Abstract
The ultimate formation of a four-chambered heart allowing the separation of the pulmonary and systemic circuits was key for the evolutionary success of tetrapods. Complex processes of cell diversification and tissue morphogenesis allow the left and right cardiac compartments to become distinct but remain poorly understood. Here, we describe an unexpected laterality in the single zebrafish atrium analogous to that of the two atria in amniotes, including mammals. This laterality appears to derive from an embryonic antero-posterior asymmetry revealed by the expression of the transcription factor gene meis2b. In adult zebrafish hearts, meis2b expression is restricted to the left side of the atrium where it controls the expression of pitx2c, a regulator of left atrial identity in mammals. Altogether, our studies suggest that the multi-chambered atrium in amniotes arose from a molecular blueprint present before the evolutionary emergence of cardiac septation and provide insights into the establishment of atrial asymmetry.
Research organism: Zebrafish
Introduction
The mature mammalian heart comprises four chambers that serve the systemic and pulmonary circuits. The parallel arrangement of the two vascular systems was a prerequisite for the evolution of tetrapods from their aquatic ancestors. Intriguingly, the emergence of the four-chambered heart of amniotes from the primitive two-chambered heart of bony fish appears not to have been a single event by axis duplication, but gradually as evidenced by the partial septation of lungfish atria and the complete septation of amphibian atria (Olson, 2006).
The formation and patterning of the tetrapod heart relies on the precise integration of cells from distinct progenitor populations during cardiogenesis. These progenitor pools include cells from the bilateral cardiogenic mesoderm, proepicardial progenitor cells, and cardiac neural crest derived cells. Progenitors from the cardiogenic mesoderm are further grouped into the so called first and second heart fields (FHF and SHF, respectively) (Buckingham et al., 2005). The integration of these spatiotemporally and molecularly distinct cellular fields is key in cardiac development, as their contributions increase heterogeneity by the addition of distinct cell types, and also define morphogenetic boundaries that shape the heart. Lineage tracing in mammals has revealed that the FHF forms most of the left ventricle, whereas the SHF contributes to the right ventricle and outflow tract (Buckingham et al., 2005; Simões-Costa et al., 2005; Koshiba-Takeuchi et al., 2009; Jensen et al., 2013) thereby establishing a left-right organ asymmetry in the ventricle that is important during ventricular septation and morphogenesis. In contrast to the ventricular chambers, both atria contain cells from both heart fields, but exhibit discrete gene expression programs. Notably, the Paired-like homeodomain transcription factor 2 c (Pitx2c) gene is exclusively expressed in the left atrial myocardium in amphibians and mammals where it regulates a wide variety of genes with left atrial expression (Figure 1a) (Ryan et al., 1998; Franco et al., 2014). Taken together, apparently distinct mechanisms of cellular heterogeneity establish ventricular and atrial laterality in mammals to allow the systemic and pulmonary compartments of the heart to be physically and molecularly distinct. Consequently, mutations interfering with FHF or SHF development or atrial asymmetry are associated with cardiac patterning and septation defects in humans and constitute the leading cause of congenital disorders (Hoffman, 1995; Lin et al., 2012a; Li et al., 2015).
Figure 1. Asymmetric expression of atrial-specific transcription factor genes in adult zebrafish.
(a) Comparison of different vertebrate hearts showing the emergence of distinct left and right chamber identities and atrial septation. (b) Schematic representation of the workflow for the identification of cardiac chamber-specific transcription factor gene expression. (c) Atrial gene expression level (signal intensity) and chamber-specific enrichment in the atrium (fold change >0) or ventricle (fold change <0). Red line marks 2.5 (log2) fold cutoff. Highly enriched atrial transcription factor genes (blue), ventricular (myh7l) and atrial (myh6) chamber-specific myosin genes (orange) are shown. (d) Atrial enriched transcription factor genes (log2 fold change >2.5). (e–f) Whole-mount in situ hybridization on adult zebrafish hearts; myh6 expression covers the entire atrium while meis2b expression is restricted to the left side of the atrium. (g–h) In situ hybridization on adult cardiac sections; myl7 is expressed in all cardiomyocytes while meis2b is expressed on the left side of the atrium in the wall and trabeculae (see Figure 1—figure Supplement 1a for higher magnification). (i–i’) Brightfield and fluorescence images of a 4 mpf Tg(meis2b-reporter) zebrafish atrium. (j–j’) High-magnification confocal projections of a 4 mpf Tg(meis2b-reporter) heart showing expression of the Tg(meis2b-reporter) in the myocardium (Tg(meis2b-reporter)+/MF20+/Fli cells) but not the endocardium (Tg(meis2b-reporter)-/MF20-/Fli1+cells). (e–f, i–i’) Red and white dotted lines delineate the atrium; (i’) yellow line delineates Tg(meis2b-reporter) expression. Scale bars: (e–i) 1 mm, (j’) 50 µm.
Figure 1—figure supplement 1. Validation of the meis2b reporter line.
Several transcription factors including Tbx5, Gata4, Nkx2.5 and Isl1 have been implicated in controlling lineage contribution from the FHF and SHF (Francou et al., 2013; Ang et al., 2016). However, it remains enigmatic how atrial left-right asymmetry is established in the developing tetrapod heart, in part because of the limitations posed by mammalian model systems when it comes to in vivo imaging. Despite its simple two-chambered heart, the zebrafish shows a surprisingly conserved cardiac ontogeny (Staudt and Stainier, 2012), including a SHF that contributes to the outer curvature of the ventricle, the outflow tract and atrium (Zhou et al., 2011; Witzel et al., 2012; Guner-Ataman et al., 2013). Hence, studies of cardiac development in zebrafish have contributed significantly to our understanding of the mechanisms underlying lineage diversity and spatiotemporal differentiation (Witzel et al., 2012; Guner-Ataman et al., 2013; Stainier et al., 1993; Lee et al., 1994; Mosimann et al., 2015; Rydeen and Waxman, 2016). Whether the single zebrafish atrium also shows myocardial asymmetry along its left-right axis has not been investigated thus far. Here, we systematically address this question using transcriptomic approaches, combined with the generation of transgenic reporter lines, high-resolution confocal microscopy, lineage tracing, as well as knock-down and loss-of-function experiments. We describe for the first time the existence of two different transcriptional compartments in the atrium of the zebrafish heart, which are analogous to the left and right atria of mammals. Furthermore, we show that this asymmetry appears to be established from two distinct antero-posterior fields during embryonic heart development. We find that the homeobox transcription factor Meis2b is important to establish and maintain left-right asymmetry in the zebrafish atrium, and that it regulates atrial morphogenetic growth and conduction. Altogether, our findings suggest that the multi-chambered heart in mammals evolved from transcriptional compartments present in the piscine heart.
Results
Identification of asymmetrically expressed transcription factor genes in the zebrafish atrium
We hypothesized that the four-chambered heart evolved from preexisting cellular compartments rather than the introduction of new structures. To test this hypothesis, we examined in zebrafish atrial-specific transcription factor genes which could potentially be involved in establishing compartments across the atrial left-right axis (Figure 1b). Microarray analysis revealed a strong enrichment of pitx2 transcripts in the zebrafish atrium, reflecting the principal expression domain of its orthologues in the three- and four-chambered hearts of amniotes (Ryan et al., 1998; Campione et al., 1999; Tessari et al., 2008) (Figure 1b–d). In humans, PITX2 mutations can cause Axenfeld-Rieger syndrome and are associated with defects in atrial conduction and septation (Mammi et al., 1998; Gudbjartsson et al., 2007). In mice, Pitx2 deficiency causes atrial and ventricular septal defects, right ventricular and right atrial enlargement, and there is strong evidence that Pitx2 is also key in maintaining left-right atrial identity and sinoatrial node formation (Franco et al., 2014; Tessari et al., 2008). In addition to pitx2, we detected a high atrial enrichment of the myeloid ecotropic integration site 2b (meis2b) transcription factor gene. This gene was an interesting candidate, as mutations in its human orthologue, MEIS2, are strongly implicated in atrial and ventricular septation defects (Louw et al., 2015). Meis2-deficient mice display outflow tract malformations and persistent truncus arteriosus (Machon et al., 2015), while zebrafish meis2b morphants reportedly show cardiac looping defects and a decreased heart rate (Paige et al., 2012). Analysis of chromatin remodeling further indicates a role for MEIS2 during myocardial differentiation of human embryonic stem cells (Paige et al., 2012). Interestingly, using in situ hybridization, we found asymmetric expression of meis2b in the adult zebrafish heart, as it is exclusively expressed in the left compartment of the mature atrium (Figure 1e–h, Figure 1—figure Supplement 1a). To investigate meis2b expression in detail using live imaging, we established a transgenic reporter system (TgBAC(meis2b:GAL4FF)bns15;Tg(5xUAS:EGFP)nkuasgfp1a) [abbreviated Tg(meis2b-reporter)], which recapitulates the embryonic and adult meis2b expression as assessed by in situ hybridization (Figure 1i–i’, Figure 1—figure Supplement 1a–f). Notably, Tg(meis2b-reporter) expression is restricted to the left compartment of the adult zebrafish atrium, where it is expressed in the myocardial wall and trabecular myocardium but not expressed in other cardiac cells (Figure 1j–j’).
Left atrial compartments share gene expression programs in fish and mammals
To better understand the myocardial compartmentalization across the atrial left-right axis in the meis2b reporter line, we compiled an expression profile of the Tg(meis2b-reporter)-positive and -negative domains. We dissected the respective domains in individual hearts (n = 6), isolated RNA and identified the differentially expressed genes (Figure 2a). Interestingly, we found a number of genes whose expression showed significant enrichment in the Tg(meis2b-reporter)-positive domain (Figure 2b). Notably, besides meis2b itself, the five most significantly enriched genes were the Pentraxin-related protein gene ptx3a, the mitochondrial creatine kinase encoding gene ckmt2a, a muscle-specific heat-shock protein hspb6a, a zebrafish orthologue of the Norepinephrine transporter encoding gene Slc6a2, si:ch211-117c9.5, and importantly pitx2c, a marker of left atrial identity in mammals (Figure 2b). If the meis2b-positive left atrial compartment of zebrafish shares an evolutionary origin with the left atrium of mammals, we speculated that genes with left atrial expression in zebrafish would show left atrial enrichment in mammalian hearts as well. We were able to identify direct mammalian orthologues of the five most significantly enriched zebrafish genes and investigated their expression in a previously published expression dataset for atrial asymmetry in mice (Kahr et al., 2011). Indeed, we found that all five orthologues were robustly expressed in the mammalian heart and showed strong enrichment within the group of genes with significantly higher expression in the left atrium of mouse hearts (Figure 2c). To validate these results, we isolated RNA from mouse left and right atria and, by performing RT-qPCR analysis, found that the expression of Pitx2c, Slc6a2, Ckmt2 and Hspb6 was significantly enriched in the left atrium (Figure 2d–h). Notably, we further found that in Xenopus tropicalis, the expression of Pitx2c was also significantly enriched in the left atrium compared to the right one (data not shown). As Pitx2c serves as a marker of left atrial identity in mice, we asked whether pitx2c and meis2b share a common expression domain within the zebrafish atrium. Hence, we performed fluorescent in situ hybridization experiments for pitx2c expression in the meis2b reporter line and found that meis2b and pitx2c expression largely overlap in the adult zebrafish atrium and are restricted to the left atrial compartment (Figure 2i–i’’’).
