Abstract.
The development of artemisinin (ART)-resistant parasites in Southeast Asia (SEA) threatens malaria control globally. Mutations in the Kelch 13 (K13)-propeller domain have been useful in identifying ART resistance in SEA. ART combination therapy (ACT) remains highly efficacious in the treatment of uncomplicated malaria in Sub-Saharan Africa (SSA). However, it is crucial that the efficacy of ACT is closely monitored. Toward this effort, this study profiled the prevalence of K13 nonsynonymous mutations in different malaria ecological zones of Kenya and in different time periods, before (pre) and after (post) the introduction of ACT as the first-line treatment of malaria. Nineteen nonsynonymous mutations were present in the pre-ACT samples (N = 64) compared with 22 in the post-ACT samples (N = 251). Eight of these mutations were present in both pre- and post-ACT parasites. Interestingly, seven of the shared single-nucleotide polymorphisms were at higher frequencies in the pre-ACT than the post-ACT parasites. The A578S mutation reported in SSA and the V568G mutation reported in SEA were found in both pre- and post-ACT parasites, with their frequencies declining post-ACT. D584Y and R539K mutations were found only in post-ACT parasites; changes in these codons have also been reported in SEA with different amino acids. The N585K mutation described for the first time in this study was present only in post-ACT parasites, and it was the most prevalent mutation at a frequency of 5.2%. This study showed the type, prevalence, and frequency of K13 mutations that varied based on the malaria ecological zones and also between the pre- and post-ACT time periods.
INTRODUCTION
There is compelling evidence that nonsynonymous mutations in the Kelch 13 (K13) propeller domain result in reduced sensitivity of Plasmodium falciparum to artemisinin (ART).1,2 ART resistance is characterized by a long parasite clearance half-life after treatment with either ART monotherapy or ART combination therapy (ACT),3,4 reduced susceptibility of ring-stage parasites in the in vitro ring-stage survival assay (RSA),5,6 and/or the presence of mutations in the K13-propeller domain.1,7 The nonsynonymous mutations in the K13 gene have only been identified to be a causal determinant of ART resistance in Southeast Asia (SEA) where increasing rates of ACTs failure are evident.6,8–11 Some of these mutations have also been observed in Sub-Saharan Africa (SSA) but are not associated with ART resistance.7,12 Presently, the World Health Organization defines suspected ART resistance as the high prevalence of the delayed parasite clearance phenotype or a high prevalence of K13-propeller mutants, whereas confirmed ART resistance is defined as the combination of delayed parasite clearance and K13 resistance-validated mutation from the same patient.13
In SEA, six nonsynonymous mutations have been validated in the K13-propeller domain as markers of ART resistance (N458Y, Y493H, R539T, I543T, R561H, and C580Y).1,13,14 These markers have been reported throughout the greater Mekong subregion with specific mutations being prevalent in particular areas.13 However, the C580Y is the most common mutation in SEA1,14,15 and has been shown to have independent origins in western and eastern Cambodia.14,15 The C580Y mutation has also been reported in SSA and appears to have emerged independently and did not migrate from SEA.16 In SSA, the A578S nonsynonymous mutation is the most predominant17–21; however, it has since been reported to have no association with ART resistance.12,22 There are other nonsynonymous K13 mutations that have been reported in SSA including the S522C found in Uganda18,23; the V520A found in west, central, and east Africa18; the N531I found in Ethiopia24; the V581F found in Ghana17; and the M579I found in the Equatorial Guinea,25 among others. These mutations, however, are at low frequencies and vary from region to region7,16 and sometimes appear to be transient where they change with seasons.19
Malaria is endemic in Kenya and although it is not homogenized across the country, around 70% of the population is at risk.26 The country is grouped into four malaria ecological zones: 1) holoendemic zones in the western lake and the coastal regions, 2) the highland epidemic zones in the western Kenya highlands, 3) semiarid seasonal zones in the northern and eastern part of the country, and 4) low-risk zones in the central highlands.26 This study sets out to analyze the prevalence of polymorphisms in the P. falciparum K13-propeller domain in Kenyan parasites. Samples were stratified based on the malaria ecological zones which they were obtained from, and whether they were obtained before or after the introduction of artemether–lumefantrine (AL) as the first-line treatment of uncomplicated malaria in 2006.27
METHODS
Samples.
