Abstract
A strategy to rapidly determine if a matched unrelated donor (URD) can be secured for allograft recipients is needed. We sought to validate the accuracy of 1) HapLogic™ match predictions, and, 2) a resultant novel Search Prognosis (SP) patient categorization that could predict 8/8 HLA-matched URD(s) likelihood at search initiation. Patient prognosis categories at search initiation were correlated with URD confirmatory typing results. HapLogic™-based SP categorizations accurately predicted the likelihood of an 8/8 HLA-match in 830 patients (1,530 donors tested). Sixty percent of patients had 8/8 URD(s) identified. Patient SP categories (217 Very Good, 104 Good, 178 Fair, 33 Poor, 153 Very Poor, 145 Futile) were associated with a marked progressive decrease in 8/8 URD identification and transplantation. Very Good-Good categories were highly predictive of identifying and receiving an 8/8 URD regardless of ancestry. Europeans in Fair/ Poor categories were more likely to identify and receive an 8/8 URD compared to non-Europeans. In all ancestries, Very Poor and Futile categories predicted no 8/8 URDs. HapLogic™ permits URD search results to be predicted once patient HLA typing and ancestry is obtained dramatically improving search efficiency. Poor-Very Poor-Futile searches can be immediately recognized thereby facilitating prompt pursuit of alternative donors.
Keywords: Allogeneic hematopoietic stem cell transplantation, unrelated donors, search categorization
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) is a potentially curative therapy for many patients with malignant and non-malignant hematologic diseases. However, only 25-30% of potential allograft recipients will have a human leukocyte antigen (HLA)-identical sibling donor. In the absence of a suitable HLA-identical sibling, an 8/8 HLA-A, -B, -C, -DRB1 allele-matched URD has priority as the next best donor option at many centers. This match requirement is consistent with current recommendations1–3 and is based on multiple single center and registry studies4-9 including those from Memorial Sloan Kettering Cancer Center (MSKCC)10.
Unfortunately, most URDs listed on global registries are not typed at high resolution. The need to request samples for confirmatory typing (CT) can cause major delays in proceeding to donor clearance and transplantation especially in patients with difficult searches. This delay can adversely affect transplant outcomes and has special relevance for patients of non-European or mixed ancestries. Such patients due to diverse HLA haplotypes, lesser registry representation, and lower rates of donor availability are far less likely to identify donors who are HLA-matched11-13. It can also adversely affect the care of patients in need of urgent transplantation as disease status at the time of transplant is an important determinant of survival in patients with hematologic malignancies (reviewed in Lown et al14 and references15-17). Moreover, patients requiring urgent transplantation are an increasingly larger proportion of transplant service referrals. Consequently, until all donors on global registries can be fully typed at high resolution, a mechanism to rapidly and accurately predict URD search outcome is a significant unmet medical need.
The NMDP facilitates access to potential URDs through their online computer system, Traxis™ and collaborates with volunteer URD registries worldwide. Traxis™ allows transplant centers to request URD DNA samples for either customized typing (individual HLA locus typing) or CT (high resolution typing of all loci of interest). In a preliminary search, after entering a patient’s typing, Traxis™ displays two lists of potential URDs. The first list includes the upfront NMDP search result (domestic U.S. and affiliated international registry donors) and the second is the BMDW list (all other international URDs not listed on the upfront NMDP list).
The inclusion of the HapLogic™ matching predictions into the upfront NMDP search was introduced in 2006, and as of 2008 it included the predictions at each of the donor’s 8 HLA loci. This technology was further refined in 2011. An example of the resultant chance of the potential donor being an 8/8 (or 10/10) HLA-allele match is shown in Figure 118. The aims of this study were to validate the HapLogic™ 8/8 and individual locus match predictions and a resultant MSKCC Search Prognosis (SP) categorization to permit rapid and accurate prediction of the outcome of URD searches at search initiation prior to donors being requested for CT.
Figure 1. Examples of HapLogic™ Match Predictions.