Figure 2. The zebrafish atrium is divided into two transcriptionally distinct domains.
(a) Schematic illustrating the dissection of a Tg(meis2b-reporter) atrium into Tg(meis2b-reporter)-positive and -negative domains for RNAseq analysis. (b) Global heatmap depicting Z-score of differentially expressed genes between Tg(meis2b-reporter)-positive and -negative domains (m2b-pos and m2b-neg, respectively). Red color indicates higher expression and blue color lower expression in the respective sample listed at the bottom of each column. (c) Schematic representation of asymmetrically expressed transcripts in mouse atria as determined by microarray analysis (Kahr et al., 2011); only significantly differentially expressed genes included (3695/25697 probes). Green: significantly left atrial enriched transcripts; grey: significantly right atrial enriched transcripts. (d–h) RT-qPCR analysis of left vs. right atria in adult mouse shows that the orthologues of the most significantly enriched genes in the Tg(meis2b-reporter)-positive domain are also significantly enriched in the left atrium in mouse. Plots show relative gene expression in left atrium with respect to right atrium (n = 5). (i–i’’’) Wholemount fluorescent in situ hybridization (RNAscope) for pitx2c expression on a Tg(meis2b-reporter) zebrafish atrium [DAPI in blue, Tg(meis2b-reporter) in white, pitx2c in red]. (b–c, e) si:ch211-117c9.5 is a zebrafish orthologue of Slc6a2. (i–i’’) Red and white dotted lines delineate the atrium; (i’) yellow line delineates Tg(meis2b-reporter) expression. (d–h) Two-tailed student t-tests were performed, *p<0.05; **p<0.005; ***p<0.0005; error bars indicate ±SD. Scale bars: (i) 100 µm, (i’’’) 25 µm.
Atrial laterality appears to derive from two distinct antero-posterior progenitor fields
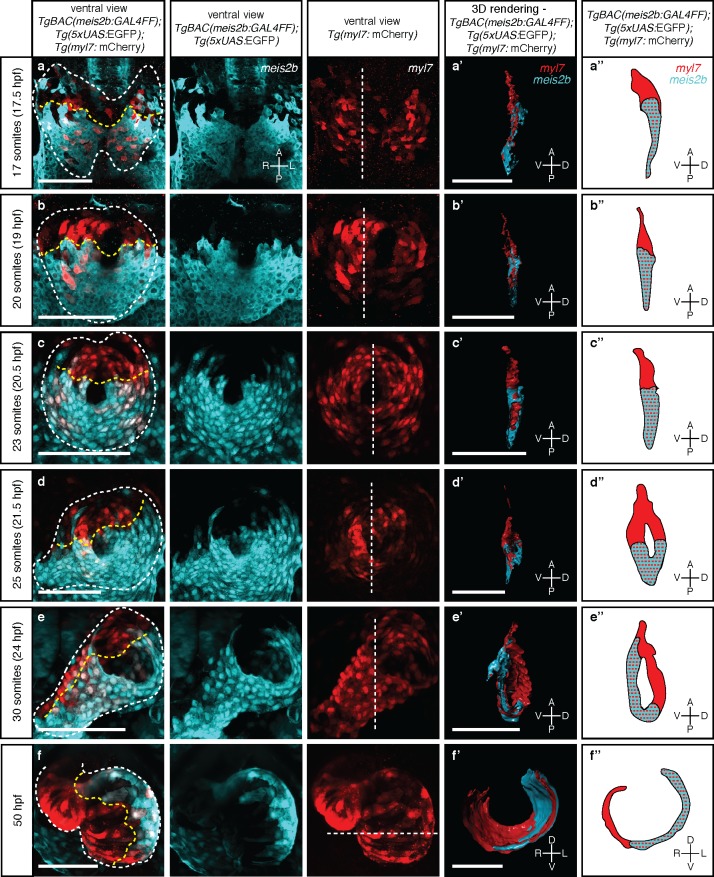
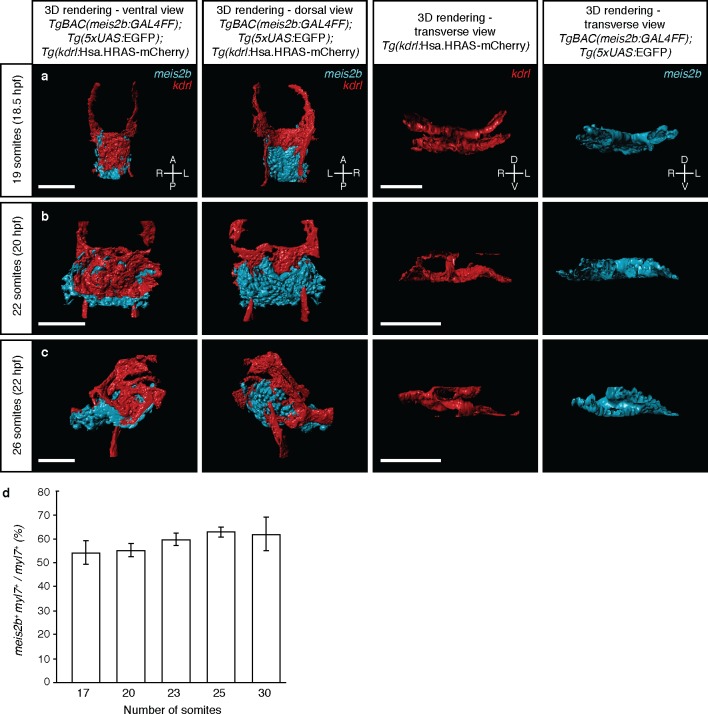
Little is known about how atrial laterality is established during development. To determine the developmental origin of the observed asymmetric meis2b expression, we first performed in situ hybridization studies. We observed that meis2b expression appears bilaterally at the 12 somite stage (ss) within the cardiogenic anterior lateral plate mesoderm (ALPM) but is restricted to its posterior aspect (Figure 3—figure supplement 1) (Paige et al., 2012). The cells of the bilateral ALPM collectively migrate towards the midline where they fuse to form the cardiac disc (Stainier et al., 1993; Trinh and Stainier, 2004). Consistently, Tg(meis2b-reporter) expression was observed only within the posterior aspect of the cardiogenic ALPM (Figure 3a, Figure 3—figure supplement 1j). Migration to the midline and fusion of the bilateral fields lead to the formation of the cardiac disc which is strikingly asymmetric based on gene expression. At the 20 to 23 ss, once the cardiac disc is formed, Tg(meis2b-reporter)-positive cells form a posterior disc compartment (PDC) establishing a sharp boundary with the anterior part of the disc (ADC) which remains Tg(meis2b-reporter) negative (Figure 3b–c). At this stage, the endocardial progenitors are located ventral to the myocardium, and pass through the central ring of the cardiac disc to connect dorsally to the aortic arches (Stainier et al., 1993; Bussmann et al., 2007) (Figure 3—figure supplement 2; Video 1). Notably, the ADC and PDC are rearranged into a dorso-ventral configuration as the heart tube starts to extend (Smith et al., 2008; Rohr et al., 2008), with the Tg(meis2b-reporter)-positive cells located on the ventral side of the tube (Figure 3d–e) (Figure 3—videos 1–5). Furthermore, we observed that the Tg(meis2b-reporter)-positive PDC comprised 54% to 62% of all cardiomyocytes from 17 to 30 ss (Figure 3a, Figure 3—figure Supplement 2d).
Figure 3. Myocardial morphogenesis establishes left-right laterality from two distinct antero-posterior fields.
Confocal projections of Tg(meis2b-reporter);Tg(myl7:mCherry) embryos between the 17 ss (16 hpf) and 50 hpf. (a-c) Myocardial progenitors migrate to the midline and form the cardiac disc, where Tg(meis2b-reporter) expression is confined to the posterior compartment of the cardiac disc (PDC). (d) Cardiomyocytes forming the cardiac disc appear to migrate clock-wise, rotating the forming heart tube. (e) Tg(meis2b-reporter)-expressing cells are located on the ventral side of the heart tube. (f) At 50 hpf, Tg(meis2b-reporter) expression appears on the left side of the atrium, and in a small part of the ventricle near the AV canal. (a’-f’) 3D rendered sections (as indicated in the third column, white dotted lines) of the hearts shown in a-f (see Figure 3—videos 1–6 for full reconstructions). (a’’-f’’) Schematic representation of Tg(meis2b-reporter);Tg(myl7:mCherry) expression from (a’-f’). White dotted lines delineate the heart, yellow dotted lines delineate Tg(meis2b-reporter) expression. White dotted lines in third column indicate the level of the sagittal and transverse views. First to third columns: ventral views, anterior up; (a’-e’, a’’-e’’) sagittal views, anterior up; (f’, f’’) transverse views, dorsal up. Scale bars: (a-e’) 100 µm, (f’) 50 µm.
Figure 3—figure supplement 1. meis2b is expressed in the cardiogenic anterior lateral plate mesoderm.

Figure 3—figure supplement 2. Tg(meis2b-reporter) expression with respect to the endothelium during the cardiac disc and heart tube stages.
Figure 3—video 1. 3D surface rendering of Tg(meis2b-reporter);Tg(myl7:mCherry) expression at 17 somite stage from Figure 3a.
Figure 3—video 2. 3D surface rendering of Tg(meis2b-reporter);Tg(myl7:mCherry) expression at 20 somite stage from Figure 3b.
Figure 3—video 3. 3D surface rendering of Tg(meis2b-reporter);Tg(myl7:mCherry) expression at 23 somite stage from Figure 3c.
Figure 3—video 4. 3D surface rendering of Tg(meis2b-reporter);Tg(myl7:mCherry) expression at 25 somite stage from Figure 3d.
Figure 3—video 5. 3D surface rendering of Tg(meis2b-reporter);Tg(myl7:mCherry) expression at 30 somite stage from Figure 3e.
Figure 3—video 6. 3D surface rendering of Tg(meis2b-reporter);Tg(myl7:mCherry) expression at 50 hpf from Figure 3f.
Video 1. 3D surface rendering of Tg(meis2b-reporter);Tg(kdrl:Hsa.HRAS-mCherry) expression at 22 somite stage from Figure 3—figure supplement 2a.
