briefly, the extraction was performed using the supernatant obtained from the suspension of 10 g of sample in 30 mL of saline solution containing 0.001% Tween 80 and 0.1% antibiotics (gentamycin and penicillin). The suspension was stirred vigorously for 1 minute and allowed to settle for 20 minutes. This procedure was repeated twice. After the last stirring, the suspension was allowed to settle only until the largest particles were settled; then, the supernatant was collected for DNA extraction. The other modification consisted of an increased contact time between the sample and kit reagents.
Hc100 nested PCR assay for H. capsulatum.
Two sets of specific primers for H. capsulatum were used for this test (a pair of external primers Hc I→ 5′-GCG TTC CGA GCC TTC CAC CTC AAC-3′ and Hc II→ 5′-ATG TTC CAT CGG GCG CCG TGT AGT-3′ and a pair of internal primers Hc III→ 5′-GAG ATC TAG TCG CGG CCA GGT TCA-3′ and Hc IV→ 5′-AGG AGA GAA CTG TAT CGG TGG CTT G-3′) that were previously designed by Bialek et al.30 These primers target a fragment of the sequence of the single copy gene encoding a 100 kDa protein that is constitutive in the H. capsulatum genome.31 The external primers amplified a 391 bp fragment within which the internal primers annealed and amplified a 210 bp fragment. The presence of this amplification product was considered positive for H. capsulatum. The primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA).
Based on the protocol proposed by Dr. Taylor in 2005,34 temperature gradients of each pair of primers (depending on the guanine-cytosine content and magnesium concentration gradients) were taken into account to define the assay conditions.
Agarose gel electrophoresis.
Agarose gels (Ref.: N605-500G; Amresco, Solon, OH) prepared at 1.5% in Tris-borate-EDTA buffer were used to visualize the amplification products of the Hc100 nested PCR. Electrophoresis was performed for 40 minutes at 80 V with 10 μL of the PCR product and 5 μL of GelRed Nucleic Acid Gel stain (Ref.: 41003; Biotum, Hayward, CA) in each lane. The bands were visualized and documented in an ultraviolet transilluminator.
Detection limit of the Hc100 nested PCR.
The detection limit was defined as the smallest amount of DNA that could be detected by the technique and visualized by electrophoresis in an agarose gel. The detection limit was tested in two ways. First, the assay was performed with DNA obtained from clinical isolate 13475 with serial dilutions starting from a concentration of 2 ng/µL up to 0.1 pg/µL. Second, the assay was performed to determine its ability to detect the minimum amount of mycelial DNA using a portion of a sample of soil fertilized with manure and inoculated with 1:10, 1:100, and 1:1,000 dilutions of a solution prepared with 3,000 colony-forming units (CFU)/mL of H. capsulatum in the mycelial phase. The DNA extraction was performed using the FastDNA Spin Kit For Soil® (MP Biomedicals).
Specificity of the Hc100 nested PCR.
To assess the capacity of the PCR to amplify only the H. capsulatum DNA, DNA samples from H. capsulatum var. capsulatum and H. capsulatum var. duboisii were used. DNA samples from fungi phylogenetically related to H. capsulatum belonging to the order Onygenales, such as Blastomyces dermatitidis, Paracoccidiodes brasiliensis, and Coccidiodes immitis, and DNA samples from fungi that shared the same habitat as H. capsulatum, such as Cryptococcus gatti and Aspergillus spp., were used.
Search for H. capsulatum in organic fertilizers by Hc100 nested PCR.
Because of the characteristically high number of PCR inhibitors in the organic fertilizer samples, each sample processed to detect the presence of H. capsulatum was accompanied by an inhibition control consisting of the addition of equal parts of the DNA obtained from the sample and H. capsulatum DNA obtained from a pure culture to the reaction mixture as a positive control. The absence of an amplification product in the inhibition control indicated the presence of inhibitors. If PCR inhibitors were found in the samples, the inhibitors were removed by washing the DNA with phenol–chloroform–isoamyl alcohol.36 In addition, a positive control and a negative control were included in all PCR assays consisting of DNA purified from a culture of H. capsulatum in yeast phase (1 ng/µL) and sterile distilled water, respectively.
Sequencing of the products amplified by Hc100 nested PCR.
Bidirectional sequencing of the amplification products obtained by the Hc100 nested PCR with the HcIII and HcIV internal primers was performed using the chain termination method. The method used ABI 3730XL DNA sequencing technology with quality criteria QV20 (Macrogen Inc., Geumcheon-gu, Korea) to elucidate whether the amplification products corresponded to the H. capsulatum genetic material. The obtained sequences were edited manually based on the chromatograms, and the mold and complementary sequences were identified using BioEdit v7.2.3. software. Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to verify that the sequenced PCR products belonged to H. capsulatum. The G186AR, H88, H147, and NAm1 strain genomes registered at the Broad Institute Fungal Genomics database (http://www.broadinstitute.org/scientific-community/science/projects/fungal-genome-initiative/fungal-genome-initiative) were used as the references. In addition, the sequences were aligned using MUSCLE software, and a distance tree was constructed using the neighbor-joining method. Four B. dermatitidis sequences were used as the outgroups because these sequences were the closest phylogenetic species to H. capsulatum.
Histoplasma capsulatum recovery in microbiological cultures from fertilizer and organic amendment samples that tested positive in the Hc100 nested PCR.
The Mycosel culture medium supplemented with 100 mg/µL oxytetracycline (Genfar, Cali, Colombia) was used at 2% to recover environmental isolates of H. capsulatum from organic fertilizer samples.
Statistical analysis.
The physicochemical and microbiological parameters according to the NTC 5167 of the 175 organic fertilizer and amendment samples were compared with the presence of H. capsulatum determined by the Hc100 nested PCR to determine their correlations. This analysis was performed with the Mann–Whitney test with GraphPad Prism version 5.00 for Windows. For this analysis, the samples were separated into two groups based on the results of the Hc100 nested PCR (presence of H. capsulatum genetic material or the absence of the fungal DNA). A total of 24 variables were analyzed in each group subdivided into the following categories: essential nutrients, heavy metals, microorganisms, and physicochemical characteristics.