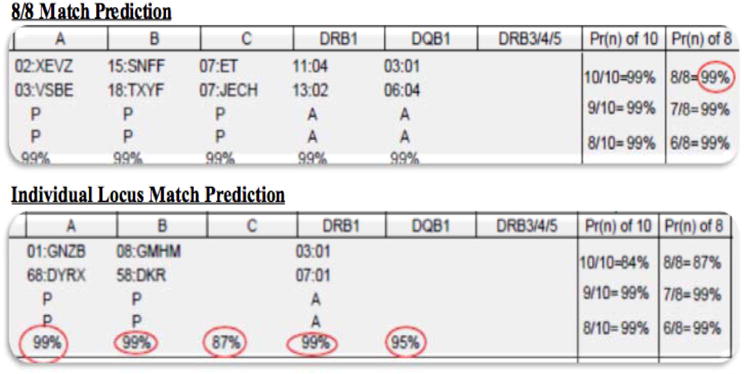
These examples were obtained from the NMDP Traxis™ system.
Methods
Patient Population
All patients (n = 830) referred to the Adult Bone Marrow Transplant Service at MSKCC who were ≥ 18 years old, diagnosed with a hematologic malignancy or aplastic anemia, and had an URD search performed from 2012-2015 were included in this analysis. Patient ancestry was prospectively obtained from detailed family history as previously described10. Patients were categorized into Europeans or non-Europeans, and ancestry subtype (northwestern European, eastern European, southern European, mixed European, African, Asian, Middle Eastern, White Hispanic, or mixed non-European) was also recorded. MSKCC IRB approval was obtained for all data collection and analysis.
Patient Searches
All patients were high resolution HLA-typed prior to commencing the search. At search initiation the results from the upfront NMDP Traxis™ search for each patient were prospectively collected and saved. Specifically, both the HapLogic™ match predictions for each HLA-locus and the predictions of the percentage likelihood of an 8/8 HLA-match for each potential donor were recorded. Up to the top 25 donors were evaluated (if available), but the number recorded and pursued varied according to how good the search was. Donor selection priority for pursuit of a donor sample was based on their likelihood of being an 8/8 HLA-match. After this, donor age was the next criterion with priority being given to younger donor age. Likelihood of donor availability (date of registration/ last contact) and donor center were also major characteristics of importance. Patients with limited donor options also underwent searches through the BMDW list. However, the BMDW list in Traxis does not include HapLogic™ match predictions.
HapLogic™ 8/8 Match Prediction Validation
Eligible URDs were defined as those that had HapLogic™ 8/8 match predictions recorded prior to any HLA typing being resulted, had at least an 1% chance of being an 8/8 match, and had a CT sample obtained and typed without any preceding customized typing requests. Donors who were matched at high resolution were included. These donors are listed as both an 8/8 HLA-match and having a HapLogic match prediction likelihood of 99%. We placed each eligible URD into one of the following groups based on their 8/8 match prediction: 99%, 90-98%, 80-89%, 70-79%, 60-69%, 50-59%, 40-49%, 30-39%, 20-29%, 10-19%, or 1-9%. The number of URDs who were confirmed by CT (once resulted) to be an 8/8 match was subsequently determined, and the frequency of 8/8 HLA-match within each group was compared to that group’s 8/8 HLA-match predicted range.
Individual Locus Match Prediction Validation
Eligible loci were those that had HapLogic™ individual locus match predictions recorded prior to any high resolution typing results being obtained and had at least a 1% chance of being a locus match. For loci that had both customized typing and full donor CT completed, only the customized typing result was included. Each eligible locus was placed into groups based on their locus match prediction: 99%, 90-98%, 80-89%, 70-79%, 60-69%, 50-59%, 40-49%, 30-39%, 20-29%, 10-19%, or 1-9%. The number of loci confirmed to be matched within each group was determined, and the frequency of locus match within each group was compared to the group’s predicted range.
Categorization of Patient Searches
Patients were placed into one of 6 SP categories devised at MSKCC as outlined in Table 1. The categories (Very Good, Good, Fair, Poor, Very Poor, or Futile) were based on the number of potential 8/8 URDs on a patient’s preliminary upfront NMDP search and the corresponding percentage chance of each URD being an 8/8 HLA-match according to HapLogic™. The aim was to segregate patient searches within a spectrum from Very Good to Futile.