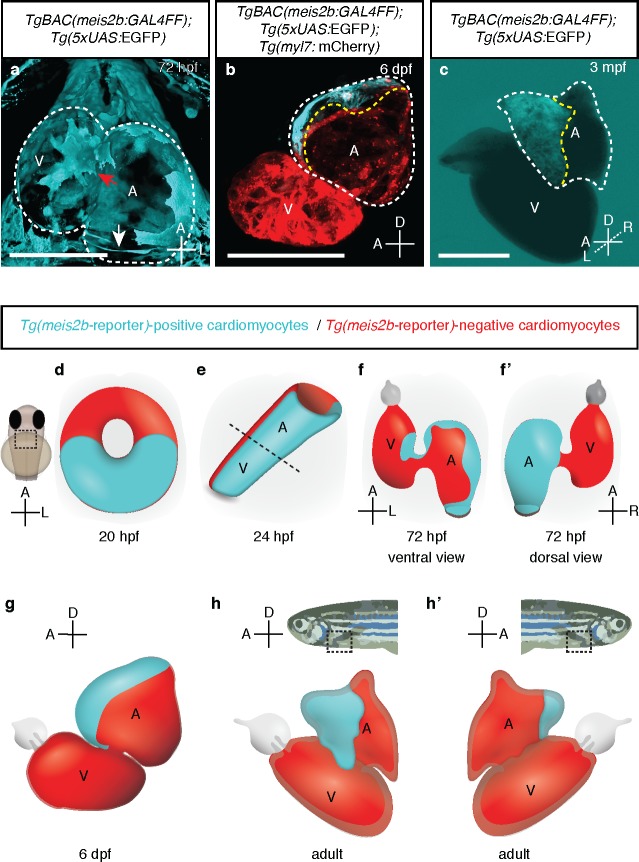
At 50 and 72 hr post fertilization (hpf), after cardiac looping and chamber ballooning have occurred, the Tg(meis2b-reporter)-positive cells reside in the dorsal part of the atrium, parts of the sinus venosus, and a small part of the ventricle near the atrio-ventricular canal (AVC) (Figure 3f; Figure 4a,f–f`; Figure 3—video 6, Figure 4—video 1). While Tg(meis2b-reporter) expression is switched off in the ventricle between 4 and 6 days post-fertilization (dpf), Tg(meis2b-reporter)-positive cells populate the left atrial compartment at this stage (Figure 4—video 2). As atrial meis2b expression appears to be stable during development, it reveals a persistent left-right asymmetry in the adult atrium (Figure 4b–c,g–h’). Together, these findings strongly support the existence in zebrafish of atrial compartmentalization from early stages and throughout life (summarized in Figure 4d–h’). To our knowledge, a molecular asymmetry in the single atrium of teleosts has not been described. Altogether, our data provide evidence that asymmetric patterns of gene expression existed in the atrium of fish hearts prior to the evolutionary emergence of the inter atrial septum.
Figure 4. meis2b expression unveils an atrial asymmetry throughout development.
(a) At 72 hpf, Tg(meis2b-reporter) expression is observed near the sinus venosus (white arrow), the dorsal and distal sides of the atrium, and a small part of the ventricle near the atrioventricular canal (red arrow) (n = 8) (Figure 4—video 1). (b) At 6 dpf Tg(meis2b-reporter) expression is found exclusively in the anterior-left compartment of the atrium at (n = 5) and in adult stages (c) in the left side of the atrium (n = 15) (Figure 4—video 2). (d-h’) Schematic of Tg(meis2b-reporter) expression throughout heart development [Tg(meis2b-reporter) expression in cardiomyocytes (cyan); meis2b-negative myocardium (red); (e) dotted line indicates the atrioventricular boundary]. (a, d–f) ventral views, anterior up; (b–c, g–h) lateral views, anterior to the left; (f’) dorsal view, anterior up; (h’) lateral view, anterior to the right. A, atrium; V, ventricle. (b–c) white dotted lines delineate the heart, yellow dotted lines delineate Tg(meis2b-reporter) expression. Scale bars: (a–b) 100 µm, (c) 500 µm.
Figure 4—video 1. 3D rendering of a 72 hpf Tg(meis2b-reporter) heart from Figure 4a.
Figure 4—video 2. 3D surface rendering of Tg(meis2b-reporter);Tg(myl7:mCherry) expression at 6 dpf from Figure 4b.
Atrial asymmetry is established independently of SHF contribution
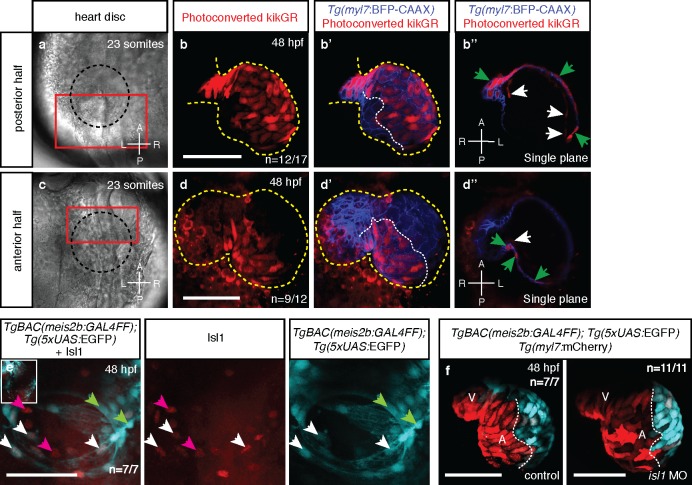
The establishment of cardiac laterality from two distinct antero-posterior fields has not been described before. To confirm our observations, we lineage traced cells within the cardiac disc by fluorescent labeling and confocal imaging. We injected one-cell stage zebrafish embryos with mRNA encoding KikGR, a photoconvertible green-to-red fluorescent protein, leading to green KikGR expression throughout the developing embryo. After locating the cardiac disc at the 23 ss, photoconversion of the PDC was performed using a confocal laser scanning microscope. Subsequent tracing of the photoconverted red cells revealed their exclusive contribution to the left side of the atrium at 48 hpf, similar to what we observed in the Tg(meis2b-reporter) line (Figure 5a–b’’). Conversely, cells of the ADC contributed to the right side of the atrium at 48 hpf (Figure 5c–d’’; Figure 5—videos 1 and 2). Together, these results reveal a process that establishes atrial left-right asymmetry from an antero-posterior embryonic asymmetry.
Figure 5. Atrial laterality is established from distinct antero-posterior fields without contribution from the second heart field.
(a, c) Cardiac disc (dotted circle) at 23 ss. (b–b’) Confocal projections of photoconverted kikGR with Tg(myl7:BFP-CAAX) hearts (yellow lines) show that cells from the PDC populate the left side of the atrium while (d–d’) cells from the ADC populate the right side of the atrium (white dotted lines). (b’’, d’’) Single optical planes from b’ and d’, respectively (cardiomyocytes, green arrows; endocardial cells, white arrows). (e) Tg(meis2b-reporter) expression and antibody staining for Isl1 in the 48 hpf heart [meis2b+/Isl1+ (white arrows), meis2b+/Isl1- (green arrows) and meis2b-/Isl1+ (pink arrows) cardiomyocytes]. (f) Confocal projections of control uninjected and isl1 MO injected 48 hpf Tg(meis2b-reporter);Tg(myl7:mCherry) hearts [white lines delineate Tg(meis2b-reporter) expression]. Scale bars: 100 µm.
Figure 5—figure supplement 1. isl1 morphants have reduced levels of Isl1 protein and phenocopy isl1 mutants.
Figure 5—video 1. 3D surface rendering of photoconverted kikGR with Tg(myl7:BFP-CAAX) expression at 48 hpf from Figure 5b’.
Figure 5—video 2. 3D surface rendering of photoconverted kikGR with Tg(myl7:BFP-CAAX) expression at 48 hpf from Figure 5d’.
Figure 5—video 3. 3D rendering of a 48 hpf Tg(meis2b-reporter);Tg(myl7:mCherry) heart from Figure 5f, control.
Figure 5—video 4. 3D rendering of a 48 hpf Tg(meis2b-reporter);Tg(myl7:mCherry) heart from Figure 5f, isl1 morphant.
The majority of cells that form the early heart tube are derived from FHF progenitors (Liu and Stainier, 2012). Subsequently, the SHF contributes cells at its cranial and caudal boundaries (Guner-Ataman et al., 2013; de Pater et al., 2009; Hami et al., 2011). As Tg(meis2b-reporter) is expressed in a subset of the atrial myocardium at 48 hpf, we tested whether the Tg(meis2b-reporter)-negative atrial myocardium was derived from the SHF. Using immunostainings for the established venous SHF marker Isl1, we tested for a potential inverse correlation between the Tg(meis2b-reporter) expression and Isl1-positive cells (Figure 5e). As previously reported (Witzel et al., 2012), we detected Isl1 expression at the venous pole of the atrium where it is essential for SHF-derived cardiomyocyte differentiation and contribution to the heart tube. However, Isl1 expression did not correlate with Tg(meis2b-reporter) expression in the heart. We could detect Isl1 in Tg(meis2b-reporter)-positive and -negative cells (Figure 5e), suggesting that asymmetric meis2b expression is not exclusively associated with the FHF or SHF. However, if a substantial contribution of the SHF was involved in the establishment of a meis2b-negative population in the atrium, a knockdown of isl1 should reduce it significantly. To test this hypothesis, we injected a previously published isl1 morpholino into the Tg(meis2b-reporter) line (Witzel et al., 2012). Interestingly, while the injection of isl1 morpholino resulted in a significant decrease in Isl1 immunostaining, and did not show any obvious toxic effects, the asymmetric expression of meis2b within the heart was unaffected at 48 hpf (Figure 5f, Figure 5—figure Supplement 1; Figure 5—videos 3 and 4), indicating that SHF contribution is not required to establish atrial asymmetry. These findings are consistent with our current understanding on the establishment of atrial asymmetry in mammals: in the mammalian heart, while the SHF is important for right ventricular formation, it contributes equally to both atrial chambers and thus is not essential to establish right atrial identity.
