Abstract.
Loop-mediated isothermal amplification (LAMP) is ideal for the detection of Leishmania DNA as it is a quick and easy-to-perform test that does not require complex or sophisticated equipment or infrastructure. However, the application of this technique in the detection of Leishmania DNA has not been comprehensively analyzed to date (analytical validation). Our objective was to evaluate the sensitivity and analytical specificity (anticipated reportable range [ARR], the limit of detection [LoD], and accuracy) of LAMP targeting the 18S rRNA gene in the diagnosis of six New World Leishmania species. We then applied the validated LAMP assay across 50 samples of sandflies and 50 direct smears from a recent outbreak of cutaneous leishmaniasis in Colombia to determine its diagnostic performance. The LAMP assay exclusively amplified the DNA of Leishmania spp., and an ARR of between 1 × 104 and 1 × 10−2 equivalent parasites/mL was determined. An LoD of 1 × 10−2 equivalent parasites/mL was established and there was no statistically significant variation in terms of accuracy. Finally, a sensitivity of 100% in direct smears and sandflies samples was calculated and a specificity of 90.9% for direct smears using microscopy as reference and 96.8% for sandflies using real-time polymerase chain reaction as reference were determined. To our knowledge, this is the first attempt to analytically validate a LAMP test to detect Leishmania DNA, which showed good diagnostic potential from sandflies and direct smear samples.
INTRODUCTION
Leishmaniasis encompasses a group of neglected tropical diseases caused by parasites of the genus Leishmania that are transmitted by the bite of female sandflies from the Phlebotominae subfamily.1 The spectrum of diseases can be classified into three clinical manifestations according to the associated signs and symptoms (cutaneous, mucocutaneous, and visceral leishmaniasis). Cutaneous leishmaniasis (CL) is considered the most common manifestation of the disease in the New World.2 The disease is prevalent in 98 countries, with around 350 million people estimated to be at risk and around 12 million cases, with an annual incidence of 0.7–1.2 million cases of CL.2,3 In the New World, leishmaniasis is endemic in many areas of North, Central, and South America, thus constituting a major public health problem.2,4,5
Molecular tools have been used for the diagnosis of leishmaniasis, owing to their high levels of sensitivity (40–92%) and specificity (57.1–100%).6,7 However, the requirement for complex equipment and infrastructure means that molecular diagnostic techniques are not suitable for use in remote areas or in the search for active cases (field work). In 2000, Notomi et al.8 designed a new technique known as loop-mediated isothermal amplification (LAMP). This method is characterized by its ability to amplify large amounts of DNA from a few copies in an average time of 30–60 minutes.9 The reaction occurs under isothermal conditions without the need for thermal cyclers.8 It has a high degree of sensitivity owing to the design and quantity of the primers used (i.e., two external primers, two internal primers, and two first loop primers).10 Some studies have demonstrated the use of LAMP against extracts from macerated sandflies8,11 and several alternatives exist for the visualization of the results including turbidity, fluorescence, and/or color changes.8–10,12–14
To date, LAMP has been used for the diagnosis of bacterial,15 fungal,16,17 and viral18,19 infections, as well as parasitic infections such as those caused by Trypanosoma brucei,20 Plasmodium falciparum,21 and Trypanosoma cruzi.22 For the diagnosis of leishmaniasis, LAMP has been used with the implementation of primers targeting the small ribosomal subunit (18S rRNA gene),10,11,23 kinetoplast DNA (kDNA) for the detection of Old World Leishmania species,24–26 and the Internal Transcribed Spacer 1 (ITS-1).12 Loop-mediated isothermal amplification has also been used in the detection of parasites in their insect vectors, as in the case of Dirofilaria immitis detected in Aedes aegypti,27 and T. cruzi and Trypanosoma rangeli.28 Importantly, there has only been one report of the analytical validation of a LAMP assay for T. cruzi.22
Although LAMP platform-based tests targeting different markers (most frequently 18S rRNA gene) have shown to be efficient for the detection of Leishmania in biological samples, a comprehensive assessment of the sensitivity and analytical specificity of these tests to elucidate their diagnostic potential is lacking, especially in endemic regions and where several species coexist.29 Therefore, the objective of this study was to determine, for the first time in Leishmania, the analytical specificity (exclusivity and inclusivity) and sensitivity (anticipated reportable range [ARR], limit of detection [LoD], and accuracy) of the LAMP assay for detecting 18S rRNA gene to evaluate its performance against sandflies and direct smears of CL lesions from a recent outbreak of CL in Colombia.
METHODS
Ethical statement.
This project has a certificate of approval from the ethics committee of the National University of Colombia, number 002-010-15, issued on February 12, 2015. The patients included in this study signed a written informed consent.
Reference strains.
Promastigote cultures of the major Leishmania reference strains frequently associated with CL and mucocutaneous leishmaniasis in Colombia were donated by the International Center for Medical Research and Training that has a preexisting collection of Leishmania species (MHOM/BR/75/M2903 Leishmania braziliensis, MHOM/PA/71/LS94 Leishmania panamensis, MHOM/BR/75/M4147 Leishmania guyanensis, MHOM/TN/80/IPT1 Leishmania infantum, IFLA/BR/67/PH8 Leishmania amazonensis and MHOM/BZ/82/BEL21 Leishmania mexicana). These strains were cloned and maintained in Novy, Nicolle, and McNeal media or Schneider medium supplemented with 20% fetal bovine serum (Microgen, Bogotá, Colombia).
DNA extraction and serial dilutions.
DNA extraction was performed according to the instructions of the High Pure PCR Template Preparation kit (Roche® Ref. 11796828001, Basel, Switzerland) from a stock that contained 1 × 105 parasite equivalents/mL (An average of 100 ng/µL was measured using Nanodrop equipment). DNA obtained from each reference strain was subsequently used to perform serial dilutions from 1 × 104 to 1 × 10−2 parasite equivalents/mL to determine the analytical performance of the LAMP test.