Table 1.
SP Category Definitions
| SP Category | Chance of Matching at 8 Alleles* |
|---|---|
|
|
|
| Very Good | ≥ 20 8/8 donors with a ≥85% chance |
|
|
|
| Good | 5-19 8/8 donors with a ≥ 85% chance |
|
| |
| ≥ 20 8/8 donors with a ≥ 70% chance | |
|
| |
| Fair | 1-4 8/8 potential donors with ≥ 85% chance |
|
| |
| 1-19 8/8 potential donors with ≥ 70% chance | |
| ≥ 5 8/8 potential donors with a 40-69% chance | |
|
| |
| Poor | 1-4 8/8 potential donors with 40-69% chance |
|
| |
| ≥ 1 8/8 potential donor with 25-39% chance | |
|
| |
| Very Poor | ≥ 1 8/8 potential donor with ≤ 24% chance |
|
|
|
| Futile | 0 8/8 potential donors |
percentage likelihoods according to HapLogic™ 8/8 match predictions.
Search Outcomes Analysis
Search outcomes were analyzed by SP category and results for European versus non-European patients within each category were compared. The search outcomes examined were: 1) whether the patient had at least one 8/8 URD confirmed by CT (i.e. identified), and, 2) whether the patient proceeded to transplant using an 8/8 URD. The frequency of potential 8/8 URD availability was also examined.
Statistical analysis
The Brier score was used to assess the accuracy of the HapLogic™ 8/8 URD match and the individual locus match predictions. The improvement in the Brier score over a null model where all URD or loci are assigned the same predicted value was assessed using a permutation test. For the locus level prediction, the permutation was done within each donor for each locus. A Fisher’s exact test or chi-square test was used to compare identification of or transplantation with an 8/8 URD based on patient SP groups. All analyses were conducted using the R statistical program (R Foundation for Statistical Computing, Vienna, Austria).
Results
Patient Characteristics and Number of Donors Tested
Of the 830 patients studied, there were 558 European patients and 268 Non-European patients with 4 patients having unknown ancestry. European patients (n = 558) included: 170 northwestern Europeans, 150 eastern Europeans, 91 southern Europeans, 144 mixed Europeans, and 3 Europeans (specific sub-group not known). Non-European patients (n = 268) included 44 Asians, 96 Africans, 52 White Hispanic, 17 Middle Eastern, and 59 Mixed Non-European.
Of the 830 patients, 718 (87%) patients had at least one potential 8/8 URD (on the upfront NMDP search or the BMDW list) whereas 112 (13%) did not. The majority of donors were low/ intermediate resolution typed at one or more loci or missing HLA-C. Of the 718 patients with at least one potential 8/8 URD, we pursued 3,432 (4.8 per patient) for CT. Of these, we obtained 2,055 donor samples. Of these 2,055 donor samples, 1,530 had HapLogic™ donor match predictions recorded. The most common reason for not having a HapLogic™ prediction recorded was due to the donor being from the BMDW.
Validation of HapLogic™ 8/8 URD and Individual Locus Match Predictions
The HapLogic™ 8/8 match prediction validation is shown in Table 2 and Figure 2. The 70-79%, 60-69%, and 1-9% URD groups had 8/8-match rates slightly outside of their respective HapLogic™ match prediction ranges at 88%, 53% and 11%, respectively. Otherwise, all other URD groups had frequencies of 8/8-match within their HapLogic™ match prediction ranges. With a Brier score of 0.035, and an associated p-value of < 0.001, there was a strong association between the estimate of the likelihood of 8/8 HLA-match and the percentage match rate for each donor group.
Table 2.
