Abstract
Background
Defining conserved molecular pathways in animal models of successful cardiac regeneration could yield insight into why adult mammals have inadequate cardiac regeneration after injury. Insight into the transcriptomic landscape of early cardiac regeneration from model organisms will shed light on evolutionarily conserved pathways in successful cardiac regeneration.
Methods
Here we describe a cross-species transcriptomic screen in three model organisms for cardiac regeneration –axolotl, neonatal mice and zebrafish. Apical resection to remove ~10 – 20% of ventricular mass was carried out in these model organisms. RNA-seq analysis was performed on the hearts harvested at three time points – 12, 24 and 48 hours post-resection. Sham surgery was used as internal control.
Results
Genes associated with inflammatory processes were found to be upregulated in a conserved manner. Complement receptors (activated by complement components, part of the innate immune system) were found to be highly upregulated in all three species. This approach revealed induction of gene expression for Complement 5a receptor1 (C5aR1) in the regenerating hearts of zebrafish, axolotls and mice. Inhibition of C5aR1 significantly attenuated the cardiomyocyte proliferative response to heart injury in all three species. Furthermore, following left ventricular apical resection, the cardiomyocyte proliferative response was abolished in mice with genetic deletion of C5aR1.
Conclusions
These data reveal that activation of C5aR1 mediates an evolutionarily conserved response that promotes cardiomyocyte proliferation following cardiac injury and identify complement pathway activation as a common pathway of successful heart regeneration.
Keywords: Complement system, C5aR1, cardiac regeneration, cross-species, axolotl, mice, zebrafish
Introduction
While some organisms like zebrafish, axolotl and newt exhibit cardiac regenerative ability throughout life1–5, mammalian hearts have limited natural regenerative potential with the exception of a narrow regenerative window demonstrated in neonatal mice6. Adult mammals, including humans, fail to regenerate significant myocardium following injury. Instead, the injured muscle is replaced with scar tissue, compromising the contractility of the remaining myocardium when the extent of injury is severe2, 7, 8. This inability of the adult mammalian heart to regenerate significant amounts of tissue can lead to subsequent heart failure, a leading cause of mortality7, 9.
Upon injury in animals that can regenerate myocardium, some cardiomyocytes undergo de-differentiation and proliferation to regenerate the lost tissue4. Lineage mapping of newly-derived cardiomyocytes in regenerating hearts has shown that new cardiomyocytes are primarily produced via cardiomyocyte cell division in zebrafish1, 10, 11 and neonatal mice6. Furthermore, it has also been shown that cardiomyocyte renewal in adult uninjured mouse hearts occurs through cardiomyocyte division12. The evidence that a significant majority of new cardiomyocytes are derived from pre-existing cardiomyocytes has focused attention on cardiomyocyte division as a key step in successful cardiac regeneration13.
In this study, we leveraged three different animal models of successful cardiac regeneration to obtain insights into early events that may activate the cardiomyocyte cell cycle after cardiac injury. We aimed to define a conserved molecular pathway that plays a critical role early in the heart regenerative response using a cross-species comparative genomic approach. We performed an RNA-sequencing screen in zebrafish (Danio rerio), axolotl (Ambystoma mexicanum) and neonatal mouse (Mus musculus), 12, 24 and 48 hours after cardiac apical resection. With the availability of recent tools to conduct such large-scale and multi-species studies, gene expression profiling in multiple species with enhanced capacity for cardiac regeneration is a powerful method to identify genes and pathways that play critical roles in cardiac regeneration. In addition to the upregulation of cell cycle and inflammatory genes across all three species, components of complement signaling were also upregulated in the regenerating hearts. Complement 5a receptor, C5aR1, was one of the most significantly upregulated genes early in the regenerating hearts of all species analyzed.
C5aR1 is a G-protein coupled receptor (GPCR) activated by complement 5a, a peptide anaphylotoxin generated by the cleavage of full length complement 5 protein14. The complement pathway is a component of the innate immune system that serves to augment the ability of the immune system to identify and initiate the clearance of foreign material15. Synthesized by the liver as zymogens, sequential proteolytic cleavage of complement proteins upon activation by external cues results in complement peptides that activate complement receptor GPCRs15, 16. Expression of complement components has previously been noted in other regenerating tissues such as the axolotl and newt limb, mammalian liver, and chick embryonic retina17–20. C5a has been shown to play a crucial role in murine liver regeneration21, and several studies implicate C5a and C5aR in ischemic injury and cardiac dysfunction after sepsis22–24. Here we show that complement signaling is required to initiate cardiomyocyte proliferation during heart regeneration.
Materials and Methods
The data and pipelines from our RNA-seq experiments have been deposited with GEO (Accession number GSE108493), the material will be publicly released on 2.1.18. Additional data and methods pertinent to the findings of this study are available from the corresponding author (Richard T Lee) upon reasonable request.
Animals
All experiments were conducted in accordance with the Guide for the Use and Care of Laboratory Animals and approved by the Harvard University Institutional Animal Care and Use Committee.
Wild-type (WT) CD-1 and C57Bl/6 mice were obtained from Charles River Laboratories. C5ar1 knock-out mice were obtained from the laboratory of Dr. Craig Gerard, Boston Children’s Hospital.
For RNASeq experiments, zebrafish of strain Wild-type AB and Wild-type TuAB were produced, maintained, and housed in the laboratory of Dr. Calum MacRae, Massachusetts General Hospital. Ventricular apical resection was performed on 9-month-old zebrafish. For inhibition of C5aR1 transgenic zebrafish Tg(cmlc2:nlsDsRed-Express)hsc4 were used25. Adult zebrafish used for experiments were grown and maintained in compliance with the Institutional Animal Care and Use Committee of Massachusetts General Hospital.
Wild-type black axolotl were maintained and housed in the laboratory of Dr. Jessica Whited, Brigham Regenerative Medicine Center. Sub-adult axolotls were used for ventricular resection, leucistic axolotls were used for C5aR1 inhibition studies.
Ventricular apical resection
Zebrafish
Ventricular apical resection was performed on zebrafish between 4–6 months of age as previously described1.
Axolotl
Axolotl resection surgeries were performed on 8- 12-months old axolotls (as previously described by Cano-Martinez et al., 2010)26. Animals were anesthetized in tricaine mesylate (MS-222) for 20 minutes. Lateral thoracotomy was then performed to expose the heart ventrally. Microsurgery scissors were used to resect approximately 10 – 20% of the ventricle. The control—or sham—animal group underwent the thoracotomy but not apical resection. After a blood clot was formed, a 6-0 prolene suture was then used to suture the incision in the thoracic cavity.
Neonatal mice
Neonatal mouse resection procedures were performed on 1-day old pups as described previously6, 27. 1-day-old mice were anesthetized on ice for 4 minutes. All mice underwent a lateral thoracotomy. Resected animals received a ~10% resection of the ventricle to expose the left ventricular chamber and the control surgery animals (“sham”) only received a thoracotomy.
RNA isolation and sequencing
Total RNA from the lower half of the ventricle of resected and sham hearts was isolated using Trizol at three different time points: 12, 24, and 48 hours post-surgery. Three biological replicates were used for each time point and condition in each species. For neonatal mice and axolotls, each replicate represents myocardium from a single animal. Due to the smaller size of the zebrafish, each biological replicate represents three pooled tissue samples. Poly-adenylated mRNA was isolated using the Wafergen #400047 protocol, and RNA-sequencing libraries were prepared with the IntegenX Directional PrepX mRNA kit as previously described28. Paired-end (50 base pairs) sequencing was performed using an Illumina HiSeq 2500. Poly-adenylated mRNA isolation, library preparation, and sequencing were performed by the Biopolymers Facility at Harvard Medical School.