RESULTS
Standardization of the Hc100 nested PCR assay to assess the presence of H. capsulatum in organic fertilizers.
The standardized conditions for the Hc100 nested PCR consist of a first reaction with 10 μL of DNA in a final volume of 50 μL per reaction and a final concentration of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1 U of Taq polymerase (Thomas Scientific, Swedesboro, NJ), 0.2 mM of each primer (HcI-HcII), and 0.2 mM deoxynucleoside triphosphates (Thomas Scientific). The nested PCR mix was similar except that 2 μL of the product of the first PCR and 0.2 mM internal primers (HcIII–HcIV) were used. The temperatures and times for the first reaction were as follows: one cycle at 94°C for 5 minutes; 35 cycles at 94°C for 30 seconds, 66°C for 1 minute, and 72°C for 1 minute; and a final cycle at 72°C for 5 minutes. The second reaction consisted of one cycle at 94°C for 5 minutes, 30 cycles at 94°C for 30 seconds, 65°C for 30 seconds, and 72°C for 1 minute, and a final extension at 72°C for 5 minutes.
Detection limit of H. capsulatum DNA amplified by Hc100 nested PCR.
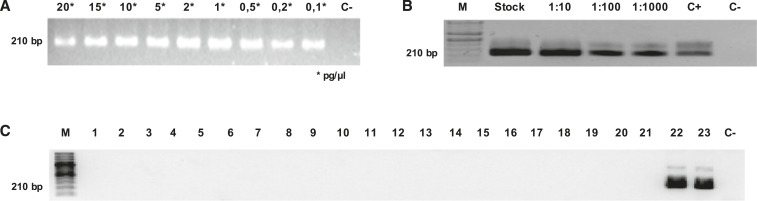
The amplification product was obtained from 0.1 pg/µL DNA using the standardized Hc100 nested PCR (Figure 1A), which corresponded to three H. capsulatum cells per 10 g of organic fertilizer and amendment sample based on the genome of the fungus weighing between 2.3 and 3.2 × 107 bp.37 These results were correlated with the assay performed with the DNA extracted from sterile compost samples infected with 1:10, 1:100, and 1:1,000 dilutions of a suspension containing 3,000 CFU/mL of H. capsulatum in the mycelial phase. The fungus was detected in all dilutions, with the 1:1,000 dilution equivalent to having three cells of H. capsulatum per 10 g of these types of samples (Figure 1B).
Figure 1.
Detection limit of Histoplasma capsulatum DNA and specificity test of the Hc100 nested polymerase chain reaction (PCR). (A) Application of the Hc100 nested PCR with DNA extracted from the yeast phase of H. capsulatum diluted from 20 to 0.1 pg/μL. (B) Amplification products of the Hc100 nested PCR from the 1:10, 1:100, and 1:1,000 serial dilutions of compost infected with 3,000 CFU/mL of H. capsulatum in the mycelial phase. (C) In the specificity test the amplification product was obtained only from the Histoplasma spp. DNA sample. The observed bands were due to the excess DNA used and was corrected when the DNA concentration was controlled (e.g., the positive control with 2 ng/μL DNA). Line: 1 Paracoccidioides brasiliensis (Pb) 01, 2: Pb03, 3: Pb18, 4: Pb60855, 5: Coccidioides immitis, 6: Blastomyces dermatitidis, 7: Emmonsia crecens, 8: Emmonsia parva, 9: Sporothryx schenckii, 10: Colletotrichum spp., 11: Trichoderma spp., 12: Pseudocercospora fijiensis, 13: Aspergillus spp., 14: Chrysosporium spp., 15: Microsporum gypseum, 16: Geotrichum spp., 17: Fusarium spp., 18: Penicillium spp., 19: Saccharomyces cerevisiae, 20: Candida albicans, 21: Cryptococcus neoformans, 22: Histoplasma duboisii, 23: H. capsulatum, 24: negative control.
Specificity of the Hc100 nested PCR to amplify H. capsulatum DNA.
The amplification product obtained from the Hc100 nested PCR with an expected molecular weight of 210 bp was detected only in the H. capsulatum var. capsulatum DNA and H. capsulatum var. duboisii DNA samples. No amplification product (Figure 1C) was found when assessing this PCR with DNA from other fungal species.
Usefulness of the Hc100 nested PCR for the detection of H. capsulatum in soil, organic fertilizers, and poultry depositions.
The Hc100 nested PCR analysis of the 239 tested samples showed that 25 samples (10.5%) were positive for the presence of genetic material from H. capsulatum based on detection of the 210 bp band in each case. These 25 samples were distributed as follows: 17 samples (7.1%) were composted fertilizers certificated according to the ICA regulation for marketing; two samples (2.1%) were fertilizers collected by the Grupo de Micología Médica (GMM); five samples (0.8%) were depositions of birds, and one sample (0.4%) was collected in a cave in the Rio Claro area in Antioquia.
Persistence of H. capsulatum in the environment.
Positive Hc100 nested PCR samples were used in a traceability study to identify areas where the presence of the fungus was detected. This analysis showed that the locations where H. capsulatum was found appeared repeatedly among the positive samples. In the 17 organic fertilizers collected by GIEM, five came from the same company that produced compost and samples studied throughout the work at different times. This company was contacted to collect new samples, and 15 samples were obtained; H. capsulatum genetic material was detected in two of these samples. In the organic fertilizer samples collected by GMM, one came from the municipality of Concordia, Antioquia, where an outbreak of histoplasmosis that affected 19 people was reported in 199323,38; the fungal genetic material was detected in a soil sample from the El Condor cave in the Rio Claro region where Dr. Moncada isolated the fungus in 1989.39
Sequencing of the amplification products obtained by Hc100 nested PCR.