HapLogic™ 8/8 Match Prediction Validation, N = 1,530 URDs
| Prediction: Chance of 8/8 Match | N URDs Tested | CT Result: N (%) URDs 8/8 Matched |
|---|---|---|
| 99% | 1218 | 1,207 (99%) |
| 90 – 98% | 92 | 85 (92%) |
| 80 – 89% | 42 | 35 (83%) |
| 70 – 79% | 24 | 21 (88%)* |
| 60 – 69% | 19 | 10 (53%)* |
| 50 – 59% | 11 | 6 (55%) |
| 40 – 49% | 14 | 6 (43%) |
| 30 – 39% | 13 | 4 (31%) |
| 20 – 29% | 23 | 6 (26%) |
| 10 – 19% | 19 | 3 (16%) |
| 1 – 9% | 55 | 6 (11%)* |
Result for URD group out of predicted range.
Figure 2. Validation of the accuracy of Haplogic™ donor predictions.
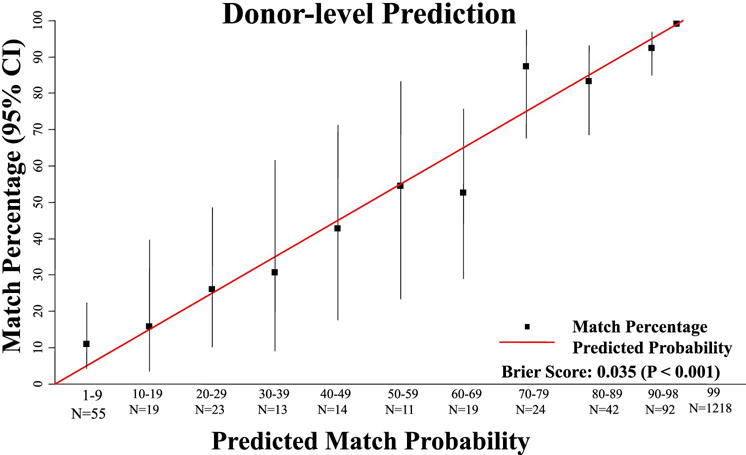
This figure shows a highly significant correlation between the predicted 8/8 HLA-match probability and the result of the donor CT.
The HapLogic™ match prediction validation for individual loci is displayed in Table 3 and Figure 3. The 70-79% and 40-49% locus groups had a locus match rate slightly higher than their respective HapLogic™ match prediction ranges at 80% and 52%, respectively, whereas the frequencies of individual locus match for all other locus groups were within their HapLogic™ match prediction ranges. Similar to the donor level results, the predictions for individual loci were overall well calibrated (Brier score 0.030, p-value < 0.001).
Table 3.
HapLogic™ Individual Locus Match Prediction Validation, N = 11,141 Loci
| Prediction: Chance of Locus Match | N Loci Tested | CT Result: N (%) Loci Matched |
|---|---|---|
| 99% | 8673 | 8627 (99%) |
| 90 – 98% | 444 | 411 (93%) |
| 80 – 89% | 161 | 135 (84%) |
| 70 – 79% | 126 | 101 (80%)* |
| 60 – 69% | 131 | 90 (69%) |
| 50 – 59% | 115 | 61 (53%) |
| 40 – 49% | 122 | 64 (52%)* |
| 30 – 39% | 150 | 46 (31%) |
| 20 – 29% | 168 | 40 (24%) |
| 10 – 19% | 224 | 35 (16%) |
| 1 – 9% | 827 | 38 (5%) |
Result for locus group out of predicted range.
Figure 3. Validation of the accuracy of Haplogic™ HLA-locus predctions.
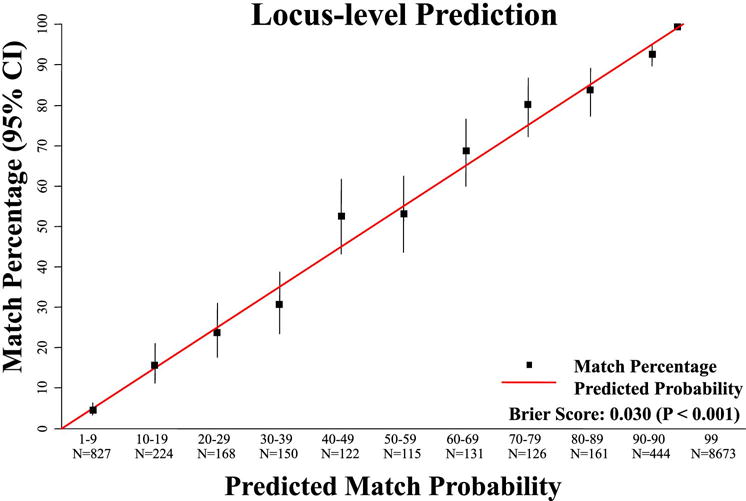
This figure shows a highly significant correlation between the predicted locus HLA-match probability and the result of the high resolution locus CT.