Plasmodium falciparum isolates used in this study were from archived DNA extracts collected from different malaria ecological zones in Kenya before (pre) and after (post) the ACT era. The pre-ACT samples (N = 64) were collected between 1995 and 2003 from the malaria holoendemic lake (Kisumu [N = 37]) and coastal endemic regions (Malindi [N = 27]). The post-ACT samples (N = 251) were collected in 2013, approximately 8 years after the implementation of ACT as the first-line treatment of malaria in Kenya. The study sites are as shown on the map (Figure 1): Kisumu and Kombewa are in the malaria holoendemic lake region of western Kenya; Kisii and Kericho are in the highland epidemic regions in western Kenya; Malindi is in the coastal holoendemic region; and Marigat is in the semiarid seasonal region in the Great Rift Valley. The study was approved by the Ethical Review Committee of the Kenya Medical Research Institute (KEMRI), Nairobi, Kenya, and Walter Reed Army Institute of Research (WRAIR) Institutional Review Board, Silver Spring, MD. The pre-ACT and post-ACT cryopreserved samples were collected under the approved study protocol KEMRI-SSC 1330/WRAIR 1384 (Epidemiology of malaria drug sensitivity patterns in Kenya).
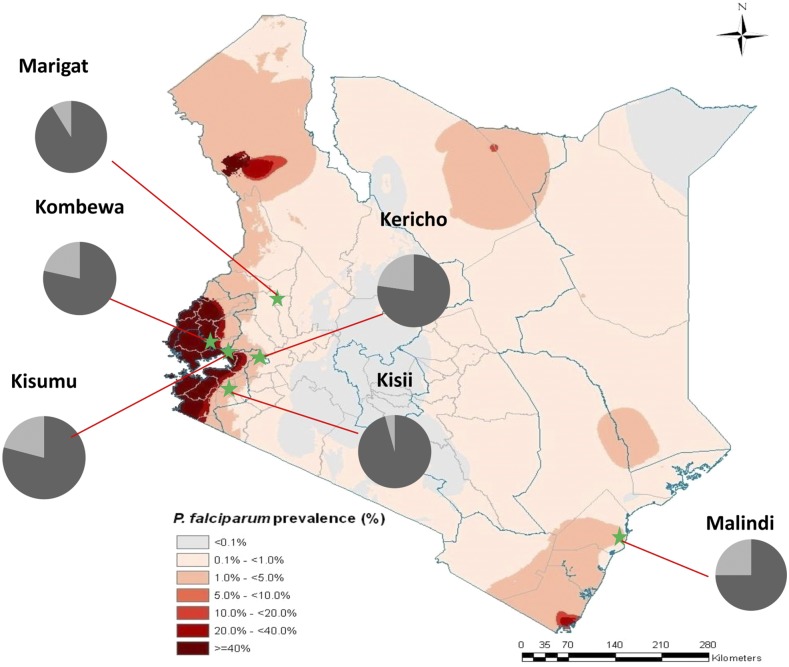
Figure 1.
Kenyan map showing the six field sites where parasites were collected. The pie chart shows the proportion of K13 mutations per site, light gray represents parasites carrying mutant alleles and dark gray the wild-type alleles. Map adapted with permission from Noor et al.49
Patient recruitment.
The samples were obtained from consenting patients presenting with uncomplicated malaria to the study site clinics, aged between 6 months and 65 years. Eligibility criteria included a history of fever (temperature of ≥ 37.5°C) within 24 hours before presentation, monoinfection with P. falciparum, and a baseline parasitemia of 2,000–200,000 asexual parasites/µL. Persons treated for malaria within the preceding 2 weeks were excluded from the study. Written informed consent and/or assent was obtained from adult subjects (> 18 years of age) or legal guardians for subjects < 18 years of age. The presence of malaria was confirmed by using microscopy and rapid diagnostic test (Parascreen®; Zephyr Biomedicals, Verna, Goa, India). Whole blood was collected and aliquots preserved for analysis as specified in the study protocol. The post-ACT study subjects were treated with oral AL (Coartem) administered over three consecutive days after the standard care for uncomplicated P. falciparum malaria in Kenya. On enrolment, the patient’s demographics (including age and gender), place of birth, place of residence, occupation, and travel history in the last 2 months were recorded on a clinical data sheet.
K13-propeller genotyping.
