Abstract.
Defining the optimal diagnostic tools for evaluating onchocerciasis elimination efforts in areas co-endemic for other filarial nematodes is imperative. This study compared three published polymerase chain reaction (PCR) methods: the Onchocerca volvulus–specific qPCR-O150, the pan-filarial qPCR melt curve analysis (MCA), and the O150-PCR enzyme-linked immunosorbent assay (ELISA) currently used for vector surveillance in skin snip biopsies (skin snips) collected from the Democratic Republic of the Congo. The pan-filarial qPCR-MCA was compared with species-specific qPCRs for Loa loa and Mansonella perstans. Among the 471 skin snips, 47.5%, 43.5%, and 27.0% were O. volvulus positive by qPCR-O150, qPCR-MCA, and O150-PCR ELISA, respectively. Using qPCR-O150 as the comparator, the sensitivity and specificity of qPCR-MCA were 89.3% and 98.0%, respectively, whereas for O150-PCR ELISA, they were 56.7% and 100%, respectively. Although qPCR-MCA identified the presence of L. loa and Mansonella spp. in skin snips, species-specific qPCRs had greater sensitivity and were needed to identify M. perstans. Most of the qPCR-MCA misclassifications occurred in mixed infections. The reduced sensitivity of O150-PCR ELISA was associated with lower microfilaria burden and with lower amounts of O. volvulus DNA. Although qPCR-MCA identified most of the O. volvulus–positive skin snips, it is not sufficiently robust to be used for stop-mass drug administration (MDA) evaluations in areas co-endemic for other filariae. Because O150-PCR ELISA missed 43.3% of qPCR-O150–positive skin snips, the qPCR-O150 assay is more appropriate for evaluating skin snips of OV-16 + children in stop-MDA assessments. Although improving the sensitivity of the O150-PCR ELISA as an alternative to qPCR might be possible, qPCR-O150 offers distinct advantages aside from increased sensitivity.
INTRODUCTION
Human onchocerciasis, or river blindness, is a neglected tropical disease caused by infection with the filarial parasite, Onchocerca volvulus, and is transmitted by Simulium blackflies that breed near fast-flowing streams or rivers. An estimated 186 million people live at risk of infection.1 Onchocerciasis can cause severe itching, disfiguring skin lesions, and eye disease, which can lead to blindness.2 Mass drug administration (MDA) of the microfilaricide, ivermectin (IVM),3 administered annually for 10–15 or more years is necessary to interrupt transmission in endemic areas.4,5 Several countries in Africa have targeted elimination of the disease by 2025, although recent modeling suggests that 2040 might be a more realistic goal for regional elimination.6
Successful elimination of onchocerciasis requires proper evaluation protocols to determine when MDA can be stopped. Laboratory tools that have been used for onchocerciasis include evaluation of skin snip biopsies (skin snips) either by microscopy or PCR testing,7–12 detection of antibody specific for OV-16, an O. volvulus–specific antigen,13,14 and blackfly surveillance using a pool screen PCR assay targeting a repeated sequence of roughly 150 base pairs (O-150 repeat) specific for O. volvulus followed by ELISA detection (O150-PCR ELISA).15 The fact that millions of people at risk for onchocerciasis live in regions in Africa co-endemic for Loa loa or Mansonella spp. poses challenges to diagnostic testing for onchocerciasis.16–19 Mansonella streptocerca microfilariae (MF) typically migrate through the skin,20 whereas MF from both L. loa and M. perstans typically circulate within blood and not skin.18,21 The presence of MF or DNA from other filarial species in skin snips could potentially lead to false positives by microscopy or PCR methods that are not species-specific for O. volvulus.12 With the exception of a study by Wilson et al.22 in Senegal, diagnostic performance of OV-16–based immunoassays has been assessed mostly in populations that are not co-endemic with other filarial species.13,14,23–27 Because cross-reactivity may occur, diagnostic tools intended for onchocerciasis stop-MDA assessments need to be evaluated within several epidemiological contexts including those co-endemic for other filariae.
The 2016 WHO guidelines for stopping MDA require measuring O. volvulus prevalence in the blackfly vector using pool screen O150-PCR ELISA and prevalence in at-risk children younger than 10 years using OV-16 serology.28 Skin snip microscopy has insufficient sensitivity to be used for stopping decisions,11,12,29 but skin snip PCR can be used in some circumstances. If fewer than 10 children are found to have positive OV-16 serology, those children may be evaluated using skin snip PCR. If all children tested have negative PCR results and continue to be negative after not receiving IVM for 1 year, the evaluation areas may stop MDA, assuming the blackfly criterion has been met.28 In this situation, skin snip PCR is used to exclude patent infection in a low-prevalence setting and, thus, test sensitivity needs to be maximized. However, these guidelines do not identify which PCR method should be implemented for this type of testing.28 This is a new procedure and to date, fewer than 10 OV-16–positive children have been tested by skin snip PCR using the O150-PCR ELISA used for blackfly vector surveillance (T. R. Unnasch, personal communication, November 2017). Because of a lack of studies that have directly compared the performance of the O150-PCR ELISA with other available PCR-based methods, there is no published evidence that the O150-PCR ELISA is the optimal PCR method for testing skin snips collected from OV-16–positive children.
Several PCR-based methods have been developed for detecting O. volvulus, including loop-mediated isothermal amplification,30 conventional PCR targeting the repeat O-150 region,7–10 and the O150-PCR ELISA used for blackfly vector surveillance.15 Within the last few years, real-time PCR (qPCR) methods have been developed for O. volvulus detection. Lloyd et al.11 described a TaqMan® (Life Technologies, Grand Island, NY) qPCR assay that targets the O-150 repeat region (qPCR-O150) and a modified conventional O150-PCR method. In addition, our group previously described the optimization of a single-reaction pan-filarial qPCR with melt curve analysis (qPCR-MCA) tool that targets a region within the first internal transcribed spacer (ITS1) of the ribosomal DNA (rDNA) and the flanking 5.8S tract12 using skin snips from sites in Ethiopia (Jimma Zone) and Uganda (Kitgum and Lamwo districts) that were not endemic for either L. loa or M. perstans (P. Cantey and V. Cama, manuscript in preparation).