Donor Identification and 8/8 HLA-Matched URD Transplantation by Patient SP Category and Ancestry
Table 4 demonstrates search outcomes by SP category with each group stratified by patient ancestry. In total, 499 (60%) of 830 patients had ≥ 1 identified 8/8 URD. Additionally, 286 (65%) of the 443 patients who had a transplant received an 8/8 URD as the graft source. SP categories from Very Good to Futile demonstrated a decreasing likelihood (100% to 6%) of identifying an 8/8 URD, and a decreasing likelihood (99% to 2%) of receiving an allograft from an 8/8 URD. Search categories Very Good and Good were highly predictive of both identified 8/8 URD and 8/8 URD transplantation regardless of patient ancestry. Notably, Europeans in search categories Fair and Poor were more likely to have 8/8 URDs identified and receive an 8/8 URD transplant compared to non-Europeans. Search categories Very Poor and Futile were highly predictive of no 8/8 URD identified and no transplant with an 8/8 URD. Non-Europeans in the Very Poor and Futile search categories almost never had an 8/8 URD identified and none received an allograft with an 8/8 URD. However, URD access for Europeans in these search categories was also very poor.
Table 4.
SP Category and Patient Ancestry* by Search Outcome, N = 830 Patients
| SP Category by Patient Ancestry | N Patients | N (%) with Identified 8/8 URD | Patient Ancestry p-value | N Received Transplant | N (%) Transplants with 8/8 URD | Patient Ancestry p-value |
|---|---|---|---|---|---|---|
| Total: | 830 | 499 (60%) | — | 443 | 286 (65%) | — |
| Very Good | 217 | 217 (100%) | — | 137 | 135 (99%) | |
| European | 188 | 188 (100%) | N/A | 119 | 118 (99%) | 0.235 |
| Non-European | 28 | 28 (100%) | 17 | 16 (94%) | ||
| Good | 104 | 104 (100%) | — | 67 | 64 (96%) | — |
| European | 86 | 86 (100%) | N/A | 55 | 53 (96%) | 0.452 |
| Non-European | 18 | 18 (100%) | 12 | 11 (92%) | ||
| Fair | 178 | 136 (76%) | — | 87 | 69 (79%) | — |
| European | 119 | 97 (82%) | 0.024 | 60 | 52 (87%) | 0.018 |
| Non-European | 58 | 38 (66%) | 26 | 16 (62%) | ||
| Poor | 33 | 16 (48%) | — | 17 | 10 (59%) | — |
| European | 19 | 14 (74%) | 0.001 | 12 | 10 (83%) | 0.003 |
| Non-European | 14 | 2 (14%) | 5 | 0 (0%) | ||
| Very Poor | 153 | 18 (12%) | — | 71 | 7 (10%) | — |
| European | 89 | 15 (17%) | 0.024 | 50 | 7 (14%) | 0.180 |
| Non-European | 63 | 3 (5%) | 20 | 0 (0%) | ||
| Futile | 145 | 8 (6%) | — | 64 | 1 (2%) | — |
| European | 57 | 6 (11%) | 0.058 | 30 | 1 (3%) | 0.476 |
| Non-European | 87 | 2 (2%) | 33 | 0 (0%) |
4 patients had missing ancestry information and are included according to their Search Prognosis category result but could not be included in the sub-analysis of European versus non-European patients within each group.