Loop-mediated isothermal amplification assays.
Primers targeting the 18S rRNA gene, reported by Nzelu et al.23 (Table 1), were used for the implementation of the LAMP test. The assays were performed at a final volume of 25 μL consisting of 40 pM of each internal primer forward inner primer and back inner primer, 15 pM of each external primer (F3 and B3), 2 × the Loopamp DNA Amplification reaction mix from Eiken® (Ref. LMP205), 8 U of Bst DNA polymerase (Eiken, Tokyo, Japan), 0.004% malachite green, and 2 μL of DNA. The mixture was incubated at 63°C for 60 minutes, with a final step at 80°C for 5 minutes for inactivation of the enzyme in a dry heating block (Labnet, Edison, NJ). At the end of the reaction, the amplifications were confirmed by visual inspection (light blue color in positive reactions and colorless in negative reactions). Then, to reconfirm the results, 2 μL of each LAMP product were subjected to 2% agarose electrophoresis plus the addition of Syber Safe (Invitrogen, Carlsbad, CA) for staining.
Table 1.
Sequences of LAMP primers used to target the 18S rRNA gene
| Target | Label | Sequence 5′–3′ |
|---|---|---|
| 18S rRNA gene | F3 | GGGTGTTCTCCACTCCAGA |
| B3 | CCATGGCAGTCCACTACAC | |
| FIP | TACTGCCAGTGAAGGCATTGGTGGCAACCATCGTCGTGAG | |
| BIP | TGCGAAAGCCGGCTTGTTCCCATCACCAGCTGATAGGGC |
LAMP = loop-mediated isothermal amplification.
Loop-mediated isothermal amplification analytical specificity.
Analytical specificity was evaluated in terms of the selectivity of the LAMP assay to distinguish blank from non-blank samples. This was analyzed on the basis of the following.
Inclusivity.
This describes the ability of the assay to detect the diversity of blank DNA, which in this case included six New World Leishmania species associated with CL. We performed LAMP on the DNA of six Leishmania reference strains within a single day using the conditions described earlier.
Exclusivity.
This describes the ability of the assay to provide a negative result when closely related but non-target sample sources are tested. In this case, we selected microorganisms that were phylogenetically related to Leishmania and also those associated with differential diagnoses of CL. DNA of parasites belonging to the order Kinetoplastida and obtained from a biological supply vendor (ATCC: The Global Bioresource Center) (ATCC PRA-330 T. cruzi and ATCC 30032 T. rangeli) and eight microorganisms with differential diagnoses of CL (ATCC 25923 Staphylococcus aureus, ATCC 12344 Streptococcus pyogenes, ATCC 26033 Histoplasma capsulatum, ATCC 27294 Mycobacterium tuberculosis, ATCC 26329 Sporothrix schenckii, and ATCC 18827 Fonsecaea pedrosoi) were subjected to the LAMP assay within a single day.
Loop-mediated isothermal amplification analytical sensitivity.
The analytical selectivity of the assay involved measuring the degree of error that can exist within specified limits, using the following parameters.
Anticipated reportable range.
The ARR refers to the range of concentrations over which an analyte can be determined with an adequate level of confidence and accuracy. To achieve this, seven serial dilutions (1 × 104 to 1 × 10−2 parasite equivalents/mL) of each DNA of the six Leishmania species were subjected to the LAMP assay. With the values obtained, consensus tables of the results were constructed.
Limit of detection.
The LoD was calculated as the lowest dilution providing 95% positive results, as established by the National Committee for Clinical Laboratory Standards.30 Five serial dilutions of each DNA of the six Leishmania species were subjected to LAMP. The amplification of each dilution was performed with eight replicates over five consecutive days. The LoD was determined using Probit Regression software (Probit Minitab 15 software, College Station, PA).
Accuracy.
Intra-assay reproducibility was assessed in terms of the accuracy of each test. A dilution above and below the LoD for each DNA of the six Leishmania species was evaluated in triplicate for 10 days (one run per day) under the same conditions. With the values obtained, consensus tables of the results were constructed.
Evaluation of LAMP in biological samples (sandflies and direct smears from skin lesions of patients with CL).
Two sets of biological samples were used to evaluate the performance of the test. The sets of samples were selected by convenience due to the lack of studies expressing values of sensitivity and specificity for LAMP tests in Leishmania. The first set consisted of 50 direct smears, collected during an outbreak of suspected CL cases that occurred in the municipality of El Chaparral in the Department of Tolima during 2003 and 2004, that had previously been tested by microscopy. The second set consisted of 50 pools of samples of female sandflies corresponding to the following: 30 pools of Psychodopygus panamensis, 12 pools of Micropygomyia cayennensis, and eight pools of Lutzomyia gomezi, which were captured in Valledupar, Cesar, in northeastern Colombia. The sandfly pools were subjected to DNA extraction using the ZR Tissue & Insect DNA Microprep kit (Zymo Research® Ref. D6016, Irving, CA) eluting in 50 µL. The direct smears of skin lesions that were fixed to glass slides, stained with Giemsa, and analyzed by microscopy for reference, were immersed in Xilol for 2 seconds to remove any remaining immersion oil. Subsequently, 200 μL of lysis buffer contained in the High Pure PCR Template Preparation kit were added and the entire slide was scraped. Then, the contents were transferred to a clean Eppendorf tube with the aid of a micropipette, and DNA was purified using the protocol described by the manufacturer eluting in 100 µL. The DNA was then subjected to real-time PCR (qPCR) and LAMP. Finally, we compared the operative capabilities of microscopy, qPCR, and LAMP.
Amplification using qPCR.