A total of 80% of the positive samples from the Hc100 nested PCR were sequenced and submitted to GenBank, these sequences can be accessed from the code MF801604 to MF801619. The sequences of the amplification products were compared with one another and with the sequences of reference strains deposited in GenBank. Blast analysis of the alignments confirmed that the amplification products corresponded to a 210 bp fragment of the 100 kDa H. capsulatum protein gene coding region with an overall value of e−7, which was a highly reliable value that allowed us to validate that the samples were positive.
Recovery of H. capsulatum in culture from Hc100 nested PCR positive samples.
The 25 positive samples from the Hc100 nested PCR were seeded into Mycosel culture medium. Despite the use of broad-spectrum antibiotics, such as oxytetracycline, and a longer observation period for the cultures (8 to 10 weeks), we were not able to obtain environmental isolates of H. capsulatum. Abundant bacterial and mold growth was obtained, such as Penicillium spp., Aspergillus spp., Trichoderma spp., and Geotrichum spp.
Correlation of physicochemical and microbiological characteristics of organic fertilizers with the presence of H. capsulatum genetic material by Hc100 nested PCR.
Essential nutrients.
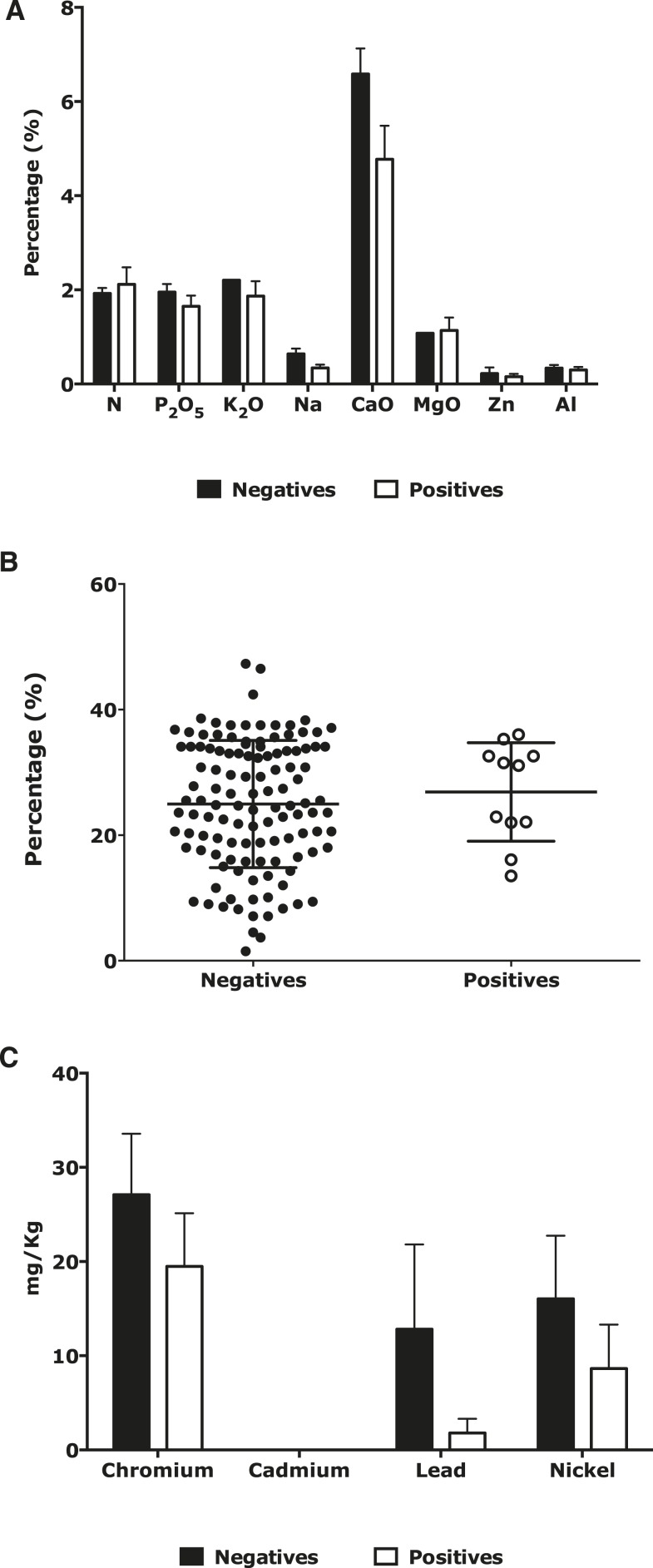
The P values for N, P2O5, and K2O between the negative and positive samples for the Hc100 nested PCR were 0.63, 0.61, and 0.47, respectively. Therefore, the differences were not significant. The mean concentrations in both the positive and negative samples were above 1%, which was considered a high value for organic fertilizers (i.e., both groups of samples were rich in these components; Figure 2A). Moreover, the standard deviations of both the positive and negative samples had a very wide range that showed large variability in the values of nutrients assessed in these types of samples. The N, P2O5, and K2O results were reviewed because the literature reported the preference of H. capsulatum for inhabiting sites rich in these three components.6,40,41
Figure 2.
Comparison of concentrations of essential nutrients (2A), organic carbon (2B), and heavy metals (2C) in organic fertilizers and amendments positive and negative for Histoplasma capsulatum by Hc100 nested polymerase chain reaction (PCR). The comparison was done with the percentage of the different nutrients and the standard error mean between negative and positives samples for Hc100 nested PCR.
Microorganisms use organic carbon (C) for metabolic oxidation. The temperature rises and the volume of the pile is reduced as a result of C consumption. The analysis of this essential nutrient showed no significant differences (P value = 0.54) between samples positive and negative for the presence of H. capsulatum by Hc100 nested PCR (Figure 2B).
Heavy metals.
The concentrations of the assessed heavy metals (chromium, cadmium, lead, and nickel) in the group of samples negative and positive for H. capsulatum by Hc100 nested PCR were below 22 mg/kg, which was lower than the limit values required by NTC 5167. Thus, both groups of samples met the regulations. Moreover, no significant differences were found when comparing the concentrations of these metals between the two groups of samples because the P values were above 0.05, as shown in Figure 2C.