Molecular tools for O. volvulus detection, including both the qPCR-O150 and qPCR-MCA assays, have shown increased sensitivity compared with parasitological detection by microscopy. However, varying sensitivities have been reported for different assays, perhaps, because of being tested in different epidemiological contexts. Therefore, it will be informative to determine the performance characteristics of qPCR methods and to conduct a side-by-side comparison of the O150-PCR ELISA and the two recently developed qPCR assays (qPCR-O150 or qPCR-MCA). This work will provide information on their relative sensitivity and specificity for detecting O. volvulus DNA in skin snips.
Thiele et al.12 demonstrated in a subset of samples (N = 248) that the qPCR-MCA had greater sensitivity than the O150-PCR ELISA, but the qPCR-0150 assay has not been compared with O150-PCR ELISA.11 The qPCR-MCA tool broadly amplifies the ITS1-5.8S region from multiple filarial species, relying on MCA for species categorization.12 Because of the way this tool was designed and because it was originally tested in two regions where L. loa and M. perstans were not endemic, an additional objective of this study was to further validate this tool with samples from settings that were co-endemic for L. loa and M. perstans.
The qPCR-O150 amplifies a repeat sequence,11 which has been reported to improve PCR detection; therefore, it is plausible that the qPCR-O150 might have increased sensitivity compared with the qPCR-MCA. A direct diagnostic comparison between qPCR-MCA, qPCR-O150, and O150-PCR ELISA was needed to determine which method had the greatest sensitivity and specificity and would be best suited for testing skin biopsies of OV-16–positive children.
The goals of this study were to 1) directly compare the performance of three PCR methods for O. volvulus detection—qPCR-O150, qPCR-MCA, and O150-PCR ELISA and 2) evaluate the qPCR-MCA tool in a setting where mixed filarial infections occurred.
MATERIALS AND METHODS
Study site and sample collection.
Specimens were collected as part of a broader study designed to support the evaluation of several different diagnostic tools for onchocerciasis and other filarial infections. Samples for the evaluations described here came from the study site in the community of Banalia, Tshopo Province, Democratic Republic of the Congo (DRC). This site is co-endemic for onchocerciasis, loiasis, and mansonellosis. Only three prior MDA distributions had occurred in this study area with the last one about 11 months before sample collection. A convenience sample of 500 participants ≥ 5 years of age who had resided within the study site for ≥ 10 years or since birth was enrolled. Consent to participate was obtained from participants aged 18 or older and from the parents of children under the age of 18 years; assent was obtained from children between the ages of 7 and 18 years. Two skin snips per participant were collected from the iliac crest and daytime blood samples were collected by venipuncture. The protocol was approved by the institutional review boards of the U.S. Centers for Disease Control and Prevention (CDC [protocol number 6196]) and the Kinshasa School of Public Health, DRC (approval number ESP/CE/032/14).
Parasitological detection by microscopy.
Microscopic evaluation of the skin snips for MF from 471 study participants was performed as previously described using a light microscope at ×200–400 magnification.12 All samples diagnosed to have MF were verified by a second microscopist. After emergence of MF from the skin snips and visualization by microscopy, the skin snips were then preserved in 400 μL of RNALater® (Life Technologies) and stored at −80°C until PCR testing was performed at the CDC laboratories in Atlanta, GA. The presence or absence of L. loa and M. perstans infection was assessed by microscopic evaluation of Giemsa-stained daytime thick blood smears on-site in DRC.
DNA extraction from skin snips.
The two skin snips from each participant, which had been preserved after MF emergence and microscopic evaluation, were pooled into one DNA extraction. Genomic DNA was extracted using the QIAmp DNA Investigator Kit (Qiagen, Valencia, CA) as previously described.12 DNA quantity and quality were assessed using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Waltham, MA).
PCR assays.
PCR assays used in this study were performed as previously described.11,12,15,31,32 Primer and probe sequences are detailed in Supplemental Table 1. The limit of detection (LOD) for the three PCR assays for O. volvulus was determined and compared using serial 10-fold dilutions of adult O. volvulus gDNA that had been quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific).
qPCR-O150.
The qPCR assay targets the O-150 repeat region of O. volvulus and was performed as previously described.11 Non-endemic Homo sapiens gDNA and a no-template reaction were used as negative controls; all assays were run in duplicate. Samples were considered positive if the cycle threshold (Ct) values were below 40 amplification cycles for both duplicates. Samples were retested if duplicate qPCR-O150 results were discordant.
qPCR-MCA.
Whole gDNA representative of single infections for O. volvulus, L. loa, Mansonella ozzardi, Dirofilaria immitis, and Brugia pahangi was used as references for MCA. Non-endemic H. sapiens gDNA and a no-template reaction were used as negative controls. All assays were run in duplicate. The melting temperature (Tm) ranges for MCA and filariae identification were as follows: O. volvulus (78.8–79.35°C, SD ± 0.21), L. loa (77.85–78.35°C, SD ± 0.24), and Mansonella spp. (76.85–77.35°C, SD ± 0.24). Previous assay optimization demonstrated that M. perstans and M. ozzardi had an indistinguishable Tm;12 therefore, M. ozzardi gDNA was used for the identification of Mansonella spp. PCR amplifications were considered positive if the Ct value was < 36 and the observed Tm fell within the specified range for O. volvulus, Mansonella spp., or L. loa. Samples with the appropriate Tm but with Ct values of 36–38 were considered indeterminate and reassayed, whereas samples with a Ct > 38 were considered negative. Samples were repeated if discordant results were observed between the experimental duplicates. Species-specific qPCRs for O. volvulus,11 M. perstans,31 and L. loa32 were used to validate qPCR-MCA results.
O150-PCR ELISA.