To compare the LAMP assay with qPCR, DNA was obtained from the direct smears, andpools of sandflies were subjected to qPCR amplification, which is considered the optimal methodology for molecular diagnosis in many cases. This test was implemented using the primers R223-TCCCATCGCAACCTCGGTT and R333-AAAGCGGGCGCGGTGCTG,31 which target the same molecular marker as the LAMP assay (18S rRNA gene). The master mix, at a final volume of 12 μL, contained 5.0 μL of Fast SYBR Green (Ref. 4385370; Applied Biosystems, Foster City, CA), 0.6 μL of each primer (5 pM), and 2 μL of DNA. The thermal profile comprised 50°C for 2 minutes followed by 40 cycles of 95°C for 10 minutes, then 95°C for 45 seconds, and 60°C for 15 seconds. The qPCR was executed using a 7,500 Fast Real-Time PCR System (Applied Biosystems).
Statistical analyses.
The rate of detection of positive samples for each test and each sample type (sandflies or direct smears) was reported in terms of the frequency and the corresponding 95% confidence interval (95% CI). The agreement between the tests (LAMP, qPCR, and microscopy in the case of the smears) was determined by calculating the concordance percentages and was supported by the Kappa coefficients with 95% CI. The operative capabilities of the LAMP test were determined through simple calculations of the sensitivity, specificity, positive and negative predictive values (PPV and NPV, respectively), and the likelihood ratio (LR), using microscopy and qPCR as reference tests for the smears, and qPCR only as a reference test for the sandfly samples. These tests were considered appropriate references because they are the only tests currently available for diagnosis in practice, in contexts similar to those in this study.
RESULTS
The implementation of the LAMP platform using primers directed to the 18S rRNA gene of Leishmania revealed adequate levels of amplification under the conditions described. The main parameters used to evaluate the analytical performance of the test are described in the following paragraphs.
Inclusivity and exclusivity.
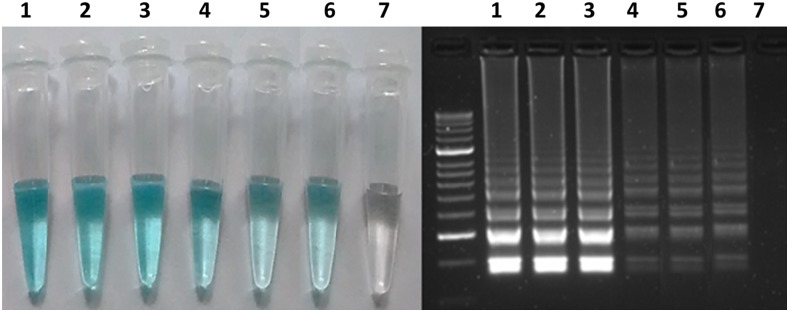
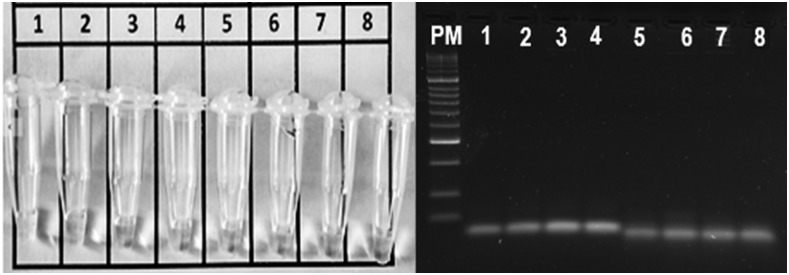
When analyzing the DNA from the six Leishmania strains using the LAMP assay, a light blue color was observed in the reaction tubes of all of the samples. The assay, therefore, appears to be inclusive when amplifying all Leishmania DNAs (Figure 1). Regarding exclusivity, the technique was found to be unique for Leishmania amplification with the 18S rRNA gene, as no color was observed in any of the reaction tubes containing DNAs from samples with a differential diagnosis (Figure 2).
Figure 1.
Inclusivity of the loop-mediated isothermal amplification test directed to the 18S rRNA gene. 1 = Leishmania amazonensis; 2 = Leishmania braziliensis; 3 = Leishmania guyanensis; 4 = Leishmania panamensis; 5 = Leishmania mexicana; 6 = Leishmania infantum; 7 = negative control. This figure appears in color at www.ajtmh.org.
Figure 2.
Exclusivity of the loop-mediated isothermal amplification test directed to the 18S rRNA gene. 1 = Trypanosoma cruzi; 2 = Trypanosoma rangeli; 3 = Staphylococcus aureus; 4 = Streptococcus pyogenes; 5 = Histoplasma capsulatum; 6 = Mycobacterium tuberculosis; 7 = Sporothrix schenckii; 8 = Fonsecaea pedrosoi.
Anticipated reportable range, LoD, and accuracy.
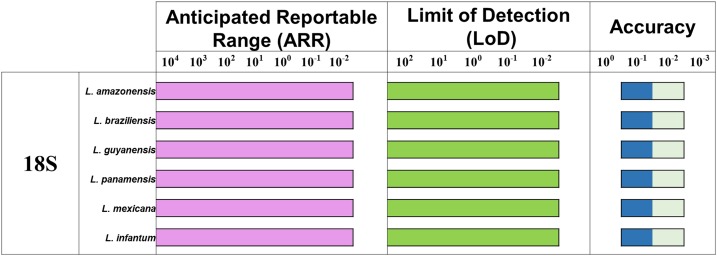
Consensus results were obtained for each dilution of each parameter evaluated (ARR, LoD, and accuracy). The ARR for the LAMP test was evaluated for dilutions 1 × 104 to 1 × 10−2 equivalent parasites/mL of DNA extracted from the six Leishmania species (Figure 3A). It was determined that the LoD was up to the 1 × 10−2 dilution (Figure 3B). These results were consistent with the ARR. The results obtained for more than 10 days of analysis indicated no statistically significant differences and thereby high levels of accuracy (Figure 3C). The findings were concordant with those observed for the ARR and LoD.