Microorganisms.
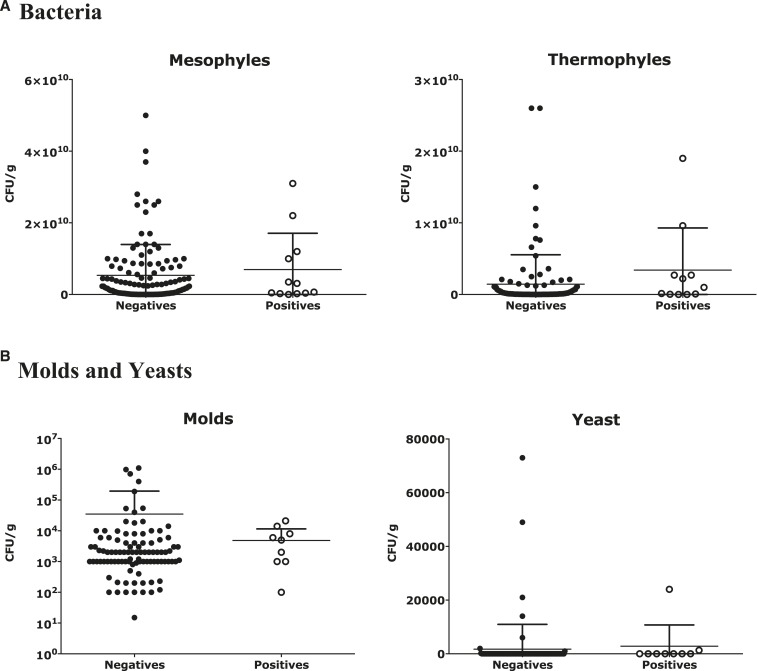
As mentioned, NTC 5167 requires the determination of mesophilic and thermophilic bacteria, mold, and yeast; nematodes and/or protozoa; enterobacteria; and Salmonella spp. The latter is tested because mature compost should not contain pathogenic microorganisms. Although NTC 5167 does not provide reference values for microorganisms, the GIEM, which is based on experience and knowledge of issues relating to composted organic fertilizers and amendments, states that the bacterial population must be reduced in the final product. The bacterial population is eliminated after the competition process between saprophytes and pathogens and the environmental saprophytic fungal population is increased because of fungal involvement in the degradation of more complex compounds such as lignin which are found at the end of the process. Figure 3 shows that both positive and negative samples for the presence of H. capsulatum by Hc100 nested PCR had a similar population of both bacteria and environmental saprophytic fungi (i.e., no significant differences were found, and the P values were > 0.05). Figure 3 also shows that the bacterial population is both mesophilic and thermophilic in both groups of samples in the order of millions of CFU/g. In the fungal population, more mold (> 107 CFU/g) was found than yeast (> 104 CFU/g), indicating that organic fertilizers generally exhibited great variability in their microorganism populations.
Figure 3.
Populations of microorganisms in the samples positive and negative for the presence of Histoplasma capsulatum by Hc100 nested polymerase chain reaction. Colony-forming units (CFU) were determined by plate cultures to determine the amount of mesophyles, termophyles, molds, and yeast recovered by gram of organic fertilizer. The data are presented as mean and standard deviation of CFUs (log10) detected on plate cultures after 8 weeks.
Physicochemical characteristics.
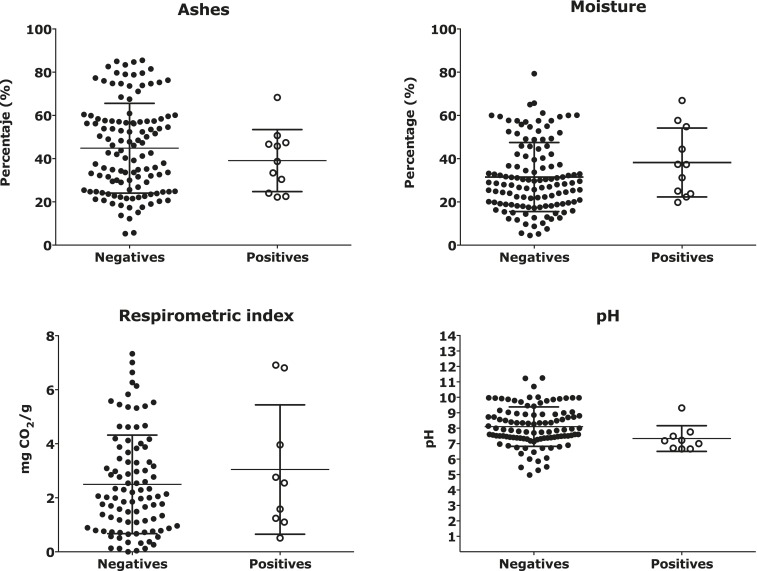
Generally, in a mature compost, the pH should be approximately 7.5 ± 0.5, the CEC should not be less than 150 meq/100 g, and the water holding capacity must be greater than 1.5 mL/g of biomass according to the NTC 5167. These conditions were met by both the positive and negative samples for H. capsulatum by Hc100 nested PCR. No differences were found in any of these parameters between organic fertilizer samples that were Hc100 nested PCR positive or negative. In evaluating the most important physicochemical characteristics in microbial metabolism (respirometric index, ash, moisture, and pH), no differences were found, and in all cases, the P value was above 0.05, which reiterates the lack of significant differences for these variables between the positive and negative samples for the presence of H. capsulatum by the Hc100 nested PCR (Figure 4).
Figure 4.
Evaluation of the respirometric index, ashes, moisture, and pH in positive and negative samples for Histoplasma capsulatum. The respirometric index, ashes, moisture, and pH were determined in their respective units for the negative and positive samples for Hc100 nested polymerase chain reaction and the results are shown by the mean in standard deviation.
