Abstract
Agrobacterium-mediated genetic transformation not only represents a technology of choice to genetically manipulate plants, but it also serves as a model system to study mechanisms employed by invading pathogens to counter the myriad defenses mounted against them by the host cell. Here, we uncover a new layer of plant defenses that is targeted by A. tumefaciens to facilitate infection. We show that the Agrobacterium F-box effector VirF, which is exported into the host cell, recognizes an Arabidopsis transcription factor VFP4 and targets it for proteasomal degradation. We hypothesize that VFP4 resists Agrobacterium infection and that the bacterium utilizes its VirF effector to degrade VFP4 and thereby mitigate the VFP4-based defense. Indeed, loss-of-function mutations in VFP4 resulted in differential expression of numerous biotic stress–response genes, suggesting that one of the functions of VFP4 is to control a spectrum of plant defenses, including those against Agrobacterium tumefaciens. We identified one such gene, ATL31, known to mediate resistance to bacterial pathogens. ATL31 was transcriptionally repressed in VFP4 loss-of-function plants and activated in VFP4 gain-of-function plants. Gain-of-function lines of VFP4 and ATL31 exhibited recalcitrance to Agrobacterium tumorigenicity, suggesting that A. tumefaciens may utilize the host ubiquitin/proteasome system to destabilize transcriptional regulators of the host disease response machinery.
Plant and animal pathogens have evolved a variety of elegant strategies to block, subvert, or redirect diverse host pathways for their own benefit. For example, host membrane trafficking, mitogen-activated protein (MAP) kinase signaling, control of cytoskeleton dynamics, and the ubiquitin-proteasome system (UPS) can each be manipulated by pathogens to facilitate infection (Alto and Orth 2012; Banfield 2015; Marino et al. 2012; Salomon and Orth 2013; Spallek et al. 2009; Trujillo and Shirasu 2010). The subversion of the UPS is of particular interest because of its potential effects on the host innate immune system, which has a key role in plant defense against pathogens (Banfield 2015; Marino et al. 2012; Spallek et al. 2009; Trujillo and Shirasu 2010). Agrobacterium tumefaciens genetically transforms its hosts in nature, eliciting neoplastic growths (Stachel and Zambryski 1989), and it was the first plant microbial pathogen shown to encode a UPS component, the F-box protein VirF (Schrammeijer et al. 2001).
A. tumefaciens infects plants by exporting into its host cells a single-stranded DNA molecule (T-DNA) with a covalently attached virulence protein effector VirD2 and, separately, additional virulence effectors VirE2, VirE3, VirD5, and VirF (Vergunst et al. 2000). VirE2 is presumed to package the T-DNA into a nucleoprotein complex (T-complex) in the host cell cytoplasm. The T-complex then associates with the host VirE2-binding protein VIP1 (Tzfira et al. 2001) and is imported into the nucleus, in which it is uncoated and the T-DNA is integrated into the plant chromatin (Citovsky et al. 2007; Tzfira et al. 2000; Zupan and Zambryski 1997; Zupan et al. 2000). This uncoating is thought to occur via the SCFVirF pathway, in which VirF binds VIP1 and targets it and the associated VirE2 for degradation by the host UPS (Tzfira et al. 2004; Zaltsman et al. 2013). Interestingly, VirF is not essential for Agrobacterium infection of some plant species (Hooykaas et al. 1984); indeed, in Arabidopsis, Agrobacterium infection induces expression of a host F-box protein, VBF, that can substitute for the VirF function (Zaltsman et al. 2010).
VirF is the only F-box protein that A. tumefaciens exports into host cells. Bacterial effectors are, in general, multifunctional proteins (Backert and Meyer 2006; Dean 2011; Galán 2009; Kenny et al. 2002). This raises the possibility that VirF may fulfill several functions during infection by interacting with multiple targets in the host cells. Here, we report one such target, VFP4, a transcription factor that both VirF and VBF target for degradation by the UPS. We further identified ATL31, which encodes a RING-type ubiquitin ligase that enhances bacterial resistance and controls carbon/nitrogen responses as one of the genes upregulated by VFP4, and showed that overexpression of either VFP4 or ATL31 renders plants less susceptible to Agrobacterium infection. These findings suggest that A. tumefaciens may utilize the host cell UPS to destabilize transcriptional regulators of components of the host disease response machinery that limit the bacterial infection.
RESULTS
VirF and its plant functional homolog VBF interact with the Arabidopsis GLABROUS1 enhancer/binding protein (GeBP)-like transcription factor VFP4
Functions of VirF in Agrobacterium infection can be revealed by identifying its potential cellular substrates. To this end, we used the yeast two-hybrid system to screen for Arabidopsis proteins that interacted with mutVirFdel1 (García-Cano et al. 2015), a mutated form of VirF that neither self-interacts (due to a 15–amino acid residue N-terminal deletion) nor interacts with the Skp1/ASK1 components of plant SCF complexes (due to two point mutations in the F-box domain) (García-Cano et al. 2015; Magori and Citovsky 2011). A cDNA library from Arabidopsis in pGAD424 (Ballas and Citovsky 1997) was probed with LexA-mutVirFdel1 as bait, as described previously (Ballas and Citovsky 1997; García-Cano et al. 2015; Hollenberg et al. 1995; Tzfira et al. 2001). Screening of approximately 3.97 × 106 transformants resulted in identification of three independent cDNA clones encoding VirF-interacting proteins (VFPs) (García-Cano et al. 2015). We focus here on one of these interacting proteins, designated VFP4, which was identified in two independent experiments.
We used bimolecular fluorescence complementation (BiFC) to confirm the interaction of full-length VirF and VFP4 and the subcellular localization of these interacting proteins in plant cells. VirF and VFP4 were tagged with carboxyl- and amino-terminal yellow fluorescent protein (cYFP and nYFP, respectively) fragments of YFP and were then transiently coexpressed in N. benthamiana leaves. Figure 1 shows that cYFP-VirF and nYFP-VFP4 interacted with each other in planta to reconstitute YFP fluorescence. As negative controls, we used two unrelated Arabidopsis control proteins Cullin 1 (CUL1) and FLD, with CUL1 representing a part of the SCF complex, similarly to VirF, and FLD representing a nuclear protein, similarly to VirF and VFP4. No fluorescent signal was produced when we coexpressed nYFP-VFP4 and cYFP-CUL1 or cYFP-VirF and nYFP-FLD (Fig. 1). The cYFP-VirF:nYFP-VFP4 complexes accumulated predominantly in the cell nucleus (Fig. 1). In contrast, YFP-tagged VFP4 (YFP-VFP4) itself, when transiently expressed in N. benthamiana leaves, was detected both in the cell nucleus and in the cytoplasm (Fig. 2A). We also examined whether VFP4 expression was induced by Agrobacterium infection. Real-time reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) analysis detected no statistically significant changes in VFP4 transcript levels in the wild-type Col-0 plants infected by A. tumefaciens as compared with mock-inoculated plants (Fig. 2B).
Fig. 1.
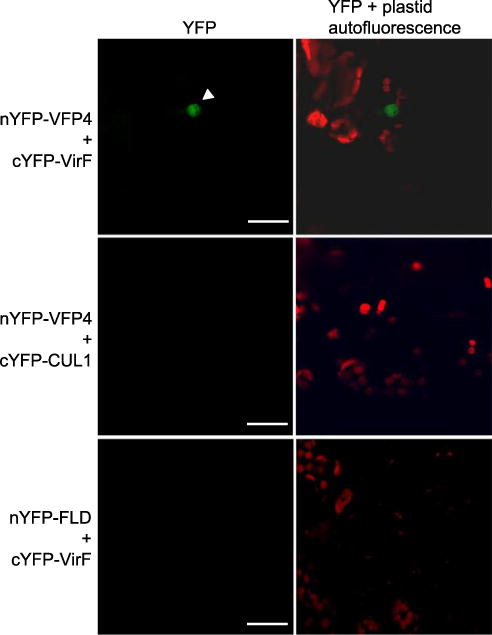
VirF interacts with VFP4 in plant cells. The interactions were visualized by BiFC. Location of the cell nucleus is indicated by a white arrowhead. All images are projections of single confocal sections, and they are representative images of three independent experiments. Scale bars = 20 μm.
Fig. 2.