RNAseq data analysis
Tophat 2 was used to align paired-end sequence reads to the GRCm30 genome for mouse and GRCz10 genome for zebrafish29. Cufflinks was used to estimate gene expression levels in aligned reads and the resulting data was processed with Cuffdiff to identify differentially expressed genes between resected and sham groups at each time point and species30. In contrast to zebrafish and the mouse, the full genome of the axolotl has not been sequenced. Thus, we took an approach were we first mapped the axolotl RNA-sequencing data to a previously published transcriptome that was constructed using the Trinity program31–33. RSEM was used to quantify gene expression34, and differential gene expression analyses were performed using EBseq35. For all species, a given gene had to be identified as differentially expressed (False Discovery Rate (FDR) adjusted p-value <0.05) by the programs described above and have a fold change of >1.2 in order to be considered in our final analyses. After identifying differentially expressed genes using the criteria described above, we constructed two datasets for each species that consisted of either genes that were 1) upregulated or 2) downregulated at 12, 24, or 48 hours post-surgery. We performed this step in order to account for species-specific time differences during early heart regeneration.
Orthology and Gene Ontology (GO) Analyses
Ensembl BioMart was used to identify the mouse orthologs of differentially expressed zebrafish genes36. Axolotl genes were annotated using Trinotate as previously described33. The Web-based Gene Set Analysis Toolkit (WebGestalt) was used to perform Gene Ontology analyses on the set of genes that were either upregulated during heart regeneration in both neonatal mice and zebrafish or downregulated during heart regeneration in both neonatal mice and zebrafish at 12, 24, or 48 hours post-surgery37, 38. M. musculus was used as the organism of interest, Overrepresentation Enrichment Analysis (ORA) was used as the method of interest, and gene ontology was used for the functional database. Ensembl gene I.D.s were used as inputs, and the genome was used as the reference set for enrichment analysis. GO terms with an FDR-adjusted p-value less than 0.05 were considered to be statistically significant.
Inhibition of C5aR1 using PMX205 following apical resection
Axolotl
0.05 mg PMX20539, 40 (Tocris, #5196) was delivered via i.p. injection for 14 days post-resection.
Zebrafish
PMX205 (R&D, 5196, 2.5 mg·kg−1) or vehicle control were injected intraperitoneally intro adult zebrafish. Injections were initiated 1 hour prior to ventricular resection and then daily for 10 days after ventricular amputation.
Neonatal mouse
On the day of surgery, 0.025 mg PMX205 was administered by a subcutaneous injection 30 minutes before resection and 0.025 mg was delivered post resection. For the next three days, 0.05 mg PMX20539, 40 (Tocris, #5196) was administered.
Histology, Immunofluorescence and western
Zebrafish
Zebrafish hearts were extracted and fixed as described41. Hearts were then processed for histological and immunofluorescence analysis as described42. Primary antibodies used were anti-PCNA (sc-56, 1:50, Santa Cruz Biotechnologies, USA), anti-DsRed (1:200, Clontech, USA) and anti-Tropomyosin (clone CH1, 1:100, Developmental Studies Hybridoma Bank, USA). Alexa (488, 568, 633)-conjugated antibodies (Invitrogen, USA) were used to detect primary antibody signal. Nuclei were stained with DAPI and slides were mounted in FluorSave (Millipore, USA). We tried two antibodies for C5aR1 localization – SantaCruz sc-56 and Thermo Fisher MA1-81761. We observed best signal with SantaCruz anti-C5aR1 in our immunostaining experiments.
Axolotl and Mouse
Hearts were fixed with 4% paraformaldehyde (PFA) overnight followed by cryoprotection through a sucrose gradient of 10%, 15%, 20%, and 30% sucrose in PBS. Samples were left in each sucrose solution till the tissue sank to the bottom of the vial, and then left in 30% sucrose overnight at 4°C. The following day samples were embedded in OCT and sectioned longitudinally (12µM). A list of antibodies used for immunostaining are listed in Table 1.
Table 1.
List of antibodies used for immunostaining
| Primary antibodies | phospho Histone 3 (Rabbit, Millipore), 1:100 |
| C5aR1 (Rabbit, SantaCruz), 1:100 | |
| Cardiac Troponin T (mouse, Abcam), 1:200 | |
| CD31 (rat, Abcam), 1:100 | |
| CD68 (rabbit, Abcam), 1:100 | |
|
| |
| Secondary antibodies | Alexa 488, 568, 647 (Invitrogen), 1:500 |
Western blot
Mouse anti C5a (Abcam) was used at a 1:1000 dilution to detect C5a levels in resected and sham hearts, 48 hours post-resection. 50ug tissue lysate was loaded per well in the western blot. Cell signaling rabbit anti GAPDH was used at 1:2000 for loading control.
EdU incorporation assay
EdU was purchased from Carbosynth, Inc. Primary cardiomyocytes were isolated from wild-type mice and cultured in fibronectin – coated dishes at a density of 300,000 cells per well. 72 hours post – plating, cells were treated with varying concentrations of C5a (saline for control) and incubated with medium containing 20 µM EdU for 24 hours. Following treatment, cells were fixed and stained with PCM-1 (Abcam), EdU (Sulfo-Cy3 azide) and DAPI and imaged using CellDiscoverer (Zeiss).
Imaging
Zebrafish
A total of 3 sections per heart were quantified for each group. DsRed+/PCNA+ cardiomyocyte nuclei were divided by the total DsRed+ cardiomyocytes to generate a proliferation index.
Axolotl and mouse
Fluorescently stained sections were imaged with Zeiss LSM 700 (mouse) and Zeiss Axiozoom (Axolotl), (HCBI, Harvard University). A total of 5 sections per heart were quantified for each group in a blinded manner.
Statistical analysis
All data are presented as mean ± standard error of the mean (SEM). All statistical analyses were performed using Prism 7 software (Graph Pad). After normality testing using the Wilk-Shapiro test; two-tailed Student’s t-test was used to compare data from individual experimental groups. A value of p<0.05 was considered significant.
Results
Transcriptomic analyses indicate a conserved role for the inflammatory response and metabolic regulation during early neonatal mouse and zebrafish heart regeneration
We utilized three model organisms capable of successful cardiac regeneration: neonatal mice, zebrafish, and axolotls. We resected ~10 – 20% of the ventricular myocardium in all three species, following which RNA-sequencing was carried out on the regenerating hearts of each species at 12, 24, and 48 hours post-resection. We undertook this approach to assess differential gene expression at three different time-points as studies have indicated that cardiac regeneration occurs at different rates across the three species; neonatal mice successfully accomplish regeneration in as few as 21 days while zebrafish and axolotl heart regeneration take 60 days or more to replace the lost myocardial tissue6, 43. RNA-sequencing on sham-operated hearts at each time point was used as control for data analysis. These internal controls were performed to account for potential systemic effects due to surgical opening of the thoracic cavity and for the rapid growth of the neonatal mouse during the first few days after birth. The zebrafish and the mouse genomes are well annotated and enabled feasibility of an RNA sequencing experiment with multiple samples. Even though the axolotl genome is incomplete, a high number of transcriptomes and various annotations exist to assist in analysis33.