Loss of meis2b causes dysmorphic atrial growth and conduction defects
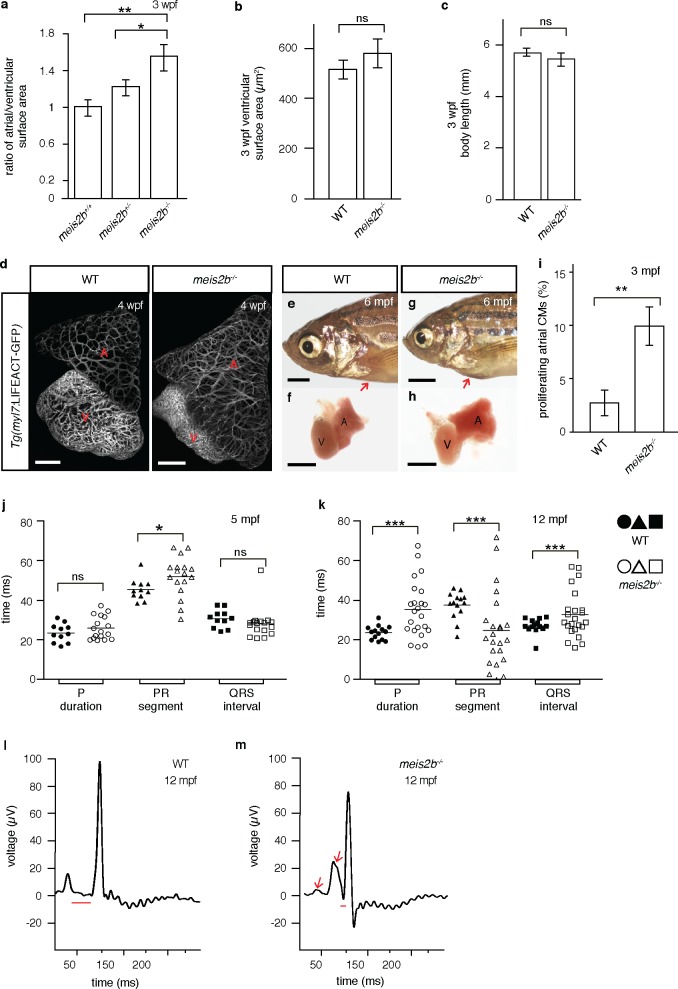
In mammals, atrial asymmetry is important for cardiac septation, morphogenesis and conduction (Koshiba-Takeuchi et al., 2009; Tessari et al., 2008; Wang et al., 2010; Campione and Franco, 2016). To determine the role of Meis2b in compartmentalization of the teleost atrium, we induced a frameshift mutation in the highly conserved Hth/Meis domain (Bürglin, 1997; Longobardi et al., 2014) of meis2b, predicted to lead to a truncated protein lacking most functional domains, including the DNA-binding homeodomain (Figure 6—figure supplement 1). In contrast to previously published knockdown experiments (Paige et al., 2012), no early cardiac morphogenetic defect was observed in meis2b-/-, and cardiac jogging did not appear to be affected (Figure 6—figure supplement 2). However, as early as 3 weeks post-fertilization (wpf), the ratio of atrial to ventricular surface areas was significantly increased in meis2b-/- animals, while body length and ventricular size were unaffected (Figure 6a–c). Using confocal imaging in the Tg(myl7:LIFEACT-GFP) background (Reischauer et al., 2014), we assessed atrial myocardial specification and overall myofibril organization. We did not detect architectural defects other than the previously noted overgrowth of the atrium (Figure 6d), which results in a dysmorphic atrium and, in some animals, in pericardial bulging (Figure 6e–h). As previous reports have suggested an important role for other Meis proteins in the regulation of the myocardial cell cycle (Mahmoud et al., 2013), we used EdU incorporation assays in the pan myocardial Tg(−5.1myl7:nDsRed2) background to test whether the observed phenotype in meis2b-/- was a consequence of atrial hypertrophy or increased myocardial proliferation. Strikingly, meis2b-/- animals showed 10.02% (±1.12) of all atrial cardiomyocytes positive for EdU incorporation after a 4-day exposure in contrast to 2.78% (±0.49) in wild-type (WT) animals, suggesting a major contribution of proliferation to the abnormal atrial growth in meis2b-/- animals (Figure 6i).
Figure 6. Loss of meis2b causes dysmorphic atrial growth and conduction defects.
(a) meis2b-/- larvae exhibit a significant increase in the ratio of atrial to ventricular surface area, while (b) ventricular surface area and (c) body length are not affected compared to WT siblings. (d) Analysis of Tg(myl7:LIFEACT-GFP) fish reveals no obvious defects in atrial myofibrillar architecture in 4 wpf meis2b-/-. (e–h) Adult meis2b-/- display pericardial bulging (arrow) due to abnormal atrial growth. (i) meis2b-/- exhibit a significant increase in atrial cardiomyocyte (CM) proliferation at 3 mpf. (j–k) Cardiac physiology assessed using electrocardiography (ECG) at 5 and 12 mpf. (j) meis2b-/- exhibit a significant increase in PR-segment variability compared to WT siblings, while P-duration and ventricular conduction (QRS) appear unaffected at 5 mpf. (k) At 12 mpf, PR-segment, P-duration and QRS interval are significantly affected in meis2b-/-. (l–m) Representative ECGs of 12 mpf animals show reduced PR-segment duration (red line) and multiple P-waves (arrow) in meis2b-/-. Scale bars: (d) 100 µm, (e–h) 1 mm. (a–c, i) One-tailed student t-tests were performed. (j–k) F-test and Bartlett’s test were performed. Significant differences compared with control are indicated (*p<0.05; **p<0.005; ***p<0.0005); error bars indicate ± SD. A, atrium; V, ventricle; ns, not significant.
Figure 6—figure supplement 1. Generation of a meis2b mutant allele by TALEN mutagenesis.
Figure 6—figure supplement 2. Cardiac positioning (jogging) appears unaffected in meis2b mutants.
We next determined how loss of Meis2b affects cardiac physiology by performing electrocardiographic (ECG) analysis and observed that adult meis2b-/- frequently display cardiac conduction defects (Figure 6j–k). In 5 mpf animals, we found a significantly increased variability in P-R segment duration while P wave duration and ventricular conduction (QRS) appeared unaffected, suggesting a functional defect of the atrial and atrio-ventricular conduction systems in the absence of Meis2b (Figure 6j). In 12 mpf fish, these defects became more severe and we observed high variability in P wave duration, extremely long or short P-R segments and, in a few cases, even prolonged ventricular conduction (Figure 6k). Most frequently, shortened P-R segments and broad, biphasic P waves (Figure 6l–m) were observed (Weinsaft et al., 2014). Taken together, these data suggest that meis2b controls atrial morphogenetic growth and directly or indirectly the patterning and function of the atrial conduction system.
Meis2b positively regulates cardiac pitx2c expression
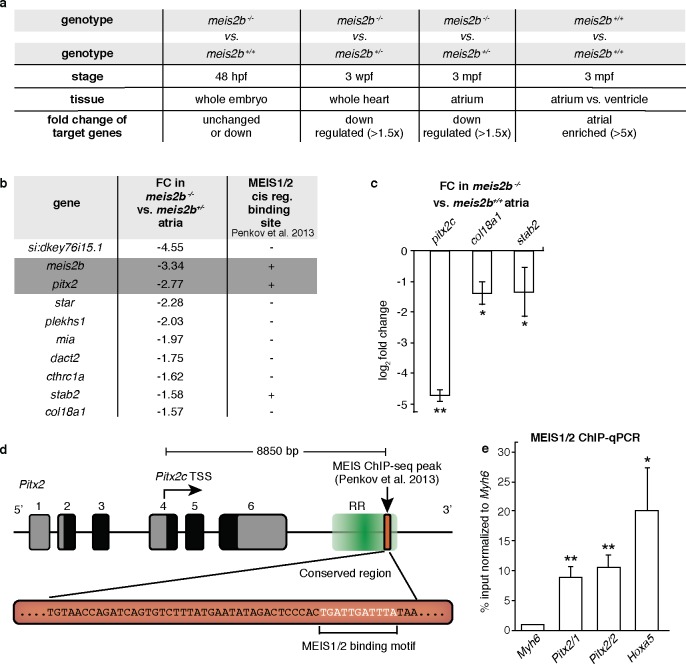
Meis proteins evolved from their invertebrate ancestor Homothorax which acts together with Extradenticle (Pbx) and a variety of Hox proteins to regulate the transcription of target genes (Merabet and Mann, 2016). In mammals, several splice variants of MEIS2 have been described including isoforms that lack a C-terminal transactivation domain (TAD), and thus can act as transcriptional repressors (Hyman-Walsh et al., 2010). To test whether the zebrafish paralogs meis2a and meis2b encode transcriptional activators or repressors, we tested their transactivation potential in cultured cells. Using PCR amplification on adult cardiac cDNA, we were able to isolate two different splice isoforms for meis2a and one for meis2b. Fusions of the C-terminus of all three cardiac meis2 isoforms to a GAL4-DNA Binding Domain conferred strong transactivation potential (Figure 7—figure supplement 1), suggesting that all cardiac Meis2b isoforms function as positive regulators of transcription. To identify potential downstream targets, we next performed expression profiling of whole larvae, entire hearts and dissected atrial chambers of meis2b-/- and meis2b+/- siblings at different developmental stages (Figure 7a), resulting in a list of 10 atrial-enriched genes with reduced expression in cardiac tissues of meis2b-/- animals at all stages when compared to meis2b+/- siblings. Notably, for three of these genes, proximal binding sites for MEIS1/2 have been identified in previous ChIP-seq experiments performed on E11.5 mouse embryos marking them as potential direct targets (Figure 7b) (Penkov et al., 2013). Notably, the list of genes with reduced expression included pitx2c, a key gene of left atrial identity in mammals. Subsequent testing using RT–qPCR confirmed a strong 4.7 (±0.18) log2 fold reduction of pitx2c transcripts in adult meis2b mutant atria compared to WT siblings (Figure 7c).
Figure 7. Meis2b positively regulates cardiac pitx2c expression.
(a) Overview of the different samples used for microarray comparison, showing the stage, type of tissue and genotype of the compared samples. The bottom row shows the expected outcome of downstream genes potentially regulated by Meis2b. (b) Table showing the 10 genes fitting all selection criteria (for genes with n > 1, the values are averaged). Cis-regulatory MEIS1/2 DNA binding sites in orthologous mouse genes were extracted from (Penkov et al., 2013). (+, proximal binding site detected; -, no proximal binding site detected)]. (c) RT-qPCR analysis in meis2b-/- vs WT sibling atria. (d) Representation of the Pitx2 locus in mouse indicating the location of a previously reported MEIS1/2 ChIP-seq peak at the 3’ end of the regulatory region (RR) (ENSMUSR00000406450) (Penkov et al., 2013). A 53 bp conserved sequence inside the ChIP-seq peak is shown together with the MEIS1/2 binding motif. (e) ChIP-qPCR analysis on E12 embryonic mouse trunks of MEIS1/2 on the Pitx2 locus shown in (d), on the Hoxa5 locus (a known MEIS1/2 direct target), and on Myh6 locus (used as a negative control). One-tailed student t-tests (c), and one-sample t-tests (e) were performed, significant differences compared with control are indicated (*p<0.05; **p<0.005); error bars indicate ±SD.
Figure 7—figure supplement 1. Transactivation potential of Meis2a/b C-terminal domains.
Figure 7—figure supplement 2. Conserved element inside ChIP-seq peak.