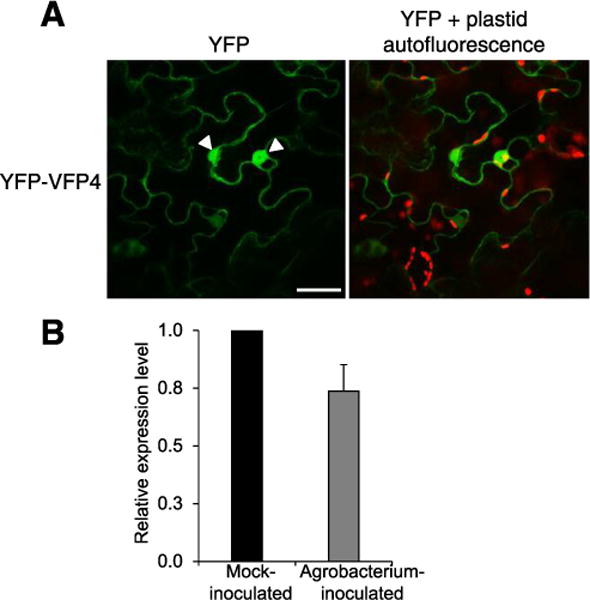
VFP4 localizes to the nucleus and cytoplasm. A, Subcellular localization of VFP4. Location of the cell nucleus is indicated by a white arrowhead. All images are projections of single confocal sections and are representative images of three independent experiments. Scale bar = 20 μm. B, Real-time quantitative polymerase chain reaction analysis of VFP4 gene expression following inoculation of wild-type plants by agrobacterium. The levels of expression were normalized to those of ACT7 and 18S RNA. The expression level of VFP4 in the mock-inoculated plants is set to 1.0, and error bars represent standard error of the mean of independent biological replicates, n = 3.
The VFP4 gene is located on chromosome 5 in the Arabidopsis genome (At5g28040). Sequence analysis of the full-length VFP4 cDNA predicted a single open reading frame (ORF) that would encode a protein of 427 amino acid residues with a molecular mass of 46.9 kDa (Fig. 3A). VFP4 is annotated as a plant-specific member of the GeBP/GeBP-like (GPL) transcription factor family in the database of Arabidopsis transcription factors. The 21 members of this family (Fig. 3B) share a central DNA-binding domain, which, in VFP4, is located between amino acid residues 122 and 223 (Chevalier et al. 2008; Curaba et al. 2003), which overlaps the DUF573 domain of unknown function (Fig. 3A). GeBP/GPL proteins are a class of leucine-zipper transcription factors that have been implicated in regulation of cell expansion, cytokinin response, and stress and defense responses (Chevalier et al. 2008; Perazza et al. 2011). For example, overexpressing Arabidopsis GPL2 enhances plant resistance to bacterial pathogens (Perazza et al. 2011). As a potential transcription factor and consistent with its ability to enter the nucleus (Fig. 2A), VFP4 is predicted to contain a monopartite nuclear localization signal (NLS) between amino acid residues 182 and 197 (Fig. 3A).
Fig. 3.
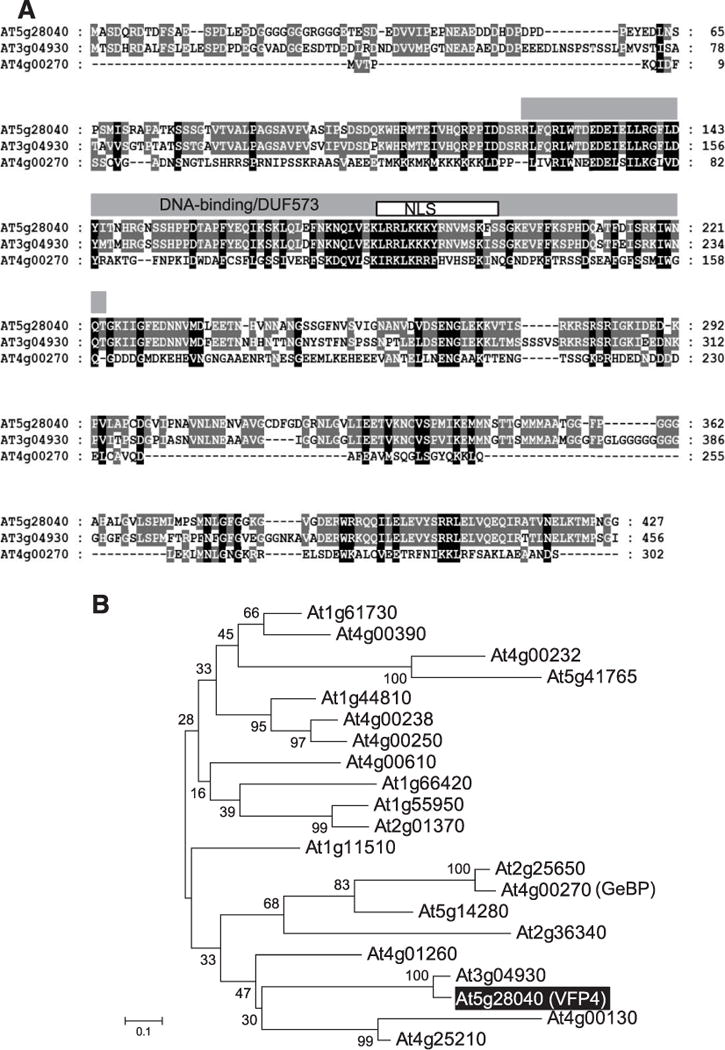
VFP4 is a member of the GeBP/GPL family of Arabidopsis transcription factors. A, Sequence alignment of VFP4 and its selected Arabidopsis homologs. The amino acid sequence of VFP4 (At5g28040) was aligned with those of proteins encoded by At3g04930 and At4g00270 (GeBP) using ClustalX (ver. 2.1). The DNA-binding domain and the overlapping DUF573 domain are delineated by a gray box. A white box delineates the putative monopartite nuclear localization signal (NLS) predicted by cNLS Mapper. Identical residues in the aligned sequences are highlighted in white letters on black background and similar residues are shaded in gray. B, Phylogenetic tree of the 21 members of the GeBP/GPL family of Arabidopsis transcription factors. VFP4 (At5g28040) is labeled with white letters in a shaded box. GeBP is indicated in parenthesis next to its locus name. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei 1987). The optimal tree with the sum of branch length of 7.00512281 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved 22 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 45 positions in the final dataset. Evolutionary analyses were conducted using the Molecular Evolutionary Genetics Analysis tool (MEGA, version 6.0.5 for Mac OS) (Tamura et al. 2013), which also generated this description of the analysis. Scale bar = 0.1 amino acid substitutions per site.
VirF and VBF destabilize VFP4 via the plant UPS
As an F-box protein, VirF is expected to promote proteasomal degradation of proteins that it specifically recognizes. For example, the interaction of VirF with host VIP1 protein destabilizes VIP1 (Tzfira et al. 2004; Zaltsman et al. 2013). We therefore used a cell-free protein degradation assay to examine the stability of VFP4 in the presence or absence of VirF (García-Cano et al. 2014). Cell extracts were prepared from N. benthamiana plants transiently expressing cyan fluorescent protein (CFP)-tagged VFP4 alone or in the presence of Myc-tagged VirF, and the kinetics of changes in the amounts of CFP-VFP4 were analyzed and quantified by Western blotting. Figure 4 shows that VFP4, when expressed alone, remained stable. In contrast, the levels of VFP4 sharply declined to 20% of the initial level when it was coexpressed with VirF (Fig. 4A and B). Consistent with the known F-box protein activity of VirF (Tzfira et al. 2004), this VirF-mediated destabilization of VFP4 was substantially reduced in the presence of the proteasomal inhibitor MG132 (Fig. 4C and D), suggesting the involvement of the UPS.
Fig. 4.
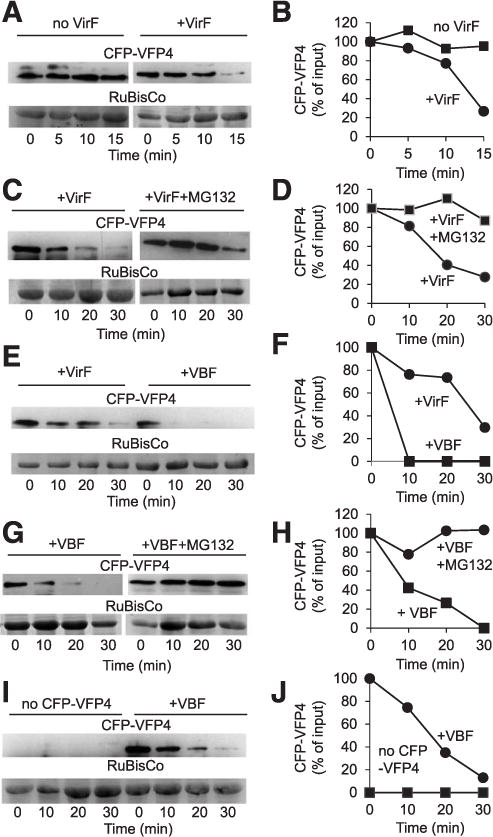
VirF and VBF (a host F-box protein) destabilize VFP4 in a cell-free degradation assay. A, Cyan fluorescent protein (CFP)-tagged VFP4 (CFP-VFP4) destabilization induced by VirF. B, Quantification of CFP-VFP4 accumulation data shown in A. C, CFP-VFP4 destabilization induced by VirF and effect of the proteasome inhibitor MG132. D, Quantification of CFP-VFP4 accumulation data shown in C. E, CFP-VFP4 destabilization induced by VirF or VBF. F, Quantification of CFP-VFP4 accumulation data shown in E. G, CFP-VFP4 destabilization induced by VBF and effect of the proteasome inhibitor MG132. H, Quantification of CFP-VFP4 accumulation data shown in G. I, Specific immunodetection of CFP-VFP4 and CFP-VFP4 destabilization induced by VBF. J, Quantification of CFP-VFP4 accumulation data shown in I. CFP-VFP4 was detected by Western blot analysis using anti-CFP antibody and RuBisCo was detected by Coomassie blue staining. When double bands were observed occasionally on some blots due to antibody cross-reactivity, only the band that corresponded to the size of VFP4 was used for quantification. The putative RuBisCo large chain was used as loading control and as reference for normalization of relative protein amounts. The data are representative of experiments conducted at least three times.