Potential 8/8 URD Availability After CT Request By Patient Ancestry
Potential 8/8 URD availability after CT request was stratified by patient ancestry (Table 5). URD availability for Europeans (64%) was greater than that of non-Europeans (48%) (p < 0.001). The ancestry subtypes with the lowest URD availability rates were the White Hispanic (46%), Asian (43%), and African (39%) patients.
Table 5.
Potential 8/8 URD Availability After CT Request Stratified by Patient Ancestry*, N = 3,353 URDs
| Patient Ancestry | N URDs Pursued for CT For Each Patient Group* | N URDs Available for CT | Patient Ancestry p-value |
|---|---|---|---|
| Total: | 3,353 | 2,040 (61%) | — |
|
European (n = 558 patients) |
2,618 | 1,682 (64%) | < 0.001 |
|
Non-European (n = 268 patients) |
726 | 352 (48%) | |
| Ancestry Subtype | — | — | — |
| NW European | 869 | 575 (66%) | — |
| Eastern European | 793 | 498 (63%) | — |
| Southern European | 277 | 186 (67%) | — |
| European Mix | 672 | 413 (61%) | — |
| Asian | 166 | 71 (43%) | — |
| African | 148 | 58 (39%) | — |
| White Hispanic | 193 | 89 (46%) | — |
| Middle Eastern | 47 | 31 (66%) | — |
| Mixed Non-European | 175 | 109 (62%) | — |
9 URDs were excluded from the European versus non-European analysis as the European versus non-European ancestry was not determined for 4 patients. Also, 13 URDs were excluded from ancestry subtype analysis due to undetermined ancestry subtype in a total of 7 patients.
BMDW 8/8 URD Search Outcomes by Patient Ancestry for Patients in Poor, Very Poor, or Futile SP Categories
Donor availability by ancestry was analyzed for the patients from the Poor, Very Poor, and Futile SP categories (n = 329) who only had potential 8/8 URD(s) identified via the BMDW list (and none in the NMDP upfront search). Once CT was resulted, Europeans (18/165, 11%) were more likely to have 8/8 URDs identified only via BMDW than non-Europeans (5/164, 3%) (p = 0.008). Furthermore, Europeans in these lower SP categories were more likely to receive an 8/8 URD allograft facilitated by the BMDW than non-Europeans (10 patients vs none) (p = 0.007).
Discussion
In this analysis we have shown for the first time that utilization of HapLogic™ by a transplant center can provide reliable 8/8 and individual locus HLA-match predictions for URDs. This is compelling support of using HapLogic™ or similar programs to derive an assessment of search prognosis. Furthermore, our SP categories reliably separate patients into a spectrum from nearly guaranteed 8/8 URD to none. Notably, however, search success differed according to patient ancestry within the Fair, Poor, and Very Poor SP categories with access to matched donors being significantly worse in non-Europeans. This is partially explained by potential 8/8 URDs for non-European patients having a worse CT availability rate than those for European patients. These disparities are also due in part to HapLogic™ match predictions only being available for potential 8/8 URDs on the upfront search in Traxis, and Europeans having a higher likelihood than non-Europeans of identifying an 8/8 URD that can only be accessed through the BMDW list. The NMDP is currently working on adding more international registries to their upfront search which in turn would include HapLogic™ match predictions for these donors. In the interim, our findings highlight the validity of HapLogic™ predictions as well as the importance of considering patient ancestry when initially assessing a patient’s search and determining the likelihood of obtaining a matched donor.
A notable value of this analysis is that it reflects a large volume of patients with highly diverse ancestries at a large U.S. transplant center. Nonetheless, our findings should be further investigated in even larger patient populations at other centers and in the future could be further refined. A transplant center validation of patient genotype frequency-based search prognostication described by Wadsworth et al19, and how this compares with our current approach of evaluating the actual searches, is also of interest. In the interim, the findings of the current study have major implications for searches. The SP categorization permits the development of an algorithm to guide search strategy so alternative graft sources (7/8 URDs, cord blood, or haploidentical donors20,21) can be immediately pursued in the event that an 8/8 URD will never be identified in the required time period, or is very unlikely. In situations in which a patient has a Fair search, alternative stem cell sources can be evaluated while potential 8/8 URDs are simultaneously requested for additional HLA typing. Conversely, patients with Very Good or Good searches will very likely have at least one 8/8 URD identified regardless of patient ancestry. Thus, pursuit of alternative graft sources and the associated costs may not be necessary unless a search is very urgent.
