To the Editor
We read with great interest the paper by Williams et al.1, who reported evidence for neutral evolution in tumors by analyzing data from The Cancer Genome Atlas (TCGA). They supported this conclusion by showing high R2 values for fits to a neutral evolutionary model predicting M ∝ 1/f, where M is the number of somatic mutations with allele frequency ≥f. However, we believe a conclusion of neutrality must be treated with caution, as high R2 values are R2 values are consistent with many evolutionary models.
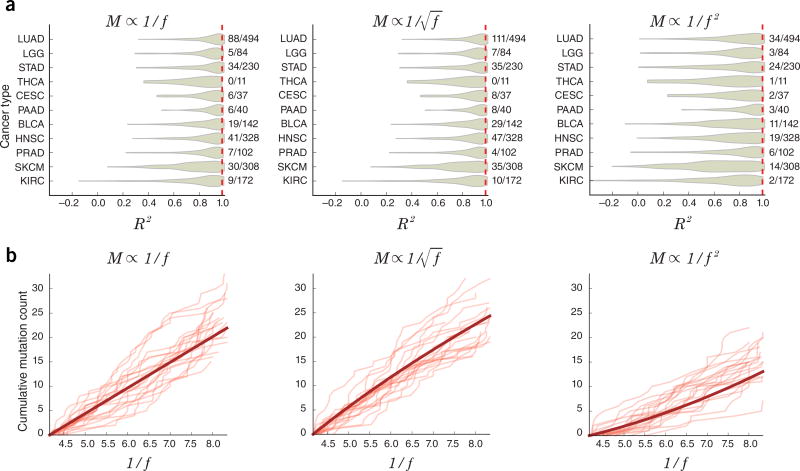
For example, we analyzed phenomenological models similar to that of ref. 1 but with parameter k, such that M ∝ 1/fk. Here k = 1 corresponds to the neutral model, k > 1 corresponds to diversifying selection (excess of rare mutations), and k < 1 corresponds to purifying selection (excess of high-frequency mutations). We reanalyzed the TCGA data to determine whether values other than k = 1 fit the data better. To reduce pipeline uncertainties, we used only tumors for which calls were made by Mutect2, and similarly to ref. 1 we only used mutations with read count ≥10 and alternative read count ≥3 and only analyzed tumors with ≥12 genes within the fitting range (0.12 < f < 0.24). We then reproduced Figure 3 from ref. 1 by fitting mutation count to 1/f (Fig. 1a). Our R2 values were high although not identical to those in ref. 1, likely owing to differences in tumor sets and perhaps as a result of insufficient information about the exact methodological details in ref. 1. To determine whether the fit was due to neutral evolution, we repeated the same analysis by fitting to the functions 1/f2 (diversifying selection) and (purifying selection) (Fig. 1a). In all cases, we were able to closely fit the TCGA data (mean R2 values were 0.84, 0.88, and 0.73 for k = 1, 0.5, and 2, respectively), but the purifying selection model in fact fit the data slightly better. Although our analysis does not clearly show a lack of neutrality, it does indicate that R2 is not a good measure for distinguishing neutral evolution.
Figure 1.
Comparison of evolutionary models for TCGA and simulated data. (a) Distribution of R2 values for fits of TCGA allele frequency distribution data to three different models. The numbers on the right side of each plot show the fraction of total tumors in each cancer type with R2 >0.98 (right side of red dashed line). (b) Simulated allele frequency distributions for different generating processes. Thin curves are individual examples of simulated M curves from the neutral (left), purifying selection (middle), and diversifying selection (right) processes, while thick curves are the ideal when no measurement noise exists. See the Supplementary Note and Supplementary Code for details.
Another consideration is that noise inherent in M(f) curves limits conclusions about neutrality. Assuming that the true allele frequency of a mutation is ftrue, the observed allele frequency fobs will be a sample from a binomial distribution with mean μ = ftrue and s.d. , given read depth n (on average, n = 102 in the TCGA samples). In the fitting range 0.12 < ftrue < 0.24, σf can take on values as large as 0.04, that is, ~30% of the fitting range. We analyzed the effect of this noise directly by simulating observed M(f) curves according to underlying neutral (k = 1), purifying (k = 0.5), and diversifying (k = 2) selection models. M(f) curves were generated by sampling values of ftrue from the underlying model and then for each value reporting an fobs generated from the binomial distribution with mean ftrue and read depth n, where n was drawn from a lognormal fit to the pooled TCGA read depth distribution. Figure 1b shows randomly generated M curves obtained by resimulating this process, suggesting that measurement uncertainty can substantially influence the shape of the observed curve and obscure the underlying evolutionary process. Moreover, we repeatedly simulated M(f) curves for each generating process (k = 0.5, 1, and 2) and tested whether the true generating process could be identified. Mean and s.d. of R2 values are shown in Table 1. R2 values to the true model (diagonal elements) were only marginally better than those to the incorrect models and in all cases these differences were less than the s.d. across replicates, suggesting that R2 is not a sensitive measure for resolving the evolutionary process.
Table 1.
Fits of simulated data from neutrality (1/f), purifying selection ( ), and diversifying selection (1/f2) to the expected M curves for all three processes
| 1/f |
|
1/f2 | |||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Generating process | Mean | s.d. | Mean | s.d. | Mean | s.d. | |
|
|
|
|
|||||
| M ∝ 1/f | 0.95 | 0.04 | 0.95 | 0.04 | 0.93 | 0.06 | |
|
|
0.94 | 0.05 | 0.95 | 0.04 | 0.90 | 0.09 | |
| M ∝ 1/f2 | 0.94 | 0.05 | 0.93 | 0.05 | 0.94 | 0.05 | |
Mean R2 values are shown along with their s.d.
The relationship M ∝ 1/f can be derived from assumptions of a homogeneously replicating population with constant mutation rate per cell division (M ∝ N) and neutral evolution: that is, a mutation that arises when the tumor is of size N will obey f ∝ N−1 at the time of measurement. Our model can be interpreted as maintaining the first assumption while replacing the second with f ∝ N−1/k to take selection into account. The described cases for k give the correct sign of the second derivative of M with respect to 1/f for purifying and diversifying selection. Still, the model is a simplification and treats selection as monotonic with N. In reality, selective pressures are likely to be spatially diverse and punctuated, although investigation of these aspects will require more extensive parameterization.
Williams et al.1 have provided a valuable conceptualization of population dynamics in tumors and have shown that neutrality is possible. However, models with selection can provide similarly good fits to the TCGA data, and TCGA data still yield substantial uncertainties about the true frequency distribution. More refined evolutionary models and further increases in sequencing depth, along with careful statistical modeling of sequencing data3, will be important to resolve what balance of selection and neutrality exists in cancer. Interestingly, even aside from the considerations we have raised, Williams et al.1 already found there to be many cases that did not fit the neutral model, and in some cases the selective processes may be resolvable. Promising areas for future investigation may include location-dependent selection, deviations from M ∝ N due to cell cycle–independent mutations, and tissue-specific selection such as differences in solid and liquid tumors.
Supplementary Material
Acknowledgments
We would like to thank H.S. Kim for helpful discussions and J. Cha for graphics design. J.H.C. was supported by the National Cancer Institute of the NIH under award R21CA191848 and supplement R21CA191848-01A1S1. Research was also partially supported by the National Cancer Institute under award P30CA034196.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.N. and J.H.C. jointly designed the study and wrote the manuscript. J.N. performed all computational data analyses.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.William MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Nat. Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cibulskis K, et al. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerstung M, et al. Nat. Commun. 2012;3:811. doi: 10.1038/ncomms1814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.