The K13-propeller domain was amplified using QuantiFast Probe PCR Kit (Qiagen, Valencia, CA) and K13-propeller–specific primers (K13_F 5′-TGG AAG ACA TCA GTC AAC CAG AGA-3′ and K13_R 5′-TTA TAT ATT TGC TAT TAA AAC GGA GTG-3′) designed using Primr3 software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Polymerase chain reaction (PCR) analyses were performed in a 25-μL reaction mixture with 2 μL of DNA template and 1 μL each of 10 μM primers. Reaction conditions consisted of an initial denaturation at 95°C for 5 minutes followed by 40 cycles of 95°C for 30 seconds, 60°C for 45 seconds, and 72°C for 3 minutes, and a final extension step of 7 minutes at 72°C. The PCR products were analyzed by electrophoresis on a 1.5% ethidium bromide–stained agarose gel to confirm amplification. Agencourt AMPure XP (Beckman Coulter, Brea, CA) bead purification was performed on the PCR products to remove excess nucleotides and primer remnants on the PCR amplicons as per the manufacturer’s instructions with minor adjustments/modification of the protocol where 80% ethanol was used instead of 70%. Sanger sequencing was performed on purified PCR products using version 3.1 of the Big Dye terminator method on the 3500xL ABI Genetic Analyzer (Applied Biosystems, Foster City, CA). The sequencing primers (K13_2F 5′-GCC AAG CTG CCA TTC ATT TG-3′ and K13_3R 5′-GCC TTG TTG AAA GAA GCA GA-3′) were designed using Primer3 software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi).
Data analysis.
Generation of consensus sequences were performed using the CLC Main workbench 7 (Qiagen). Nucleotide basic local alignment search tool (BLASTn) (https://blast.ncbi.nlm.nih.gov) analysis was carried out to ascertain the sequences were K13 and to determine whether the sequences were in the forward or reverse orientation. Alignment of these sequences was performed using Muscle v3.8,28 and PF3D7_1343700 retrieved from PlasmoDB (www.plasmodb.org) was used as the reference sequence. BioEdit was then used to visualize, edit, and call out single-nucleotide polymorphisms (SNPs). Mixed alleles were considered as mutants. Odds ratio (OR) was used to assess mutations in the parasite population in pre- and post-ACT samples. The prevalence of mutations was also determined.
RESULTS
K13-propeller polymorphisms before and after the introduction of ACT.
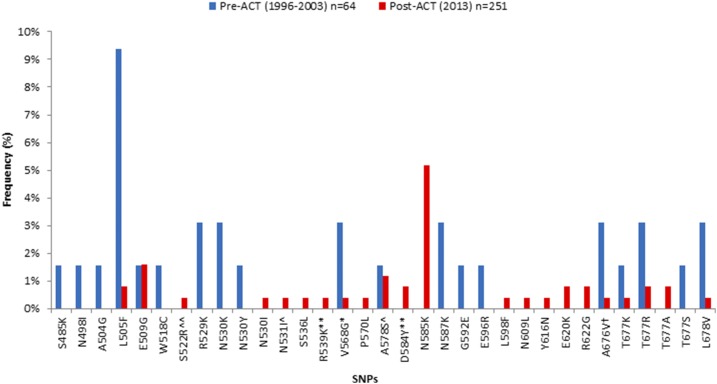
Of the 315 samples successfully analyzed for K13-propeller polymorphism, 64 were pre-ACT and 251 were post-ACT. There were 19 nonsynonymous K13 mutations in the pre-ACT compared with 22 in the post-ACT parasites, with 8 present in both pre- and post-ACT populations (Figure 2). There were 11 private mutations (found in only one sample) in the pre-ACT and 13 in the post-ACT period (Table 1). Most of the private mutations in the post-ACT parasites (N = 8) were found in Malindi. Mutations such as the V568G reported in SEA1 and A578S reported in SSA,19,20 Bangladesh,29,30 and India31 were found both in pre- and post-ACT parasites, but the frequencies declined from 3.2% to 0.4% (for V568G) and 1.6% to 1.2% (for A578S) (Figure 2) in the pre-ACT era compared with the post-ACT era. Nonsynonymous mutations at positions 539 and 584 reported in SEA1 were present in post-ACT parasites but at low frequencies (< 1%) and contained different amino acids compared with those reported in SEA.1 The N585K mutation reported for the first time in this study was only present in the post-ACT parasites. Interestingly, it was the most frequent mutation in post-ACT parasites (5.2%) and was present in all the study sites. A578S was the third most frequent mutation at 1.2%, present only in two study sites (Kombewa and Kisii), whereas the rest of the nonsynonymous mutations were less than 1%. The frequency of nonsynonymous mutations pre-ACT ranged between 1.6% and 9.4%, with L505F (9.4%) showing the highest frequency. However, the frequency of L505F fell from 9.4% pre-ACT to 0.8% post-ACT (Figure 2).