Penkov et al. (2013) performed a ChIP-seq experiment using antibodies that recognize MEIS1 and MEIS2 isoforms. They found in addition to genes in the Hoxa cluster, a region annotated as regulatory (Ensembl, ENSMUSR00000406450), located downstream of the Pitx2 locus with the closest transcriptional start site belonging to Pitx2c. Interestingly, this 187 bp ChIP-seq peak contains a highly conserved 53 bp long element, which includes the MEIS1/2 binding motif (Figure 7d, Figure 7—figure Supplement 2). To validate these results, we performed a ChIP-qPCR analysis on E12 embryonic mouse trunks using the antibodies employed by Penkov et al. (2013). We performed qPCR on two regions of the Pitx2 element flanking the MEIS1/2-binding motif, on the Hoxa5 locus (a known direct target of MEIS1/2 (Penkov et al., 2013)), and on Myh6 (a region where no MEIS1/2 binding site is present). Interestingly, we observed that MEIS1/2 binds Pitx2 8.87 (±1.92) to 10.56 (±2.20) fold higher than Myh6, while enrichment for the MEIS1/2-binding motif within the Hoxa5 locus was 20.21 fold (±7.21) (Figure 7e). These data suggest that MEIS1/2 can directly bind the regulatory region next to Pitx2 in mouse. Taken together, our data showing the overlapping expression patterns of meis2b and pitx2 along with the reduced pitx2c expression in meis2b mutants suggest that meis2b acts upstream of pitx2c in the heart. These findings are further supported by our ChIP-qPCR data showing that MEIS1/2 directly binds the mouse Pitx2 locus in vivo as well as the similar MEIS2 and PITX2 mutant phenotypes in human and mouse.
Discussion
Mechanisms that establish cellular heterogeneity during development fulfill two principal roles. First, they increase organ complexity, allowing the emergence of specialized cellular subtypes from a single progenitor field. Second, they define tissue compartments that can serve morphogenetic or functional roles. In the developing vertebrate heart, myocardial cells from the FHF establish a tube that is rapidly compartmentalized into atrial and ventricular domains. Subsequently, a spatiotemporally and molecularly distinct SHF contributes additional myocardial cells to both ends of the tube. At the arterial pole, these cells form the right ventricle, and a septum is formed at the interface between the ventricular descendants of the FHF and SHF (Buckingham et al., 2005; Lin et al., 2012a). Mutations affecting genes that regulate SHF integration consequently impair right ventricular development and can cause septal defects (Koshiba-Takeuchi et al., 2009; Bruneau, 2008; Kelly et al., 2014). In the venous pole, the second heart field contributes cells to both atria, including functionally important subtypes of cardiomyocytes such as the pacemaker cells and cells that contribute to the atrial septum. The identification of a SHF in zebrafish contributing myocardium to both the ventricle and atrium (Guner-Ataman et al., 2013; de Pater et al., 2009; Hami et al., 2011), was possibly unexpected given the single chamber configuration of its ventricle and the absence of a pulmonary circuit. However, its presence in teleosts suggests that the two-chambered ventricle of amniotes evolved from a preexisting pattern rather than the addition of novel structures. We now report the identification of atrial compartmentalization in the teleost species zebrafish. Moreover, our data suggest that the left atrial compartment in zebrafish shares an evolutionarily conserved origin with the left atrium of tetrapods including the left-specific expression of several genes. Amongst the most significantly left atrial expressed genes in the zebrafish atrium are the transcription factor genes meis2b and pitx2c, which show overlapping expression patterns in the myocardium of the mature zebrafish heart. Pitx2c is a well-established marker of the left atrium in amniotes and a key determinant of left atrial identity (Tessari et al., 2008; Campione et al., 2001). Furthermore, PITX2C mutations have been associated with atrial septation and conduction defects in mammals (Franco et al., 2014; Mammi et al., 1998; Gudbjartsson et al., 2007). meis2b, a transcription factor gene of the TALE family is one of two highly conserved zebrafish orthologues of MEIS2, a gene that has also been strongly linked to congenital septation defects in humans (Louw et al., 2015). Canonically, embryonic left-right development is regulated through the well studied nodal signaling pathway (Bakkers et al., 2009). Using a transgenic reporter line for meis2b expression, we were able to track the establishment of atrial asymmetry in zebrafish back to early embryonic stages. Surprisingly, in contrast to components of the nodal pathway, meis2b is expressed bilaterally and restricted to the posterior domain of the cardiogenic ALPM. Using in vivo confocal microscopy, we observed that this posteriorly restricted expression of meis2b leads to the establishment of two compartments within the cardiac disc, a meis2b-positive PDC and an anterior, meis2b-negative ADC. Subsequently, a previously reported nodal dependent rotation of the cardiac disc through cell migration breaks lateral symmetry (Bakkers et al., 2009), leading to the observed left-right pattern. Using an isl1 knockdown strategy, we further found that the restricted expression pattern of meis2b does not depend upon the integration of cells from the SHF. Meis proteins typically form trimers with their interaction partners, Pre-B-cell leukemia transcription factor Pbx as well as different Hox Proteins, to bind DNA and regulate gene expression (Merabet and Mann, 2016). This trimeric mode of action allows highly complex target gene regulation through spatially overlapping transcriptional fields. An important aspect of future studies will be to identify the specific elements that regulate meis2b expression and to identify further transcriptional compartments within the cardiac mesoderm to better understand the mechanisms that control cell diversification.
To investigate the role of meis2b in cardiac development and asymmetry, we generated a meis2b allele predicted to encode a non-functional protein (Figure 6—figure supplement 1) (Longobardi et al., 2014). Surprisingly, zebrafish meis2b homozygous mutants did not exhibit the severe cardiac defects previously reported in a Meis2b knockdown study using splice and translation blocking morpholinos (Paige et al., 2012). Notably, maternal zygotic meis2b mutants did not exhibit obvious developmental defects either (data not shown). However, meis2b homozygous mutants develop atrial overgrowth as well as conduction defects, and these phenotypes become more severe with age. These results suggest that zebrafish meis2b does not regulate cardiac looping but atrial growth and conduction.
In mouse, the meis2b homologue Meis1 is important for cardiomyocyte cell cycle regulation, whereby lack of Meis1 function is sufficient to extend the cardiomyocyte proliferation window in neonatal hearts and induce cardiomyocyte mitosis in adult hearts (Mahmoud et al., 2013). Similarly, lack of meis2b function in zebrafish could lead to the increase in myocardial proliferation observed in the mutant atria. It will be interesting to further investigate this phenotype including the cell-autonomy of meis2b in regulating cardiomyocyte proliferation.
Intriguingly, pitx2c, the evolutionary conserved marker for left atrial identity is strongly downregulated in meis2b mutants, suggesting that meis2b functions upstream of myocardial pitx2c expression in zebrafish. Furthermore, we have shown that mouse MEIS1/2 directly binds a conserved regulatory region proximal to the Pitx2 locus, suggesting a direct regulation of myocardial Pitx2 expression by MEIS2. These data are particularly interesting, as mutations affecting PITX2 function have been linked to conduction and septation defects in humans (Mammi et al., 1998; Gudbjartsson et al., 2007), similar to what has been reported for cases of MEIS2 haploinsufficiency (Louw et al., 2015). Altogether, our findings create new opportunities for the study of atrial compartmentalization and myocardial heterogeneity in zebrafish. It is likely that further investigation of the intrinsic and extrinsic signals that regulate antero-posterior patterning of the cardiac disc will lead to additional insights into the control of cardiac mesoderm diversification and patterning as well as the establishment of atrial asymmetry.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers |
|---|---|---|---|
| Strain (Danio rerio) | Tg(5xUAS:EGFP)nkuasgfp1a | doi: 10.1073/pnas0.0704963105 | ZFIN_ZDB-GENO-080528–1 |
| Strain (Danio rerio) | Tg(myl7:mCherry)chb1 | doi: 10.1016/j.ydbio.2011.12.004 | ZFIN_ZDB-GENO-110720–1 |
| Strain (Danio rerio) | meis2bs988 | This paper | ZFIN: ZDB-ALT-180327-7 |
| Strain (Danio rerio) | TgBAC(meis2b:Gal4FF)bns15 | This paper | ZFIN: ZDB-ALT-180330-1 |
| Strain (Danio rerio) | Tg(myl7:BFP-CAAX)bns193 | This paper | ZFIN: ZDB-ALT-180327-8 |
| Antibody | anti-Fli1 Rabbit Polyclonal | Abcam, UK. | ab133485; RRID: AB_2722650 |
| Antibody | anti-MF20 Mouse Monoclonal | Developmental Studies Hybridoma Bank, US. | MF20-s; RRID: AB_2147781 |
| Antibody | Anti-MEIS1/2 | doi: 10.1016/j.celrep.2013.03.029 | Produced at CNIC. K830; K844 |
| Sequence-based reagent | isl1 ATG morpholino | doi: 10.1242/dev.02355; Gene Tools, LLC, US. | ZFIN_ZDB-MRPHLNO-060728–3 |
| Commercial assay or kit | RNAscope | Advanced Cell Diagnostics, US. | Multiplex Fluorescent kit |
| Software | ZEN Black and Blue 2012 | Zeiss, Germany. | |
| Software | NIS-Elements | Nikon Instruments Inc. | |
| Software | Imaris - Version 8.4.0. | Bitplane, UK. | |
| Software | LabChart 8 | ADInstruments, Australia. | |
| Software | PerlPrimer - Version 7 | doi: 10.1093/bioinformatics/bth254 | |
| Other | Dynabeads Protein G | Invitrogen, Germany. | 10003D |
Zebrafish husbandry and strains
All zebrafish husbandry was performed under standard conditions in accordance with institutional (MPG) and national ethical and animal welfare guidelines. The following transgenic and mutant lines were used in this study: Tg(myl7:LIFEACT-GFP)s974 (Reischauer et al., 2014), Tg(−5.1myl7:nDsRed2)f2 (Mably et al., 2003), Tg(5xUAS:EGFP)nkuasgfp1a (Asakawa et al., 2008), Tg(myl7:mCherry)chb1 (Langenbacher et al., 2012), Tg(myl7:EGFP-Hsa.HRAS)s883 (D'Amico et al., 2007), Tg(kdrl:Hsa.HRAS-mCherry)s896 (Chi et al., 2008), and isl1sa0029 (de Pater et al., 2009).
Generation of transgenic and mutant lines
TALEN technology was used to target meis2b. TALEN-binding sites were selected using TALEN Targeter 2.0 (tale-nt.cac.cornell.edu) for exon 4 of meis2b (NM_130910.1), and constructs were assembled as previously described (Cermak et al., 2011; Bedell et al., 2012) to bind the target sequences 5’-TTGAGAAGTGCGAGCT-3’ and 5’-AGAGAGCCGGGAGTCGCTGGA-3’ for the right and left arms, respectively. mRNA encoding the TALEN arms was synthesized using the mMESSAGE mMachine Kit (Ambion), and 100 pg per TALEN arm were injected into one-cell stage embryos to generate potential founders (F0). To assess the efficacy of the TALEN injections, DNA was extracted from injected embryos at 48 hr post fertilization (hpf) and subjected to high-resolution melt analysis (HRMA, Illumina Eco Real-Time PCR System). F0 founders were identified by outcrossing to wild-type (WT) fish. Genomic DNA was extracted from individual F1 embryos and analyzed for induced genomic mutations by PCR amplification of the TALEN target region and subsequent HRMA (Supplementary file 1a). Using the genomic DNA of F1 fin clipped adult fish as a template, PCR products were obtained with the primers shown in Supplementary file 1b and sequenced in order to determine the specific alleles generated. An allele containing a 10 bp deletion with a 3 bp insertion leading to a frameshift mutation and a premature stop codon was identified and designated meis2bs988. Genotyping of meis2bs988 allele was carried out by PCR using allele specific primer combinations (Supplementary file 1c-d).