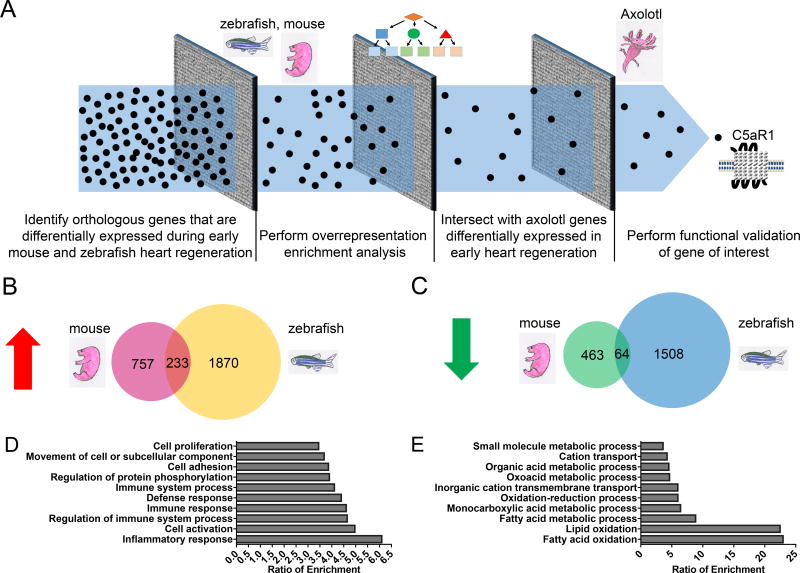
Following acquisition of the RNA-sequencing data, we found that 1433 genes were differentially expressed in neonatal mice, 4502 genes were differentially expressed in zebrafish, and 4639 gene contigs were differentially expressed in axolotl heart regeneration across all time points. We undertook a systematic computational approach to refine the dataset in order to identify genes that are functionally important for heart regeneration across all three species (Figure 1A). First, we identified orthologous mouse and zebrafish genes that were consistent in terms of their differential expression (i.e. both upregulated or both downregulated) during early regeneration (Figure 1A). We found that 233 genes were commonly upregulated during the first two days of heart regeneration (Figure 1B). These genes represented 23.5% of upregulated mouse genes (233/990 genes) and 11.1% of mouse genes (233/2103) with an orthologous relationship to upregulated zebrafish genes. Furthermore, we identified 64 genes common to zebrafish and mice that were downregulated during this early time period (Figure 1C). These genes constitute 12.1% of mouse genes that were downregulated within the first two days of heart regeneration (64/527) and 4.1% (64/1572) of genes corresponding to the mouse orthologs of downregulated zebrafish genes.
Figure 1. RNA-seq analysis pipeline to identify evolutionarily conserved genes involved in early heart regeneration.
Ventricular myocardium was resected, and RNA-sequencing was performed at 12, 24, and 48 hours post-injury in neonatal mice, axolotl, and zebrafish. Each time point was internally controlled with sham surgery. (A) Schematic of transcriptomic approach for identifying genes whose expression was conserved during early heart regeneration. Black dots represent genes, and gray squares represent computational filtration steps. (B) Intersection of genes that were upregulated at 12, 24, or 48 hours relative to sham controls in neonatal mice and zebrafish. (C) Intersection of genes that are downregulated at 12, 24, or 48 hours relative to sham controls in neonatal mice and zebrafish. (D) Gene ontology (GO) analysis of commonly upregulated neonatal mouse and zebrafish genes. (E) Gene ontology (GO) analysis of commonly downregulated neonatal mouse and zebrafish genes.
Next, we attempted to identify conserved gene expression modules by performing overrepresentation analyses on the common differentially expressed genes (Figure 1A). Using WebGestalt38, we found that the set of genes commonly upregulated during mouse and zebrafish heart regeneration were significantly enriched for Gene Ontology (GO) terms such as “Inflammatory Response”, “Regulation of Immune System Process”, and “Immune System Process” (FDR <0.05; Figure 1D). These data suggest that regulation of the inflammatory response during early heart regeneration is a conserved process across species and is consistent with previous studies demonstrating the functional importance of the early immune response to regeneration44–46. Similarly, we also observed significant enrichment (FDR <0.05) for “Cell proliferation” (Figure 1D), which is consistent with cardiomyocyte division as well as possibly non-myocytes during heart regeneration6, 7, 10, 11. Biological processes such as “Fatty Acid Oxidation”, “Lipid Oxidation”, and “Fatty Acid Metabolic Process” had the highest ratio of enrichment among significant GO terms (FDR <0.05) in the set of genes commonly downregulated in neonatal mouse and zebrafish heart regeneration (Figure 1E). These data suggest that fundamental changes in metabolic processes during early heart regeneration are evolutionarily conserved across species.
Cross-species gene expression profiling indicates a conserved role for complement components during early heart regeneration
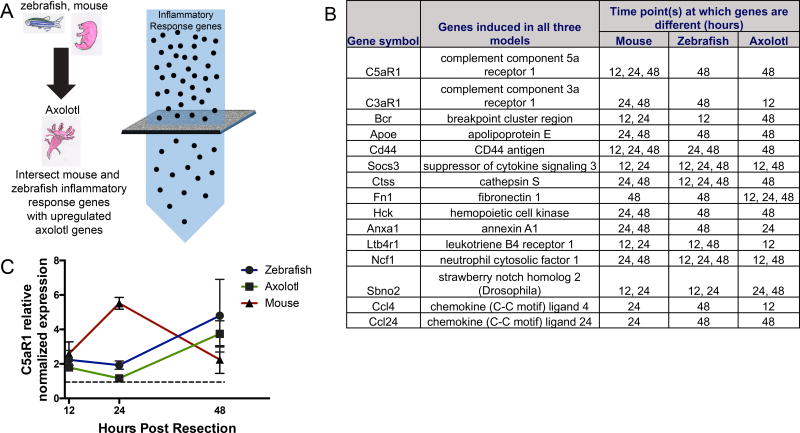
Motivated by our ability to uncover broader biological processes that are known to be essential for regeneration, we focused our attention on genes belonging to the “Inflammatory Response” classification that are commonly upregulated during mammalian and zebrafish heart regeneration (Figure 1D). This particular category comprised 38 genes and had the highest enrichment ratio relative to other GO terms in the upregulated gene set (Figure 1D). We then identified 15 inflammatory response genes that were also upregulated during early axolotl heart regeneration (Figure 2A); these comprised the highest confidence set of upregulated genes during early regeneration across the three species (Figure 2B). The 15 inflammatory genes identified to be upregulated in early cardiac regeneration is listed in Figure 2B. It is important to note that the top two inflammatory genes found to be upregulated in a conserved manner are both complement receptors, C5aR1 and C3aR1.
Figure 2. Schematic of analysis pipeline used to identify upregulated inflammatory genes in mouse, zebrafish an axolotl.
(A). Inflammatory response genes including complement components and associated proteins that are upregulated in early cardiac regeneration in an evolutionarily conserved manner. The list is ordered according to fold change; genes with higher fold change in mouse are first, followed by genes in decreasing order. Note that the top two genes are receptors for activated complement components, C5aR1 and C3aR1. (B). Complement 5a receptor 1 (C5aR1) is upregulated in early cardiac regeneration in zebrafish, axolotl and mouse at the timepoints shown, in hours (C), expression of C5aR1 is normalized to sham levels (dotted line).
The 15 genes included complement component 5a receptor (C5aR1) and complement component 3a receptor (C3aR1), which serve as the receptors for complement components C5a and C3a, respectively, and have been shown to mediate complement signaling in macrophages47 (Figure 2A). The complement cascade is an ancient component of the innate immune system. Recent studies have shown that complement may also mediate cross-talk between the innate and adaptive immune response48. Upon binding to their corresponding receptors, complement peptides C5a and C3a act as potent inflammatory mediators that can trigger the degranulation of endothelial cells, mast cells, and phagocytes, and they function as chemoattractants for neutrophils, monocytes, and macrophages49. Complement proteins have been noted in several regenerating tissues such as newt lens, axolotl limb and mouse liver19, 20. Interestingly, the expression of C5 and C3 was observed in regenerating limb tissues such as blastema and wound epidermis, suggesting that the complement system could play functional roles in other regenerative contexts beyond heart regeneration20. Although most of the evidence supporting the expression and role for the complement system in regeneration is mainly observational, Strey et.al. showed mechanistic evidence supporting the crucial role of C5a signaling in murine liver regeneration after partial hepatectomy21.
The evolutionarily conserved upregulation of complement components observed during early heart regeneration in neonatal mouse, zebrafish, and axolotl, along with studies implicating the complement system in regenerative processes outside of the heart, suggest a role of complement receptors in cardiac regeneration. Of the two receptors identified, C5aR1 was upregulated during regeneration (Figure 2A). When normalized to expression levels in sham-operated animals, C5aR1 expression in all three species was upregulated significantly, peaking at slightly different time points within the first 48 hours (Figure 2B). Specifically, we observed the highest fold-change in C5aR1 expression at 24 hours post-injury in mice and 48 hours post-injury in zebrafish and axolotls. We speculate that the time differences in expression may be attributed to species-specific differences in the heart regeneration response and could possibly reflect the different rates at which heart regeneration occurs in these species. Taken together, our analyses suggested that conserved activation of C5aR1 during early heart regeneration could play a role in cardiac regeneration.