Overall, the use of this algorithm has dramatically improved search efficiency at our center as it has facilitated efficient patient triage to cord blood or haplo-identical donor transplantation in the absence of an 8/8 matched URD, or the acceptance of a 7/8 HLA-matched URD. This can speed access to transplant and almost guarantees a donor for all patients. This approach can also facilitate cost savings from streamlining the process. Optimal efficiency of this approach to searches is contingent upon: 1) a clearly accepted definition of urgency and time frame for transplantation and 2) what alternative graft source is the priority (and available) if an 8/8 URD is not feasible in the required time period.
Recognition of futile searches should provide robust evidence that would argue against waiting for donors that will never likely eventuate. Our findings have major relevance to centers who would prefer to use an alternative donor (haplo-identical or cord blood) if a matched URD cannot be secured. Furthermore, the institution of alerts to transplant centers early in the search process notifying them of the search prognosis is appropriate, especially in difficult searches. Additionally, our data would also strongly argue against conducting futile donor drives that will not help the patient. At the very least the patient and family should be informed that such donor drives will have negligible impact on their search success if the search has already shown to be very poor or futile. That only two-thirds of donors for European patients and less than half for those of Asian, African and white Hispanic patients are available for CT should also be recognized. This is especially important to efficiently triage search strategy to successful alternative donors.
Highlights.
HapLogic™ provides reliable 8/8 and individual locus HLA-match predictions for unrelated donors (URDs).
A novel Search Prognosis patient categorization can accurately predict 8/8 HLA-matched URD likelihood at search initiation.
Rapid recognition of poor, very poor or futile searches can facilitate immediate pursuit of alternative graft sources.
Acknowledgments
This research was supported in part by P30 CA008748.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributions
Eric Davis, Courtney Byam, and Juliet N. Barker designed and performed the research, and analyzed the data. Sean Devlin analyzed the data. All authors wrote the paper.
Conflicts of Interest
The authors have no relevant conflicts of interest to declare.
References
- 1.Spellman SR, Eapen M, Logan BR, et al. A perspective on the selection of unrelated donors and cord blood units for transplantation. Blood. 2012;120(2):259–65. doi: 10.1182/blood-2012-03-379032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howard A, Fernandez-Vina MA, Appelbaum FR, et al. Recommendations for Donor HLA Assessment and Matching for Allogeneic Stem Cell Transplantation: Consensus Opinion of the Blood and Marrow Transplant Clinical Trials Network (BMT CTN) Biol Blood Marrow Transplant. 2015;21(1):4–7. doi: 10.1016/j.bbmt.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kollman C, Spellman SR, Zhang MJ, et al. The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood. 2016;127(2):260–7. doi: 10.1182/blood-2015-08-663823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flomenberg N, Baxter-Lowe LA, Confer D, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–1930. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 5.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 6.Woolfrey A, Klein JP, Haagenson M, et al. HLA-C antigen mismatch is associated with worse outcome in unrelated donor peripheral blood stem cell transplantation. Biology of Blood and Marrow Transplantation. 2011;17:885–892. doi: 10.1016/j.bbmt.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunstein CG, Eapen M, Ahn KW, et al. Reduced-intensity conditioning transplantation in acute leukemia: the effect of source of unrelated donor stem cells on outcomes. Blood. 