DISCUSSION
The search for H. capsulatum in environmental samples has traditionally been performed using mouse inoculations. Therefore, animal models have been considered the gold standard to retrieve the fungus, but problems such as the expensive cost and the skill and time required for its performance have led to the search for alternatives among molecular biology techniques, especially PCR.42 These techniques have been widely used in recent years for diagnostic microbiology because of their capacity to solve sample problems, such as the low microorganism inoculum, poor quality, and abundant contamination with other microorganisms, and because molecular biology techniques shorten the time required for diagnosis.29,42
Several PCR-based assays have been designed for the diagnosis of H. capsulatum in clinical samples.30,32,33,43–45 These PCRs target gene sequence fragments that have been described within the H. capsulatum genome, including the genes encoding the 100 kDa protein, the M33 and H32 antigens, and more recently SCAR sequences.45 However, to date, most studies have used Hc100-PCR to target the gene encoding the 100 kDa protein of H. capsulatum as a molecular diagnostic tool.34,46–50
The Hc100 nested PCR designed by Bialek for diagnosis in clinical samples was optimized during the development of this work to search for H. capsulatum in organic fertilizers, poultry excrement, and soil. The assay obtained a sensitivity similar to the report by the original design by Bialek et al.30 The authors detected genetic material from five fungal cells, whereas our study detected DNA from three H. capsulatum cells in the samples. Moreover, our method did not show cross-reactivity with DNA from the other assessed fungi. The ability to detect the fungi in very small quantities is of great importance because H. capsulatum is immersed with other soil microorganisms in environmental samples.16,51,52 This study was the first report in which Hc100 nested PCR was used to search for H. capsulatum in organic fertilizers, soil, and bird droppings. In the other studies, the technique was applied to clinical but not environmental samples.34,48,53
Although Hc100 nested PCR has ideal characteristics for application in these samples, it is important to note that organic fertilizers contain a large number of PCR reaction inhibitors, including humic acid products and a large number of metabolites produced by the organisms present in them.16,51,52,54,55 Therefore, the “Fast DNA SPIN Kit for Soil” instructions were optimized to extract nucleic acids from organic fertilizers that had high quality, a high concentration, and were free of PCR inhibitors. The coupling of the DNA extraction method with Hc100 nested PCR allowed us to develop a protocol to detect the presence of H. capsulatum genetic material in poultry excrement, soil, organic fertilizers, and amendments and different types of environmental origin samples.
The assessment of 239 poultry excrement, cave soil, organic fertilizer, and amendment samples with the Hc100 nested PCR allowed the detection of H. capsulatum DNA in 10.5% of the samples. From the results, two very interesting findings are observed. First, when discriminating the results by sample grouping, higher positivity was found in samples that were not subjected to the composting process (i.e., groups or poultry depositions with 16.7% positive samples and cave soils where the fungus was detected in 12.5% samples compared with composted organic fertilizers where the amplification product of H. capsulatum was detected in 9.7% of samples). This finding suggests that the materials subjected to the complete composting process achieved greater sanitization in terms of the presence of H. capsulatum.
The second finding was related to the persistence of H. capsulatum in places where the fungus was previously detected by PCR during the development of this work or was isolated in previous studies. As mentioned, the amplification product from the Hc100 nested PCR was obtained in 25 organic fertilizer samples. Five samples were sent by the same supplier at different times, and the presence of the fungus was detected again in new samples collected from the same production center. In addition, the fungus was detected in samples collected from two sites where it was retrieved in previous studies. In 1993, there was an outbreak in the municipality of Concordia, Antioquia, in which 19 people were infected after handling contaminated chicken manure.23,24 During the development of this study, a positive sample was obtained from the same municipality with weather characteristics56 that might be favorable for the persistence of the fungus in the area (i.e., an average temperature of 24°C and humidity above 56%). These characteristics have been described for soil inhabited by H. capsulatum.57 Similarly, in 1989, Dr. Moncada and her team isolated the fungus in the Rio Claro region from the soil of the El Condor cave. Here, 25 years later, we detected the presence of H. capsulatum in a soil sample from the Los Guacharos cave in the same region.39 The detection of the fungus repeatedly and after long periods of time reaffirms the need to more deeply study the factors associated with the persistence of the fungus in substrates and materials that favor its survival.
The recovery of H. capsulatum in culture from positive Hc100 nested PCR samples was not possible despite the use of Mycosel, which contains cycloheximide to inhibit the growth of saprophytic fungi. However, the growth of numerous environmental molds was observed, including Penicillium spp., Aspergillus spp., Trichoderma spp., and Geotrichum spp. These fungi were reported by Brandsberg et al.,16,58 who demonstrated that they shared a microhabitat with H. capsulatum and acted as strong competitors for space and nutrients, which are factors that affect the survival of fungi in soil.9,16 This finding is also associated with the stabilization of organic matter by composting because composting is a decomposition process in which multiple microorganisms, such as fungi, actinomycetes, and bacteria,51,59 take part. Thus, this process does not allow the easy recovery of H. capsulatum from these samples.
The difficulty of H. capsulatum detection and isolation is a feature shared with other thermally dimorphic fungi that reproduce by blastoconidia formation and are human pathogens, such as B. dermatitidis and Paracoccidiodes spp. Despite having data that guide the environmental search, attempts to isolate these fungi from soils have not been successful. Knowledge of the meteorological parameters and soil characteristics that define their habitat60,61 or mark a place as a possible source of infection when the fungus has the ability to generate outbreaks in humans and animals60,61 and the availability of epidemiological data obtained from contact studies by intradermoreaction tests20 have not guaranteed the recovery of these fungi from environmental samples.
For example, B. dermatitidis has been isolated from soil only 20 times60,62 since its discovery in 1894, and the search has been so difficult that in a study conducted by Denton et al.60 in 1958, only one sample of 600 was positive. Despite attempts to isolate Paracoccidiodes spp. from environmental samples, the isolation has only been achieved six times. However, PCR-based techniques have also been developed for these fungi to facilitate the detection of their presence and thus delimit areas that constitute their habitats to allow the implementation of prevention programs.
