Summary
Adenosine (A) to inosine (I) RNA editing introduces many nucleotide changes in cancer transcriptomes. However, due to the complexity of posttranscriptional regulation, the contribution of RNA editing to proteomic diversity in human cancers remains unclear. Here we performed an integrated analysis of TCGA genomic data and CPTAC proteomic data. Despite of limited site diversity, we demonstrate that A-to-I RNA editing contributes to proteomic diversity in breast cancer through changes in amino acid sequences. We validate the presence of editing events at both RNA and protein levels. The edited COPA protein increases proliferation, migration, and invasion of cancer cells in vitro. Our study suggests an important contribution of A-to-I RNA editing to protein diversity in cancer and highlights its translational potential.
Graphical abstract

By an integrated analysis of TCGA genomic data and CPTAC proteomic data, Peng et al. show that A-to-I RNA editing contributes to proteomic diversity in breast cancer through changes in amino acid sequences. The edited COPA protein increases proliferation, migration, and invasion of cancer cells in vitro.
Introduction
A-to-I RNA editing is the most prevalent RNA editing mechanism in humans, where ADAR enzymes convert adenosine (A) to inosine (I) at specific nucleotide sites of select transcripts without affecting the DNA sequence identity (Bass, 2002). Although the vast majority of A-to-I editing events occur in non-coding regions, the absolute number of high-confidence missense RNA editing sites in humans is large (>1,000) (Bazak et al., 2014; Peng et al., 2012; Ramaswami et al., 2012; Ramaswami et al., 2013). Intriguingly, several individual editing events have been reported to play critical roles in tumorigenesis, such as AZIN1 editing in liver cancer (Chen et al., 2013), CDC14B editing in glioblastoma (Galeano et al., 2013), RHOQ editing in colorectal cancer (Han et al., 2014), SLC22A3 and IGFBP7 editing in esophageal cancer (Chen et al., 2017; Fu et al., 2017), PODXL editing in gastric cancer (Chan et al., 2016), and GABRA3 editing in breast cancer (Gumireddy et al., 2016). Using RNA-sequencing data from The Cancer Genome Atlas (TCGA), recent studies have detected a large number of A-to-I editing events in cancer transcriptomes, many of which show clinically relevant patterns (Fumagalli et al., 2015; Han et al., 2015; Paz-Yaacov et al., 2015).
However, the surveys on the patterns of RNA editing in human cancer have so far focused on the RNA level. Given the tremendous complexity of posttranscriptional regulation (Moore, 2005), we aimed to address to what extent genetic information engendered by missense A-to-I editing is translated to protein sequences, thereby contributing to proteomic diversity in cancer. The mass spectrometry (MS) data recently available from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) (Mertins et al., 2016; Zhang et al., 2014; Zhang et al., 2016) provide an opportunity to address this question since they were generated from patient sample cohorts with parallel genomic and transcriptomic data from TCGA (The Cancer Genome Atlas Research Network, 2011; The Cancer Genome Atlas Research Network, 2012a; The Cancer Genome Atlas Research Network, 2012b).
Results
Relative contributions of RNA editing and somatic mutations to cancer proteomic diversity
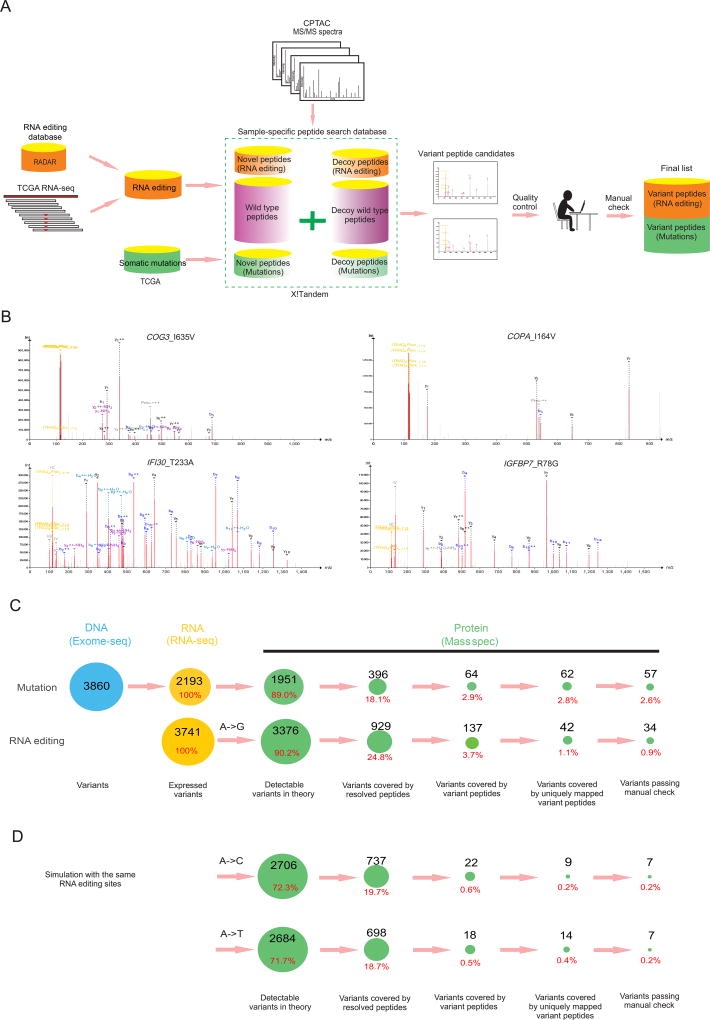