Figure 2.
Frequency of K13-propeller mutations pre- and post-ACT in Kenya. Most mutations had frequencies ≤ 2%. L505F was highest in frequency pre-ACTs (9.4%), whereas N585K was highest in frequency post-ACTs (5.2%). * SNP reported in SEA, ** SNP reported in SEA but with a different amino acid, ^ SNP reported in SSA, ^^ SNP reported in SSA but with a different amino acid, and † SNP reported in both SSA and SEA. ACT = artemisinin combination therapy; SEA = Southeast Asia; SNP = single-nucleotide polymorphism; SSA = Sub-Saharan Africa.
Table 1.
Frequency of K13 polymorphisms pre- and post-ACT
| Mutation | Pre-ACT (1996–2003) | Post-ACT (2013) | ||||||
|---|---|---|---|---|---|---|---|---|
| Kisumu* (N = 37) | Malindi† (N = 27) | Kisumu* (N = 41) | Kombewa* (N = 47) | Kericho‡ (N = 43) | Malindi† (N = 39) | Marigat§ (N = 34) | Kisii‡ (N = 47) | |
| S485K | 1 (2.7%) | – | – | – | – | – | – | – |
| N498I | 1 (2.7%) | – | – | – | – | – | – | – |
| A504G | – | 1 (3.7%) | – | – | – | – | – | – |
| L505F | 4 (10.8%) | 2 (7.4%) | – | – | – | 1 (2.6%) | 1 (2.9%) | – |
| E509G | – | 1 (3.7%) | 1 (2.4%) | – | 3 (7.0%) | – | 1 (2.9%) | – |
| W518C | 1 (2.7%) | – | – | – | – | – | – | – |
| R529K | 2 (5.4%) | – | – | – | – | – | – | – |
| S522R‖ | – | – | – | – | – | 1 (2.6%) | – | – |
| N530K | 2 (5.4%) | – | – | – | – | – | – | – |
| N530Y | – | 1 (3.7%) | – | – | – | – | – | – |
| N530I | – | – | – | – | – | 1 (2.6%) | – | – |
| N531I¶ | – | – | – | – | – | 1 (2.6%) | – | – |
| S536L | – | – | – | – | – | 1 (2.6%) | – | – |
| R539K# | – | – | – | – | – | 1 (2.6%) | – | – |
| V568G** | 2 (5.4%) | – | 1 (2.4%) | – | – | – | – | |
| P570L | – | – | – | – | – | 1 (2.6%) | – | – |
| A578S¶ | 1 (2.7%) | – | – | 2 (4.3%) | – | – | – | 1 (2.1%) |
| D584Y# | – | – | 2 (4.9%) | – | – | – | – | – |
| N585K | – | – | 4 (9.8%) | 5 (10.6%) | 1 (2.3%) | 1 (2.6%) | 1 (2.9%) | 1 (2.1%) |
| N587K | 2 (5.4%) | – | – | – | – | – | – | – |
| G592E | – | 1 (3.7%) | – | – | – | – | – | – |
| E596R | – | 1 (3.7%) | – | – | – | – | – | – |
| L598F | – | – | – | – | – | 1 (2.6%) | – | – |
| N609L | – | – | – | 1 (2.1%) | – | – | – | – |
| Y616N | – | – | – | – | 1 (2.3%) | – | – | – |
| E620K | – | – | – | – | 2 (4.7%) | – | – | – |
| R622G | – | – | – | – | 2 (4.7%) | – | – | – |
| A676V†† | 1 (2.7%) | 1 (3.7%) | – | 1 (2.1%) | – | – | – | – |
| T677K | 1 (2.7%) | – | – | – | – | 1 (2.6%) | – | – |
| T677R | 2 (5.4%) | – | 1 (2.4%) | 1 (2.1%) | – | – | – | – |
| T677A | – | – | – | – | – | 1 (2.6%) | – | – |
| T677S | 1 (2.7%) | – | – | – | – | – | – | – |
| L678V | 1 (2.7%) | 1 (3.7%) | – | 1 (2.1%) | – | – | – | – |
ACT = artemisinin combination therapy; SEA = Southeast Asia; SSA = Sub-Saharan Africa.
Holoendemic lake region.
Holoendemic coastal region.
Highland epidemic.
Semiarid seasonal region.
Reported in SSA but with a different amino acid.
Reported in SSA.
Reported in SEA but with a different amino acid.
Reported in SEA.
Reported in SSA and SEA but with different amino acid.