Inhibition of C5aR1 severely attenuates cardiomyocyte proliferation following apical resection
Based on our finding that upregulation of complement signaling is an evolutionarily conserved mechanism in early cardiac regeneration, we sought to test the hypothesis that C5aR1 signaling is required for successful cardiac regeneration after apical resection in zebrafish, axolotl and mice. We utilized a peptide C5aR1 antagonist, PMX20539, 40 to inhibit C5aR1 activity across the three species. PMX205 is a highly selective C5aR1 antagonist with an IC50 of 31nM50. Although studies have focused on the immunomodulatory role of C5aR1, it is known to be expressed in other tissues including liver, kidney and heart14. We tested the hypothesis that C5aR1 receptor function is required for effective cardiac regeneration by administering PMX20539, 40 after apical resection in zebrafish, axolotl and neonatal mice. PMX205 was administered to the animals following cardiac apical resection (vehicle treatment of resected animals and sham surgery with vehicle / PMX205 administration for control). Cardiomyocyte proliferation was quantified by immunostaining for proliferation markers phospho-Histone 3 (pH3, mouse and axolotl) and PCNA (zebrafish). The total number of proliferating cardiomyocytes per field of view was quantified after imaging of immunostained heart sections.
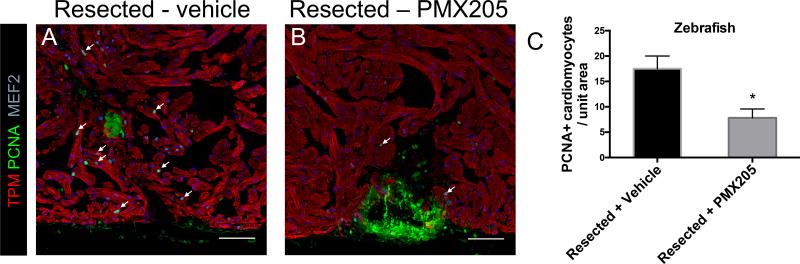
Due to differences in the rate at which cardiac regeneration occurs in the three species, different lengths of treatment with PMX205 were employed for each species studied. Apical resection was performed on zebrafish (sham for control), following which PMX205 was administered for 10 days at 0.05mg / zebrafish by i.p. injection. Zebrafish hearts were harvested 14 days post-resection and stained with proliferation marker PCNA. A significant reduction in the number of proliferating cardiomyocytes per unit area was observed in resected zebrafish hearts that were treated with PMX205 (Figure 3A,B). Quantification of proliferating cardiomyocytes from the experimental groups showed that PMX205 effected ~ 2-fold reduction in cardiomyocyte proliferative response in zebrafish following apical resection (Figure 3C).
Figure 3. Inhibition of C5aR1 decreases proliferating cardiomyocytes after apical resection in zebrafish.
(A) resected zebrafish treated with vehicle, representative image, (B) resected zebrafish treated with PMX205, representative image, cardiomyocyte marker tropomyosin (red), PCNA (green), cardiomyocyte nuclei (MEF2, blue); scale bar - 50µm (C) quantification of proliferating cardiomyocytes in vehicle and PMX205 treated hearts of zebrafish, * p<0.01, n=5 (vehicle), 8 (PMX205).
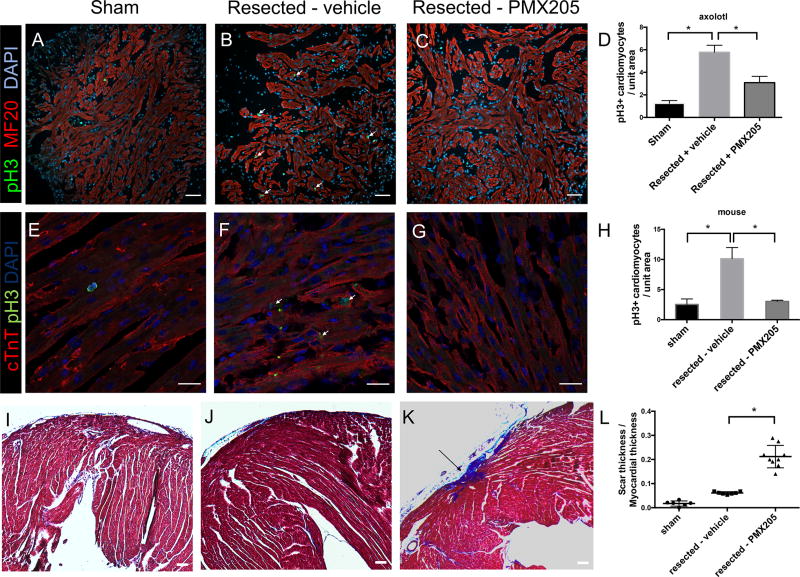
Axolotls received 14 days of PMX205 administration (0.05mg/kg) following apical resection. Axolotl hearts were harvested following PMX205 treatment and stained for pH3 and the cardiomyocyte marker MF20 to quantify cardiomyocyte proliferation. Inhibition of C5aR1 significantly decreased cardiomyocyte cell cycle activity after apical resection in axolotl (Figure 4A–C), revealed by a reduction of pH3 positive cardiomyocytes per field of view in resected axolotl hearts treated with PMX205. Quantification of the number of proliferating cells per field of view showed a ~ 2-fold reduction in the number of pH3 positive cardiomyocytes in axolotl hearts treated with PMX205 (quantified blindly in Figure 4D).
Figure 4. Inhibition of C5aR1 in axolotl and mouse.
Inhibition of C5aR1 results in a reduction of proliferating cardiomyocytes in axolotl hearts. cardiomyocyte marker MF20 is shown in red, phospho Histone 3 (pH3) is shown in green and nuclei are in blue. (A) Sham surgery of axolotl hearts, (B) resected – vehicle treatment and (C) resected – PMX205 treatment; scale bar - 100µm. Quantification of proliferating cardiomyocytes is summarized in (D), * p<0.05, n=4.
Effect of C5aR1 inhibition on cardiomyocyte proliferation in mouse (E–H). Cardiac Troponin T (cTnT, cardiomyocyte marker) is shown in red, phosphoHistone 3 staining is in green and nuclei are in blue. (E) sham surgery of mouse hearts, (F) resected – vehicle treatment and (G) resected – PMX205 treatment, scale bar - 20µm. Quantification of cardiomyocyte proliferation is summarized in (H), * p<0.05, n=4.
Effect of C5aR1 inhibition on scar formation after apical resection (I–L). Masson trichrome images of hearts 21 days post-resection show an increase in scar formation in PMX205 treatment after apical resection. Representative images of sham surgery (I), resected – vehicle treatment, resected – PMX205 treatment. Scar infiltration is quantified in (L). * p<0.05, n≥6, scale bar 100 µm.
As cardiac regeneration in neonatal mice is faster than in axolotl and zebrafish, we assessed the effect of PMX205 treatment for 4 days after apical resection on neonatal mice (P1). Hearts from experimental mice were harvested 7 days post-resection and proliferation was assessed by staining for pH3. We observed a significant reduction of pH3 positive cardiomyocytes in the PMX205 treated animals compared to vehicle treatment, consistent with the reduction of cardiomyocyte proliferation we observe in axolotl and zebrafish (Figure 4E–H). As the mouse model is the most relevant for mammalian heart regeneration among the three model organisms used, the effect of C5aR1 inhibition was then assessed at the histological level by quantifying scarring of the murine heart in the different experimental groups, 21 days post injury by Masson Trichrome staining. An increase in scarring of the ventricular apex was observed in the group of animals treated with PMX205, further demonstrating that C5aR1 signaling is crucial for the initiation of effective cardiac regeneration (Figure 4I–L). While minimal scarring was observed in resected hearts treated with vehicle (Figure 4J, L), we observed small but consistent scars in the ventricular apex of mice treated with PMX205 after resection (Figure 4K,L). To further investigate the role of C5aR1 on cardiomyocyte proliferation, we quantified proliferation in primary cardiomyocytes treated with C5aR1 agonist C5a for a period of 24 hours by measuring EdU incorporation. We did not observe a significant increase in cardiomyocyte proliferation upon C5a treatment (Figure S1). This indicates that the proliferative effect elicited by C5a – C5aR1 signaling is potentially invoked after cardiac injury. Overall, our results across the three species support the hypothesis that C5aR1 plays an important role in heart regeneration and is essential for the initiation of a cardiomyocyte proliferative response required for successful heart regeneration. Our findings also indicate that one possible function of the C5aR1 pathway is to inhibit collagen deposition and scarring in mammalian heart regeneration.