We next examined whether the plant F-box protein VBF, which can substitute for the VirF function during Agrobacterium infection (Zaltsman et al. 2010), would also destabilize VFP4. We found that coexpression of His-tagged VBF promoted rapid and virtually complete destabilization of CFP-VFP4, which was more efficient and rapid than VFP4 destabilization by VirF (Fig. 4E and F). Similarly to VirF, this VFP4 destabilization of VBF was blocked by treatment with MG132 (Fig. 4G and H), again suggesting that it occurred via UPS. Note that MG132 reduced destabilization rather than blocking it completely, consistent with previously observed partial effects of this proteasomal inhibitor on both VirF (Tzfira et al. 2004) and VBF (Zaltsman et al. 2010). In negative control experiments, no immunosignal was observed in the absence of the CFP-VFP4 expression, whereas, as expected, coexpression of CFP-VFP4 and VBF resulted in VBF destabilization (Fig. 4I and J). Taken together, these results suggest that VirF and VBF destabilize approximately 70 to 90% of VFP4, respectively (Supplementary Fig. S1).
Interestingly, unlike the VirF-VFP4 interaction (Fig. 1), our initial BiFC experiments did not detect consistent interaction between VBF and VFP4 (data not shown). The rapid VFP4 destabilization by VBF, however, suggested it may hinder detection of the BiFC signal in interaction experiments. Thus, we performed the BiFC assay in the presence of MG132 to impede degradation and allow detection of the BiFC signal. Under these conditions, cYFP-VBF interacted with nYFP-VFP4 in planta (Fig. 5). In a negative control, cYFP-VBF did not interact with an unrelated Agrobacterium VirE2 protein (Zaltsman et al. 2010) tagged with nYFP (Fig. 5).
Fig. 5.
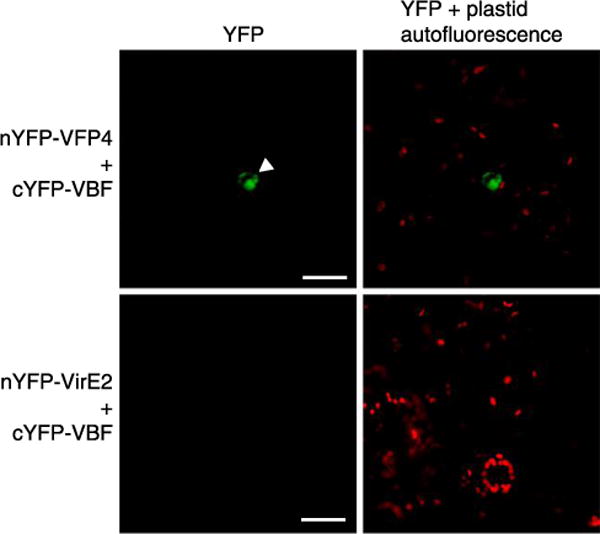
VFP4 interacts with VBF (a host F-box protein). The interaction was visualized by bimolecular fluorescence complementation in the presence of MG132. Location of the cell nucleus is indicated by a white arrowhead. All images are projections of single confocal sections and are representative images of three independent experiments. Scale bars = 20 μm.
Effects of VFP4 on Agrobacterium infection
If VirF-mediated destabilization of VFP4 acts to facilitate Agrobacterium infection, then one would expect wild-type Col-0 and VPF4 loss-of-function plants to be equivalently susceptible to Agrobacterium infection but VPF4 gain-of-function plants to exhibit reduced susceptibility to Agrobacterium infection. We, therefore, investigated whether VFP4 affected the efficiency of Agrobacterium infection, using two different transgenic plant lines with altered VFP4 gene expression, i.e., the loss-of-function homozygous T-DNA insertional mutant line vfp4-1 and the gain-of-function overexpressing lines VFP4 OE-6 and VFP4 OE-14.
Based on qualitative RT-PCR analysis, we found that, whereas levels of VFP4 transcripts were readily detected in wild-type Col-0 plants, VFP4 transcripts were not detected in vfp4-1 loss-of-function plants (Fig. 6A). Control reactions, using constitutively expressed actin (ACT2), confirmed equal input of RNA and reaction efficiency. We then inoculated vfp4-1 plants with a tumorigenic strain of A. tumefaciens to examine their susceptibility to Agrobacterium infection, using the root-tumor assay (Nam et al. 1999) and found that wild-type Col-0 and vfp4-1 plants exhibited similar levels of susceptibility to A. tumefaciens (Fig. 6B), with both lines developing comparable numbers of tumors on inoculated roots. Analysis by the Student’s t test confirmed that Agrobacterium tumorigenicity in Col-0 plants was not significantly different from that in the vfp4-1 plants.
Fig. 6.
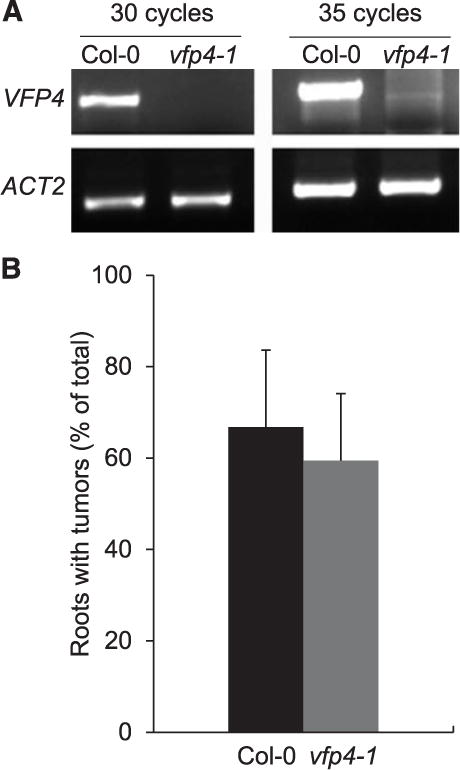
Loss-of-function vfp4-1 plants exhibit no detectable changes in susceptibility to Agrobacterium tumorigenicity. A, Qualitative polymerase chain reaction analysis of VFP4 transcript levels in leaves of the wild-type Col-0 and vfp4-1 plants. ACT2 was used as internal reference. B, Quantification of tumorigenicity in root explants inoculated with A. tumefaciens. Error bars represent standard error of the mean of independent biological replicates, n = 3.
To examine whether VFP4 gain-of-function plants were less susceptible to Agrobacterium infection than wild-type Col-0, we generated several transgenic lines that constitutively expressed the VFP4 cDNA from a Cauliflower mosaic virus (CaMV) 35S promoter. RT-qPCR analysis of two independent T2 transgenic lines, designated VFP4 OE-6 and VFP4 OE-14, showed that these moderate expressor lines accumulated 1.5- to 1.8-fold higher levels of VFP4 transcripts in their roots than did wild-type Col-0 plants (Fig. 7A). Our analyses, using the root-tumor assay, demonstrated that the number of neoplastic growths elicited by A. tumefaciens in both VFP4 OE-6 and VFP4 OE-14 plants was lower than that elicited in the Col-0 roots (Fig. 7B). We quantified the difference in tumor formation between the wild-type Col-0 plants and the two VFP4 gain-of-function lines and found it to be statistically significant (P values < 0.05); in contrast, the differences in tumor formation between VFP4 lines OE-6 and OE-14 lines were not significant (Fig. 7C). Thus, VFP4 appears to be a limiting factor for Agrobacterium infection. Notably, we did not detect any overt changes in morphology or developmental phenotypes in any of the VFP4 loss-of-function or gain-of-function lines as compared with the wild-type Col-0 plants, although the VFP4 gain-of-function plants did appear to develop more root hairs (data not shown).