Figure 3.
Analytical sensitivity for loop-mediated isothermal amplification. For each concentration, a positive result is determined by the presence of a light blue color in the reaction tube. (A) Anticipated reportable range determined from seven serial dilutions analyzed; (B) Limit of detection (LoD) as a consensus of five serial dilutions and (C) accuracy, including dilution above and below the LoD (box with lighter color). This figure appears in color at www.ajtmh.org.
Leishmania DNA detection from biological samples.
To analyze the ability of the tests to detect Leishmania DNA from biological samples, the tests were first applied to a set of 50 CL smears. Previously, by traditional microscopic diagnosis, 72.0% positivity was detected for Leishmania DNA using this set of smears (n = 36; 95% CI: 57.5–83.8). This frequency of infection was lower than that detected by molecular tests, which in the case of qPCR was 78.0% (n = 39; 95% CI: 64.0–88.5) and 80.0% for the LAMP test (n = 40; 95% CI: 60.3–90.0). The second set of biological samples corresponded to 50 pools of sandflies, for which an infection rate of 40.0% was detected (n = 20; 95% CI: 26.4–54.8) by the same two molecular tests (qPCR and LAMP) (Supplemental Table 1). The results obtained for these two sets of samples are presented in Table 2A. The evaluated tests showed high concordance (> 90% in all cases), with high kappa coefficients. The results of comparisons between the tests are shown in Table 2B.
Table 2.
Comparison of the detection of Leishmania DNA by a range of tests
| Smears | Sandflies | ||||||
|---|---|---|---|---|---|---|---|
| Microscopy | qPCR | qPCR | |||||
| A. | Positive, n (%) | Negative, n (%) | Positive, n (%) | Negative, n (%) | Positive, n (%) | Negative, n (%) | |
| LAMP test | Positive | 36 (72.0) | 4 (8.0) | 39 (78.0) | 1 (2.0) | 20 (40.0) | 0 |
| Negative | 0 | 10 (20.0) | 0 | 10 (20.0) | 0 | 30 (60.0) | |
| B. | Concordance (%) | 92.0 | 98.0 | 100 | |||
| Kappa index | 0.7826 | 0.9398 | 1.000 | ||||
| 95% confidence interval | (0.5832–0.9821) | (0.8231–1.000) | (1.000–1.000) | ||||
qPCR = real-time polymerase chain reaction.
The operative capabilities of the LAMP test were determined using the techniques currently used to detect infection in each of the biological sample sources (smears and pools) evaluated as reference tests. The LAMP test was found to display 100% sensitivity for the detection of Leishmania DNA for the different sample sources. Although the sensitivity was lower (71.4%) compared with that of microscopy, the values remained greater than 90% for both types of samples when compared with qPCR. In agreement with these findings, the NPVs were 100% in all cases and the PPVs were higher than 90%. In the case of the LR (useful for predicting risk in clinical practice), the LR (+) were higher than 3.0 and the LR (−) were not quantifiable because in none of the cases was a positive result obtained for the reference and a negative result obtained for the LAMP test. The results of the operative capabilities of the LAMP test versus the reference test used are described in Table 3.
Table 3.
Operative capabilities of the LAMP test compared with microscopy and qPCR
| Smears | Sandflies | ||
|---|---|---|---|
| Reference test | Microscopy % (95% CI) | qPCR % (95% CI) | qPCR % (95% CI) |
| Sensitivity | 100 (98.6–100.0) | 100 (98.7–100.0) | 100 (97.5–100.0) |
| Specificity | 71.4 (44.2–98.7) | 90.9 (69.4–100.0) | 96.8 (88.9–100.0) |
| PPV | 90.0 (79.5–100.0) | 97.5 (91.4–100.0) | 95.2 (83.8–100.0) |
| NPV | 100.0 (95.0–100.0) | 100.0 (95.0–100.0) | 100.0 (98.3–100.0) |
| LR (+) | 3.5 (1.5–8.0) | 11.0 (1.7–71.3) | 31.0 (4.5–213.8) |
| LR (−) | – | – | – |
LAMP = loop-mediated isothermal amplification; LR = likelihood ratio; NPV = negative predictive value; PPV = positive predictive value; qPCR = real-time polymerase chain reaction. The estimator could not be calculated because one of the fields contained no data during the dispersion analysis.
DISCUSSION
Routine diagnosis of leishmaniasis has been developed based on the direct demonstration of the causative agent from lesions and/or aspirates depending on the clinical manifestation.32 However, these procedures are invasive and often uncomfortable for the patient.33,34 In the case of CL, microscopy remains the “gold standard” in primary health care centers in endemic regions. This test is characterized by its high specificity (100%), ease to perform, and low cost. However, it can present low and variable levels of sensitivity (between 40% and 74.4%) depending on the number and dispersion of parasites in the sample, the sampling procedure, and the ability and expertise of the person preparing and reading the sample smear.35–39 The disadvantages presented by parasitological tests prompted the search for molecular techniques that would be effective for the diagnosis and surveillance of CL.