C5aR1 is primarily expressed in cardiomyocytes following apical resection
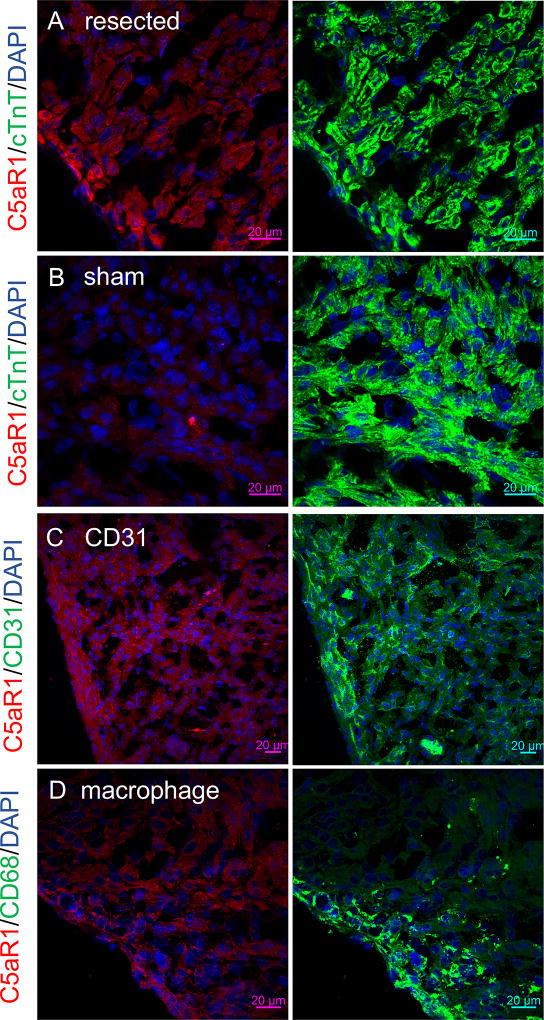
The observed upregulation of C5aR1 expression in the early regenerating myocardium could be due to immune infiltration of the heart following injury, since C5aR1 is expressed in myeloid cells51. Macrophages play an essential role in cardiac regeneration, and depletion of macrophages in neonatal mice results in incomplete regeneration46. However, despite extensive scarring and inhibition of microvasculature formation in the regenerating heart, macrophage depletion does not appear to cause a decrease in cellular proliferation following cardiac injury46. This suggests that decreased cardiomyocyte proliferation upon C5aR1 inhibition may not be due to the contribution of C5aR1 from macrophages. C5aR1 expression has been noted in cardiomyocytes24, 52, 53 as well as endothelial cells, where the C5a component of complement has been shown to activate the endothelium54–56. C5aR1 expression has been shown to be upregulated in ischemic cardiomyocytes in adult mice following ischemia/reperfusion injury24. To identify the cell types expressing C5aR1 in the neonatal mouse heart during regeneration, we conducted extensive immunostaining analyses in the neonatal mouse heart following apical resection (sham for control). We found that C5aR1 was primarily expressed in cardiomyocytes (Figure 5A, seen by co-localization with cardiac Troponin T), 48 hours post-resection in neonatal mice. Hearts from littermate sham controls did not have expression of C5aR1 (Figure 5B). We validated the antibody by staining sections from hearts from C5aR1 neonates, 48 hours post-resection. We do not observe any signal from the antibody in C5aR1 knock-out sham and resected hearts (Figure S2). Furthermore, we also observed co-localization of C5aR1 with the endothelial marker CD31 in the injured hearts (Figure 5C). However, we did not observe localization of C5aR1 staining with the macrophage marker CD68 in injured neonatal hearts (Figure 5D). In-depth immunostaining and imaging analyses of C5aR1 expression pattern in the injured neonatal heart show expression of C5aR1 in the injured zone at the apex (site of resection, Figure S3A), while, no significant expression was observed in the border and the remote zones (Figure S3 B,C respectively). Expression of C5aR1 in cardiomyocytes following cardiac injury circumstantially supports the concept that C5aR1 plays an important role in cardiomyocyte proliferation after injury and suggests that either cardiomyocytes or cardiac endothelial cells—or both—are cells likely to be receiving the C5a signal during heart regeneration.
Figure 5. Localization of C5aR1 to cardiomyocytes and endothelial cells after apical resection in neonatal mice.
Localization of C5aR1 to cardiomyocytes and endothelial cells after apical resection in neonatal mice. C5aR1 (red) localizes with cardiomyocyte marker Troponin T (green) in neonatal mouse hearts, 48 hours after apical resection (nuclei are in blue), scale bar 20µm (A). C5aR1 upregulation is absent in hearts from littermate sham controls (B). C5aR1 (red) expression localizes with endothelial marker CD-31 (green) (C), scale bar - 20µm. C5aR1 (red) does not localize with macrophage marker CD68 (green)in resected hearts (D), scale bar - 20µm.
A majority of the complement components are secreted proteins, synthesized mainly by the liver as zymogens. Complement component 5, upon activation by complement 5 convertase, is cleaved to form C5a and C5b in target tissues. Therefore, we carried out western analysis of neonatal murine hearts 48 hours post-injury to detect activation of C5 and cleavage to C5a. (sham for control). We observed a significant increase in C5a production in the injured heart 48 hours post resection in mice, in comparison with sham hearts of littermate animals (Figure S4), n=3 (all groups); *p<0.05. This increase of C5a levels upon cardiac resection accompanied the upregulation of C5aR1 observed in our RNA-seq screen, supporting a role for C5a-C5aR1 signaling in early cardiac regeneration. This data also gives further confidence in the approach of pursuing the consequences of genetic ablation of the C5aR1 receptor in the process of heart regeneration.
Genetic deletion of C5aR1 abolishes cardiomyocyte proliferation following apical resection
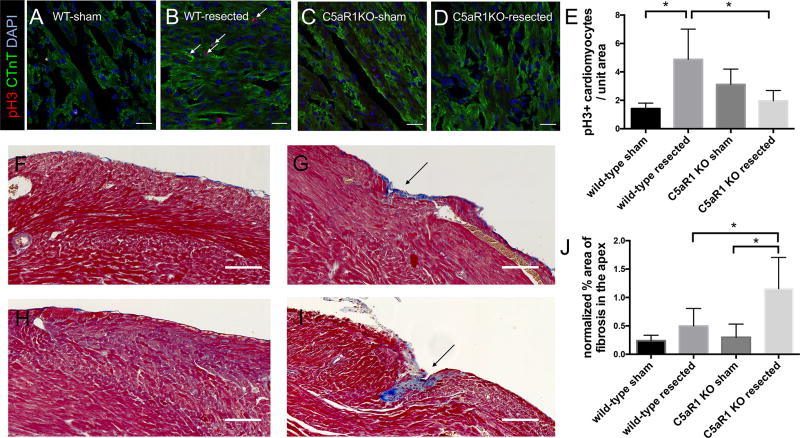
To further evaluate the role of C5aR1 in cardiac regeneration, a murine global genetic deletion model of C5aR1 was used. Cardiomyocyte proliferation was assessed in C5aR1 knock-out mice and wild-type mice after apical resection. Our hypothesis was that the cardiomyocyte proliferative response 7 days after apical resection would be reduced in C5aR1 KO mice in comparison to C5aR1 wild-type (WT) mice. Consistent with the regenerative response observed by Porrello et al. in neonatal mice6, the littermate WT mice had an increase in the fraction of proliferating cardiomyocytes 7 days post resection (Figure 6 A–E), while in the C5aR1 KO mice, no significant induction of cardiomyocyte proliferation is observed. This is consistent with the results of the receptor inhibition experiments, showing an essential role of C5aR1 in cardiomyocyte proliferation during regeneration.