The TgBAC(meis2b:Gal4FF)bns15 line was generated by rapid modification of a Bacterial Artificial Chromosome (BAC) clone using a recombination-based protocol (Bussmann and Schulte-Merker, 2011). The modified clone (CH211-128L12) contains ~139 kb of genomic sequence upstream of the meis2b start codon and ~20 kb downstream its stop codon. Briefly, after Tol2 transposon LTR insertion into the vector backbone, the recombination was performed by amplifying a Gal4FF-Kan(R) cassette using primers with 50 bp of homology flanking the start codon of GAL4FF (Supplementary file 1e). Transgenesis was performed in the Tg(5xUAS:EGFP) background, following a previously described protocol (Kawakami et al., 2004).
Generation of Tg(myl7:BFP-CAAX)bns193: Blue fluorescent protein (BFP) coding sequence was fused to a CAAX membrane targeting motif (Lin et al., 2012b) using Cold Fusion technology (System Biosciences, CA) and cloned under the control of the myl7 promoter (Reischauer et al., 2014) to obtain myl7:BFP-CAAX. 20 pg of the resulting plasmid along with 15 pg Tol2 mRNA were injected in one-cell stage embryos to generate Tg(myl7:BFP-CAAX)bns193.
RNA in situ hybridization
RNA in situ hybridization experiments on embryos were performed as described (Thisse and Thisse, 2008). For whole-mount adult hearts, in situ hybridization was performed with the following modifications: permeabilization of the tissue was achieved by incubating the hearts in proteinase-K (10 µg/mL) for 30 min on ice and then for 40 min at 37°C with gentle agitation.
In situ hybridization on 12-µm-thick paraffin sections of adult hearts: briefly, after rehydration, the slides were incubated with pre-hybridization solution (50% formamide, 5X SSC pH 4,5, 50 µg/mL yeast tRNA, 1% SDS, 50 µg/mL Heparin, 5X Denhardt’s solution) for 1 hr at 65°C. Subsequently, the probe was added to a final concentration of 1 µg/mL to hybridize overnight at 65°C. The next day, samples were washed with a series of SSC to MBST (100 mM Maleic acid, 150 mM NaCl, 0.1% Tween 20, pH 7.5) and blocked for 2 hr using MBST with 2% Blocking Reagent (Roche) and 5% sheep serum. Subsequently, anti-DIG-AP Fab fragments (Roche) were added and incubated overnight at 4°C. Staining was performed with BM Purple (Roche). Samples were imaged using a Zeiss Axiocam mrc5 camera or Nikon SMZ25. RNA probes were synthesized from cDNA from 52 hpf zebrafish embryos and adult zebrafish hearts using the combination of primers listed in Supplementary file 1.
RNAscope (Advanced Cell Diagnostics, Hayward, CA) was performed on 5 weeks post-fertilization (wpf) Tg(meis2b-reporter) hearts, following a published protocol (Gross-Thebing et al., 2014) for whole-mount RNAscope. Advanced Cell Diagnostics designed the pitx2c probe used in this study.
Morpholino injections
isl1 ATG morpholino from Gene Tools, LLC (5’-CCCATGTCAAGAAAGTAAGGCGGTG-3’) (Hutchinson and Eisen, 2006) was prepared at a stock concentration of 1 mM and diluted to the desired concentration for microinjection. 5 ng of the isl1 morpholino were injected into one-cell stage Tg(meis2b-reporter);Tg(myl7:mCherry), Tg(myl7:EGFP-Hsa.HRAS) and isl1 mutant embryos. Uninjected embryos and embryos injected with control morpholino served as controls. Embryos injected with control morpholino did not show any detectable phenotype. The embryos were imaged at 48 hpf using a Zeiss LSM 700 confocal microscope.
kikGR injection and photoconversion
100 pg of kikGR mRNA were injected into one-cell stage Tg(myl7:BFP-CAAX) embryos. Dechorionated embryos of 20–23 somite stage (ss) were mounted in 0.8% agarose with the dorsal side up. Using a Zeiss LSM 880 confocal microscope, the cardiac disc was identified, and the anterior or posterior half of the disc was selectively photoconverted using a 405 nm wavelength laser (7 pulses of 1 s each and 15% laser power).
After photoconversion, the embryos were transferred to egg water and raised until 48 hpf. Then, the embryos were mounted in 0.8% agarose and the hearts were imaged by confocal microscopy (Zeiss LSM 880). Each heart was imaged twice: first, the green kikGR was imaged together with the photoconverted kikGR. In a second step, Tg(myl7:BFP-CAAX) expression was visualized by excitation with a 405 nm wavelength laser. The resulting images containing the different channels were merged using ImageJ.
Embryonic and juvenile heart imaging and measurements
Offspring of a Tg(myl7:LIFEACT-GFP);meis2b+/- incross were grown until 3 and 4 wpf. The juvenile fish were fixed in Fish-Fix (0.1 M phosphate buffer, 4% PFA, 4% sucrose, 120 µM CaCl2) for 1 hr at room temperature. The hearts were dissected and individually mounted in 1% low-melt agarose. After imaging with a Zeiss LSM 780 confocal microscope, the fish were genotyped by PCR using the above-mentioned set of allele specific primers.
Zen Microscope Software (Zeiss, Black Edition 2012) was used to create maximum intensity projections. Ventricular and atrial maximal surface area was defined as the outer boundary of each compartment, as assessed by manually examining each z-stack. ImageJ was used to compute the surface area.
For imaging at early stages, embryos were mounted in 1% low-melt agarose and imaged using a Zeiss LSM 700 confocal microscope. ImageJ was used to count the cells.
For determining the heart rate at 48 hpf, the embryos were embedded in 1% low-melt agarose containing 0.004% tricaine (Chan et al., 2009). Videos of 1 min duration were acquired using a Nikon SMZ25 and the heart rate was measured.
Adult zebrafish electrocardiograms were performed as described (Orr et al., 2016). Signals were amplified using the Animal BioAmp FE136, acquired using the PowerLab 4/30, and analyzed using LabChart 8, all from ADInstruments (Sydney, Australia). 40 meis2b-/- and 25 WT siblings were tested. For each fish, a minimum of 20 heartbeats with a clearly identifiable baseline were used to analyze ECG tracings.
Image processing
Figure 5B was processed with PhotoFiltre seven for noise reduction (pixel dimension, two and threshold, 0). Figure 4B was processed using Imaris 8.4.0 for cropping areas outside of the heart and to enhance brightness. Adjustments for brightness and contrasts of other confocal projections and brightfield pictures were performed with Zen Black and Zen Blue editions (Zeiss), NIS-Elements (Nikon) and ImageJ (NIH).
Gene expression profiling
To identify chamber-specific transcripts, 12 adult zebrafish hearts (8 months post-fertilization, mpf) were isolated as described (Arnaout et al., 2014), and bisected into atria and ventricles. The expression profile of the pooled atria was compared to the expression profile of the pooled ventricles. Briefly, total RNA was isolated using TRIzol (Sigma), treated with DNase (Promega, Germany), and purified using an RNA Clean and Concentrator kit (Zymo Research). Gene expression profiles were established using dual color microarray analysis by MOgene (St. Louis, MO) using the Agilent Zebrafish V3 44K platform.
To determine the expression profiles of the Tg(meis2b-reporter)-positive vs -negative atrial compartments, a total of 6 hearts of 3 mpf Tg(meis2b-reporter) zebrafish were micro-dissected. A total of 4 pools were made: the first two pools each contained 3 Tg(meis2b-reporter)-positive atrial compartments, and the other two contained the Tg(meis2b-reporter)-negative parts of the atria. RNA was isolated using the miRNeasy micro Kit (Qiagen) combined with on-column DNase digestion (DNase-Free DNase Set, Qiagen) to avoid contamination by genomic DNA. RNA and library preparation integrity were verified with a BioAnalyzer 2100 (Agilent) or LabChip Gx Touch 24 (Perkin Elmer). 50–100 ng of RNA were used as input for Truseq Stranded mRNA Library preparation following the low sample protocol (Illumina). Sequencing was performed on a NextSeq500 instrument (Illumina) using v2 chemistry, resulting in a minimum of 27 M reads per library with a 2 × 75 bp paired-end setup. The resulting raw reads were assessed for quality, adapter content and duplication rates with FastQC. Trimmomatic version 0.33 was employed to trim the reads after a quality drop below a mean of Q18 in a window of 5 nucleotides (Bolger et al., 2014). Only reads above 30 nucleotides were cleared for further analyses. Trimmed and filtered reads were aligned versus the Ensembl Zebrafish genome version DanRer10 (GRCz10.87) using STAR 2.4.0a with the parameter ‘--outFilterMismatchNoverLmax 0.1’ to increase the maximum ratio of mismatches to mapped length to 10%. The number of reads aligning to genes was counted with featureCounts 1.4.5-p1 tool from the Subread package. Only reads mapping at least partially inside exons were admitted and aggregated per gene. Reads overlapping multiple genes or aligning to multiple regions were excluded. Differentially expressed genes were identified using DESeq2 version 1.62. Only genes with a minimum fold change of ±1.5 (log2 ±0.59), a maximum Benjamini-Hochberg corrected p-value of 0.05, and a minimum combined mean of 5 reads were deemed to be significantly differentially expressed. The Ensembl annotation was enriched with UniProt data (release 06.06.2014) based on Ensembl gene identifiers.
For the study of possible downstream targets of Meis2b, we compared the following expression profiles: 48 hpf whole larvae of meis2b-/- to meis2b+/+ siblings, whole hearts of 3 wpf meis2b-/- to meis2b+/- siblings, whole atria of 3 mpf meis2b-/- to meis2b+/- siblings, and ventricles to atria of 3 mpf WT zebrafish. Total RNA was isolated using TRIzol and purified using column-based affinity purification (RNA Clean and Concentrator kit, Zymo Research). Gene expression profiles were established using single color microarray analysis OakLabs (Hennigsdorf, Germany) using the ArrayXS Zebrafish 8 × 60K platform (OakLabs).
RT-qPCR analysis
To study gene expression in the adult mouse heart, the left and right atria of 5 adult hearts were dissected. RNA was isolated using TRIzol and cDNA was synthesized using a Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following manufacturer’s instructions. Plots show the result of five different experiments (one atrium per experiment). Primers were designed using PerlPrimer (Marshall, 2004), and Actb was used for normalization. Primer sequences are listed in Supplementary file 1. The Ct values obtained for every gene are shown in Figure 2—source data 1.
For RT-qPCR expression analysis of potential meis2b downstream targets, total RNA was isolated from the atria of 3 mpf (adult) meis2b-/- and meis2b+/+ siblings using TRIzol and cDNA was synthesized as described above. Plots show the result of three different experiments, representing three pools of six atria each. Primers were designed using PerlPrimer (Marshall, 2004), and rpl13 was used for normalization. Primer sequences are listed in Supplementary file 1. The Ct values obtained for every gene are shown in Figure 7—source data 1.