The O150-PCR ELISA assay was performed as previously described.15 Briefly, 5 μL of purified gDNA was used as template for PCR amplification. Assays included one high and one low-concentration positive control and 10 no-template negative control reactions. Conventional PCR for the O150 PCR–ELISA was performed in a T-100 thermal cycler (Bio-Rad, Hercules, CA), and ELISA detection of amplicons was measured with a SpectraMax 190 reader using SoftMax Pro v5.4.1 for data capture and analyses (Molecular Devices, Downingtown, PA). Samples were classified as positive using one of two cutoff values, always selecting the higher of the two, as previously described15: 1) the mean plus three standard deviations of the 10 negative controls if this value exceeded 0.1 or 2) 0.1. Samples that were initially positive were retested beginning with a new PCR reaction. Any sample with values above the cutoff in two independent PCRs was confirmed positive.15
Species-specific TaqMan qPCRs for M. perstans and L. loa.
Mansonella perstans was detected using a published protocol targeting the ITS1 region of the rDNA,31 whereas L. loa was detected using a species-specific TaqMan qPCR targeting the predicted ORF LLMF72 as previously described.32 qPCR results were considered positive if the Ct values were below 40 amplification cycles for both duplicates. Samples were retested if duplicate results were discordant.
Sanger sequencing.
Species specificity of TaqMan qPCRs was verified by Sanger sequencing of the ITS1 region of the rDNA in a subset of samples that was found to be either L. loa (N = 16) or M. perstans (N = 22) positive, but not mixed.12 DNA sequencing chromatograms were analyzed, trimmed, and assembled using Geneious version R8 (Biomatters, Inc., Auckland, New Zealand). Filariae species were determined using the National Center for Biotechnology Information’s nucleotide database via the Basic Local Alignment Search Tool’s MegaBlast algorithm (https://blast.ncbi.nlm.nih.gov). To distinguish O. volvulus and O. ochengi sequences for the five samples that were qPCR-MCA(+) but qPCR-O150(−), whole ITS1 sequences were compared by BLAST analysis with the O. volvulus and O. ochengi genomic sequences from the WormBase ParaSite (http://parasite.wormbase.org/index.html).33,34
Statistical analyses.
Data analysis and statistical tests were performed using Epi Info (CDC), SPSS v. 24 (IBM SPSS Statistics, Armonk, NY), and GraphPad Prism version 7 (GraphPad Software, La Jolla, CA). Diagnostic test performance was compared using the McNemar’s test for paired nominal data, and P values were corrected for multiple comparisons using the Bonferroni correction method. A two-sided χ2 test was performed to assess the role of non-single peak melt curve profiles in relation to the presence of mixed filarial DNA. Non-normally distributed continuous variables, as determined by D’Agostino and Pearson omnibus and Shapiro–Wilk tests for normality, were analyzed using the nonparametric Spearman’s rank correlation test.
RESULTS
Filarial detection by microscopy.
Samples from 189 (40.1%) study participants were positive for O. volvulus, whereas none were positive for M. streptocerca by microscopic evaluation of skin snips. Microscopic evaluation of daytime thick blood smears detected 104 (22.2%) individuals with L. loa and 257 (54.7%) individuals with M. perstans, whereas 83 (17.6%) were positive for both.
Comparison of three PCR-based methods for detection of O. volvulus.
Limit of detection.
Overall, the qPCR-O150 had the lowest LOD at 10 fg/μL genomic DNA (Table 1). Controlling for differences in the volume of extracted DNA used in each PCR protocol, qPCR-O150 had a 2.5× lower LOD than qPCR-MCA and > 100× lower LOD when compared with O150-PCR ELISA (Table 1).
Table 1.
Limit of detection (LOD) for three PCR-based methods for Onchocerca volvulus diagnosis
| Detection method | DNA added (μL) | Reaction volume (μL) | LOD | LOD normalized (by vol. DNA) |
|---|---|---|---|---|
| qPCR-O150 | 2.0 | 10 | 10 fg/μL | 20 fg |
| qPCR-MCA | 0.5 | 10 | 100 fg/μL | 50 fg |
| O150-PCR ELISA | 5.0 | 50 | 1 pg/μL | 5 pg |
Detection of O. volvulus DNA in skin snip biopsies.
PCR and microscopy results for O. volvulus detection in skin snips are presented in Table 2. The qPCR-O150 assay detected O. volvulus DNA in significantly more skin snips (224) than qPCR-MCA (205; P = 0.003) or O150-PCR ELISA (127; P = 0.0003). Both qPCR methods detected more O. volvulus–positive skin snips than microscopy; however, the O150-PCR ELISA detected fewer positive skin snips than microscopy.
Table 2.
Performance of three PCR-based methods for Onchocerca volvulus diagnosis in skin snips
| Detection method | # Positive | % Positive (%) | P value* |
|---|---|---|---|
| qPCR-O150 | 224 | 47.5 | – |
| qPCR-MCA | 205 | 43.5 | 0.003 |
| O150-PCR ELISA | 127 | 27.0 | 0.0003 |
| Microscopy | 189 | 40.1 | 0.0003 |
McNemar’s test with Bonferroni correction for multiple comparisons.
The sensitivity and specificity of each assay were compared relative to qPCR-O150, which was positive in the greatest number of O. volvulus–positive skin snips (Table 3). The qPCR-MCA performed similarly to the comparator, with a sensitivity and specificity of 89.3% and 98.0%, respectively. The O150-PCR ELISA had a sensitivity of 56.7% and a specificity of 100%. Sequences of the five skin snips that were negative by qPCR-O150 but positive by qPCR-MCA for O. volvulus all had amplicons with ≥ 99% sequence identity (615/619 bases) to O. volvulus sequences but only 96–98% sequence identity to O. ochengi genome (102/105 bases) by BLAST analysis.
Table 3.