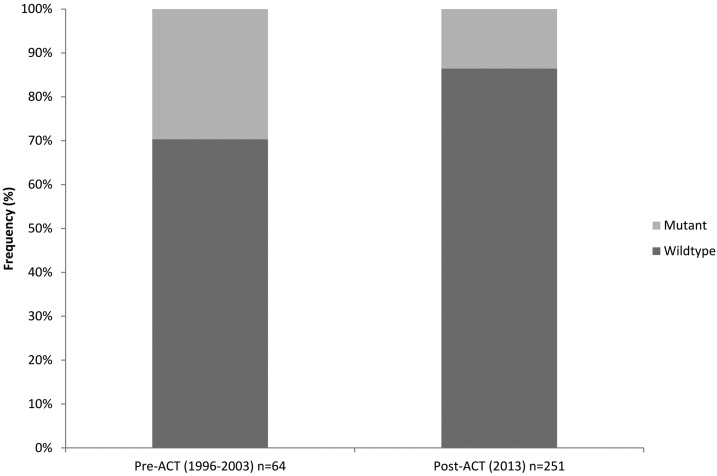
The percentage of samples with nonsynonymous mutations was higher in the pre-ACT (29.7%; 19 of 64) than the post-ACT parasites (13.5%; 34 of 251) (Figure 3). Of interest, more individual parasites carried more than one mutation pre-ACT compared with post-ACT. The OR was calculated to estimate the effect of AL on the introduction of mutations; OR = 0.3711 (95% confidence interval = 0.1944–0.7085) with a P value of 0.0027. The odds of having a mutation post-ACT was 62.9% less likely to occur compared with the pre-ACT period.
Figure 3.
Bar graph showing frequency of K13-propeller mutations pre- and post-ACT including the wild-type in Kenya. ACT = artemisinin combination therapy.
Frequency of K13-propeller polymorphisms in Kenya post-ACT parasites.
Figure 1 is a cumulative summary of all SNPs included in the post-ACT parasites per study site. Some individual K13 nonsynonymous mutations were present in all the study sites, whereas others were private showing restricted geographic localization, all with varying frequencies. The Malindi parasite population had the most polymorphic K13-propeller gene with a total of 10 different SNPs, followed by Kombewa and Kericho with a total of 6 SNPs each, whereas Kisii had the least with a total of 2 SNPs. When analyzed per study site, the frequencies of nonsynonymous mutations ranged from 2.1% to 10.6% (Table 1) with N585K having the highest frequency in Kombewa. The cumulative frequency of mutations per study area was also highest in Malindi (28.2%), followed by Kombewa (23.4%), whereas Kisii had the lowest frequency (4.3%). There were several nonsynonymous mutations reported for the first time in this study: N585K was present in all study sites and its frequency was highest in Kombewa at 10.6%, N531I (2.6%) was exclusive to Malindi, L678V (2.1%) and N609G (2.1%) were exclusive to Kombewa, and E620K (4.7%) was exclusive to Kericho (Table 1). The A578S mutation was present in Kisii and Kombewa at frequencies of 2.1% and 4.3%, respectively. Kisumu and Malindi had mutations at positions D584Y (4.9%) and R539K (2.6%), respectively; mutations at these positions have been reported in SEA1 but with different amino acids, D584V and R539T.
DISCUSSION
In this study, we profiled the distribution of K13 nonsynonymous mutations in pre- and post-ACT parasites and in different malaria ecological zones of Kenya. There were 19 nonsynonymous mutations present in the pre-ACT compared with 22 in the post-ACT parasites; eight of these SNPs were present in both pre- and post-ACT parasites. Interestingly, seven of the shared SNPs had frequencies that were higher in the pre-ACT than the post-ACT parasites. Furthermore, the odds of having a mutation in K13 was more likely to occur in the pre-ACT than the post-ACT parasites. We detected 11 novel nonsynonymous mutations that were private and four SEA nonsynonymous mutations, including mutation at codon 539, a validated marker of ART resistance, albeit with a different amino acid substitution. In SEA, R539T is a nonconservative mutation, whereas in Kenyan parasites, R539K is a conservative mutation that may not change the function of the protein.