Figure 6. Mice lacking C5aR1 mice have a reduction in cardiomyocyte proliferation after apical resection compared to wild-type littermate controls.
Representative images from C5aR1 wild-type sham surgery (A), C5aR1 wild-type resected (B), C5aR1 knock-out sham surgery (C) and C5aR1 knock-out resected (D). Cardiac troponin T (green), phosphoHistone 3 (red) and nuclei (blue), scale bar - 20µm. Quantification of proliferating cardiomyocytes shows the absence of proliferative response upon resection in C5aR1 knock-out hearts (E). *p <0.05, n≥4. Measurement of fibrosis 21 days after apical resection (F–J). Representative Masson Trichrome stained images from C5aR1 wild-type sham surgery (F), C5aR1 wild-type resected (G), C5aR1 knock-out sham surgery (H) and C5aR1 knock-out resected (I), scale bars, 100µm. Quantification of fibrosis in cardiac apex after resection (J), *p<0.05, n=8.
Furthermore, we analyzed scarring in the hearts of C5aR1 KO and littermate WT mice 21 days post-resection to investigate the effect of C5aR1 in cardiac regeneration at a longer time-point. We observed moderate scarring of the apex 21 days post-resection in C5aR1 KO mice, while no significant differences in scarring were observed after resection in matched wild-type control mice (Figure 6 F–J). Although we did not observe a complete inhibition of apical regeneration of the hearts of C5aR1 knock-out mice, an increase in fibrosis observed indicates that the overall regenerative process is partially hampered upon loss of C5aR1 (Figure 6 F–J). This increase of fibrosis was not observed in littermate wild-type hearts after resection, further supporting a role for C5aR1 in effective cardiac regeneration and minimization of fibrosis following an injury.
Expression of C5aR1 has been reported mainly in myeloid cells and macrophages51; therefore, we quantified macrophage infiltration after apical resection in C5aR1 wild-type and knock-out hearts, 48 hours post-resection, by immunostaining. While we saw a significant increase in the macrophage infiltration of the injured zone in C5aR1 wild-type mice 48 hours post-resection; we did not see any significant difference in macrophage infiltration between C5aR1 knock-out sham and resected hearts (Figure S5). Since we observed localization of C5aR1 with the endothelial marker CD31, we analyzed the number of blood vessels in the apex, 7 days post-resection, in C5aR1 knock-out and wild-type mice. The total number of blood vessels per field of view was quantified. We did not see a significant difference in the number of blood vessels between sham and resected hearts of both C5aR1 knock-out and wild-type mice (Figure S6). Therefore, further downstream experiments need to be carried out with endothelial cell-specific deletion for estimating the effect of C5aR1 on angiogenesis after cardiac injury.
Discussion
Achieving cardiac regeneration in adult mammals is an important goal for cardiovascular research. Although it is now known that adult mammalian cardiomyocytes can undergo cell proliferation to generate new cardiomyocytes, the rate of proliferation is insufficient to replace lost cardiomyocytes and restore function after a cardiac injury57, 58. Previous work has established multiple models of successful heart regeneration, including neonatal mice, zebrafish, and axolotl1–4. Over the past decade, studies of cardiac regeneration in different experimental models has shed light on several shared mechanisms across the species including stimulation of an essential immune response, a role for nerves and a critical role of cardiomyocyte division4, 28, 46. However, we still do not understand the barriers to heart regeneration that lead to extensive scarring and eventual heart failure in humans who have major cardiac injuries. This study leveraged similarities among model organisms that have substantial heart regenerative capacity to define a regenerative molecular pathway in the heart across species. The use of three unique model systems to identify the transcriptomic landscape of early cardiac regeneration provided insight into an evolutionarily conserved pathway underlying regeneration.
Our observation that C5aR1 is important for effective cardiac regeneration in an evolutionarily-conserved manner fits with the existing body of literature supporting a role for the complement system in mediating regenerative responses19. Although the role of macrophages in cardiac regeneration has been previously examined46, the role played by the immune system in initiating the cardiac regenerative response is not clearly understood. C5a and its receptor C5aR1 have been previously studied in the adult murine heart after septic shock, ischemia-reperfusion and in hypertension24, 52, 53, 59, 60. Activation of C5aR1 due to excessive C5a generation in sepsis has been shown to contribute to cardiac dysfunction as excessive C5a results in increased ROS and impaired Ca2+ handling in the cardiomyocytes52, 53. However, it has also been shown that C5a - C5aR1 signaling decreases hypertension-induced fibrosis in an angiotensin II – induced hypertension model61. Our finding that C5aR1 knock-out mice have increased scarring of the heart following apical resection agrees with the observations made by Weiss et al., who found that loss of C5aR1 results in increased cardiac fibrosis upon stress or cardiac injury. Furthermore, C5aR1 has been shown to be upregulated in the adult mouse heart; following ischemia / reperfusion, C5aR1 upregulation was evident in ischemic cardiomyocytes24. However, the role of C5aR1 and complement signaling in initiating cardiomyocyte proliferation and cardiac regeneration has not been investigated.
Following apical resection, we observe an upregulation of C5aR1 expression in cardiomyocytes and endothelial cells in the neonatal murine heart (Figure 5). This suggests a role for non-myeloid C5aR1 in heart regeneration. Although we did not observe localization of C5aR1 with the macrophage marker, differences in macrophage infiltration of the injured zone between C5aR1 knock-out and wild-type mice necessitate further downstream studies into the interplay of immune cells and complement components in early cardiac regeneration. It should be noted that differences in macrophage infiltration were not quantified by flow cytometry. Cell-specific conditional C5aR1 deletion models in cardiomyocytes, endothelial cells and myeloid cells need to be examined in future experiments to define the cell-type(s) expressing C5aR1 that contributes to cardiomyocyte proliferative response following injury. Furthermore, the intracellular signaling mechanisms by which cardiomyocyte division is initiated after an injury by C5a signaling in the adult heart may provide new insights. Although we did not observe a significant increase in cardiomyocyte proliferation upon treatment with C5a in vitro, it is possible that the effect of C5a – C5aR1 on cardiomyocyte proliferation could be mediated by paracrine signals from non-myocytes.
A pilot experiment was conducted to analyze the therapeutic potential of C5aR1 activation in adult murine cardiac regeneration, following an ischemia – reperfusion (I/R) injury . A pilot dose of C5a was administered for four days after I/R. We observed a marginal, non-significant increase in cardiomyocyte proliferation in the adult mouse heart after I/R and treatment with C5a. Future analyses of the mechanistic processes underlying the proliferative effect seen by C5aR1 activation need to be conducted. Additionally, elucidation of the mechanisms that lead to the transcriptional upregulation of the receptor gene, C5aR1, following injury should provide important details for how the system is regulated. Finally, we have not yet defined the dynamics of the entire complement cascade in these models in comparison to myocardium that fails to regenerate, such as in adult mammals, and these studies might reveal that complement activation is different or identical in these settings.
Currently, treatment for myocardial infarction focuses on reperfusion and limiting ventricular remodeling60; therapy targeted at replenishing lost cardiomyocytes has not entered the clinical arena. Our findings uncover a molecular pathway involving the interplay of the complement system and cardiac regeneration. We suggest that defining the molecular events of successful heart regeneration could eventually allow us to understand what events are absent in myocardium that cannot regenerate.
Supplementary Material
Clinical perspective.
What is new?
RNA-seq analysis of models of successful cardiac regeneration identified evolutionarily conserved mechanisms that are active in early cardiac regeneration.
Genes associated with inflammatory pathways are upregulated in all three species.
Complement receptors (C5aR1 and C3aR1) were highly upregulated in a conserved manner during early cardiac regeneration.