Performance of three diagnostic tests relative to qPCR-O150 for Onchocerca volvulus detection in skin snips
| qPCR-O150 | |||
|---|---|---|---|
| Positive | Negative | Total | |
| qPCR-MCA | |||
| qPCR-MCA positive | 200 | 5 | 205 |
| qPCR-MCA negative | 24 | 242 | 266 |
| Total | 224 | 247 | 471 |
| Sensitivity | Specificity | ||
| 89.3% | 98.0% | ||
| O150-PCR ELISA | |||
| O150-PCR ELISA positive | 127 | 0 | 127 |
| O150-PCR ELISA negative | 97 | 247 | 344 |
| Total | 224 | 247 | 471 |
| Sensitivity | Specificity | ||
| 56.7% | 100% | ||
| Microscopy | |||
| Microscopy positive | 179 | 10 | 189 |
| Microscopy negative | 45 | 237 | 282 |
| Total | 224 | 247 | 471 |
| Sensitivity | Specificity | ||
| 80% | 96% | ||
Evaluation of qPCR-MCA.
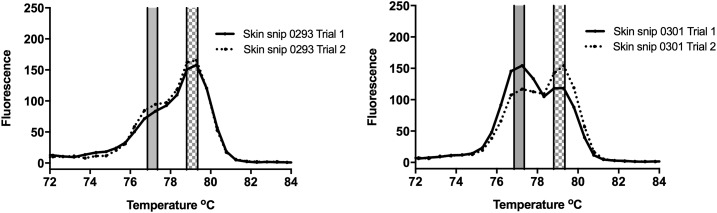
There were 266/471 skin snips (56.5%) that were positive by qPCR-MCA, indicating the presence of at least one filarial species. The Tm and dissociation curve allowed further categorization into 205 (43.5%) O. volvulus, 19 (4.0%) L. loa, 15 (3.2%) Mansonella spp., and 27 (5.7%) as non-categorized (Table 4). qPCR-MCA results were classified as non-categorized if they displayed dissociation curves that contained more than one peak, a peak with a shoulder, or a peak that was broader than expected, as shown in Figure 1, possibly indicating the presence of more than one filarial species. Of the 24 samples that were positive by qPCR-O150 but negative by qPCR-MCA for O. volvulus in Table 3, one was positive for L. loa by MCA, two were negative by MCA, and 21 were non-categorized by MCA.
Table 4.
Performance of the pan-filarial qPCR-MCA tool compared with species-specific qPCRs for Onchocerca volvulus, Loa loa, and Mansonella perstans
| qPCR-O150 | L. loa qPCR | M. perstans qPCR | ||||||
|---|---|---|---|---|---|---|---|---|
| qPCR-MCA result | Total | % | Positive | Negative | Positive | Negative | Positive | Negative |
| O. volvulus | 205 | 43.5 | 200 | 5 | 7 | 198 | 15 | 190 |
| L. loa | 19 | 4.0 | 1 | 18 | 19 | 0 | 5 | 14 |
| Mansonella spp. | 15 | 3.2 | 0 | 15 | 1 | 14 | 15 | 0 |
| Non-categorized* | 27 | 5.7 | 21 | 6 | 13 | 14 | 15 | 12 |
| Negative | 205 | 43.5 | 2 | 203 | N/A | N/A | N/A | N/A |
| Total | 471 | 100.0 | 224 | 247 | 40 | 226 | 50 | 216 |
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |||
| 89.3% | 98% | 47.5% | 100% | 30% | 100% | |||
MCA dissociation curve did not display a single peak
Figure 1.
Representative melt curve profiles from two different skin snip DNA samples that displayed non-single peak profiles indicative of mixed template DNA. Species-specific qPCR results for these samples indicated the presence of both Mansonella perstans and Onchocerca volvulus. The Tm range for O. volvulus and Mansonella spp.–positive controls is indicated with checkered gray and solid gray bars, respectively. The melt curve profile from each skin snip sample is shown in black (duplicate replicates in solid vs. dashed line) (A) melt curve profile with a main peak and a secondary smaller peak. (B) Melt curve profile with two distinct peaks.
The qPCR-MCA species categorizations were compared with species-specific qPCR results (Table 4). For O. volvulus, qPCR-MCA and qPCR-O150 results were concordant in 200/224 (89.3%) of the skin snips. Most of the samples non-categorized by MCA were positive for O. volvulus by qPCR-O150, with more than half of these also positive for L. loa and/or M. perstans. When comparing the qPCR-MCA categorizations to species-specific qPCR results for L. loa or M. pertans, qPCR-MCA demonstrated 100% specificity; however, the relative sensitivity for detecting L. loa DNA (47.5%) and M. perstans DNA (30%) in skin snips was low.
A total of 46 (9.8%) samples had DNA from multiple filarial species detected. qPCR-MCA non-categorized skin snips (N = 27) were more likely to contain DNA from multiple filarial species—20/46 mixed infections had non-categorized results and 7/215 single infections had non-categorized results (P < 0.0001, two-sided χ2). Mixed filarial DNA, however, was detected in 26 cases when the dissociation curve was a single peak.
The frequency of the different filarial species DNA present in the skin snips defined by species-specific qPCR results is presented in Table 5. Overall, 208 (44.2%) of the skin snips were negative, 224 (47.5%) had O. volvulus, 40 (8.5%) had L. loa, and 50 (10.6%) had M. perstans. Most of the individuals with skin snips that were L. loa qPCR positive were also positive for this parasite by blood smear microscopy (35/40 or 87.5%). Likewise, a majority of individuals with skin snips that were M. perstans qPCR positive were also positive by blood smear microscopy (48/50 or 96%).
Table 5.