A recent study that sequenced the entire genome of more than 3,000 clinical samples of P. falciparum from SEA and SSA showed that SSA parasites had an excess of polymorphisms that were at low frequencies and emerged independently. Some of these mutations are known to cause ART resistance in SEA.16 The proportion of K13 nonsynonymous mutations have been shown to be heterogeneous in SEA, ranging from fixed (> 95%) to very high (80% to 94%) in western Cambodia, to low (< 5%) in other regions.12 In this study, we obtained 33 different nonsynonymous mutations from 315 samples. The frequency of mutations was higher in the pre-ACT (all the frequencies were ≥ 1.6%) than the post-ACT samples (19 of 22 mutations were < 1%). All but one of the K13 nonsynonymous mutations that were present in both pre- and post-ACT parasites declined in frequency, with L505F declining from 9.4% in pre-ACT to 0.8% in post-ACT. The low frequencies of K13 mutations in SSA parasites is suggestive of much less pressure for evolutionary selection than what is present in SEA.16 It is likely that the K13 gene in Kenya might have been under slightly higher evolutionary selection pre-ACT than post-ACT or it is simply the polymorphic nature of the K13 protein.
Individual K13 nonsynonymous mutations have shown restricted geographic localization.12 Although ART resistance has not been reported in Kenya or SSA, and ACTs remain highly efficacious,21,32–38 we found geographic disparity in the proportion of polymorphisms in parasites from different malaria ecological zones of Kenya. Malindi, which is the coastal holoendemic region, had the most polymorphic K13 gene in the post-ACT parasites with 7 of the 10 nonsynonymous mutations being private to this region. Overall, the K13 polymorphisms were present at low frequencies, study site specific, and private. This corroborates previous studies that observed rare and highly diverse K13 mutations in SSA parasites.12,16,18–21,23,39–43 In a study by Ménard et al.,12 where more than 9,000 sequences were analyzed and 150 distinct alleles were found in the K13 gene in SSA parasites, none of the SEA ART-resistant alleles were present. The MalariaGEN study,16 however, found seven nonsynonymous mutations in SSA parasites that have been associated with ART resistance in SEA, including C580Y, the most common allele in resistant parasites.1,14,15 Studies are yet to link these mutations with increased parasite half-life or clinical failures in SSA parasites. We observed four nonsynonymous mutations that have been reported in SEA, but three of these mutations carried a different amino acid compared with what has been reported in SEA. Two of the mutations, R539K and D584Y, were present only in the post-ACT parasites, whereas V568G and A676V were present in both pre- and post-ACT parasites. Interestingly, the V568G mutation, which is a candidate marker associated with ART resistance, was at higher frequency in the pre-ACT than the post-ACT parasites.
A578S is the most commonly reported K13 mutant allele in SSA.17–21 In this study, we found A578S in post-ACT parasites in two different study sites, Kombewa (at a frequency of 4.3%) and Kisii (at a frequency of 2.1%); Kombewa is located in the holoendemic lake region and Kisii is in the highland epidemic region. In our previous study, we obtained A578S at a frequency of 2.8% in 108 samples from Kisumu, which is also in the holoendemic lake region of western Kenya.20 Clinical and in vitro studies suggest that this mutation is not associated with ART resistance.12,31,39,40 Our study further corroborates this finding because A578S was also present in the Kenyan parasites before the introduction of ACTs.
The N585K allele was only present post-ACT in all the study sites, with the highest frequencies in Kombewa (10.6%) and Kisumu (9.8%). This suggests that this mutation might be under pressure for evolutionary selection either by antimalarial drugs or some other factors; usage of AL is high in Kisumu and Kombewa because of high malaria transmission. The N585K mutation is located in Kelch domain 4, and the mutation introduces a positively charged amino acid, which might impact the functioning of the domain. Additional studies such as genome editing44 and RSAs5 might elucidate the role of this and other mutations in ART resistance, if any, in the Kenyan parasite population.
This study had several limitations. First, the pre-ACT samples were collected from only two field sites (Kisumu and Malindi) as opposed to all field sites where post-ACT samples were collected. As such, the changes reported might not be a true reflection of the change in parasite genotypes between the two periods (pre- and post-ACT) because individual K13 nonsynonymous mutations show restricted geographic localization.12 However, previous studies of parasite populations in Kenya have shown gene flow between the different regions,45 and there was no difference in statistical analysis when samples were stratified per site (pre-ACT Kisumu versus post-ACT Kisumu and pre-ACT Malindi versus post-ACT Malindi). Another limitation was the sample size; a larger sample size would have provided a more comprehensive analysis including the impact of the introduction of AL on the parasite population. Although other studies have used similar sample sizes,5,19–21,23,24,42,43,46,47 more recent studies have used large sample sizes providing a more comprehensive genetic analysis at a global scale.12,16
In conclusion, this study compared the P. falciparum parasites collected in the pre- and post-ACT era in Kenya and elucidated differences in K13 polymorphic profiles from the different geographic and malaria ecological zones of Kenya. In Africa, parasites have a large excess of polymorphisms with minor allele frequencies, which are evenly distributed across the genome.16 This is likely to be the result of different demographic histories and epidemiological characteristics, such as changes in effective population size,48 rather than differences in selective pressures that might be transient and may not necessarily be as a result of ART drug pressure. Whether polymorphisms in the K13-propeller domain will have any impact on ART resistance in SSA still remains to be studied.