Several molecular tests have been used to improve the diagnosis of CL40,41; however, PCR has been found to offer high sensitivity and specificity compared with traditional parasitological methods37 and can detect parasite DNA in a variety of clinical samples,6,38 including direct smears,42–44 Flinders Technology Associates filter paper,45 and swabs from the lesion.46 In many cases, the efficacy of PCR depends on the DNA extraction process, the number of copies of the molecular marker selected, and the type of PCR used (PCR-restriction fragment length polymorphism, conventional PCR, or qPCR).47,48 According to the literature, the molecular markers predominantly used to detect parasite DNA by conventional PCR and qPCR are kDNA with a sensitivity of 88.2–97% and a specificity of 57.1–87%,49–51 the 70 kDa heat shock protein 70 with a 90–95% sensitivity and a specificity of 95–100%,4,52 transcribed ITS-1 with 40–91% sensitivity and 96% specificity33,53 and finally the 18S rRNA gene, for which the results were similar to those of kDNA.10 However, the requirement for complex equipment and infrastructure demonstrate that these tests are not an option as diagnostic tools in remote areas or in the search of active cases (field work). In 2000, Notomi and collaborators designed LAMP, which is characterized as an isothermic technique with a fast reaction time and high sensitivity that offers several alternatives for the visualization of results including turbidity, fluorescence, and/or color changes.8–10,12–14 In 2016, Mondal et al.54 reported the use of a recombinant polymerase amplification assay for the detection of Leishmania donovani; this technique offers a shorter reaction time compared with LAMP. Despite advances in the development of new diagnostic techniques, studies often only report certain aspects relating to the sensitivity and specificity of these techniques. A comprehensive evaluation of the analytical performance of these tests and the molecular markers used has not been carried out to date. Evaluation of the analytical performance is vital to determine the relative analytical specificity and sensitivity of the various molecular methods. Such evaluations have been performed for qPCR and LAMP in Chagas disease22,55,56 and conventional PCR and qPCR in toxoplasmosis,57 but not for the LAMP assay in leishmaniasis.
Herein, we evaluated the analytical sensitivity and specificity of a LAMP test targeting the 18S rRNA gene in six Leishmania species circulating in Colombia and determined the operational capabilities of the technique in direct smears from patients with presumptive CL and in sandflies. Our study was the first to include microorganisms with differential diagnoses of CL and two parasites of the order Kinetoplastida to evaluate the exclusivity of the technique. Our findings confirmed the exclusivity of the LAMP assay (Figure 2). Nzelu et al.23 included trypanosome DNA of anurans in their assays and found no cross-reactivity with these parasites. Contrary to the findings of Adams et al.10 who used DNA from differentially diagnosed microorganisms for visceral leishmaniasis and CL (P. falciparum, Escherichia coli, S. aureus, M. tuberculosis, and Mycobacterium leprae) and T. brucei and T. cruzi DNA, and found cross-reactivity with these last two parasites using PCR. In our study, we also found that the assay was inclusive for at least the six Leishmania species tested (Figure 1). However, future studies including a larger range of human infective Leishmania species are needed to confirm the inclusivity of this test. To our knowledge, this is the first time that a large number of microorganisms have been used to determine the exclusivity of the LAMP test, showing its tremendous potential for field implementation in the diagnosis of CL.
We also determined the ARR of the test over a range of dilutions (1 × 104 to 1 × 10−2 equivalent parasites/mL) that were appropriate to test the LoD. The LoD was established to be 1 × 10−2 equivalent parasites/mL (Figure 2), similar to that reported in the Nzelu study using serial dilutions of L. mexicana.23,40 Our results differed from those reported by Adams et al.10 with a LoD of 1 × 102 parasites/mL for L. donovani and Sriworarat et al.14 with a LoD of 1 × 103 parasites/mL for Leishmania siamensis in whole blood samples. In addition, a report by Tiwananthagorn et al.58 concluded that a PCR-kDNA assay was more sensitive than PCR-ITS and LAMP with the 18S rRNA gene.42 Therefore, a comprehensive study to evaluate these parameters in a vast set of Leishmania species causing CL in the New World is needed.
Finally, we evaluated the diagnostic performance of the LAMP test using DNA from sandflies and, for the first time, DNA extracted from direct smear slides that had previously been stained with Giemsa and diagnosed by microscopy. Comparison of the results of the LAMP test with other tests, considered as reference tests in this study, revealed high concordance (> 90%, with kappa coefficients > 0.78) and no cases in which a positive test result was obtained for the reference test that was then found to be negative for the LAMP test (Table 2). This may indicate the high analytical sensitivity of the LAMP test for the detection of Leishmania DNA. Analysis of the operative capabilities of this technique revealed 100% sensitivity and NPVs, and greater than 90% specificity and PPVs, with the exception of comparisons with microscopy where the sensitivity was reduced to 71.4% (Table 3). These findings may be related to the fact that different types of techniques are being compared, which together with the sensitivity and analytical specificity data for the LAMP assay, indicate that the cases of non-concordance between the tests may correspond to failures in baseline tests and not limitations of the LAMP assay. However, field studies should be considered where the diagnostic performance of the technique is evaluated over a representative number of samples. Positive samples were obtained for LAMP from P. panamensis DNA, a sandfly known to be a vector of parasites of the subgenus Viannia in the country,59,60 further indicating that LAMP could be of potential use for the entomological surveillance of CL. Our results clearly depict the potential use of this LAMP assay in remote or rural areas, especially owing to the possibility of amplifying DNA from smears collected on slides that had previously been stained with Giemsa, indicating the potential to conduct retrospective studies and plausibly identify infective species. Future studies need to standardize a method of DNA extraction that could be conducted in the field, which is a current limitation for the implementation of a LAMP test or other type of molecular test such as a recombinant polymerase amplification assay. In terms of cost effectiveness, Adams et al.10 performed a comparison of LAMP versus qPCR and PCR-RFLP. Loop-mediated isothermal amplification using the Eiken fluorescent reagent was estimated to cost approximately $3.5 per reaction, compared with $12 for qPCR and $2.5 for PCR. The use of malachite green to visualize the results, as reported in the current study, may reduce the cost per reaction considerably. These findings differ from our previous results that revealed better analytical performance for LAMP than PCR-based tests.61
In conclusion, LAMP is a valuable diagnostic tool that offers advantages compared with qPCR and microscopy such as a LoD of 1 × 10−2 parasites/mL, exclusivity for Leishmania amplification, lower cost, and ease to perform and interpret. One of the most important advantages is its ability to detect Leishmania DNA from direct smears and sandflies, which indicate its potential application in primary care centers and in the diagnostic and entomological surveillance of leishmaniasis in endemic countries. Field trials will confirm the potential of this LAMP assay for the timely diagnosis of leishmaniasis in communities vulnerable to infection.