Overall prevalence of filarial DNA in skin snips as determined by species-specific qPCRs
| Filarial species | Total # | Total (%) |
|---|---|---|
| Total positive | 263 | 55.8 |
| Onchocerca volvulus* | 224 | 47.5 |
| Loa loa | 40 | 8.5 |
| Mansonella perstans | 50 | 10.6 |
| Total single infections | 217 | 46.1 |
| O. volvulus* | 185 | 39.3 |
| L. loa | 16 | 3.4 |
| M. perstans | 16 | 3.4 |
| Total mixed infections | 46 | 9.8 |
| O. volvulus* + L. loa | 12 | 2.5 |
| O. volvulus* + M. perstans | 22 | 4.7 |
| O. volvulus* + L. loa + M. perstans | 5 | 1.1 |
| L. loa + M. perstans | 7 | 1.5 |
| Negative | 208 | 44.2 |
| Total | 471 | 100.0 |
As defined by qPCR-O150.
Comparison of O150-based PCR methods.
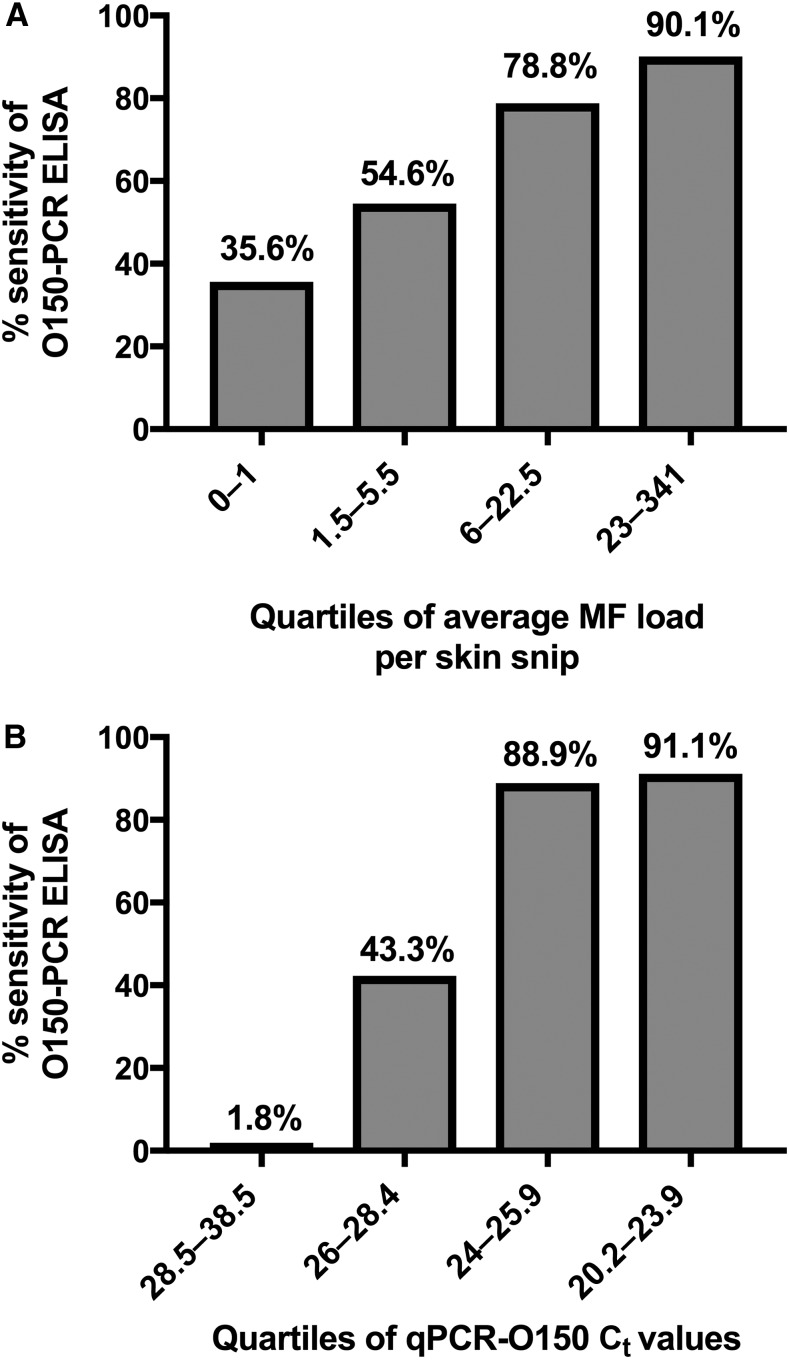
The O150-PCR ELISA was less sensitive than either of the two qPCR methods tested. Therefore, potential factors influencing reduced O150-PCR ELISA sensitivity were investigated. A total of 161/471 (34.2%) samples were initially positive by O150-PCR ELISA. However, 34/161 (21% of the initial positives) were negative on repeat testing, so the final result was negative (per protocol). Twenty-four of those 34 negatives on repeat testing (70.6%) were qPCR-O150(+). Receiver operator characteristics analysis for O150-PCR ELISA showed the optimal threshold at OD = 0.055. Compared with the standard threshold at 0.1, the use of this optimal threshold would increase the sensitivity from 56.7% to 83%, but specificity would be reduced from 100% to 90.7% (Supplemental Figure 1). Comparison of the sensitivity of the qPCR-O150 and O150-PCR ELISA methods showed decreased O150-PCR ELISA sensitivity in skin snips with lower MF loads (Figure 2A). Because the qPCR-O150 Ct values are semiquantitative and highly correlated with MF load (Supplemental Figure 2; Spearman’s rank ρ = −0.74, P < 0.0001), the O150-PCR ELISA sensitivity was compared with quartiles of Ct values (Figure 2B); this showed a decline in O150-PCR ELISA sensitivity with higher Ct values.
Figure 2.
O150-PCR ELISA demonstrates reduced sensitivity in low-intensity Onchocerca volvulus infections. (A) Decreased O150-PCR sensitivity relative to qPCR-O150 is observed in skin biopsy samples with lower MF loads. Bar graph depicting the percent sensitivity of O150-PCR ELISA relative to qPCR-O150 on the y axis and MF load group on the x axis; samples were divided into quartiles based on MF load. (B) Decreased O150-PCR sensitivity relative to qPCR-O150 is observed in skin biopsy samples with lower concentration of O. volvulus DNA. Bar graph depicting the percent sensitivity of O150-PCR ELISA relative to qPCR-O150 on the y axis and on the x axis; samples were divided into quartiles based on qPCR-O150 cycle threshold (Ct) values.