Acknowledgments:
We thank the patients and the clinical and other support staff at the study sites. We are grateful to our colleagues in the Malaria Drug Resistance laboratory for their technical and moral support. We are grateful to the previous and current director of Centre for Biotechnology and Bioinformatics (CEBIB), University of Nairobi, for supporting this work. We also thank the Director of KEMRI for permission to publish this work. This work was supported by the Armed Forces Health Surveillance Center, Division of Global Emerging Infections Surveillance and Response System Operations. The opinions and assertions contained herein are private opinions of the authors and are not to be construed as official or reflecting the views of the U.S. Department of the Army, the U.S. Department of Defense, or the Walter Reed Army Institute of Research. The identification of specific products, scientific instrumentation, or organization is considered an integral part of the scientific endeavor and does not constitute endorsement or implied endorsement on the part of the author, DoD, or any component agency.
REFERENCES
- 1.Ariey F, et al. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straimer J, et al. 2014. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347: 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dondorp AM, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaratunga C, Witkowski B, Dek D, Try V, Khim N, Miotto O, Ménard D, Fairhurst RM, 2014. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. 58: 4935–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witkowski B, et al. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13: 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amaratunga C, Neal AT, Fairhurst RM, 2014. Flow cytometry-based analysis of artemisinin-resistant Plasmodium falciparum in the ring-stage survival assay. Antimicrob Agents Chemother 58: 4938–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley EA, et al. 2014. Spread of artemisinin resistance in Plasmodium falciparum Malaria. N Engl J Med 371: 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrara VI, et al. 2009. Changes in the treatment responses to artesunate-mefloquine on the northwestern border of Thailand during 13 years of continuous deployment. PLoS One 4: e4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaratunga C, et al. 2012. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet Infect Dis 12: 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, et al. 2015. In vivo monitoring of dihydroartemisinin-piperaquine sensitivity in Plasmodium falciparum along the China–Myanmar border of Yunnan Province, China from 2007 to 2013. Malar J 14: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miotto O, et al. 2013. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ménard D, et al. 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374: 2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO , 2016. Artemisinin and Artemisinin-Based Combination Therapy Resistance Available at: http://apps.who.int/iris/bitstream/10665/208820/1/WHO_HTM_GMP_2016.5_eng.pdf?ua=1. Accessed June 27, 2017.
- 14.Miotto O, et al. 2015. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 47: 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takala-Harrison S, et al. 2015. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis 211: 670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MalariaGEN Plasmodium falciparum Community Project , 2016. Genomic epidemiology of artemisinin resistant malaria. eLife 5: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouattara A, et al. 2015. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg 92: 1202–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor SM, et al. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211: 680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH, Kaneko A, 2015. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis 21: 490–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamau E, et al. 2015. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis 211: 1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, Dorsey G, Rosenthal PJ, 2014. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One 9: e105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muwanguzi J, Henriques G, Sawa P, Bousema T, Sutherland CJ, Beshir KB, 2016. Lack of K13 mutations in Plasmodium falciparum persisting after artemisinin combination therapy treatment of Kenyan children. Malar J 15: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ocan M, Bwanga F, Okeng A, Katabazi F, Kigozi E, Kyobe S, Ogwal-Okeng J, Obua C, 2016. Prevalence of K13-propeller gene polymorphisms among Plasmodium falciparum parasites isolated from adult symptomatic patients in northern Uganda. BMC Infect Dis 16: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuchert A, Abduselam N, Zeynudin A, Eshetu T, Löscher T, Wieser A, Pritsch M, Berens-Riha N, 2015. Molecular markers of anti-malarial drug resistance in southwest Ethiopia over time: regional surveillance from 2006 to 2013. Malar J 14: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu F, et al. 2017. Emergence of indigenous artemisinin-resistant Plasmodium falciparum in Africa. N Engl J Med 376: 991–993. [DOI] [PubMed] [Google Scholar]
- 26. National Malaria Control Programme (NMCP), Kenya National Bureau of Statistics (KNBS) and ICF International, 2016. Kenya Malaria Indicator Survey 2015. Nairobi, Kenya, and Rockville, Maryland NMCP, KNBS, and ICF International.