We obtained three CPTAC MS datasets (breast cancer [BRCA], ovarian cancer [OV], and colorectal cancer [CRC]) and focused respectively on 101 samples, 90 samples and 84 samples, with parallel RNA-seq and somatic mutation data in these datasets for subsequent analyses (Table S1). We combined this information trove with another LC-MS/MS-based dataset of the NCI60 cell line collection (Moghaddas Gholami et al., 2013). We developed a sample-customized search strategy to identify variant peptides caused by A-to-I RNA editing or somatic mutations (Figure 1A, STAR Methods). Briefly, for each cancer sample in a MS set, we first obtained somatic mutation data and detected missense RNA editing events using RNA-seq data based on well-annotated, literature-curated RNA editing sites (1,369 sites) in the RADAR database (Ramaswami and Li, 2014). We then constructed a customized peptide database by adding variant peptides resulting from the mutations and RNA editing events, and employed the X!Tandem algorithm (Wen et al., 2014) to search the corresponding MS data to identify variant peptide candidates at FDR = 0.01 (Table 1). In the analysis, we considered fixed and variable amino acid modifications as in the original CPTAC papers (Mertins et al., 2016; Zhang et al., 2014; Zhang et al., 2016). We implemented a series of quality control steps to rule out possibilities that the resolved peptide comes from a homologous protein or that the detected RNA editing event is due to variants at the DNA level such as mutations or SNPs. Finally, to reduce false positives to a minimal level, we manually reviewed all the candidate spectra identified from variant peptides to obtain the final list attributable to RNA editing or somatic mutations for each MS set. We also removed variants leading to N-to-D amino acid changes in the manual check step since they may be due to post-translational deamidation. Figure S1A–E summarizes the characteristics of the datasets surveyed and resolved peptides.
Figure 1. Relative contributions of RNA editing and somatic mutations to proteomic diversity in cancer.
(A) The flow chart of variant peptide identification using MS data. For each MS set, all missense RNA editing sites and somatic mutations from the corresponding samples were pooled to construct a sample-set-specific database. After the quality control steps, only variants with a uniquely mapped variant peptide were considered as candidates. We manually reviewed each candidate and generated the final lists. (B) Four representative peptide spectrum matches of variant peptides due to RNA editing (Upper: COG3_I635V, COPA_I164V; Lower: IFI30_T233A, IGFBP7_R78G). (C) Information flow during the analysis of CPTAC breast cancer samples. We dissected the genetic information flow from DNA to RNA to protein into seven steps: the number of variants at the DNA level, somatic mutations (variants); number of variants with evidence of expression, both RNA editing and mutations (expressed variants); number of MS detectable variants in theory; number of variants covered by resolved peptides (wild-type or variant); number of variants covered by variant peptides; number of variants covered by uniquely mapped variant peptides; and number of variants passing the manual check. (D) Information flow of simulation analysis with the same editing sites but different nucleotide changes (A-to-C and A-to-T). See also Tables S1–3, Figure S1.
Table 1.