DISCUSSION
We directly compared the performance of three PCR-based methods for O. volvulus detection—two real-time methods (qPCR-O15011 and qPCR-MCA12) and one conventional PCR-based method (O150-PCR ELISA15)—using skin snips from an onchocerciasis endemic region co-endemic for L. loa and M. perstans. We also evaluated the qPCR-MCA tool in a setting co-endemic for several filariae. The results show that qPCR-O150 detected O. volvulus DNA in significantly more skin snips than either of the other PCR methods tested. Notably, the O150-PCR ELISA currently in use by onchocerciasis programs detected only 56.7% of the skin snips in which O. volvulus DNA was detected by qPCR-O150. The reduced sensitivity of both tests compared with qPCR-O150 was because of differences in limits of detection among the assays. As a result, the O150-PCR ELISA was less sensitive in samples with lower counts of MF. However, specific issues with the qPCR-MCA leading to reduced sensitivity also included the misclassifications of filarial species in the cases of mixed infections.
There were 10 individuals who were microscopy positive but negative by qPCR-O150 and O150-PCR ELISA. The MF loads in these 10 samples were low (median 1.5 MF/2 snips; range 1.0–22), and we cannot exclude the possibility that PCR amplifications were negative because of our use of skin snips that had been incubated overnight to allow the emergence of O. volvulus MF for microscopy. However, two of these 10 individuals were skin snip positive by qPCR for M. perstans, one of which was also positive for L. loa. It may be that there was misclassification of these skin snips by microscopy. This observation highlights the importance of PCR-based methods in assessments for the elimination of transmission of onchocerciasis, as microscopy misdiagnosis in field settings can occur, despite the expertise of the diagnostician.
The qPCR-MCA tool detected skin snips that displayed a Tm consistent with either O. volvulus, L. loa, or Mansonella spp., accurately detecting 89.3% of O. volvulus–positive skin snips. However, this tool was unable to categorize 27 samples because of their irregular melt curve profiles. Compared with species-specific qPCRs, the qPCR-MCA had lower sensitivity for detecting L. loa and M. perstans in skin snips than for detecting O. volvulus. It is plausible that in an onchocerciasis hyperendemic setting such as the one tested here, the relative abundance of DNA from M. perstans and L. loa, which are not typically found in the skin, would be lower than that from O. volvulus DNA and, thus, might not always be detected by the MCA tool. Another point to consider is if O. volvulus MF loads are suppressed by multiple rounds of IVM, the ability of the qPCR-MCA to detect O. volvulus might decrease even further in the context of coinfections with other filariae.
The use of species-specific qPCRs was imperative to evaluate the performance of qPCR-MCA because initial assay optimization demonstrated that Tm profiles could not discriminate between closely related species, limiting its discrimination to the genus level.12 qPCR-MCA alone cannot discriminate M. streptocerca, which typically circulates within the skin,20 from M. perstans. Furthermore, another potential limitation of qPCR-MCA might be its inability to distinguish between O. volvulus and other closely related Onchocerca species such as O. ochengi and Onchocerca sp. “Siisa,” which can be found in African blackflies,35–37 but we did not directly test this hypothesis. Interestingly, five skin snips that were categorized as qPCR-MCA(+) for O. volvulus were qPCR-O150(−), with all five demonstrating ITS1 sequence similarity to O. volvulus. These five qPCR-O150 false negatives might be because of suboptimal primer/probe binding. Although the qPCR-MCA identified five positive results that were not detected by the qPCR-O150, this assay nevertheless was less sensitive than qPCR-O150. A more extensive validation of qPCR-MCA for detection of filarial species within other sample types derived from blood is necessary to assess the utility of this tool within other contexts.
Despite the limitations of qPCR-MCA, its use in DRC allowed for the unexpected identification of L. loa and M. perstans DNA in skin snips samples. The presence of DNA from these two species might be more common in skin snips than previously recognized. Whether MF are present and thus could be sources of false positives by microscopy remains unclear. However, MF from both L. loa and M. perstans are not usually found circulating in the skin.18,21 Therefore, one plausible explanation for the presence of DNA of these species in skin snips could be the unintentional capture of blood during the biopsy; however, this seems unlikely given the fact that there were blood smear negative individuals that were skin snip PCR positive. Alternatively, the PCR assays might be detecting cells or circulating DNA from either M. perstans or L. loa because adults migrate through subcutaneous tissue.21
Currently, the O150-PCR ELISA test is the standard tool used for detection of O. volvulus in Simulium blackflies, and thus far, it has been used for confirmation of patent infection in less than 10 children with positive OV-16 antibody results (T. R. Unnasch, personal communication, Nov. 2017). As mentioned previously, the sensitivity of this method as tested was low and decreased as O. volvulus MF and DNA decreased, corroborating the observations of Thiele et al.12 As PCR will be used to exclude patent infection in children positive for OV-16 serologic test, sensitivity is very important and efforts should be made to maximize the sensitivity of the PCR used by programs. We showed that the qPCR-O150 assay would be the most appropriate test for this based on its LOD and high sensitivity.