Supplementary Material
Acknowledgments:
We thank Kate Fox, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1.Akhoundi M, et al. 2017. Leishmania infections: molecular targets and diagnosis. Mol Aspects Med 57: 1–29. [DOI] [PubMed] [Google Scholar]
- 2.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M; WHO Leishmaniasis Control Team , 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO)/Department of Control of Neglected Tropical Diseases , 2017. Global leishmaniasis update, 2006–2015: a turning point in leishmaniasis surveillance. Weekly Epidemiological Record 38: 557–565. [Google Scholar]
- 4.Montalvo AM, Fraga J, Tirado D, Blandón G, Alba A, Van der Auwera G, Vélez ID, Muskus C, 2017. Detection and identification of Leishmania spp.: application of two Hsp70-based PCR-RFLP protocols to clinical samples from the New World. Parasitol Res 116: 1843–1848. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez JD, Hernandez C, Leon CM, Ayala MS, Florez C, González C, 2016. Taxonomy, diversity, temporal and geographical distribution of cutaneous leishmaniasis in Colombia: a retrospective study. Sci Rep 6: 28266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohammadiha A, Mohebali M, Haghighi A, Mahdian R, Abadi AR, Zarei Z, Yeganeh F, Kazemi B, Taghipour N, Akhoundi B, 2013. Comparison of real-time PCR and conventional PCR with two DNA targets for detection of Leishmania (Leishmania) infantum infection in human and dog blood samples. Exp Parasitol 133: 89–94. [DOI] [PubMed] [Google Scholar]
- 7.Munoz EB, Santander S, Rojas-Silva P, Cardenas PA, Fornasini M, Cifuentes SC, Salvador D, Baldeón ME, 2016. Diagnostic efficacy of molecular techniques for detection and identification of Leishmania species in human whole blood and skin samples from Ecuador. Am J Trop Med Hyg 95: 803–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T, 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomita N, Mori Y, Kanda H, Notomi T, 2008. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3: 877–882. [DOI] [PubMed] [Google Scholar]
- 10.Adams ER, Gomez MA, Scheske L, Rios R, Marquez R, Cossio A, Albertini A, Schallig H, Saravia NG, 2014. Sensitive diagnosis of cutaneous leishmaniasis by lesion swab sampling coupled to qPCR. Parasitology 141: 1891–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikita K, Maeda T, Yoshikawa S, Ono T, Miyahira Y, Kawana A, 2014. The Direct Boil-LAMP method: a simple and rapid diagnostic method for cutaneous leishmaniasis. Parasitol Int 63: 785–789. [DOI] [PubMed] [Google Scholar]
- 12.Abbasi I, Kirstein OD, Hailu A, Warburg A, 2016. Optimization of loop-mediated isothermal amplification (LAMP) assays for the detection of Leishmania DNA in human blood samples. Acta Trop 162: 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghasemian M, Gharavi MJ, Akhlaghi L, Mohebali M, Meamar AR, Aryan E, Oormazdi H, 2014. Development and assessment of loop-mediated isothermal amplification (LAMP) assay for the diagnosis of human visceral leishmaniasis in Iran. Iran J Parasitol 9: 50–59. [PMC free article] [PubMed] [Google Scholar]
- 14.Sriworarat C, Phumee A, Mungthin M, Leelayoova S, Siriyasatien P, 2015. Development of loop-mediated isothermal amplification (LAMP) for simple detection of Leishmania infection. Parasit Vectors 8: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao YM, Guo YC, Zhang XE, Zhou YF, Zhang ZP, Wei HP, Yang RF, Wang DB, 2007. Loop-mediated isothermal amplification for rapid detection of Bacillus anthracis spores. Biotechnol Lett 29: 1939–1946. [DOI] [PubMed] [Google Scholar]
- 16.Villari C, Tomlinson JA, Battisti A, Boonham N, Capretti P, Faccoli M, 2013. Use of loop-mediated isothermal amplification for detection of Ophiostoma clavatum, the primary blue stain fungus associated with Ips acuminatus. Appl Environ Microbiol 79: 2527–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inacio J, Flores O, Spencer-Martins I, 2008. Efficient identification of clinically relevant Candida yeast species by use of an assay combining panfungal loop-mediated isothermal DNA amplification with hybridization to species-specific oligonucleotide probes. J Clin Microbiol 46: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wozniakowski G, Samorek-Salamonowicz E, Kozdrun W, 2013. Comparison of loop-mediated isothermal amplification and PCR for the detection and differentiation of Marek’s disease virus serotypes 1, 2, and 3. Avian Dis 57: 539–543. [DOI] [PubMed] [Google Scholar]
- 19.Hong TC, Mai QL, Cuong DV, Parida M, Minekawa H, Notomi T, Hasebe F, Morita K, 2004. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol 42: 1956–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, Sugimoto C, Igarashi I, 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol 41: 5517–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poon LL, et al. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem 52: 303–306. [DOI] [PubMed] [Google Scholar]