To determine whether there was a correlation between the physicochemical and microbiological characteristics evaluated according to NTC 5167 and detection of the presence of H. capsulatum genetic material in organic fertilizers by Hc100 nested PCR, a statistical analysis was performed between positive and negative samples for this assay.
No significant differences in the evaluated parameters were found between the positive and negative samples. This result was in contrast to reports in the literature that stated that the soils where H. capsulatum was isolated had an acidic pH, concentrations of nutrients (particularly phosphorus [P] and nitrogen [N]) above 3%, and an average relative moisture of 70%.9,10,16,57,58 With the premise that the samples assessed for these parameters were the result of a complete composting process that led to a mature and stabilized compost, the presence of H. capsulatum in the final product could have several explanations: first, the persistence of the fungus in locations where composting plants are located; second, raw materials are contaminated with a large inoculum of the fungus that is not eliminated during the composting process; third, incomplete composting processes do not achieve adequate sanitization of the final product; and fourth, if uncomposted raw materials are mixed with composted material, a reinfestation of the composted end product may occur. To test these hypotheses, it is necessary to evaluate the presence of H. capsulatum in the raw materials to be composted at the zero time point (i.e., before starting the composting process) and in the final product obtained after the time needed to complete the process of transformation of organic material (final time = between 45 and 60 days) using both the Hc100 nested PCR assay standardized in this study and inoculation in mice. The mouse model used for the inoculation of environmental samples could obtain pure isolates of the fungus and is the most widely used method to find H. capsulatum in these samples.9,10,16,34,58,63,64 If the fungus were isolated using the mouse model, these results could let us conclude that the detection of DNA in positive samples by Hc100 nested PCR corresponds to a viable fungus and not only its genetic material. However, it should be noted that even without H. capsulatum isolation from the studied environmental samples, the detection of its genetic material demonstrates its presence in the raw material or in some stage of the composting process.
In conclusion, this work appeals to producers of composted organic fertilizers and regulators for the need to include the Hc100 nested PCR standardized in this work within the screening parameters of NTC 5167 to determine the presence of H. capsulatum genetic material in organic fertilizers. Because of the scope of this work, it was not possible to verify the viability of the fungus in the samples or the dynamics of infection by H. capsulatum during the composting process. Thus, we cannot rule out the presence of H. capsulatum in any stage of the process. To avoid causing outbreaks and isolated cases of infection with this fungus, it is important to implement the preventive measures published by the Center for Disease Control and Prevention41 for all those who produce, handle, or use organic fertilizers to protect workers at risk.
REFERENCES
- 1.Kauffman CA , 2007. Histoplasmosis: a clinical and laboratory update. Clin Microbiol Rev 20: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aidé MA, 2009. Continuing education course—mycoses. Chapter 4: histoplasmosis. J Bras Pneumol 35: 1145–1151. [DOI] [PubMed] [Google Scholar]
- 3.Porta A, Eletto A, Török Z, Franceschelli S, Glatz A, Vígh L, Maresca B, 2010. Changes in membrane fluid state and heat shock response cause attenuation of virulence. J Bacteriol 192: 1999–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garfoot AL, Rappleye CA, 2016. Histoplasma capsulatum surmounts obstacles to intracellular pathogenesis. FEBS J 283: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knox KS, Hage CA, 2010. Histoplasmosis. Proc Am Thorac Soc 7: 169–172. [DOI] [PubMed] [Google Scholar]
- 6.Gómez BL, 2011. Histoplasmosis: epidemiology in Latin America. Curr Fungal Infect Rep 5: 199–205. [Google Scholar]
- 7.Nieto-Ríos JF, Serna-Higuita LM, Guzman-Luna CE, Ocampo-Kohn C, Aristizabal-Alzate A, Ramírez I, Velez-Echeverri C, Vanegas-Ruiz JJ, Zuleta JJ, Zuluaga-Valencia GA, 2014. Histoplasmosis in renal transplant patients in an endemic area at a reference hospital in Medellin, Colombia. Transplant Proc 46: 3004–3009. [DOI] [PubMed] [Google Scholar]
- 8.Emmons CW, 1949. Isolation of Histoplasma capsulatum from soil. Public Health Rep 64: 892–896. [PubMed] [Google Scholar]
- 9.Emmons CW, 1950. Histoplasmosis: animal reservoirs and other sources in nature of pathogenic fungus, Histoplasma. Am J Public Health Nations Health 40: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emmons CW, 1958. Association of bats with histoplasmosis. Public Health Rep 73: 590–595. [PMC free article] [PubMed] [Google Scholar]
- 11.Hoff GL, Bigler WJ, 1981. The role of bats in the propagation and spread of histoplasmosis: a review. J Wildl Dis 17: 191–196. [DOI] [PubMed] [Google Scholar]
- 12.Manolakos JJ, Cooray M, Patel A, Haider S, 2013. Bats, fever and adenopathy–what is the link? Can J Infect Dis Med Microbiol 24: 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suárez-Alvarez RO, Pérez-Torres A, Taylor ML, 2010. Adherence patterns of Histoplasma capsulatum yeasts to bat tissue sections. Mycopathologia 170: 79–87. [DOI] [PubMed] [Google Scholar]