Although qPCR technology is currently available in many onchocerciasis endemic countries, some onchocerciasis programs might not have access to qPCR but would have access to the equipment used for O150-PCR ELISA testing of blackflies. A detailed comparison of the accuracy and sustainability of qPCR versus O150-PCR ELISA would be necessary to decide which PCR method would be the best to properly inform the onchocerciasis elimination programs. In the event that qPCR-O150 could not be implemented by a program, consideration will need to be given for maximizing the sensitivity of the O150-PCR ELISA to improve its detection performance, especially when MF loads are low. Adjusting the O150-PCR ELISA OD cutoff could improve sensitivity, but at a cost to specificity. Assay sensitivity is also important for blackfly surveillance, but for blackfly testing, lower sensitivity can be circumvented to an extent by increasing the numbers of blackflies tested. This cannot be performed when testing OV-16–positive children. However, increased sensitivity with loss to specificity could be important in posttreatment surveillance of blackflies to maximize early recognition of recrudescence. Testing an additional locus such as cytochrome oxidase I may also help to overcome the potential loss to specificity.38 An oligonucleotide-based magnetic bead capture method for purification of O. volvulus DNA from blackfly vectors method was previously shown to increase detection sensitivity.15 Extracting DNA from skin snips using this method might further increase the sensitivity of the O150-PCR ELISA in skin snips; however, additional studies are needed to evaluate this.
Even if the O150-PCR ELISA protocol for PCR-testing of skin snips could be adapted to augment sensitivity, the qPCR-O150 assay still affords several advantages over the O150-PCR ELISA method. qPCR techniques do not require any post-amplification processing, thus reducing the possibility for cross-contamination and need for extensive quality control measures. The assay time for the qPCR-O150 assay is less than 2 hours, much shorter than for O150-PCR ELISA, which can take up to 2 days. Moreover, the qPCR-O150 assay requires less equipment compared with the O150-PCR ELISA, which uses a regular PCR thermocycler, ELISA plate reader, incubators, and possibly an ELISA plate washer. The qPCR is also simpler to perform because there are fewer reagents and protocol steps involved. This may reduce interoperator assay variability and would make it more amenable to standardization across diverse laboratory environments. These same advantages would also apply to qPCR testing of blackflies; thus, comparing the sensitivity and specificity of qPCR-O150 versus O150-PCR ELISA in blackflies could be important, particularly for posttreatment surveillance, when early detection of recrudescence will be important. It would also be worthwhile to conduct a comprehensive comparison between the two assays, both for testing of blackflies and skin snips, to determine if it would be beneficial to programmatically implement the qPCR-O150 assay both for the evaluation of blackflies and skin snips.
In summary, we found the qPCR platform to be more sensitive than O150-PCR ELISA. Increasing the sensitivity of the currently available PCR technique or using real-time PCR technology, if logistically and technically feasible, should be considered. As increasing the sensitivity for O. volvulus detection in blackflies could enhance earlier detection of recrudescence, an evaluation of these PCR methods in blackflies could be an important next step.
Supplementary Material
Supplemental Figures And Table.
Acknowledgments:
We thank the people of the communities of Banalia, Tshopo Province; field teams for their generous participation and support for this project; and Dr. Mark Eberhard for his helpful insight and for providing O. volvulus microfilariae samples.
Note: Supplemental figures and table appear at www.ajtmh.org.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
REFERENCES
- 1.WHO , 2016. Progress report on the elimination of human onchocerciasis, 2015–2016. Wkly Epidemiol Rec 91: 505–514. [PubMed] [Google Scholar]
- 2.WHO , 2011. African programme for onchocerciasis control: meeting of National Task Forces, September 2011. Wkly Epidemiol Rec 86: 541–549. [PubMed] [Google Scholar]
- 3.Cupp EW, Sauerbrey M, Richards F, 2011. Elimination of human onchocerciasis: history of progress and current feasibility using ivermectin (Mectizan((R))) monotherapy. Acta Trop 120 (Suppl 1): S100–S108. [DOI] [PubMed] [Google Scholar]
- 4.Roberts JM, Neumann E, Gockel CW, Highton RB, 1967. Onchocerciasis in Kenya 9, 11 and 18 years after elimination of the vector. Bull World Health Organ 37: 195–212. [PMC free article] [PubMed] [Google Scholar]
- 5.Turner HC, Walker M, Churcher TS, Basanez MG, 2014. Modelling the impact of ivermectin on River Blindness and its burden of morbidity and mortality in African Savannah: EpiOncho projections. Parasit Vectors 7: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner HC, Walker M, Churcher TS, Osei-Atweneboana MY, Biritwum NK, Hopkins A, Prichard RK, Basanez MG, 2014. Reaching the london declaration on neglected tropical diseases goals for onchocerciasis: an economic evaluation of increasing the frequency of ivermectin treatment in Africa. Clin Infect Dis 59: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unnasch TR, Meredith SE, 1996. The use of degenerate primers in conjunction with strain and species oligonucleotides to classify Onchocerca volvulus. Methods Mol Biol 50: 293–303. [DOI] [PubMed] [Google Scholar]
- 8.Fink DL, Fahle GA, Fischer S, Fedorko DF, Nutman TB, 2011. Toward molecular parasitologic diagnosis: enhanced diagnostic sensitivity for filarial infections in mobile populations. J Clin Microbiol 49: 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerman PA, Guderian RH, Aruajo E, Elson L, Phadke P, Kubofcik J, Nutman TB, 1994. Polymerase chain reaction-based diagnosis of Onchocerca volvulus infection: improved detection of patients with onchocerciasis. J Infect Dis 169: 686–689. [DOI] [PubMed] [Google Scholar]
- 10.Toe L, Boatin BA, Adjami A, Back C, Merriweather A, Unnasch TR, 1998. Detection of Onchocerca volvulus infection by O-150 polymerase chain reaction analysis of skin scratches. J Infect Dis 178: 282–285. [DOI] [PubMed] [Google Scholar]