2012;119(23):5591–5598. doi: 10.1182/blood-2011-12-400630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horan J, Wang T, Haagenson M, et al. Evaluation of HLA matching in unrelated hematopoietic stem cell transplantation for nonmalignant disorders. Blood. 2012;120(14):2918–24. doi: 10.1182/blood-2012-03-417758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milano F, Gooley T, Wood B, et al. Cord-Blood Transplantation in Patients with Minimal Residual Disease. N Engl J Med. 2016;375(10):944–953. doi: 10.1056/NEJMoa1602074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponce DM, Hilden P, Devlin SM, et al. High Disease-Free Survival with Enhanced Protection against Relapse after Double-Unit Cord Blood Transplantation When Compared with T Cell-Depleted Unrelated Donor Transplantation in Patients with Acute Leukemia and Chronic Myelogenous Leukemia. Biol Blood Marrow Transplant. 2015;21(11):1985–1993. doi: 10.1016/j.bbmt.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson JA, Gibbens Y, Tram K, et al. Identification of a 10/10 matched donor for patients with an uncommon haplotype is unlikely. HLA. 2017;89(2):77–81. doi: 10.1111/tan.12946. [DOI] [PubMed] [Google Scholar]
- 12.Gragert L, Eapen M, Williams E, et al. HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N Engl J Med. 2014;371(4):339–348. doi: 10.1056/NEJMsa1311707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker JN, Byam CE, Kernan NA, et al. Availability of Cord Blood Extends Allogeneic Hematopoietic Stem Cell Transplant Access to Racial and Ethnic Minorities. Biol Blood Marrow Transplant. 2010;16(11):1541–1548. doi: 10.1016/j.bbmt.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lown RN, Shaw BE. Beating the odds: factors implicated in the speed and availability of unrelated haematopoietic cell donor provision. Blood Marrow Transplantation. 2013;48(2):210–219. doi: 10.1038/bmt.2012.54. [DOI] [PubMed] [Google Scholar]
- 15.Grubovikj RM, Alavi A, Koppel A, Territo M, Schiller GJ. Minimal Residual Disease as a Predictive Factor for Relapse after Allogeneic Hematopoietic Stem Cell Transplant in Adult Patients with Acute Myeloid Leukemia in First and Second Complete Remission. Cancers (Basel) 2012;4(2):601–617. doi: 10.3390/cancers4020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giralt SA, Horowitz M, Weisdorf D, Cutler C. Review of Stem-Cell Transplantation for Myelodysplastic Syndromes in Older Patients in the Context of the Decision Memo for Allogeneic Hematopoietic Stem Cell Transplantation for Myelodysplastic Syndrome Emanating From the Centers for Medicare and Medicaid Services. J Clin Oncol. 2011;29(5):566–572. doi: 10.1200/JCO.2010.32.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarina B, Castagna L, Farina L, et al. Allogeneic transplantation improves the overall and progression-free survival of Hodgkin lymphoma patients relapsing after autologous transplantation: a retrospective study based on the time of HLA typing and donor availability. Blood. 2010;115(18):3671–3677. doi: 10.1182/blood-2009-12-253856.. [DOI] [PubMed] [Google Scholar]
- 18.Dehn J, Setterholm M, Buck K, et al. HapLogic: A Predictive Human Leukocyte Antigen-Matching Algorithm to Enhance Rapid Identification of the Optimal Unrelated Hematopoietic Stem Cell Sources for Transplantation. Biol Blood Marrow Transplant. 2016;22(11):2038–2046. doi: 10.1016/j.bbmt.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Wadsworth K, Albrecht M, Fonstad R, et al. Unrelated donor search prognostic score to support early HLA consultation and clinical decisions. Bone Marrow Transplant. 2016;51(11):1476–1481. doi: 10.1038/bmt.2016.162. [DOI] [PubMed] [Google Scholar]
- 20.Anasetti C, Aversa F, Brunstein CG. Back to the Future: Mismatched Unrelated Donor, Haploidentical Related Donor, or Unrelated Umbilical Cord Blood Transplantation? Biology of Blood and Marrow Transplantation. 2012;18(1):S161–S165. doi: 10.1016/j.bbmt.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Ballen KK, Koreth J, Chen Y-B, Dey BR, Spitzer TR. Selection of optimal alternative graft source: mismatched unrelated donor, umbilical cord blood, or haploidentical transplant. Blood. 2012;119(9):1972–1980. doi: 10.1182/blood-2011-11-354563.. [DOI] [PubMed] [Google Scholar]