Fig. 7.
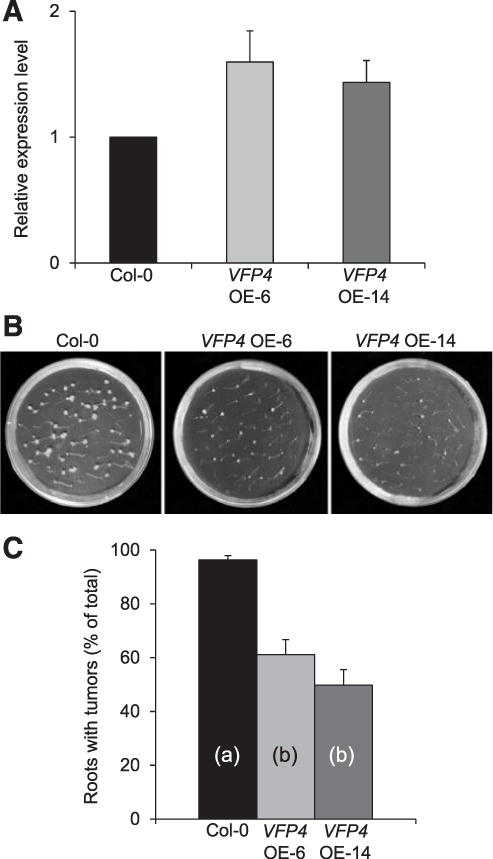
Gain-of-function VFP4 OE-6 and VFP4 OE-14 plants exhibit reduced susceptibility to Agrobacterium tumorigenicity. A, Real-time quantitative polymerase chain reaction analysis of VFP4 gene expression in roots of wild-type Col-0, VFP4 OE-6, and VFP4 OE-14 plants. The levels of expression were normalized to the internal reference genes ACT7 and 18S RNA. The expression level of VFP4 in the wild-type Col-0 was set to 1.0, and error bars represent standard error of the mean (SEM) of independent biological replicates, n = 3. B, Tumors developed on root explants inoculated with A. tumefaciens. C, Quantification of tumorigenicity. Error bars represent SEM of n = 3 biological replicates. Differences in tumorigenicity values between wild-type Col-0 and VFP4 OE-14 plants indicated by different letters are statistically significant (P values < 0.05) and by the same letter are not statistically significant.
VFP4 regulates transcription of Arabidopsis defense response genes
As a transcription factor, VFP4 most likely affected plant susceptibility to A. tumefaciens by regulating the expression of factors that would interfere with the Agrobacterium–plant cell interactions. As an approach to identify these factors, we assessed the global effects of VFP4 loss-of-function on the transcription of defense response genes, using RNA-seq analysis of the RNA samples characterized in Figure 6A. The resulting data were analyzed by MapMan, an ontology technique designed to analyze plant-specific transcriptional profiles in a variety of species, including Arabidopsis (Johnston et al. 2014; Kumar et al. 2015; Rotter et al. 2007; Urbanczyk-Wochniak et al. 2006). We used DESeq (Anders and Huber 2010) to identify differentially expressed genes (DEGs) and found statistically significant changes in the expression of 479 genes between wild-type Col-0 and the vfp4-1 loss-of-function line (false discovery rate [FDR] < 0.001 and log2 fold change [FC] > 2) (Supplementary Table S1). This represented 2.2% of the 21,519 expressed genes that had mapped reads of >5 in at least one sample. From these DEGs, MapMan annotation (TAIR10) assigned 181 genes to functional categories related to different aspects of response to either pathogen or pest attack or both (Fig. 8).
Fig. 8.
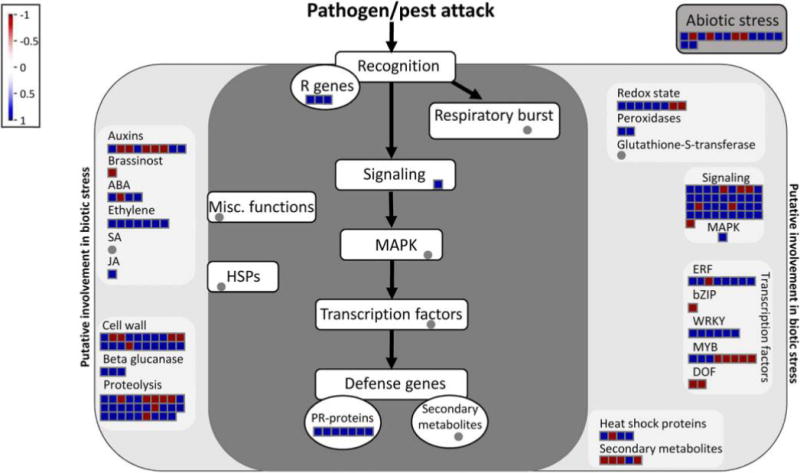
Overview of genes involved in biotic stress that are differentially expressed in vfp4-1 plants. The analysis utilized the MapMan software and values of log2 fold changes in the vfp4-1 mutant versus wild-type plants with P values < 0.001.
A VFP4-controlled gene, ATL31, negatively regulates Agrobacterium tumorigenicity
To explore whether any of the identified VFP4-regulated genes might be involved in Agrobacterium infection, we selected two DEGs for further analysis. One, ATL31/CNI1 (At5g27420), encodes a RING-type ubiquitin ligase (Serrano et al. 2006) shown to promote resistance to the bacterial pathogen Pseudomonas syringae (Maekawa et al. 2012). The second DEG, At2g32030, encodes an acyl-CoA N-acyltransferase superfamily protein involved in abscisic acid response (Xin et al. 2005). The latter has no known involvement in bacterial infection and, thus, represents a useful control. We then used RT-qPCR to examine directly the effect of VFP4 loss-of-function on expression of ATL31 and At2g32030, finding that transcript levels of ATL31 and At2g32030 in the vfp4-1 line were reduced fourfold and twofold, respectively, as compared with wild-type Col-0 plants (Fig. 9A). In reciprocal experiments, we examined the effect of VFP4 gain-of-function on expression of ATL31 and At2g32030, finding that transcript levels of ATL31 and At2g32030 were elevated 2.5-fold and threefold, respectively, in the VFP4 OE-6 line as compared with Col-0 plants (Fig. 9B). In both experiments, the data were statistically significant, with P values < 0.05.
Fig. 9.
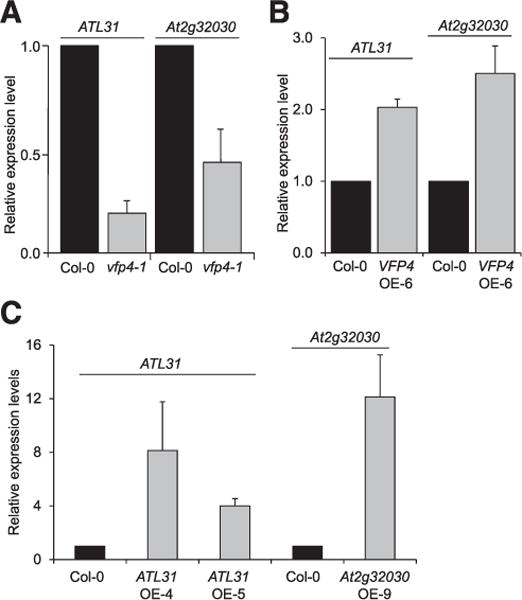
VFP4-controlled genes ATL31 and At2g32030. Real-time quantitative polymerase chain reaction (RT-qPCR) analysis of ATL31 and At2g32030 expression in roots of A, wild-type Col-0 and vfp4-1 plants and B, wild-type Col-0 and VFP4 OE-6 plants. C, RT-qPCR analysis of ATL31 and At2g32030 expression in roots of the wild-type Col-0, ATL31 OE-4, and ATL31 OE-5 and Col-0 and At2g32030 OE-9 plants, respectively. The levels of expression were normalized to the internal reference genes ACT7 and 18S RNA. The expression level of each tested gene in the wild-type Col-0 is set to 1.0, and error bars represent standard error of the mean of independent biological replicates, n = 3.
We propose that A. tumefaciens may employ VirF and its cellular functional homolog VBF to destabilize VFP4 in order to reduce the capacity of VFP4 to activate potential disease response genes, such as ATL31, that could negatively affect the infection process. According to this hypothesis, elevating the cellular levels of ATL31 would be expected to reduce Agrobacterium tumorigenicity. To test this, we produced both ATL31 and At2g32030 transgenic gain-of-function lines by expressing each coding sequence from a CaMV 35S promoter and identified several independent transgenic lines. We selected two gain-of-function ATL31 lines—one highly and one moderately expressing line designated ATL31 OE-4 and ATL31 OE-5, respectively—for further analyses. We also selected an At2g32030 gain-of-function line designated At2g32030 OE-9. RT-qPCR analysis showed that the high-expressing ATL31 OE-4 plants accumulated 11-fold higher levels of ATL31 transcript in their roots and the moderately expressing ATL31 OE-5 plants accumulated 4.4-fold higher transcript levels as compared with wild-type Col-0 plants (Fig. 9C). The At2g32030 OE-9 plants accumulated up to 16-fold higher levels of the At2g32030 transcript than did wild-type plants (Fig. 9C).