- 22.Besuschio SA, et al. 2017. Analytical sensitivity and specificity of a loop-mediated isothermal amplification (LAMP) kit prototype for detection of Trypanosoma cruzi DNA in human blood samples. PLoS Negl Trop Dis 11: e0005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nzelu CO, Gomez EA, Caceres AG, Sakurai T, Martini-Robles L, Uezato H, Mimori T, Katakura K, Hashiguchi Y, Kato H, 2014. Development of a loop-mediated isothermal amplification method for rapid mass-screening of sand flies for Leishmania infection. Acta Trop 132: 1–6. [DOI] [PubMed] [Google Scholar]
- 24.Khan MG, Bhaskar KR, Salam MA, Akther T, Pluschke G, Mondal D, 2012. Diagnostic accuracy of loop-mediated isothermal amplification (LAMP) for detection of Leishmania DNA in buffy coat from visceral leishmaniasis patients. Parasit Vectors 5: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takagi H, Itoh M, Islam MZ, Razzaque A, Ekram AR, Hashighuchi Y, Noiri E, Kimura E, 2009. Sensitive, specific, and rapid detection of Leishmania donovani DNA by loop-mediated isothermal amplification. Am J Trop Med Hyg 81: 578–582. [DOI] [PubMed] [Google Scholar]
- 26.Verma S, Avishek K, Sharma V, Negi NS, Ramesh V, Salotra P, 2013. Application of loop-mediated isothermal amplification assay for the sensitive and rapid diagnosis of visceral leishmaniasis and post-kala-azar dermal leishmaniasis. Diagn Microbiol Infect Dis 75: 390–395. [DOI] [PubMed] [Google Scholar]
- 27.Aonuma H, Yoshimura A, Perera N, Shinzawa N, Bando H, Oshiro S, Nelson B, Fukumoto S, Kanuka H, 2009. Loop-mediated isothermal amplification applied to filarial parasites detection in the mosquito vectors: Dirofilaria immitis as a study model. Parasit Vectors 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thekisoe OM, Rodriguez CV, Rivas F, Coronel-Servian AM, Fukumoto S, Sugimoto C, Kawazu S, Inoue N, 2010. Detection of Trypanosoma cruzi and T. rangeli infections from Rhodnius pallescens bugs by loop-mediated isothermal amplification (LAMP). Am J Trop Med Hyg 82: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pourmohammadi B, Motazedian M, Hatam G, Kalantari M, Habibi P, Sarkari B, 2010. Comparison of three methods for diagnosis of cutaneous leishmaniasis. Iran J Parasitol 5: 1–8. [PMC free article] [PubMed] [Google Scholar]
- 30.NCCLS , 2004. Protocols for Determination of Limits of Detection and Limits of Quantification; Approved Guideline, Vol. 24(10). Wayne, Pennsylvania: NCCLS. [Google Scholar]
- 31.Cruz IC, Cañavate JM, Rubio MA, Morales C, Chicharro F, Laguna F, Jiménez-Mejías M, Sirera G, Videla S, Alvar J, 2012. A nested polymerase chain reaction (Ln-PCR) for diagnosing and monitoring Leishmania infantum infection in co-infected patients with human immunodeficiency virus. Trans R Soc Trop Med Hyg 96: 185–189. [DOI] [PubMed] [Google Scholar]
- 32.Bezerra-Vasconcelos DR, Melo LM, Albuquerque ES, Luciano MC, Bevilaqua CM, 2011. Real-time PCR to assess the Leishmania load in Lutzomyia longipalpis sand flies: screening of target genes and assessment of quantitative methods. Exp Parasitol 129: 234–239. [DOI] [PubMed] [Google Scholar]
- 33.Marfurt J, Nasereddin A, Niederwieser I, Jaffe CL, Beck HP, Felger I, 2003. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J Clin Microbiol 41: 3147–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wortmann G, Sweeney C, Houng HS, Aronson N, Stiteler J, Jackson J, Ockenhouse C, 2001. Rapid diagnosis of leishmaniasis by fluorogenic polymerase chain reaction. Am J Trop Med Hyg 65: 583–587. [DOI] [PubMed] [Google Scholar]
- 35.Bensoussan E, Nasereddin A, Jonas F, Schnur LF, Jaffe CL, 2006. Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J Clin Microbiol 44: 1435–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goto H, Lauletta Lindoso JA, 2012. Cutaneous and mucocutaneous leishmaniasis. Infect Dis Clin North Am 26: 293–307. [DOI] [PubMed] [Google Scholar]
- 37.Szargiki R, Castro EA, Luz E, Kowalthuk W, Machado AM, Thomaz-Soccol V, 2009. Comparison of serological and parasitological methods for cutaneous leishmaniasis diagnosis in the state of Parana, Brazil. Braz J Infect Dis 13: 47–52. [DOI] [PubMed] [Google Scholar]
- 38.Singh S, Dey A, Sivakumar R, 2005. Applications of molecular methods for Leishmania control. Expert Rev Mol Diagn 5: 251–265. [DOI] [PubMed] [Google Scholar]
- 39.Brito ME, Mendonca MG, Gomes YM, Jardim ML, Abath FG, 2001. Dynamics of the antibody response in patients with therapeutic or spontaneous cure of American cutaneous leishmaniasis. Trans R Soc Trop Med Hyg 95: 203–206. [DOI] [PubMed] [Google Scholar]
- 40.Marco JD, Bhutto AM, Soomro FR, Baloch JH, Barroso PA, Kato H, Uezato H, Katakura K, Korenaga M, Nonaka S, 2006. Multilocus enzyme electrophoresis and cytochrome B gene sequencing-based identification of Leishmania isolates from different foci of cutaneous leishmaniasis in Pakistan. Am J Trop Med Hyg 75: 261–266. [PubMed] [Google Scholar]
- 41.Wincker P, Ravel C, Britto C, Dubessay P, Bastien P, Pagès M, Blaineau C, 1997. A direct method for the chromosomal assignment of DNA markers in Leishmania. Gene 194: 77–80. [DOI] [PubMed] [Google Scholar]