Of note, reference genes do not show different expression levels between the left and right components of the atria in fish or mice (Kahr et al., 2011).
ChIP-qPCR
ChIP experiments were performed as described (Penkov et al., 2013) with minor modifications using WT C57BL/6 E12 embryonic mouse trunks (without head, tail or legs). The protocol was started with approximately 5 × 107 cells (equivalent to three trunks). The Meis antibodies were kindly provided by the Torres lab (K830, recognizing Meis1a and Meis2a isoforms, and K844, recognizing Meis1a and Meis1b isoforms; both produced at the CNIC, Madrid, Spain).
Briefly, single-cell suspensions were prepared in cold DMEM +10% FBS by crushing embryonic mouse trunks against a 100 µm cell strainer. Cross-linking was performed with 1% formaldehyde (Thermo Fisher Scientific) for 10 min, and the reaction was quenched by addition of 125 mM glycine. The cells were harvested and washed three times with 30 mL cold PBS and finally resuspended in 2.5 mL LB1 for 20 min at 4°C. The nuclei were washed with LB2 for 10 min at 4°C, and resuspended in 3 mL of LB3. At this point, the samples were divided in half to continue the protocol with approximately 2.5 × 107 cells for each immunoprecipitation. Chromatin was sonicated three times (12 min cycles; 10 s pulse, 50 s rest) at 20% amplitude, using a Bandelin Sonopuls ultrasonic homogenizer to generate 200–1000 bp chromatin fragments.
After centrifugation and verification of the size of the sonicated chromatin, the sonicated lysates were incubated with antibody-bound Dynabeads protein G (Invitrogen). Rabbit IgG IP was used as a negative control. After overnight incubation at 4°C, three buffers were used (WB1, WB2 and WB3) and the immunoprecipitated material was washed twice at 4°C with 1 mL of each buffer. Finally, the elution of the immunoprecipitated material was done at 37°C with TE buffer +2% SDS for 30 min. After removing the magnetic beads, the eluted material was reverse cross-linked overnight at 65°C. Three volumes of TE buffer were added to dilute the SDS in the solution. After treatment with RNase-A and proteinase-K, the DNA was extracted twice with phenol-chloroform and purified with a MinElute PCR Purification Kit (Qiagen). Four biological samples were used for the ChIP. The qPCRs were done in triplicates, using 2X SYBR green real-time PCR master mix (Thermo Fisher Scientific) in a CFX Connect Real-Time PCR detection system (BioRad) with 39 cycles. Percentage input was calculated using the DDCT method. The list of primers employed for the qPCR can be found in Supplementary file 1. The Ct values obtained in the qPCR can be found in Figure 7—source data 2.
EdU injections
Eight meis2b-/- fish and 8 WT siblings at 3 mpf were anesthetized in 0.02% tricaine. EdU (Life Technologies, # A10044) was injected intraperitoneally (200 µg/g of body weight) and incubated for 4 days. The EdU was detected using a Click-iT EdU Alexa Fluor 647 Imaging Kit (Life Technologies, # C10340).
Antibody staining and tissue clearing
Whole-mount staining of Isl1 on 48 hpf Tg(meis2b-reporter) embryos was performed as previously described (Dong et al., 2007). With the exception that the washing solution contained 0.1% Triton X-100 to reduce background.
Tg(−5.1myl7:nDsRed2) adult zebrafish hearts were extracted and fixed in 4% paraformaldehyde overnight. Hearts were gradually dehydrated in methanol and the tissue was permeabilized by incubating the hearts for 1 hr at −80°C and then 1 hr at room temperature, repeating this process 6 to 8 times. The hearts were rehydrated in PBS and blocked in PBS + 1% BSA+1% DMSO+0.5% Triton 100x + 2% Goat Serum for 5 hr at room temperature. Rehydrated hearts were incubated for 5 days at 4°C with anti-DsRed Rabbit Polyclonal Antibody (Clontech, # 632496) diluted 1:500 in PBDT (PBS + 1% BSA+1% DMSO+0.5% Triton 100x). After incubation with the primary antibody, the hearts were washed with PBDT and incubated for 5 days at 4°C with Alexa Fluor 488 Donkey-anti-rabbit (Life Technologies, #A21206), diluted to 1:500 in PBDT. At this point, the EdU was detected following the company’s protocol.
For antibody staining on cryosections, adult hearts were fixed in 4% paraformaldehyde overnight, and washed with PBS + 0.1% Tween. Fixed samples were incubated with 1 mL of 10% sucrose in PBS (w/v) at room temperature until they sank to the bottom of the tube, and then transferred to 30% sucrose in PBS overnight at 4°C. The samples were mounted in OCT (Tissue-Tek), and 12-µm-thick sections were cut. The sections were washed first in water and then three times in PBS. After blocking the sections (with PBS + 2% sheep serum +0.2% triton X-100 +1% DMSO) for 2 hr at room temperature, the sections were incubated with anti-Fli1 Rabbit polyclonal antibody (ab133485, Abcam. Diluted 1:100), MF20 mouse monoclonal antibody (DSHB, diluted 1:200), Chicken anti-GFP (Aves Labs, GFP-1020. Diluted 1:500) overnight. The samples were washed three times with PBS + 0.1% Triton-X100 and incubated 3 hr with secondary antibodies (goat anti-rabbit Alexa Fluor 647; goat anti-mouse Alexa Fluor 568; goat anti-chicken Alexa Fluor 488, diluted 1:500) at room temperature. Finally, the samples were incubated with DAPI (diluted 1:10,000) in PBS + 0.1% Triton-X100 for 5 min, mounted in Mowiol (Sigma) and imaged with a Zeiss LSM 800 confocal microscope.
The tissues were cleared using iDISCO (Renier et al., 2014) and imaged using a Zeiss LSM 800 confocal microscope. IMARIS was used to count the number of cardiomyocyte nuclei and the number of proliferating cells. The raw numbers of proliferating atrial cardiomyocytes can be found in Figure 6—source data 1.
Luciferase assays
HEK293T cells were obtained from ATCC (CRL-3216) and authenticated by Short Tandem Repeat Analysis matching 100% with the database profile (ATCC Cell Line Authentication Service). Cells were tested using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza, Walkersville, Inc.) and tested negative for mycoplasma contamination (ratio of 0.24). Cells were transfected with Lipofectamine 2000 (Life Technologies, Carlsbad, CA) 24 hr after seeding in a 24-well plate and were harvested 48 hr post transfection. Assays were performed in quadruplicates, and the ratio of activities of firefly luciferase (expressed from a UAS-Luc2 reporter plasmid) to Renilla luciferase was determined for each well using the Dual-Luciferase Reporter Assay System (Promega). GAL4 and Meis2-GAL4 fusions were expressed under a CMV promoter in pCS2+ vector. The following amounts of plasmid DNA were transfected per well: Renilla luciferase (pTK-RL) (50 ng), GAL4 or GAL4 fusions (500 ng), UAS-Luc2 (450 ng).
Statistical analyses
No statistical tests were used to predetermine sample size. Several independent experiments were performed to ensure reproducibility. The investigators were inherently blinded by the experimental design during the experiment shown in Figure 6—figure supplement 2, until the genotyping was performed. Statistical analyses were performed by paired students t-test with one tail distribution, with the exception of the RT-qPCR analyses comparing left vs. right atria in mouse, where paired students t-test with two tailed was used. For the adult zebrafish electrocardiograms, the statistical differences were calculated using F-test and Bartlett’s test. For all bar graphs, data are represented as mean ± SD. p-Values were considered significant at p<0.05.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (SFB/TR23) (S Reischauer), the Hartstichting (Dutch Heart Foundation) and the Netherlands Organization for International Cooperation in Higher Education (Nuffic) (RFV Germano), the NIH (K08HL125945) and American Heart Association (15GPSPG238300004) (Rima Arnaout), the Leducq Foundation (V. Uribe), the NIH (HL54737) and Max Planck Society (DYR Stainier).We thank Miguel Torres for providing the MEIS antibodies, Arica Beisaw for help with ChIP-qPCR experiments, Vasanth Vedantham for the use of electrocardiography equipment, Michelle Collins for discussion and reagents, Jenny Pestel, Radhan Ramadass, Simon Howard, and Hans-Martin Maischein for technical assistance.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Sven Reischauer, Email: sven.reischauer@mpi-bn.mpg.de.
Margaret Buckingham, Institut Pasteur, France.
Funding Information
This paper was supported by the following grants:
Deutsche Forschungsgemeinschaft SFB/TR23 to Sven Reischauer.
National Institutes of Health K08HL125945 to Rima Arnaout.
American Heart Association 15GPSPG238300004 to Rima Arnaout.
Max-Planck-Gesellschaft Open-access funding to Didier YR Stainier.
National Institutes of Health HL54737 to Didier YR Stainier.
Hartstichting to Raoul F. V. Germano.
Netherlands Organization for International Cooperation in Higher Education NL.11/379 to Raoul F. V. Germano.
Fondation Leducq to Verónica Uribe.
Additional information
Competing interests
Senior Editor, eLife.
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing.
Resources, Formal analysis, Investigation, Visualization, Methodology, Writing—review and editing.
Resources, Writing—review and editing.
Investigation, Writing—review and editing.
Data curation, Formal analysis, Investigation.
Resources, Established the Tg(myl7:BFP-CAAX)bns193 zebrafish transgenic line.
Resources, Established the Tg(myl7:BFP-CAAX)bns193 zebrafish transgenic line.
Investigation, Writing—review and editing.
Supervision, Funding acquisition, Methodology, Project administration, Writing—review and editing.
Conceptualization, Resources, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Methodology, Project administration, Writing—review and editing.
Ethics
Animal experimentation: All animal experiments were done in accordance with institutional (MPG) and national ethical and animal welfare guidelines approved by the ethics committee for animal experiments at the Regierungspräsidium Darmstadt, Germany (permit numbers B2/1041, B2-1023 and B2/Anz. 1007).
Additional files
Set of primers used to clone in situ hybridization probes. Set of primers employed for RT-qPCR and qPCR analyses.