We then used the root-tumor assay to analyze these transgenic lines for their susceptibility to Agrobacterium-mediated genetic transformation. Figure 10A shows representative data for one line for each of the genes, i.e., ATL31 OE-4 and At2g32030 OE-9, and Figure 10B quantifies all tested lines, showing a statistically significant (P values < 0.05) twofold decrease in susceptibility to Agrobacterium tumorigenicity for both ATL31 OE-4 and ATL31 OE-5 plants, as compared with wild-type Col-0. This observed inhibitory effect on tumorigenicity was comparable in both ATL31 OE-4 and ATL31 OE-5 lines irrespective of their levels of ATL31 expression. In contrast, At2g32030 OE-9 plants were fully susceptible to A. tumefaciens, exhibiting no statistically significant differences from the wild-type plants (Fig. 10B). Interestingly, both ATL31 gain-of-function lines were indistinguishable from the wild-type plants in their growth but exhibited four- to eightfold reduction in seed yield and slightly impaired root gravitropism.
Fig. 10.
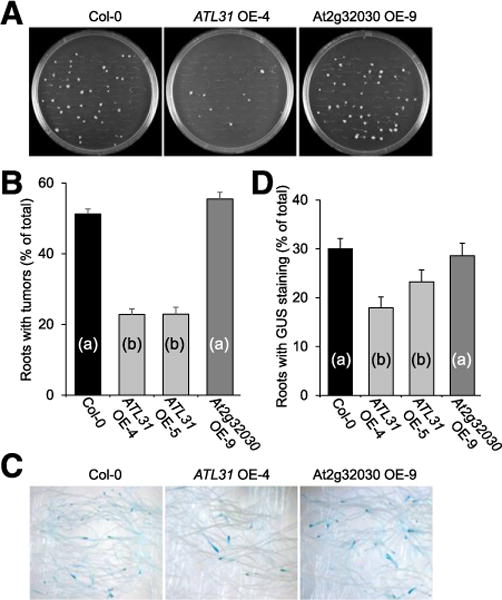
ATL31 OE-4, ATL31 OE-5, VFP4 OE9, and VFP4 OE-Y plants exhibit reduced susceptibility to stable but not to transient genetic transformation by Agrobacterium tumefaciens. Stable and transient genetic transformation was assessed by tumor formation and expression of a β-glucuronidase (GUS) reporter. A, Tumor development in root explants inoculated with A. tumefaciens. B, Quantification of tumorigenicity. C, Transient expression of GUS reporter. D, Quantification of transient GUS expression. Error bars represent standard error of the mean of independent biological replicates, n = 3. Differences in tumorigenicity values between wild-type Col-0 and mutated lines indicated by different letters are statistically significant (P values < 0.05) and by the same letter are not statistically significant.
Finally, we examined whether ATL31 affects Agrobacterium–host cell interaction only at the late, tumor-inducing stages of genetic transformation or also acts early in this process. The earlier events are detected, at 2 to 6 days after infection, as transient transgene expression that occurs prior to T-DNA integration into the host genome, whereas late events require T-DNA integration and are detected as tumor formation several weeks after infection (Nam et al. 1999). To assess transient transformation, root segments of ATL31 OE-4, ATL31 OE-5, or At2g32030 OE-9 plants were inoculated with an Agrobacterium strain carrying a gus gene for the β-glucuronidase (GUS) reporter in its T-DNA, and GUS activity was detected histochemically at 6 days after inoculation. Figure 10C shows representative data for one line for each of the genes. Quantification of GUS activity data for all tested lines (Fig. 10D) shows that both ATL31 OE-4 and ATL31 OE-5 lines exhibited slight but statistically significant reductions in transient transformation. In contrast, the At2g32030 OE-9 plants showed no statistically significant differences from the wild-type Col-0 in their ability to support transient T-DNA expression (Fig. 10D). Collectively, our findings suggest that ATL31 mainly compromises plant susceptibility to stable genetic transformation, yet, its action likely begins at earlier stages of the transformation process.
DISCUSSION
Interactions of A. tumefaciens with its host cells is both a technology of choice to genetically manipulate plants and several other eukaryotes such as fungi (Abuodeh et al. 2000; de Groot et al. 1998; Grimaldi et al. 2005; Lacroix et al. 2006) for research and biotechnology and a paradigm to study mechanisms evolved by invading pathogens to thwart the variety of defenses mounted against them by the host cell. Indeed, previous studies have identified several aspects of general plant defense that A. tumefaciens counters or subverts during infection. For example, A. tumefaciens has been shown to suppress the RNA silencing defense (Dunoyer et al. 2006). It has also been suggested that A. tumefaciens utilizes host MAP kinase defense signaling to help target its T-DNA into the plant cell nucleus (Djamei et al. 2007) and subverts the plant defense response–related UPS pathways for uncoating the associated proteins from the invading T-DNA (Zaltsman et al. 2010, 2013). In this study, we uncovered yet another aspect of plant defenses that is targeted by A. tumefaciens to facilitate genetic colonization of the host cell. We show that the Agrobacterium effector VirF, an F-box protein that is exported into host cells, and its functional cellular homolog, the Arabidopsis F-box protein VBF (Zaltsman et al. 2010), recognize and target the plant protein VFP4 for proteasomal degradation via the SCFVirF/SCFVBF pathway. VFP4 is a hitherto unknown transcription factor that is plant-specific and a member of the GeBP/GPL transcription factor family. Loss of function of VFP4 in Arabidopsis resulted in differential expression of a substantial number of biotic stress– response genes, suggesting that one of the functions of VFP4 is to control a broad spectrum of plant defenses. Based on this hypothesis and our demonstrating that Agrobacterium VirF interacts with and destabilizes VFP4, we propose that VFP4 may act to negatively affect Agrobacterium infection and that the bacterium utilizes its VirF effector to mitigate the VFP4-based defense by directly degrading VFP4 via the SCFVirF pathway. This hypothesis is consistent with the inability of A. tumefaciens to suppress VFP4 on the transcriptional level, taking instead a different route to destabilize the VFP4 protein itself. Indeed, our observations suggest that VirF as well as VBF, which is known to be induced by A. tumefaciens (Zaltsman et al. 2010), both promote proteasomal degradation of VFP4. Thus, A. tumefaciens likely has evolved to produce an F-box protein that it secretes into the host cell to subvert the host’s own UPS to target and destabilize defense response components of the host itself. That VFP4 represents one such component is supported by the decreased susceptibility to Agrobacterium tumorigenicity in VFP4 gain-of-function Arabidopsis lines.
Among the numerous defense response genes regulated by VFP4 are genes that might interfere with the Agrobacterium infection process. We identified one such gene, ATL31, which belongs to an 80-member family of RING-H2 finger ubiquitin ligases (Serrano et al. 2006) and is involved in the carbon/nitrogen response (Maekawa et al. 2012). Importantly, ATL31 is also a known defense-response gene that mediates resistance to P. syringae (Maekawa et al. 2012). Transcription of ATL31 is repressed in VFP4 loss-of-function plants and is activated in VFP4 gain-of-function plants. Consistent with the notion that VFP4 and VFP4-controlled ATL31 can act as negative regulators of Agrobacterium infection, gain-of-function lines of both genes exhibited decreased susceptibility to Agrobacterium tumorigenicity. This effect of VFP4 and ATL31 appeared to mainly target the late stages of the Agrobacterium-infection process manifested as tumor formation, yet the early stages, as characterized by transient expression of a transgene, also were affected, albeit slightly, in our gain-of-function lines. It is noteworthy that the natural outcome of Agrobacterium infection is tumor production and that A. tumefaciens has evolved different mechanisms to optimize this tumorigenicity, including, most likely, the one that targets VFP4 and the defense response genes that it controls.
Most bacterial effectors are multifunctional (Backert and Meyer 2006; Dean 2011; Galán 2009; Kenny et al. 2002). Similarly, VirF may perform multiple tasks and, by implication, recognize numerous substrates in the host cell. VFP4 represents the second VirF substrate, in addition to VIP1, identified to date (Tzfira et al. 2004). Our identification of this new substrate for the Agrobacterium VirF F-box effector and our demonstration that this substrate—a transcriptional activator of biotic stress genes that include antibacterial resistance genes—represents another line of defense of the host cell against bacterial pathogens opens a new page in the story of the Agrobacterium–plant host arms race. Considering that highly diverse pathogens utilize F-box proteins for infection, the ability of such pathogen-encoded F-box proteins to target the host transcription machinery that activates defense responses may have evolved as a widespread strategy to evade host defenses.