- 14.Stobierski MG, Hospedales CJ, Hall WN, Robinson-Dunn B, Hoch D, Sheill DA, 1996. Outbreak of histoplasmosis among employees in a paper factory–Michigan, 1993. J Clin Microbiol 34: 1220–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockwood GF, Garrison RG, 1968. The possibel role of uric acid in the ecology of Histoplasma capsulatum. Mycopathol Mycol Appl 35: 377–388. [DOI] [PubMed] [Google Scholar]
- 16.Brandsberg JW, 1968. Fungi found in association with Histoplasma capsulatum in a naturally contaminated site in Clarksburg, Maryland, USA. Sabouraudia 6: 246–254. [PubMed] [Google Scholar]
- 17.Huhn GD, et al. 2005. Two outbreaks of occupationally acquired histoplasmosis: more than workers at risk. Environ Health Perspect 113: 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz B, Martínez MA, Palma G, Ramírez A, Frías MG, Reyes MR, Taylor ML, Higuera AL, Corcho A, Manjarrez ME, 2010. Molecular characterization of Histoplasma capsulatum isolated from an outbreak in treasure hunters Histoplasma capsulatum in treasure hunters. BMC Infect Dis 10: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashford DA, Hajjeh RA, Kelley MF, Kaufman L, Hutwagner L, McNeil MM, 1999. Outbreak of histoplasmosis among cavers attending the National Speleological Society Annual Convention, Texas, 1994. Am J Trop Med Hyg 60: 899–903. [DOI] [PubMed] [Google Scholar]
- 20.Restrepo A, Robledo M, Ospina S, Restrepo M, Correa A, 1968. Distribution of paracoccidioidin sensitivity in Colombia. Am J Trop Med Hyg 17: 25–37. [PubMed] [Google Scholar]
- 21.Carmona Fonseca J, 1971. Análisis estadístico y ecológico-epidemiológico de la sensibilidad a la histoplasmina en Colombia 1950–1968. Antioquia Med 21: 109–154. [Google Scholar]
- 22.Arango M, Castañeda E, Agudelo CI, De Bedout C, Agudelo CA, Tobón A, Linares M, Valencia Y, Restrepo A, 2011. Histoplasmosis: results of the Colombian national survey, 1992–2008. Biomedica 31: 344–356. [DOI] [PubMed] [Google Scholar]
- 23.Ordóñez N, Tobón A, Arango M, Tabares A, De Bedout C, Gómez B, Castañeda E, Restrepo A, 1997. Brotes de histoplasmosis registrados en el área andina colombiana. Biomedica 17: 105. [Google Scholar]
- 24.Jiménez RA, Urán ME, de Bedout C, Arango M, Tobón AM, Cano LE, Restrepo A, 2002. Outbreak of acute histoplasmosis in a family group: identification of the infection source. Biomedica 22: 155–159. [PubMed] [Google Scholar]
- 25.Hoyos JL, Vargas CA, Velasco RJ, 2010. Evaluación de compost obtenido en pila movil empleando mezclas de gallinaza de jaula con material celulósico. Fac Ciencias Agropecu 8: 7. [Google Scholar]
- 26.Corantioquia, GIEM GI de EM- Tropicales CA para el estudio de P , 2003. Cartilla Técnica: Manejo Y Evaluación de La Porquinaza Mediante Procesos de Compostación. Medellín, Colombia: Corantioquia.
- 27.Instituto Colombiano Agropecuario , 2003. “ICA.” Resolución No. 00150 Bogotá, Colombia: Ministerio de Agricultura y Desarrollo Rural, 1–18.
- 28.Instituto Colombiano de Normas Tecnicas y Certificación I , 2011. Norma Técnica Colombiana NTC 5167 (Segunda Actualización) Bogotá, Colombia: Instituto Colombiano de Normas, Técnicas y Certificación, 1–51.
- 29.Reid TM, Schafer MP, 1999. Direct detection of Histoplasma capsulatum in soil suspensions by two-stage PCR. Mol Cell Probes 13: 269–273. [DOI] [PubMed] [Google Scholar]
- 30.Bialek R, Feucht A, Aepinus C, Just-Nübling G, Robertson VJ, Knobloch J, Hohle R, 2002. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J Clin Microbiol 40: 1644–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porta A, Colonna-Romano S, Callebaut I, Franco A, Marzullo L, Kobayashi GS, Maresca B, 1999. An homologue of the human 100-kDa protein (p100) is differentially expressed by Histoplasma capsulatum during infection of murine macrophages. Biochem Biophys Res Commun 254: 605–613. [DOI] [PubMed] [Google Scholar]
- 32.Bracca A, Tosello ME, Girardini JE, Amigot SL, Gomez C, Serra E, 2003. Molecular detection of Histoplasma capsulatum var. capsulatum in human clinical samples. J Clin Microbiol 41: 1753–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Matos Guedes HL, Guimaraes AJ, De Medeiros Muniz M, Vera Pizzini C, Hamilton AJ, Peralta JM, Deepe GS, Zancope-Oliveira RM, 2003. PCR assay for identification of Histoplasma capsulatum based on the nucleotide sequence of the M antigen. J Clin Microbiol 41: 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor ML, et al. 2005. Identification of the infectious source of an unusual outbreak of histoplasmosis, in a hotel in Acapulco, state of Guerrero, Mexico. FEMS Immunol Med Microbiol 45: 435–441. [DOI] [PubMed] [Google Scholar]
- 35.Ohno H, Tanabe K, Umeyama T, Kaneko Y, Yamagoe S, Miyazaki Y, 2013. Application of nested PCR for diagnosis of histoplasmosis. J Infect Chemother 19: 999–1003. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Russell DW, 2001. Molecular Cloning: A Laboratory Manual, 3rd edition. New York, NY: Cold Spring Harbor Laboratory Press, 2025. [Google Scholar]
- 37.Carr J, Shearer G, 1998. Genome size, complexity, and ploidy of the pathogenic fungus Histoplasma capsulatum. J Bacteriol 180: 6697–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castañeda E, De Coppiano CI, Raad J, Ajello L, Weeks R, Marin H, Coppiano T, Jimenez H, 1983. Brote epidemico de histoplasmosis asociado con exposicion a un arbol hueco. Acta Med Colomb 8: 17–22. [Google Scholar]