- 42.Eroglu F, Uzun S, Koltas IS, 2014. Comparison of clinical samples and methods in chronic cutaneous leishmaniasis. Am J Trop Med Hyg 91: 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motazedian H, Karamian M, Noyes HA, Ardehali S, 2002. DNA extraction and amplification of Leishmania from archived, Giemsa-stained slides, for the diagnosis of cutaneous leishmaniasis by PCR. Ann Trop Med Parasitol 96: 31–34. [DOI] [PubMed] [Google Scholar]
- 44.Yokota M, Tatsumi N, Tsuda I, Yano I, 1995. DNA extraction and amplification from Giemsa-stained blood smears. J Clin Lab Anal 9: 387–391. [DOI] [PubMed] [Google Scholar]
- 45.Caicedo L, Márquez P, Sánchez M, Ortíz Arauz A, Solorzano L, Castro G, Pozo W, 2016. Comparison of the sensitivity of PCR to other laboratory techniques used for the diagnosis of cutaneous leishmaniasis in Ecuador. Centro de Biotecnología 5: 80–90. [Google Scholar]
- 46.Mimori T, Matsumoto T, Calvopina MH, Gomez EA, Saya H, Katakura K, Nonaka S, Shamsuzzaman SM, Hashiguchi Y, 2002. Usefulness of sampling with cotton swab for PCR-diagnosis of cutaneous leishmaniasis in the New World. Acta Trop 81: 197–202. [DOI] [PubMed] [Google Scholar]
- 47.Cortes S, Rolao N, Ramada J, Campino L, 2004. PCR as a rapid and sensitive tool in the diagnosis of human and canine leishmaniasis using Leishmania donovani s.l.-specific kinetoplastid primers. Trans R Soc Trop Med Hyg 98: 12–17. [DOI] [PubMed] [Google Scholar]
- 48.Tsokana CN, Athanasiou LV, Valiakos G, Spyrou V, Manolakou K, Billinis C, 2014. Molecular diagnosis of leishmaniasis, species identification and phylogenetic analysis. Claborn D, ed. Leishmaniasis—Trends in Epidemiology, Diagnosis and Treatment. London, United Kingdom: InTech. [Google Scholar]
- 49.Jara M, Adaui V, Valencia BM, Martinez D, Alba M, Castrillon C, Cruz M, Cruz I, Van der Auwera G, Llanos-Cuentas A, 2013. Real-time PCR assay for detection and quantification of Leishmania (Viannia) organisms in skin and mucosal lesions: exploratory study of parasite load and clinical parameters. J Clin Microbiol 51: 1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marquez LM, Lampo M, Rinaldi M, Lau P, 2001. Gene flow between natural and domestic populations of Lutzomyia longipalpis (Diptera: Psychodidae) in a restricted focus of American visceral leishmaniasis in Venezuela. J Med Entomol 38: 12–16. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez N, Bailey BN, Martin MB, Oldfield E, Urbina JA, Docampo R, 2002. Radical cure of experimental cutaneous leishmaniasis by the bisphosphonate pamidronate. J Infect Dis 186: 138–140. [DOI] [PubMed] [Google Scholar]
- 52.Garcia AL, Tellez T, Parrado R, Rojas E, Bermudez H, Dujardin JC, 2007. Epidemiological monitoring of American tegumentary leishmaniasis: molecular characterization of a peridomestic transmission cycle in the Amazonian lowlands of Bolivia. Trans R Soc Trop Med Hyg 101: 1208–1213. [DOI] [PubMed] [Google Scholar]
- 53.Ovalle C, Porras L, Muvdi S, Rios M, 2007. Polymerase chain reaction with two molecular targets in mucosal leishmaniasis’ diagnosis: a validation study. Mem Inst Oswaldo Cruz 102: 549–554. [DOI] [PubMed] [Google Scholar]
- 54.Mondal D, Ghosh P, Khan MA, Hossain F, Bohlken-Fascher S, Matlashewski G, Kroeger A, Olliaro P, Abd El Wahed A, 2016. Mobile suitcase laboratory for rapid detection of Leishmania donovani using recombinase polymerase amplification assay. Parasit Vectors 9: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duffy T, Cura CI, Ramirez JC, Abate T, Cayo NM, Parrado R, Bello ZD, Velazquez E, Muñoz-Calderon A, Juiz NA, 2013. Analytical performance of a multiplex Real-Time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl Trop Dis 7: e2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramirez JD, Hernandez C, 2017. Trypanosoma cruzi I: towards the need of genetic subdivision? Part II. Acta Trop pii: S0001-706X(17)30250-4. [DOI] [PubMed] [Google Scholar]
- 57.Sterkers Y, et al. 2010. Multicentric comparative analytical performance study for molecular detection of low amounts of Toxoplasma gondii from simulated specimens. J Clin Microbiol 48: 3216–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiwananthagorn S, Kato H, Yeewa R, Muengpan A, Polseela R, Leelayoova S, 2017. Comparison of LAMP and PCR for molecular mass screening of sand flies for Leishmania martiniquensis infection. Mem. Inst. Oswaldo Cruz 112: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santamaria E, Ponce N, Zipa Y, Ferro C, 2006. Presence of infected vectors of Leishmania (V.) panamensis within dwellings in two endemic foci in the foothill of the middle Magdalena valley, western Boyaca, Colombia [Spanish]. Biomedica 26 (Suppl 1): 82–94. [PubMed] [Google Scholar]
- 60.Vivero RJ, Quintero LS, Pena HC, Alvar-Beltran J, Tovar C, Atencia CM, Vélez ID, 2017. Composition and distribution of medically important phlebotomines (Diptera: Psychodidae) in the municipalities of Tierralta and Valencia (Cordoba, Colombia). J Vector Borne Dis 54: 87–95. [PubMed] [Google Scholar]
- 61.León CM, Muñoz M, Hernández DC, Teherán A, Ayala MS, Florez C, Ramírez JD, 2017. Analytical performance of four polymerase chain reaction (PCR) and real time PCR (qPCR) assays for the detection of six Leishmania species DNA in Colombia. Front Microbiol 8: 1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.