MATERIALS AND METHODS
Plant growth
The homozygous Arabidopsis Col-0 SALK_129879C line, corresponding to the vfp4-1 mutant, was obtained from the Arabidopsis Biological Resource Center. Wild-type Arabidopsis thaliana Col-0 plants, vfp4-1 plants, and Nicotiana benthamiana seedlings were germinated on aseptic Murashige and Skoog (MS) medium (Murashige and Skoog 1962) and, after 2 weeks, were transferred to soil and were maintained in an environment-controlled chamber at 22 to24°C under a 16-h light (70 to 80 μmol photons m−2 s−1) and 8-h dark cycle.
Agroinfiltration and microbombardment
For agroinfiltration, A. tumefaciens EHA105 (Hood et al. 1993), containing each test construct, was grown overnight at 28°C in Luria Bertani medium with 100 μg of spectinomycin per milliliter. Cells were harvested by centrifugation, were resuspended to adsorbance at 600 nm (A600) = 0.1 in infiltration buffer (10 mM MgCl2, 10 mM MES [pH 5.5], 100 μM acetosyringone), were incubated for 2 h at 25°C, and were infiltrated into the abaxial side of intact leaves on 3- to 4-week-old N. benthamiana plants, using a 1-ml needleless syringe. Plants were grown for 48 to 72 h, as described above, before being harvested.
For biolistic delivery, test constructs were mixed at a 1:1 wt/wt ratio. The DNA mixture (100 μg) was then adsorbed onto 10 mg of 1-μm gold particles (Bio-Rad) and were bombarded into the leaf epidermis of N. benthamiana using a portable Helios gene gun system (Model PDS-1000/He; Bio-Rad) at a pressure of 90 to 150 psi, and tissues were analyzed 48 h after microbombardment.
BiFC and subcellular localization
For BiFC, the coding sequence of VFP4 was amplified using the primer pair 5′ATGCAAGCTTCGATGGCATCGGATCAACGTGA3′/5′ATGCGGTACCTCATCCTCCATTAGGCATTG3′ and was cloned into the HindIII-KpnI sites of pSAT6-nEYFP-C1 (Citovsky et al. 2006). The constructs expressing cYFP-VirF, cYFP-VBF, nYFP-VirE2, and nYFP-FLD were described previously (Krichevsky et al. 2011; García-Cano et al. 2015; Zaltsman et al. 2010). The coding sequence of CUL1 (At4g02570) was amplified using the primer pair 5′AGGATCCTAAGCCAAGTACCTAAACATGTTAGG3/5′TCTCGAGACATGGAGCGCAAGACTATTGAC3′ and was cloned into the BamHI-XhoI sites of pSAT4-cYFP-C1 (Citovsky et al. 2006). Each construct or test pair of constructs was transiently expressed in N. benthamiana leaves by microbombardment and was observed at 72 h postbombardment.
For BiFC in the presence of MG132, the VBF coding sequence was amplified using the primer pair 3′ATAAAGCTTCGATGATGATGTTACCAGAAG/5′TAGGATCCTTATGTTTTAGGCCTCACTTCAATAC3′ and was cloned into the HindIII-BamHI sites of pSAT1-cEYFP-C1 (Citovsky et al. 2006), and the nYFP-VFP4 expression cassette was transferred from pSAT6-nEYFP-C1 into the HindIII-KpnI sites of pSAT4-nEYFP-C1 (Tzfira et al. 2005). Each of the expression cassettes was then excised with I-SceI and AscI, respectively, and was inserted into the same sites in the binary vector pPZP-RCS2 (Goderis et al. 2002; Tzfira et al. 2005). The tested combination of constructs was transiently expressed in N. benthamiana leaves using agroinfiltration. The tissues were infiltrated with 10 μM MG132 at 72 h postagroinfiltration and were then incubated for 4 h, before being examined.
To determine subcellular localization, the VFP4 coding sequence was amplified with the primer pair 5′ATGCAAGCTTCGATGGCATCGGATCAACGTGA3′/5′ATGCGGTACCTCATCCTCCATTAGGCATTG3′ and was cloned into the HindIII-KpnI sites of pSAT5-EYFP-C1, which is identical to pSAT5-EGFP-C1 (Tzfira et al. 2005), except that it expresses the YFP reporter. The resulting expression cassette was excised with I-CeuI, was inserted into the pPZP-RCS1 binary vector (Goderis et al. 2002; Tzfira et al. 2005), was transiently expressed in N. benthamiana leaves by agroinfiltration, and was examined at 72 h postinfiltration. YFP fluorescence was detected using a Zeiss LSM 5 Pascal confocal microscope. All experiments were repeated at least three times in independent biological replicates, i.e., independently grown plants.
Protein destabilization in a cell-free system
The binary construct expressing Myc-VirF was described previously (García-Cano et al. 2015; Magori and Citovsky 2011). The VFP4 and VBF coding sequences were amplified with the primer pairs 5′ATGCAAGCTTCGATGGCATCGGATCAACGTGA3′/5′ATGCGGTACCTCATCCTCCATTAGGCATTG3′ and 5′TTTCTCGAGCATTATCGATGAGAAGAGA3′/5′TAGCGGCCGCTTATGTTTTAGGCCTCACTTCAATAC3′, respectively, and were cloned into the HindIII-KpnI sites of pSAT5-ECFP-C1 (García-Cano et al. 2015) or PstI-SalI sites of pSAT4-HIS-C1 (provided by A. Zaltsman, Plant Genetic Engineering, Inc., Stony Brook, NY, U.S.A.), respectively. The resulting expression cassettes were excised with I-CeuI or I-SceI, respectively, and were inserted into pPZP-RCS1 separately or together, so that each combination of the tested proteins would be expressed from the same vector. Each expression construct was then agroinfiltrated into N. benthamiana leaves and was transiently expressed for 72 h, after which the leaves were harvested, extracted, and incubated in degradation buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 5 mM dithiothreitol [DTT], 5 mM ATP, and 1× plant protease inhibitor cocktail [Sigma-Aldrich]) at 25°C for the times indicated, as described (García-Cano et al. 2014; Magori and Citovsky 2011). The levels of VFP4 protein were analyzed on Western blots, using anti–green fluorescent protein antibody (Clontech) and secondary antibody conjugated to horseradish peroxidase, as described (Magori and Citovsky 2011; García-Cano et al. 2014), detecting the approximately 77-kDa CFP-VFP4 fusion protein. For control reactions in the absence of CFP-VFP4, we expressed CFP-VFP3 as described previously (García-Cano et al. 2015). For treatment with MG132, leaves were infiltrated with 10 μM MG132 or mock-treated with 0.1% dimethyl sulfoxide and were incubated for 4 h before harvesting. For the sample loading control, we used an approximately 50-kDa major protein band, presumably representing the large chain of ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCo) (Magori and Citovsky 2011), detected on Coomassie blue-stained gels. Protein amounts were assessed by scanning densitometry of the corresponding Western blot bands, using the ImageJ software (version 1.50, National Institutes of Health), and were normalized to the loading controls for each sample.
Qualitative RT-PCR
The mutagenic T-DNA insertion into the VFP4 gene of the homozygous vfp4-1 mutant line was confirmed by PCR using the T-DNA left border–specific forward primer SALK LBb1.3 5′ATTTTGCCGATTTCGGAAC3′ (Salk Institute Genomic Analysis Laboratory) and the VFP4-specific reverse primer 5′AGTCATACGGTGCCATTTCTG3′. The wild-type copy of the VFP4 gene was detected with the primer pair 5′TCCACTGCGTTTAAACCAGTC3′/5′AGTCATACGGTGCCATTTCTG3′.
For RT-PCR analysis of VFP4 expression in vfp4-1 plants, total RNA was extracted from leaves of the wild-type Col-0 and vfp4-1 plants using TRIzol (Invitrogen) and was purified with the SV total RNA isolation system (Promega). RNA quality and quantity were assessed using Biospec-Nano (Shimadzu). RT reactions were performed with 0.5 mg of the total RNA, using oligo-dT and the RevertAid RT kit (Thermo Scientific), and the resulting cDNA was amplified for 30 or 35 cycles using the primer pairs 5′CTGGATCTGATTCGATCTACCAG3′/5′GTGATGTAATCGAGAAAGCCACG3′ specific for VFP4, and 5′AGAGATTCAGATGCCCAGAAGTCTTGTTCC3′/5′AACGATTCCTGGACCTGCCTCATCATACTC3′ specific for ACTIN2 (ACT2), a constitutively expressed gene used as an internal loading control. The PCR conditions were: 1 cycle at 94°C for 3 min, 1 cycle at 94°C for 30 s; 1 cycle at 55°C for 30 s, 1 cycle at 55°C for 30 s, the indicated number cycles (i.e., 30 or 35) at 72°C for 1 min, and 1 cycle at 72°C for 5 min.