Inhibition of C5aR1 inhibits cardiac regeneration, reducing the number of proliferating cardiomyocytes and increasing scar formation.
Genetic deletion of C5aR1 results impairs cardiac regeneration after apical resection.
What are the clinical implications?
Identification of complement receptors induced in early cardiac regeneration provides insight into how successful heart regeneration can be initiated.
Understanding how organisms achieve heart regeneration may reveal why adult humans fail to regenerate myocardium.
Acknowledgments
Supported by HL117986, HL119230, and the Leducq Foundation (RTL); National Institute of Health Common Fund/ National Institute of Child Health and Human Development 1DP2HD087953 (JLW); RO1 AI121386 (CJG); NIH (R01 HL109264, R24 OD017870 (CAM) Howard Hughes Medical Institute Gilliam Fellowship (DMB); National Science Foundation Graduate Research Fellowship DGE1144152 (MS); Eurpoean Molecular Biology Organization Long-Term Fellowship (ALTF 253-2014) and a Fund for Medical Discovery Award from the Executive Committee on Research at Massachusetts General Hospital (J.M.G.-R). The zebrafish regeneration studies were supported by National Institute of Health award R01 HL127067 to CGB and CEB and by Hassenfeld and d’Arbeloff Massachusetts General Hospital Research Scholar Awards to CEB.
We thank the Harvard Center for Biological Imaging for infrastructure and support.
We would like to thank Lingsheng Dong and Harvard Medical School's Research Computing group for their helpful advice and guidance with the analysis of the RNA-seq data presented in this study. Portions of this research were conducted on the Orchestra High Performance Compute Cluster at Harvard Medical School. This NIH supported shared facility consists of thousands of processing cores and terabytes of associated storage and is partially provided through grant NCRR 1S10RR028832-01. See http://rc.hms.harvard.edu for more information.
Abbreviations
- GPCR
G-protein coupled receptor
- C5aR1
Complement 5a receptor 1
- pH3
phospho Histone 3
- PCNA
Proliferating cell nuclear antigen
- C5a
Complement 5a
Footnotes
Author contributions: Niranjana Natarajan, Aysu Uygur, Donald Bryant, Craig Gerard, Calum Macrae, Caroline Burns, C. Geoffrey Burns, Jessica Whited and Richard Lee participated instudy design and supervision of the project. Donald Bryant and Aysu Uygur performed zebrafish and axolotl experiments in the Macrae and Whited laboratories. Juan Manuel González and Michka Sharpe performed zebrafish experiments in the Burns’ laboratory. Niranjana Natarajan, Yamen Abbas, Nhi Ho, Aysu Uygur and Lucas Cocco performed mouse experiments in the Lee laboratory. Donald Bryant and Nhi Ho conducted analyses on the RNA-seq data for cross-species comparison of gene expression. All authors reviewed and approved the final manuscript, which was drafted by Niranjana Natarajan and Richard Lee.
Conflict of interest disclosures: None
References
- 1.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 2.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gamba L, Harrison M, Lien CL. Cardiac regeneration in model organisms. Curr Treat Options Cardiovasc Med. 2014;16:288. doi: 10.1007/s11936-013-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uygur A, Lee RT. Mechanisms of Cardiac Regeneration. Dev Cell. 2016;36:362–374. doi: 10.1016/j.devcel.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon HG, Odelberg S. Assessing cardiomyocyte proliferative capacity in the newt heart and primary culture. Methods Mol Biol. 2015;1290:227–240. doi: 10.1007/978-1-4939-2495-0_18. [DOI] [PubMed] [Google Scholar]
- 6.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foglia MJ, Poss KD. Building and re-building the heart by cardiomyocyte proliferation. Development. 2016;143:729–740. doi: 10.1242/dev.132910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porrello ER, Olson EN. A neonatal blueprint for cardiac regeneration. Stem Cell Res. 2014;13:556–570. doi: 10.1016/j.scr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P American Heart Association Statistics C and Stroke Statistics S. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eschenhagen T, Bolli R, Braun T, Field LJ, Fleischmann BK, Frisen J, Giacca M, Hare JM, Houser S, Lee RT, Marban E, Martin JF, Molkentin JD, Murry CE, Riley PR, Ruiz-Lozano P, Sadek HA, Sussman MA, Hill JA. Cardiomyocyte Regeneration: A Consensus Statement. Circulation. 2017;136:680–686. doi: 10.1161/CIRCULATIONAHA.117.029343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerard C, Gerard NP. C5A anaphylatoxin and its seven transmembrane-segment receptor. Annu Rev Immunol. 1994;12:775–808. doi: 10.1146/annurev.iy.12.040194.004015. [DOI] [PubMed] [Google Scholar]
- 15.Sarma JV, Ward PA. The complement system. Cell Tissue Res. 2011;343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajic G, Degn SE, Thiel S, Andersen GR. Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J. 2015;34:2735–2757. doi: 10.15252/embj.201591881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutkowski MJ, Sughrue ME, Kane AJ, Ahn BJ, Fang S, Parsa AT. The complement cascade as a mediator of tissue growth and regeneration. Inflamm Res. 2010;59:897–905. doi: 10.1007/s00011-010-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes T, Luz-Madrigal A, Reis ES, Echeverri Ruiz NP, Grajales-Esquivel E, Tzekou A, Tsonis PA, Lambris JD, Del Rio-Tsonis K. Complement anaphylatoxin C3a is a potent inducer of embryonic chick retina regeneration. Nat Commun. 2013;4:2312. doi: 10.1038/ncomms3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mastellos DC, Deangelis RA, Lambris JD. Complement-triggered pathways orchestrate regenerative responses throughout phylogenesis. Semin Immunol. 2013;25:29–38. doi: 10.1016/j.smim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura Y, Madhavan M, Call MK, Santiago W, Tsonis PA, Lambris JD, Del Rio-Tsonis K. Expression of complement 3 and complement 5 in newt limb and lens regeneration. J Immunol. 2003;170:2331–2339. doi: 10.4049/jimmunol.170.5.2331. [DOI] [PubMed] [Google Scholar]
- 21.Strey CW, Markiewski M, Mastellos D, Tudoran R, Spruce LA, Greenbaum LE, Lambris JD. The proinflammatory mediators C3a and C5a are essential for liver regeneration. J Exp Med. 2003;198:913–923. doi: 10.1084/jem.20030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mastellos D, Papadimitriou JC, Franchini S, Tsonis PA, Lambris JD. A novel role of complement: mice deficient in the fifth component of complement (C5) exhibit impaired liver regeneration. J Immunol. 2001;166:2479–2486. doi: 10.4049/jimmunol.166.4.2479. [DOI] [PubMed] [Google Scholar]
- 23.Niederbichler AD, Hoesel LM, Westfall MV, Gao H, Ipaktchi KR, Sun L, Zetoune FS, Su GL, Arbabi S, Sarma JV, Wang SC, Hemmila MR, Ward PA. An essential role for complement C5a in the pathogenesis of septic cardiac dysfunction. J Exp Med. 2006;203:53–61. doi: 10.1084/jem.20051207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Qin G, Liang G, Li J, Barrington RA, Liu DX. C5aR-mediated myocardial ischemia/reperfusion injury. Biochem Biophys Res Commun. 2007;357:446–452. doi: 10.1016/j.bbrc.2007.03.152. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, Zhou YQ, Yeh RF, Henkelman RM, Harvey RP, Metzger D, Chambon P, Stainier DY, Pollard KS, Scott IC, Bruneau BG. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cano-Martinez A, Vargas-Gonzalez A, Guarner-Lans V, Prado-Zayago E, Leon-Oleda M, Nieto-Lima B. Functional and structural regeneration in the axolotl heart (Ambystoma mexicanum) after partial ventricular amputation. Arch Cardiol Mex. 2010;80:79–86. [PubMed] [Google Scholar]