- 39.Moncada LH, Muñoz J, Pineda F, Ferreira G, 1989. Estudio de la presencia de Histoplasma capsulatum en la tierra de 4 cuevas localizadas de la región de Río Claro (Antioquia). Iatreia 2: 195–200. [Google Scholar]
- 40.Vining LK, Weeks RJ, 1974. A preliminary chemical and physical comparison of blackbird-starling roost soils which do or do not contain Histoplasma capsulatum. Mycopathol Mycol Appl 54: 541–548. [DOI] [PubMed] [Google Scholar]
- 41.Lenhart SW, Schafer MP, Singal M, Hajjeh RA, 2004. Histoplasmosis; Protecting Workers at Risk Washington, DC: National Institute for Occupational Safety and Health, 1–39. [Google Scholar]
- 42.Muñoz CO, Cano LE, González A, 2010. Detección e identificación de Histoplasma capsulatum por el laboratorio: de los métodos convencionales a las pruebas moleculares. Infectio 14: 145–158. [Google Scholar]
- 43.Bialek R, Fischer J, Feucht A, Najvar LK, Dietz K, Knobloch J, Graybill JR, 2001. Diagnosis and monitoring of murine histoplasmosis by a nested PCR assay. J Clin Microbiol 39: 1506–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maubon D, Simon S, Aznar C, 2007. Histoplasmosis diagnosis using a polymerase chain reaction method. Application on human samples in French Guiana, South America. Diagn Microbiol Infect Dis 58: 441–444. [DOI] [PubMed] [Google Scholar]
- 45.Frías De León MG, Arenas López G, Taylor ML, Acosta Altamirano G, Reyes-Montes MDR, 2012. Development of specific sequence-characterized amplified region markers for detecting Histoplasma capsulatum in clinical and environmental samples. J Clin Microbiol 50: 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muñoz-Cadavid C, Rudd S, Zaki SR, Patel M, Moser SA, Brandt ME, Gómez BL, 2010. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J Clin Microbiol 48: 2147–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babady NE, Buckwalter SP, Hall L, Le Febre KM, Binnicker MJ, Wengenack NL, 2011. Detection of Blastomyces dermatitidis and Histoplasma capsulatum from culture isolates and clinical specimens by use of real-time PCR. J Clin Microbiol 49: 3204–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dantas KC, Freitas RS, Moreira APV, da Silva MV, Benard G, Vasconcellos C, Criado PR, 2013. The use of nested polymerase chain reaction (nested PCR) for the early diagnosis of Histoplasma capsulatum infection in serum and whole blood of HIV-positive patients. An Bras Dermatol 88: 141–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guarner J, Brandt ME, 2011. Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev 24: 247–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koepsell SA, Hinrichs SH, Iwen PC, 2012. Applying a real-time PCR assay for Histoplasma capsulatum to clinically relevant formalin-fixed paraffin-embedded human tissue. J Clin Microbiol 50: 3395–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryckeboer J, Mergaert J, Coosemans J, Deprins K, Swings J, 2003. Microbiological aspects of biowaste during composting in a monitored compost bin. J Appl Microbiol 94: 127–137. [DOI] [PubMed] [Google Scholar]
- 52.Hyde KD, Bussaban B, Paulus B, Crous PW, Lee S, Mckenzie EHC, Photita W, Lumyong S, 2007. Diversity of saprobic microfungi. Biodivers Conserv 16: 7–35. [Google Scholar]
- 53.Sampaio IDL, Freire AKL, Ogusko MM, Salem JI, De Souza JVB, 2011. Selection and optimization of PCR-based methods for the detection of Histoplasma capsulatum var. capsulatum. Rev Iberoam Micol 29: 34–39. [DOI] [PubMed] [Google Scholar]
- 54.Tsai YL, Olson BH, 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol 58: 2292–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baar C, d’Abbadie M, Vaisman A, Arana ME, Hofreiter M, Woodgate R, Kunkel TA, Holliger P, 2011. Molecular breeding of polymerases for resistance to environmental inhibitors. Nucleic Acids Res 39: e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.AccuWeather Enterprise Solutions Inc , 2015. Concordia, Colombia Local Weather Available at: http://www.accuweather.com/. Accessed October 8, 2015.
- 57.Ulloa M, Lappe P, Aguilar S, Park H, Pérez Mejía A, Toriello C, Taylor ML, 2006. Contribution to the study of the mycobiota present in the natural habitats of Histoplasma capsulatum: an integrative study in Guerrero, Mexico. Rev Mex Biodivers 77: 153–168. [Google Scholar]
- 58.Brandsberg JW, Weeks RJ, Hill WB, Piggott WR, 1969. A study of fungi found in association with Histoplasma capsulatum: three bird roosts in S. E. Missouri, USA. Mycopathol Mycol Appl 38: 71–81. [DOI] [PubMed] [Google Scholar]
- 59.Mehta CM, Palni U, Franke-Whittle IH, Sharma AK, 2014. Compost: its role, mechanism and impact on reducing soil-borne plant diseases. Waste Manag 34: 607–622. [DOI] [PubMed] [Google Scholar]
- 60.Denton JF, McDonough ES, Ajello L, Ausherman RJ, 1961. Isolation of Blastomyces dermatitidis from soil. Science 133: 1126–1127. [DOI] [PubMed] [Google Scholar]
- 61.Borelli D, 1961. Hipótesis sobre ecología de Paracoccidioides. Dermatol Venez 3: 130–132. [Google Scholar]
- 62.Baumgardner DJ, Laundre B, 2001. Studies on the molecular ecology of Blastomyces dermatitidis. Mycopathologia 152: 51–58. [DOI] [PubMed] [Google Scholar]
- 63.Ajello L, 1964. Relationship of Histoplasma capsulatum with hens and other birds. Bol Oficina Sanit Panam 56: 232–236. [PubMed] [Google Scholar]
- 64.Negroni R, Duré R, Ortiz Nareto Á, Arechavala A, Maiolo E, Santiso G, Iovannitti C, Ibarra-Camou B, Canteros C, 2010. Brote de histoplasmosis en la Escuela de Cadetes de la Base Aérea de Morón, Provincia de Buenos Aires, República Argentina. Rev Argent Microbiol 42: 254–260. [DOI] [PubMed] [Google Scholar]