RT-qPCR
A total of 0.5 μg of DNA-free total RNA was extracted from the leaves or roots of the indicated plant lines as described above and was reverse-transcribed with oligo-dT and the RevertAid RT kit. The resulting cDNA samples were then amplified using the following primer pairs: VFP4 specific, 5′CAGCTTGTGGAGAAGCTAAGG3′/5′CCAGTTTGATTCCAAATCTTCC3′ ATL31-specific, 5′TGACCCGTATGCTTACAGCG3′/5′ACACTCCAACGCTCCTTTAC3′; At2g32030-specific, 5′TCCGACGTCGACGATTTCAT3′/5′TCCTCTGATCTCATCGACTG3′; ACTIN7 (ACT7)-specific (internal control for a constitutively expressed gene), 5′CATTCAATGTCCCTGCCATGT3′/5′GGTTGTACGACCACTGGCATAG3′; and 18S ribosomal RNA (18S RNA)-specific (independent internal control gene), 5′GGTCTGTGATGCCCTTAGATGTT3′/5′GGCAAGGTGTGAACTCGTTGA3′. RT-qPCR was performed using LightCycler 480 with SYBR Green I Master (Roche Diagnostic). The PCR conditions were: 1 cycle at 95°C for 5 min, 40 cycles at 95°C for 10 s, 1 cycle at 57°C for 10 s, and 1 cycle at 72°C for 15 min. Three technical replicates from three biological replicates, i.e., leaves or roots from independently grown pools of 4 to 5 or 10 to 14 plants, respectively, were performed for each gene assayed. Relative gene expression levels were calculated using the cycle threshold (2−ΔΔCT) method (Livak and Schmittgen 2001). All quantitative data were analyzed by the Student’s t test; P values <0.05, corresponding to a statistical probability of greater than 95%, were considered statistically significant. Standard error of the mean and t test calculations were performed using Excel 2010 (Microsoft Inc.).
Generation of transgenic plants
The coding sequences of VFP4, ATL31, and At2g32030 genes were amplified using the Pfu high-fidelity DNA polymerase and the following primers: VFP4, 5′ATGCAAGCTTCGATGGCATCGGATCAACGTGA3′/5′ATGCGGTACCTCATCCTCCATTAGGCATTG3′; ATL31, 5′ATATAAGCTTCATGGATCCCATAA3′/5′ATATGTCGACCTAAACCGGTAGC3′; and, At2g32030, 5′TACCTCGAGCTGTAAGAATTGAGAGAGAT3′/5′GAAGGATCCGAGCATCACATCAAATTATAC3′. The amplified VFP4 and At2g32030 DNAs were then cloned between the HindIII-KpnI or the HindIII-SalI sites, respectively, of pSAT5A-MCS (Chung et al. 2005) and that of ATL31 was cloned between the XhoI-BamHI sites of pSAT4-MCS (Tzfira et al. 2005). The resulting expression cassettes were each excised with I-CeuI, inserted into pPZP-RCS2, and were confirmed by DNA sequencing. Transgenic Arabidopsis Col-0 plants were generated using these binary constructs and A. tumefaciens EHA105, as described previously (Bent 2006; Kim et al. 2003). Transgene expression was confirmed by RT-qPCR in T2 and T3 lines.
Agrobacterium inoculation, tumorigenesis, and GUS staining
For tumorigenesis assays, root explants from aseptically grown 15- to 20-day-old wild-type Col-0 and vfp4-1 plants (50 to 70 explants per plant) were submerged in a 0.9% saline suspension of the oncogenic A. tumefaciens LBA1010 (Koekman et al. 1982) (A600 = 0.1) and were incubated for 10 min at 25°C. These were then cultivated for 48 h at 25°C in hormone-free MS (HFMS) medium, were washed, and were then cultured for an additional 2 to 3 weeks in HFMS in the presence of 100 μg of timentin per milliliter and were scored for tumors. For transient T-DNA expression and histochemical GUS staining, the inoculation protocol utilized the same oncogenic LBA1010 strain harboring a pBISN1 plasmid with an expression cassette for a gus reporter gene with a plant intron sequence (gus-int) (Narasimhulu et al. 1996). The root explants were harvested after 4 days of culture, were stained with 5-bromo-4-chloro-3-indolyl β-D-glucuronide (BioVectra), as described (Li et al. 2005; Nam et al. 1999), and were scored for the GUS-specific indigo color. For analysis of VFP4 expression following bacterial challenge, the roots were inoculated with LBA1010 or mock-inoculated with the bacterial growth medium as described above, and total RNA was extracted from tissue samples at 24 h after inoculation and was subjected to the RT-qPCR analysis. Each experiment was performed in three biological replicates, each containing pools of 150 to 210 roots explants from 50 different plants, and statistical significance of the data were evaluated by the Student’s t test as described above.
High-throughput cDNA sequencing (RNA-seq), read mapping, and data analysis
Experiments were performed exactly as described previously (García-Cano et al. 2015). Briefly, total RNA was extracted from leaves of wild-type Col-0 and vfp4-1 mutant plants. Polyadenylated RNA was then isolated on oligo-dT-magnetic beads, fragmented and primed for cDNA synthesis (García-Cano et al. 2015). These RNA preparations (1.2 ng) were used for RNA-seq library construction according to the manufacturer’s recommendations (Illumina). cDNA fragments of approximately 200 to 500 bp were isolated by gel electrophoresis, were amplified by 15 cycles of PCR, and were sequenced on the Illumina NextSeq500 platform (García-Cano et al. 2015). Three biological replicates, i.e., independently grown pools of plants, were used for all RNA-seq experiments.
For read mapping, adapters were removed from raw reads with FASTX toolkit pipeline v0.0.13, sequence quality determined with FastQC (Babraham Bioinformatics), and low quality reads were removed with FASTX toolkit set to retain reads with 1% sequencing error rate (García-Cano et al. 2015). These reads were then mapped to the Arabidopsis genome (TAIR10.22) from EnsemblPlants, using TopHat v2.0.10 (Trapnell et al. 2009). Raw count data were obtained by Cuffdiff embedded in Cufflinks pipeline v2.1.1 (Trapnell et al. 2012). DEGs were identified by DESeq (Anders and Huber 2010) using Bioconductor, setting the FDR as <0.001 and absolute value of log2 FC > 2 (García-Cano et al. 2015).
Data analysis classified Arabidopsis loci using the MapMan functional classification system (Thimm et al. 2004). Categories with gene number <10 were not included in the presented data. overrepresented functional categories enrichments were also conducted based on Fisher’s exact test (Li et al. 2010). Overview of biotic stress DEGs was visualized using MapMan version 3.5.1 (Thimm et al. 2004).
Supplementary Material
Acknowledgments
RNA-seq experiments and their analyses were performed in the Cornell University Biotechnology Resource Center. E. García-Cano was supported in part by the Marie Curie U-Mobility Postdoctoral Fellowship. H. Hagit was supported by the Vaadia-Binational Agricultural Research and Development Fund (BARD) Postdoctoral Fellowship. The work in the V. Citovsky laboratory is supported by grants from the National Institutes of Health (NIH), National Science Foundation, United States Department of Agriculture National Institute of Food and Agriculture, and BARD to V. Citovsky, and the S. G. Lazarowitz laboratory is supported by NIH and funds from the Departments of Plant Pathology and Plant-Microbe Biology to S. G. Lazarowitz.
Funding: This work was funded by the National Institute of Food and Agriculture grant 2013-02918, the Directorate for Biological Sciences grant MCB 1118491, the National Institute of General Medical Sciences grant GM50224, and BARD grant IS-4605-13C.
Footnotes
The e-Xtra logo stands for “electronic extra” and indicates that one supplementary figure and one supplementary table are published online.