- 27.Mahmoud AI, Porrello ER, Kimura W, Olson EN, Sadek HA. Surgical models for cardiac regeneration in neonatal mice. Nat Protoc. 2014;9:305–311. doi: 10.1038/nprot.2014.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahmoud AI, O'Meara CC, Gemberling M, Zhao L, Bryant DM, Zheng R, Gannon JB, Cai L, Choi WY, Egnaczyk GF, Burns CE, Burns CG, MacRae CA, Poss KD, Lee RT. Nerves Regulate Cardiomyocyte Proliferation and Heart Regeneration. Dev Cell. 2015;34:387–399. doi: 10.1016/j.devcel.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo TH, Davis FG, Bateman J, Bryant S, Guzikowski AR, Tsai SL, Coyne S, Ye WW, Freeman RM, Jr, Peshkin L, Tabin CJ, Regev A, Haas BJ, Whited JL. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017;18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leng N, Li Y, McIntosh BE, Nguyen BK, Duffin B, Tian S, Thomson JA, Dewey CN, Stewart R, Kendziorski C. EBSeq-HMM: a Bayesian approach for identifying gene-expression changes in ordered RNA-seq experiments. Bioinformatics. 2015;31:2614–2622. doi: 10.1093/bioinformatics/btv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, Bardou P, Beck T, Blake A, Bonierbale M, Brookes AJ, Bucci G, Buetti I, Burge S, Cabau C, Carlson JW, Chelala C, Chrysostomou C, Cittaro D, Collin O, Cordova R, Cutts RJ, Dassi E, Di Genova A, Djari A, Esposito A, Estrella H, Eyras E, Fernandez-Banet J, Forbes S, Free RC, Fujisawa T, Gadaleta E, Garcia-Manteiga JM, Goodstein D, Gray K, Guerra-Assuncao JA, Haggarty B, Han DJ, Han BW, Harris T, Harshbarger J, Hastings RK, Hayes RD, Hoede C, Hu S, Hu ZL, Hutchins L, Kan Z, Kawaji H, Keliet A, Kerhornou A, Kim S, Kinsella R, Klopp C, Kong L, Lawson D, Lazarevic D, Lee JH, Letellier T, Li CY, Lio P, Liu CJ, Luo J, Maass A, Mariette J, Maurel T, Merella S, Mohamed AM, Moreews F, Nabihoudine I, Ndegwa N, Noirot C, Perez-Llamas C, Primig M, Quattrone A, Quesneville H, Rambaldi D, Reecy J, Riba M, Rosanoff S, Saddiq AA, Salas E, Sallou O, Shepherd R, Simon R, Sperling L, Spooner W, Staines DM, Steinbach D, Stone K, Stupka E, Teague JW, Dayem Ullah AZ, Wang J, Ware D, Wong-Erasmus M, Youens-Clark K, Zadissa A, Zhang SJ, Kasprzyk A. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain U, Woodruff TM, Stadnyk AW. The C5a receptor antagonist PMX205 ameliorates experimentally induced colitis associated with increased IL-4 and IL-10. Br J Pharmacol. 2013;168:488–501. doi: 10.1111/j.1476-5381.2012.02183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staab EB, Sanderson SD, Wells SM, Poole JA. Treatment with the C5a receptor/CD88 antagonist PMX205 reduces inflammation in a murine model of allergic asthma. Int Immunopharmacol. 2014;21:293–300. doi: 10.1016/j.intimp.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Rosa JM, Mercader N. Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat Protoc. 2012;7:782–788. doi: 10.1038/nprot.2012.025. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 43.Vivien CJ, Hudson JE, Porrello ER. Evolution, comparative biology and ontogeny of vertebrate heart regeneration. Npj Regenerative Medicine. 2016;1:16012. doi: 10.1038/npjregenmed.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavine KJ, Epelman S, Uchida K, Weber KJ, Nichols CG, Schilling JD, Ornitz DM, Randolph GJ, Mann DL. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci U S A. 2013;110:9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan MA, Assiri AM, Broering DC. Complement and macrophage crosstalk during process of angiogenesis in tumor progression. J Biomed Sci. 2015;22:58. doi: 10.1186/s12929-015-0151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett KM, Rooijakkers SH, Gorham RD., Jr Let's Tie the Knot: Marriage of Complement and Adaptive Immunity in Pathogen Evasion, for Better or Worse. Front Microbiol. 2017;8:89. doi: 10.3389/fmicb.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marder SR, Chenoweth DE, Goldstein IM, Perez HD. Chemotactic responses of human peripheral blood monocytes to the complement-derived peptides C5a and C5a des Arg. J Immunol. 1985;134:3325–3331. [PubMed] [Google Scholar]
- 50.March DR, Proctor LM, Stoermer MJ, Sbaglia R, Abbenante G, Reid RC, Woodruff TM, Wadi K, Paczkowski N, Tyndall JD, Taylor SM, Fairlie DP. Potent cyclic antagonists of the complement C5a receptor on human polymorphonuclear leukocytes. Relationships between structures and activity. Mol Pharmacol. 2004;65:868–879. doi: 10.1124/mol.65.4.868. [DOI] [PubMed] [Google Scholar]
- 51.Karsten CM, Laumonnier Y, Eurich B, Ender F, Broker K, Roy S, Czabanska A, Vollbrandt T, Figge J, Kohl J. Monitoring and cell-specific deletion of C5aR1 using a novel floxed GFP-C5aR1 reporter knock-in mouse. J Immunol. 2015;194:1841–1855. doi: 10.4049/jimmunol.1401401. [DOI] [PubMed] [Google Scholar]
- 52.Fattahi F, Ward PA. Complement and sepsis-induced heart dysfunction. Mol Immunol. 2017;84:57–64. doi: 10.1016/j.molimm.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalbitz M, Fattahi F, Herron TJ, Grailer JJ, Jajou L, Lu H, Huber-Lang M, Zetoune FS, Sarma JV, Day SM, Russell MW, Jalife J, Ward PA. Complement Destabilizes Cardiomyocyte Function In Vivo after Polymicrobial Sepsis and In Vitro. J Immunol. 2016;197:2353–2361. doi: 10.4049/jimmunol.1600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahajan SD, Tutino VM, Redae Y, Meng H, Siddiqui A, Woodruff TM, Jarvis JN, Hennon T, Schwartz S, Quigg RJ, Alexander JJ. C5a induces caspase-dependent apoptosis in brain vascular endothelial cells in experimental lupus. Immunology. 2016;148:407–419. doi: 10.1111/imm.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahajan SD, Parikh NU, Woodruff TM, Jarvis JN, Lopez M, Hennon T, Cunningham P, Quigg RJ, Schwartz SA, Alexander JJ. C5a alters blood-brain barrier integrity in a human in vitro model of systemic lupus erythematosus. Immunology. 2015;146:130–143. doi: 10.1111/imm.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skeie JM, Fingert JH, Russell SR, Stone EM, Mullins RF. Complement component C5a activates ICAM-1 expression on human choroidal endothelial cells. Invest Ophthalmol Vis Sci. 2010;51:5336–5342. doi: 10.1167/iovs.10-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eschenhagen T, Bolli R, Braun T, Field LJ, Fleischmann BK, Frisen J, Giacca M, Hare JM, Houser SR, Lee RT, Marban E, Martin JF, Molkentin JD, Murry CE, Riley PR, Ruiz-Lozano P, Sadek HA, Sussman MA, Hill JA. Cardiomyocyte Regeneration: A Consensus Statement. Circulation. 2017 doi: 10.1161/CIRCULATIONAHA.117.029343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalbitz M, Fattahi F, Grailer JJ, Jajou L, Malan EA, Zetoune FS, Huber-Lang M, Russell MW, Ward PA. Complement-induced activation of the cardiac NLRP3 inflammasome in sepsis. FASEB J. 2016;30:3997–4006. doi: 10.1096/fj.201600728R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiss S, Rosendahl A, Czesla D, Meyer-Schwesinger C, Stahl RA, Ehmke H, Kurts C, Zipfel PF, Kohl J, Wenzel UO. The complement receptor C5aR1 contributes to renal damage but protects the heart in angiotensin II-induced hypertension. Am J Physiol Renal Physiol. 2016;310:F1356–1365. doi: 10.1152/ajprenal.00040.2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.