- 27.Amin AA, Zurovac D, Kangwana BB, Greenfield J, Otieno DN, Akhwale WS, Snow RW, 2007. The challenges of changing national malaria drug policy to artemisinin-based combinations in Kenya. Malar J 6: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alam MS, Mohon AN, Bayih A, Folefoc A, Pillai D, 2014. Mutations in P. falciparum K13 propeller gene from Bangladesh: emerging resistance? Malar J 13 (Suppl 1): 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohon AN, Alam MS, Bayih AG, Folefoc A, Shahinas D, Haque R, Pillai DR, 2014. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013). Malar J 13: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra N, et al. 2015. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the Kelch 13 molecular marker. Antimicrob Agents Chemother 59: 2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogutu BR, et al. 2014. Efficacy and safety of artemether-lumefantrine and dihydroartemisinin-piperaquine in the treatment of uncomplicated Plasmodium falciparum malaria in Kenyan children aged less than five years: results of an open-label, randomized, single-centre study. Malar J. 13: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abuaku B, Duah N, Quaye L, Quashie N, Malm K, Bart-Plange C, Koram K, 2016. Therapeutic efficacy of artesunate-amodiaquine and artemether–lumefantrine combinations in the treatment of uncomplicated malaria in two ecological zones in Ghana. Malar J 15: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espié E, Lima A, Atua B, Dhorda M, Flévaud L, Sompwe EM, Palma Urrutia PP, Guerin PJ, 2012. Efficacy of fixed-dose combination artesunate–amodiaquine versus artemether–lumefantrine for uncomplicated childhood Plasmodium falciparum malaria in Democratic Republic of Congo: a randomized non-inferiority trial. Malar J 11: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ndounga M, Mayengue PI, Casimiro PN, Loumouamou D, Basco LK, Ntoumi F, Brasseur P, 2013. Artesunate-amodiaquine efficacy in Congolese children with acute uncomplicated falciparum malaria in Brazzaville. Malar J 12: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogouyèmi-Hounto A, Azandossessi C, Lawani S, Damien G, de Tove YS, Remoue F, Kinde Gazard D, 2016. Therapeutic efficacy of artemether–lumefantrine for the treatment of uncomplicated falciparum malaria in northwest Benin. Malar J 15: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schramm B, et al. 2013. Efficacy of artesunate–amodiaquine and artemether–lumefantrine fixed-dose combinations for the treatment of uncomplicated Plasmodium falciparum malaria among children aged six to 59 months in Nimba County, Liberia: an open-label randomized non-inferiority. Malar J 12: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirima SB, et al. 2016. Comparison of artesunate–mefloquine and artemether–lumefantrine fixed-dose combinations for treatment of uncomplicated Plasmodium falciparum malaria in children younger than 5 years in sub-Saharan Africa: a randomised, multicentre, phase 4 trial. Lancet Infect Dis 16: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashley EA, et al. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371: 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper RA, Conrad MD, Watson QD, Huezo SJ, Ninsiima H, Tumwebaze P, Nsobya SL, Rosenthal PJ, 2015. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother 59: 5061–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang B, et al. 2015. Polymorphisms of the artemisinin resistant marker (K13) in Plasmodium falciparum parasite populations of Grande Comore Island 10 years after artemisinin combination therapy. Parasit Vectors 8: 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouattara A, et al. 2015. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg 92: 1202–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torrentino-madamet M, et al. 2014. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J 13: 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio J-J, 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32: 819–821. [DOI] [PubMed] [Google Scholar]
- 45.Ingasia LA, Cheruiyot J, Okoth SA, Andagalu B, Kamau E, 2016. Genetic variability and population structure of Plasmodium falciparum parasite populations from different malaria ecological regions of Kenya. Infect Genet Evol 39: 372–380. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Shrestha S, Li X, Miao J, Yuan L, Cabrera M, Grube C, Yang Z, Cui L, 2015. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China–Myanmar border in 2007–2012. Malar J 14: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talundzic E, et al. 2015. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog 11: e1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joy DA, et al. 2003. Early origin and recent expansion of Plasmodium falciparum. Science 300: 318–321. [DOI] [PubMed] [Google Scholar]
- 49.Noor AM, et al. 2009. The risks of malaria infection in Kenya in 2009. BMC Infect Dis 9: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]