AUTHOR-RECOMMENDED INTERNET RESOURCES
Arabidopsis Biological Resource Center: http://www.arabidopsis.org/abrc/
Bioconductor software: http://www.bioconductor.org
ClustalX (ver. 2.1): http://www.clustal.org/clustal2
cNLS Mapper: nls-mapper.iab.keio.ac.jp
Cufflinks pipeline: http://cufflinks.cbcb.umd.edu
Database of Arabidopsis transcription factors (DATF): http://atrm.cbi.pku.edu.cn
EnsemblPlants database: http://plants.ensembl.org
FASTX toolkit: http://hannonlab.cshl.edu/fastx_toolkit
Babraham Bioinformatics: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Molecular Evolutionary Genetics Analysis tool: http://www.megasoftware.net
LITERATURE CITED
- Abuodeh RO, Orbach MJ, Mandel MA, Das A, Galgiani JN. Genetic transformation of Coccidioides immitis facilitated by Agrobacterium tumefaciens. J Infect Dis. 2000;181:2106–2110. doi: 10.1086/315525. [DOI] [PubMed] [Google Scholar]
- Alto NM, Orth K. Subversion of cell signaling by pathogens. Cold Spring Harb Perspect Biol. 2012;4:a006114. doi: 10.1101/cshperspect.a006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield MJ. Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell Microbiol. 2015;17:18–25. doi: 10.1111/cmi.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A. Arabidopsis thaliana floral dip transformation method. Methods Mol Biol. 2006;343:87–103. doi: 10.1385/1-59745-130-4:87. [DOI] [PubMed] [Google Scholar]
- Chevalier F, Perazza D, Laporte F, Le Hénanff G, Hornitschek P, Bonneville JM, Herzog M, Vachon G. GeBP and GeBP-like proteins are noncanonical leucine-zipper transcription factors that regulate cytokinin response in Arabidopsis. Plant Physiol. 2008;146:1142–1154. doi: 10.1104/pp.107.110270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T. A versatile vector system for multiple gene expression in plants. Trends Plant Sci. 2005;10:357–361. doi: 10.1016/j.tplants.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Kozlovsky SV, Lacroix B, Zaltsman A, Dafny-Yelin M, Vyas S, Tovkach A, Tzfira T. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 2007;9:9–20. doi: 10.1111/j.1462-5822.2006.00830.x. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol. 2006;362:1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Curaba J, Herzog M, Vachon G. GeBP, the first member of a new gene family in Arabidopsis, encodes a nuclear protein with DNA-binding activity and is regulated by KNAT1. Plant J. 2003;33:305–317. doi: 10.1046/j.1365-313x.2003.01622.x. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Bundock P, Hooykaas PJJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev. 2011;35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat Genet. 2006;38:258–263. doi: 10.1038/ng1722. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Galán JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cano E, Magori S, Sun Q, Ding Z, Lazarowitz SG, Citovsky V. Interaction of Arabidopsis trihelix-domain transcription factors VFP3 and VFP5 with Agrobacterium virulence protein VirF. PLoS One. 2015;10:e0142128. doi: 10.1371/journal.pone.0142128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cano E, Zaltsman A, Citovsky V. Assaying proteasomal degradation in a cell-free system in plants. J Vis Exp. 2014;85:e51293. doi: 10.3791/51293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goderis IJ, De Bolle MF, François IE, Wouters PF, Broekaert WF, Cammue BP. A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol. 2002;50:17–27. doi: 10.1023/a:1016052416053. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, de Raaf MA, Filetici P, Ottonello S, Ballario P. Agrobacterium-mediated gene transfer and enhanced green fluorescent protein visualization in the mycorrhizal ascomycete Tuber borchii: A first step towards truffle genetics. Curr Genet. 2005;48:69–74. doi: 10.1007/s00294-005-0579-z. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers S, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2:208–218. [Google Scholar]
- Hooykaas PJJ, Hofker M, den Dulk-Ras H, Schilperoort RA. A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid, and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid. 1984;11:195–205. doi: 10.1016/0147-619x(84)90026-x. [DOI] [PubMed] [Google Scholar]
- Johnston R, Wang M, Sun Q, Sylvester AW, Hake S, Scanlon MJ. Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell. 2014;26:4718–4732. doi: 10.1105/tpc.114.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny B, Ellis S, Leard AD, Warawa J, Mellor H, Jepson MA. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol Microbiol. 2002;44:1095–1107. doi: 10.1046/j.1365-2958.2002.02952.x. [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130:4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- Koekman BP, Hooykaas PJJ, Schilperoort RA. A functional map of the replicator region of the octopine Ti plasmid. Plasmid. 1982;7:119–132. doi: 10.1016/0147-619x(82)90072-5. [DOI] [PubMed] [Google Scholar]
- Krichevsky A, Zaltsman A, Lacroix B, Citovsky V. Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc Natl Acad Sci USA. 2011;108:11157–11162. doi: 10.1073/pnas.1014030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Datta R, Hazra S, Sultana A, Mukhopadhyay R, Chattopadhyay S. Transcriptomic profiling of Arabidopsis thaliana mutant pad2.1 in response to combined cold and osmotic stress. PLoS One. 2015;10:e0122690. doi: 10.1371/journal.pone.0122690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Tzfira T, Vainstein A, Citovsky V. A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet. 2006;22:29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA. 2005;102:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, Reidel EJ, Turgeon R, Liu P, Sun Q, Nelson T, Brutnell TP. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maekawa S, Sato T, Asada Y, Yasuda S, Yoshida M, Chiba Y, Yamaguchi J. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol Biol. 2012;79:217–227. doi: 10.1007/s11103-012-9907-0. [DOI] [PubMed] [Google Scholar]
- Magori S, Citovsky V. Agrobacterium counteracts host-induced degradation of its effector F-box protein. Sci Signal. 2011;4:ra69. doi: 10.1126/scisignal.2002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, Peeters N, Rivas S. Ubiquitination during plant immune signaling. Plant Physiol. 2012;160:15–27. doi: 10.1104/pp.112.199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nam J, Mysore KS, Zheng C, Knue MK, Matthysse AG, Gelvin SB. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol Gen Genet. 1999;261:429–438. doi: 10.1007/s004380050985. [DOI] [PubMed] [Google Scholar]
- Narasimhulu SB, Deng XB, Sarria R, Gelvin SB. Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell. 1996;8:873–886. doi: 10.1105/tpc.8.5.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Laporte F, Balagué C, Chevalier F, Remo S, Bourge M, Larkin J, Herzog M, Vachon G. GeBP/GPL transcription factors regulate a subset of CPR5-dependent processes. Plant Physiol. 2011;157:1232–1242. doi: 10.1104/pp.111.179804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter A, Usadel B, Baebler S, Stitt M, Gruden K. Adaptation of the MapMan ontology to biotic stress responses: Application in solanaceous species. Plant Methods. 2007;3:10. doi: 10.1186/1746-4811-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salomon D, Orth K. What pathogens have taught us about posttranslational modifications. Cell Host Microbe. 2013;14:269–279. doi: 10.1016/j.chom.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuïnk TJG, Crosby WL, Hooykaas PJJ. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol. 2001;11:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- Serrano M, Parra S, Alcaraz LD, Guzmán P. The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J Mol Evol. 2006;62:434–445. doi: 10.1007/s00239-005-0038-y. [DOI] [PubMed] [Google Scholar]
- Spallek T, Robatzek S, Göhre V. How microbes utilize host ubiquitination. Cell Microbiol. 2009;11:1425–1434. doi: 10.1111/j.1462-5822.2009.01346.x. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Zambryski PC. Generic trans-kingdom sex? Nature. 1989;340:190–191. doi: 10.1038/340190a0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Shirasu K. Ubiquitination in plant immunity. Curr Opin Plant Biol. 2010;13:402–408. doi: 10.1016/j.pbi.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Rhee Y, Chen MH, Kunik T, Citovsky V. Nucleic acid transport in plant-microbe interactions: The molecules that walk through the walls. Annu Rev Microbiol. 2000;54:187–219. doi: 10.1146/annurev.micro.54.1.187. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Bläsing O, Kowalczyk M, Weicht D, Polinceusz A, Meyer S, Stitt M, Fernie AR. Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: Effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol. 2006;60:773–792. doi: 10.1007/s11103-005-5772-4. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CMT, Regensburg-Tuïnk TJ, Hooykaas PJJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Xin Z, Zhao Y, Zheng ZL. Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol. 2005;139:1350–1365. doi: 10.1104/pp.105.068064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Krichevsky A, Loyter A, Citovsky V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe. 2010;7:197–209. doi: 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Lacroix B, Gafni Y, Citovsky V. Disassembly of synthetic Agrobacterium T-DNA-protein complexes via the host SCF(VBF) ubiquitin-ligase complex pathway. Proc Natl Acad Sci USA. 2013;110:169–174. doi: 10.1073/pnas.1210921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Academic Press; New York: 1965. pp. 97–166. [Google Scholar]
- Zupan J, Muth TR, Draper O, Zambryski P. The transfer of DNA from agrobacterium tumefaciens into plants: A feast of fundamental insights. Plant J. 2000;23:11–28. doi: 10.1046/j.1365-313x.2000.00808.x. [DOI] [PubMed] [Google Scholar]
- Zupan J, Zambryski PC. The Agrobacterium DNA transfer complex. Crit Rev Plant Sci. 1997;16:279–295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