Major datasets
The following dataset was generated:
Guerra A, author; Germano RFV, author; Stone O, author; Arnaout R, author; Vanhollebeke B, author; Guenther S, author; Ahuja S, author; Stainier DYR, author; Reischauer S, author. Atrial Molecular Asymmetry Precedes the Emergence of Cardiac Septation. 2017 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94492 Publicly available at the NCBI Gene Expression Omnibus (accession no: GSE94492)
The following previously published datasets were used:
Kahr PC, author; Piccini I, author. Genome-wide analysis of atrial left-right differences in two different mouse strains. 2011 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29500 Publicly available at the NCBI Gene Expression Omnibus (accession no: GSE29500)
Penkov D, author; Mateos San Martín D, author; Fernandez-Díaz LC, author; Rosselló CA, author. TALE transcription factors binding and transcriptional effect. 2013 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39609 Publicly available at the NCBI Gene Expression Omnibus (accession no: GSE39609)
References
- Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, Tippens ND, Li M, Narasimha A, Radzinsky E, Moon-Grady AJ, Yu H, Pruitt BL, Snyder MP, Srivastava D. Disease model of gata4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167:1734–1749. doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R, Reischauer S, Stainier DYR. Recovery of adult zebrafish hearts for high-throughput applications. Journal of Visualized Experiments. 2014;94:e52248. doi: 10.3791/52248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. PNAS. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkers J, Verhoeven MC, Abdelilah-Seyfried S. Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Developmental Biology. 2009;330:213–220. doi: 10.1016/j.ydbio.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nature Reviews Genetics. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Bussmann J, Bakkers J, Schulte-Merker S. Early endocardial morphogenesis requires Scl/Tal1. PLoS Genetics. 2007;3:e140. doi: 10.1371/journal.pgen.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. 2011;138:4327–4332. doi: 10.1242/dev.068080. [DOI] [PubMed] [Google Scholar]
- Bürglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Research. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione M, Franco D. Current perspectives in cardiac laterality. Journal of Cardiovascular Development and Disease. 2016;3:34. doi: 10.3390/jcdd3040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, Blum M, Franco D, Moorman AF. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Developmental Biology. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PK, Lin CC, Cheng SH. Noninvasive technique for measurement of heartbeat regularity in zebrafish (Danio rerio) embryos. BMC Biotechnology. 2009;9:11. doi: 10.1186/1472-6750-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, Stainier DY. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes & Development. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico L, Scott IC, Jungblut B, Stainier DY. A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Current Biology. 2007;17:252–259. doi: 10.1016/j.cub.2006.12.023. [DOI] [PubMed] [Google Scholar]
- de Pater E, Clijsters L, Marques SR, Lin YF, Garavito-Aguilar ZV, Yelon D, Bakkers J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nature Genetics. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- Franco D, Christoffels VM, Campione M. Homeobox transcription factor Pitx2: The rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends in Cardiovascular Medicine. 2014;24:23–31. doi: 10.1016/j.tcm.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Francou A, Saint-Michel E, Mesbah K, Théveniau-Ruissy M, Rana MS, Christoffels VM, Kelly RG. Second heart field cardiac progenitor cells in the early mouse embryo. Biochimica Et Biophysica Acta (BBA) - Molecular Cell Research. 2013;1833:795–798. doi: 10.1016/j.bbamcr.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Gross-Thebing T, Paksa A, Raz E. Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biology. 2014;12:55. doi: 10.1186/s12915-014-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- Guner-Ataman B, Paffett-Lugassy N, Adams MS, Nevis KR, Jahangiri L, Obregon P, Kikuchi K, Poss KD, Burns CE, Burns CG. Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development. 2013;140:1353–1363. doi: 10.1242/dev.088351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011;138:2389–2398. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JI. Incidence of congenital heart disease: I. Postnatal incidence. Pediatric Cardiology. 1995;16:103–113. doi: 10.1007/BF00801907. [DOI] [PubMed] [Google Scholar]
- Hutchinson SA, Eisen JS. Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development. 2006;133:2137–2147. doi: 10.1242/dev.02355. [DOI] [PubMed] [Google Scholar]
- Hyman-Walsh C, Bjerke GA, Wotton D. An autoinhibitory effect of the homothorax domain of Meis2. FEBS Journal. 2010;277:2584–2597. doi: 10.1111/j.1742-4658.2010.07668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen B, Wang T, Christoffels VM, Moorman AF. Evolution and development of the building plan of the vertebrate heart. Biochimica Et Biophysica Acta (BBA) - Molecular Cell Research. 2013;1833:783–794. doi: 10.1016/j.bbamcr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Kahr PC, Piccini I, Fabritz L, Greber B, Schöler H, Scheld HH, Hoffmeier A, Brown NA, Kirchhof P. Systematic analysis of gene expression differences between left and right atria in different mouse strains and in human atrial tissue. PLoS One. 2011;6:e26389. doi: 10.1371/journal.pone.0026389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Developmental Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Buckingham ME, Moorman AF. Heart fields and cardiac morphogenesis. Cold Spring Harbor Perspectives in Medicine. 2014;4:a015750. doi: 10.1101/cshperspect.a015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Mori AD, Kaynak BL, Cebra-Thomas J, Sukonnik T, Georges RO, Latham S, Beck L, Beck L, Henkelman RM, Black BL, Olson EN, Wade J, Takeuchi JK, Nemer M, Gilbert SF, Bruneau BG. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature. 2009;461:95–98. doi: 10.1038/nature08324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbacher AD, Huang J, Chen Y, Chen JN. Sodium pump activity in the yolk syncytial layer regulates zebrafish heart tube morphogenesis. Developmental Biology. 2012;362:263–270. doi: 10.1016/j.ydbio.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RK, Stainier DY, Weinstein BM, Fishman MC. Cardiovascular development in the zebrafish. II. Endocardial progenitors are sequestered within the heart field. Development. 1994;120:3361–3366. doi: 10.1242/dev.120.12.3361. [DOI] [PubMed] [Google Scholar]
- Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, Chang C, Yagi H, San Agustin JT, Thahir M, Anderton S, Lawhead C, Vescovi A, Pratt H, Morgan J, Haynes L, Smith CL, Eppig JT, Reinholdt L, Francis R, Leatherbury L, Ganapathiraju MK, Tobita K, Pazour GJ, Lo CW. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Lin CY, Chen CH, Zhou B, Chang CP. Partitioning the heart: mechanisms of cardiac septation and valve development. Development. 2012a;139:3277–3299. doi: 10.1242/dev.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Swinburne I, Yelon D. Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Developmental Biology. 2012b;362:242–253. doi: 10.1016/j.ydbio.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stainier DY. Zebrafish in the study of early cardiac development. Circulation Research. 2012;110:870–874. doi: 10.1161/CIRCRESAHA.111.246504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longobardi E, Penkov D, Mateos D, De Florian G, Torres M, Blasi F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Developmental Dynamics. 2014;243:59–75. doi: 10.1002/dvdy.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw JJ, Corveleyn A, Jia Y, Hens G, Gewillig M, Devriendt K. MEIS2 involvement in cardiac development, cleft palate, and intellectual disability. American journal of medical genetics. Part A. 2015;167A:1142–1146. doi: 10.1002/ajmg.a.36989. [DOI] [PubMed] [Google Scholar]
- Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Current Biology. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- Machon O, Masek J, Machonova O, Krauss S, Kozmik Z. Meis2 is essential for cranial and cardiac neural crest development. BMC Developmental Biology. 2015;15:40. doi: 10.1186/s12861-015-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, Porrello ER, Sadek HA. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammi I, De Giorgio P, Clementi M, Tenconi R. Cardiovascular anomaly in rieger syndrome: heterogeneity or contiguity? Acta Ophthalmologica Scandinavica. 1998;76:509–512. doi: 10.1034/j.1600-0420.1998.760424.x. [DOI] [PubMed] [Google Scholar]
- Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
- Merabet S, Mann RS. To be specific or not: the critical relationship between hox and tale proteins. Trends in Genetics. 2016;32:334–347. doi: 10.1016/j.tig.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Panáková D, Werdich AA, Musso G, Burger A, Lawson KL, Carr LA, Nevis KR, Sabeh MK, Zhou Y, Davidson AJ, DiBiase A, Burns CE, Burns CG, MacRae CA, Zon LI. Chamber identity programs drive early functional partitioning of the heart. Nature Communications. 2015;6:8146. doi: 10.1038/ncomms9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr N, Arnaout R, Gula LJ, Spears DA, Leong-Sit P, Li Q, Tarhuni W, Reischauer S, Chauhan VS, Borkovich M, Uppal S, Adler A, Coughlin SR, Stainier DY, Gollob MH. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nature Communications. 2016;7:11303. doi: 10.1038/ncomms11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, Moon RT, Stamatoyannopoulos J, Murry CE. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkov D, Mateos San Martín D, Fernandez-Díaz LC, Rosselló CA, Torroja C, Sánchez-Cabo F, Warnatz HJ, Sultan M, Yaspo ML, Gabrieli A, Tkachuk V, Brendolan A, Blasi F, Torres M. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Reports. 2013;3:1321–1333. doi: 10.1016/j.celrep.2013.03.029. [DOI] [PubMed] [Google Scholar]
- Reischauer S, Arnaout R, Ramadass R, Stainier DY. Actin binding GFP allows 4D in vivo imaging of myofilament dynamics in the zebrafish heart and the identification of Erbb2 signaling as a remodeling factor of myofibril architecture. Circulation Research. 2014;115:845–856. doi: 10.1161/CIRCRESAHA.115.304356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Rohr S, Otten C, Abdelilah-Seyfried S. Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circulation Research. 2008;102:e12–e19. doi: 10.1161/CIRCRESAHA.107.165241. [DOI] [PubMed] [Google Scholar]
- Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Peña J, Sabbagh W, Greenwald J, Choe S, Norris DP, Robertson EJ, Evans RM, Rosenfeld MG, Izpisúa Belmonte JC. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- Rydeen AB, Waxman JS. Cyp26 enzymes facilitate second heart field progenitor addition and maintenance of ventricular integrity. PLOS Biology. 2016;14:e2000504. doi: 10.1371/journal.pbio.2000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa MS, Vasconcelos M, Sampaio AC, Cravo RM, Linhares VL, Hochgreb T, Yan CY, Davidson B, Xavier-Neto J. The evolutionary origin of cardiac chambers. Developmental Biology. 2005;277:1–15. doi: 10.1016/j.ydbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Smith KA, Chocron S, von der Hardt S, de Pater E, Soufan A, Bussmann J, Schulte-Merker S, Hammerschmidt M, Bakkers J. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Developmental Cell. 2008;14:287–297. doi: 10.1016/j.devcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Lee RK, Fishman MC. Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development. 1993;119:31–40. doi: 10.1242/dev.119.1.31. [DOI] [PubMed] [Google Scholar]
- Staudt D, Stainier D. Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annual Review of Genetics. 2012;46:397–418. doi: 10.1146/annurev-genet-110711-155646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessari A, Pietrobon M, Notte A, Cifelli G, Gage PJ, Schneider MD, Lembo G, Campione M. Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circulation Research. 2008;102:813–822. doi: 10.1161/CIRCRESAHA.107.163188. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Developmental Cell. 2004;6:371–382. doi: 10.1016/S1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. PNAS. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinsaft JW, Kochav JD, Kim J, Gurevich S, Volo SC, Afroz A, Petashnick M, Kim A, Devereux RB, Okin PM. P wave area for quantitative electrocardiographic assessment of left atrial remodeling. PLoS One. 2014;9:e99178. doi: 10.1371/journal.pone.0099178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzel HR, Jungblut B, Choe CP, Crump JG, Braun T, Dobreva G. The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Developmental Cell. 2012;23:58–70. doi: 10.1016/j.devcel.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, Heideman W, Burns CE, Burns CG. Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474:645–648. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]