- 11.Lloyd MM, Gilbert R, Taha NT, Weil GJ, Meite A, Kouakou IM, Fischer PU, 2015. Conventional parasitology and DNA-based diagnostic methods for onchocerciasis elimination programmes. Acta Trop 146: 114–118. [DOI] [PubMed] [Google Scholar]
- 12.Thiele EA, Cama VA, Lakwo T, Mekasha S, Abanyie F, Sleshi M, Kebede A, Cantey PT, 2016. Detection of Onchocerca volvulus in skin snips by microscopy and real-time polymerase chain reaction: implications for monitoring and evaluation activities. Am J Trop Med Hyg 94: 906–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipner EM, Dembele N, Souleymane S, Alley WS, Prevots DR, Toe L, Boatin B, Weil GJ, Nutman TB, 2006. Field applicability of a rapid-format anti-Ov-16 antibody test for the assessment of onchocerciasis control measures in regions of endemicity. J Infect Dis 194: 216–221. [DOI] [PubMed] [Google Scholar]
- 14.Lobos E, Weiss N, Karam M, Taylor HR, Ottesen EA, Nutman TB, 1991. An immunogenic Onchocerca volvulus antigen: a specific and early marker of infection. Science 251: 1603–1605. [DOI] [PubMed] [Google Scholar]
- 15.Gopal H, Hassan HK, Rodriguez-Perez MA, Toe LD, Lustigman S, Unnasch TR, 2012. Oligonucleotide based magnetic bead capture of Onchocerca volvulus DNA for PCR pool screening of vector black flies. PLoS Negl Trop Dis 6: e1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downes BL, Jacobsen KH, 2010. A systematic review of the epidemiology of mansonelliasis. Afr J Infect Dis 4: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawking F, 1977. The distribution of human filariasis throughout the world. Part III. Africa. Trop Dis Bull 74: 649–679. [PubMed] [Google Scholar]
- 18.Simonsen PE, Onapa AW, Asio SM, 2011. Mansonella perstans filariasis in Africa. Acta Trop 120 (Suppl 1): S109–S120. [DOI] [PubMed] [Google Scholar]
- 19.Zoure HG, Wanji S, Noma M, Amazigo UV, Diggle PJ, Tekle AH, Remme JH, 2011. The geographic distribution of Loa loa in Africa: results of large-scale implementation of the Rapid Assessment Procedure for Loiasis (RAPLOA). PLoS Negl Trop Dis 5: e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer P, Buttner DW, Bamuhiiga J, Williams SA, 1998. Detection of the filarial parasite Mansonella streptocerca in skin biopsies by a nested polymerase chain reaction-based assay. Am J Trop Med Hyg 58: 816–820. [DOI] [PubMed] [Google Scholar]
- 21.Padgett JJ, Jacobsen KH, 2008. Loiasis: African eye worm. Trans R Soc Trop Med Hyg 102: 983–989. [DOI] [PubMed] [Google Scholar]
- 22.Wilson NO, et al. 2016. Evaluation of lymphatic filariasis and onchocerciasis in three senegalese districts treated for onchocerciasis with ivermectin. PLoS Negl Trop Dis 10: e0005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steel C, Golden A, Stevens E, Yokobe L, Domingo GJ, de los Santos T, Nutman TB, 2015. Rapid point-of-contact tool for mapping and integrated surveillance of Wuchereria bancrofti and Onchocerca volvulus infection. Clin Vaccine Immunol 22: 896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moya L, et al. 2016. Evidence for suppression of onchocerciasis transmission in Bioko Island, Equatorial Guinea. PLoS Negl Trop Dis 10: e0004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katabarwa MN, et al. 2012. Transmission of onchocerciasis in wadelai focus of northwestern Uganda has been interrupted and the disease eliminated. J Parasitol Res 2012: 748540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golden A, et al. 2013. Extended result reading window in lateral flow tests detecting exposure to Onchocerca volvulus: a new technology to improve epidemiological surveillance tools. PLoS One 8: e69231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans DS, et al. 2014. Status of onchocerciasis transmission after more than a decade of mass drug administration for onchocerciasis and lymphatic filariasis elimination in central Nigeria: challenges in coordinating the stop MDA decision. PLoS Negl Trop Dis 8: e3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.WHO , 2016. Guidelines for Stopping Mass Drug Administration and Verifying Elimination of Human Onchocerciasis: Criteria and Procedures. Geneva, Switzerland: World Health Organization. [PubMed] [Google Scholar]
- 29.Taylor HR, Munoz B, Keyvan-Larijani E, Greene BM, 1989. Reliability of detection of microfilariae in skin snips in the diagnosis of onchocerciasis. Am J Trop Med Hyg 41: 467–471. [DOI] [PubMed] [Google Scholar]
- 30.Alhassan A, Makepeace BL, LaCourse EJ, Osei-Atweneboana MY, Carlow CK, 2014. A simple isothermal DNA amplification method to screen black flies for Onchocerca volvulus infection. PLoS One 9: e108927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mourembou G, et al. 2015. Mansonella, including a potential new species, as common parasites in children in Gabon. PLoS Negl Trop Dis 9: e0004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fink DL, Kamgno J, Nutman TB, 2011. Rapid molecular assays for specific detection and quantitation of Loa loa microfilaremia. PLoS Negl Trop Dis 5: e1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe KL, et al. 2016. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res 44: D774–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M, 2017. WormBase ParaSite—a comprehensive resource for helminth genomics. Mol Biochem Parasitol 215: 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenbarth A, Achukwi MD, Renz A, 2016. Ongoing transmission of Onchocerca volvulus after 25 years of annual ivermectin mass treatments in the Vina du Nord River Valley, in North Cameroon. PLoS Negl Trop Dis 10: e0004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenbarth A, Ekale D, Hildebrandt J, Achukwi MD, Streit A, Renz A, 2013. Molecular evidence of ‘Siisa form’, a new genotype related to Onchocerca ochengi in cattle from north Cameroon. Acta Trop 127: 261–265. [DOI] [PubMed] [Google Scholar]
- 37.Krueger A, Fischer P, Morales-Hojas R, 2007. Molecular phylogeny of the filaria genus Onchocerca with special emphasis on Afrotropical human and bovine parasites. Acta Trop 101: 1–14. [DOI] [PubMed] [Google Scholar]
- 38.Lagatie O, Merino M, Batsa Debrah L, Debrah AY, Stuyver LJ, 2016. An isothermal DNA amplification method for detection of Onchocerca volvulus infection in skin biopsies. Parasit Vectors 9: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures And Table.