Identified A-to-I RNA editing events with variant peptide support
| Protein | Position (hg19) | Strand | AA Change |
MS dataset | Variant Peptide | |||
|---|---|---|---|---|---|---|---|---|
| BRCA | OV | CRC | NCI60 | |||||
|
| ||||||||
| COG3 | chr13: 46,090,371 | + | I635V | 2 | 0 | 0 | 0 | EVSLDLKK |
|
| ||||||||
| COPA | chr1: 160,302,244 | − | I164V | 11 | 1 | 2 | 3 | VWDVSGLR |
| VWDVSGLRK | ||||||||
|
| ||||||||
| EEF1A1 | chr6:74,229,074 | − | T104A | 0 | 0 | 0 | 3 | NMIAGTSQADCAVLIVAAGVG |
| EFEAGISK | ||||||||
|
| ||||||||
| EEF1A1 | chr6: 74,229,116 | − | I90V | 1 | 0 | 0 | 0 | YYVTIVDAPGHR |
|
| ||||||||
| FLNB | chr3: 58,141,801 | + | Q2327R | 1 | 0 | 0 | 0 | RLTVMSLR |
|
| ||||||||
| HSP90AB1 | chr6: 44,219,922 | + | K550R | 5 | 0 | 0 | 0 | EGLELPEDEEER |
|
| ||||||||
| IFI30 | chr19: 18,288,551 | + | T223A | 2 | 0 | 0 | 0 | PLEDQTQLLALVCQLYQGK |
|
| ||||||||
| IGFBP7 | chr4: 57,976,286 | − | R78G | 11 | 0 | 0 | 0 | GEGEPCGGGGAGGGYCAPGM |
| ECVK | ||||||||
|
| ||||||||
| SERPINB6 | chr6:2,949,167 | − | E237G | 1 | 0 | 0 | 0 | ELNMIIMLPDGTTDLR |
Through this analysis, we detected a considerable number of RNA editing events with confident variant peptide support (Table 1, and some representative MS shown in Figure 1B). For BRCA, our analysis included, on average, 104 editing-introduced variants and 107 mutation-introduced variants per MS set of 3 cancer samples, and identified 34 RNA editing events and 57 mutations with variant peptide evidence across the 36 BRCA MS sets (note that editing or mutation variants at the same site but detected in different MS sets were counted as independent events). For OV, our analysis included, on average, 93 editing-introduced variants and 54 mutation-introduced variants per MS set of 3 cancer samples, and identified 1 RNA editing event and 25 somatic mutations with variant peptide support across the 64 OV MS sets. For CRC, our analysis included, on average, 17 editing-introduced variants and 342 mutation-introduced variants per cancer sample and identified 2 RNA editing events and 84 somatic mutations with variant peptide evidence over the 89 CRC MS sets. (See details in Table S2, Table S3) Given the same patient samples, our pipeline detected similar numbers of somatic mutations to those reported in previous studies (Mertins et al., 2016; Zhang et al., 2014), supporting the sensitivity of our approach. In addition to the three patient proteomic datasets, we detected another RNA editing site with supported variant peptides from the MS dataset of 34 NCI60 cell lines.
Among the three cancer patient cohorts surveyed, OV and CRC showed much lower numbers of variant peptides caused by RNA editing than somatic mutations, but strikingly, the BRCA dataset showed just over half as many variant peptides (60%) per patient sample as caused by mutations. Therefore, we performed a more detailed analysis of BRCA to dissect the information flow of RNA editing and somatic mutations (Figure 1C, Figure S1F). For the 101 BRCA samples, we started with 3,741 RNA editing events and 3,860 somatic mutations and identified 2,193 mutations with expression evidence at the RNA level. For both RNA editing and expressed mutations, ~90% of them were detectable by MS in theory (i.e., not Ile->Leu; and length of variant peptide 8~35 amino acids) (Swaney et al., 2010). However, only 24.8% of the RNA editing sites and 18.1% of the somatic mutations were covered by a resolved peptide (wild-type or variant), and ~3% of editing sites or somatic mutations were covered by a variant peptide, which was mainly due to the low coverage of CPTAC datasets. Among variants covered by variant peptides, almost all the mutation cases (97%: 2.8%/2.9%) were associated with uniquely mapped variant peptides, whereas this proportion was much lower (30%: 1.1%/3.7%) for RNA editing, which may be due to sequence similarity of ADAR targets. After careful manual review, our final list included 0.9% of the editing events and 2.6% of expressed somatic mutations as “confident hits” (STAR Methods). Although we employed FDR = 0.01 in variant peptide search, this “decoy peptides”-based FDR could underestimate the noise rate. To estimate the false positive rate more conservatively, we performed a simulation analysis through creating the same number of artificial RNA editing events with the nucleotide changes of A-to-C or A-to-T at the same sites. Using the same analytic procedure, the false positive rate for detected editing sites was estimated to be 20%, indicating that the true positive rate of the approach is likely 80% and that the majority of the detected editing events are “real” (Figure 1D). Of note, the site diversity of RNA editing for the patient cohort was much lower than that of mutations, as most of the observed variant peptides resulted from recurrent RNA editing sites across tumor samples (Table 1, Figure S1G). In addition, we analyzed correlations between RNA-editing level detected by RNA-seq and ion intensity of variant peptides in MS and found that across 34 MS sets, the sample with the highest variant peptide intensity in each dataset tended to be that with the highest RNA editing level (Binominal test, p < 0.05). These results indicate that RNA editing can introduce amino acid changes into proteins, contributing to protein diversity at least in breast cancer.
Independent validation of RNA editing events with variant peptide support
Across the four different MS datasets, we identified 9 unique RNA editing sites with variant peptide evidence (Table 1). Among them, editing at COPA_I164V and IGFBP_R78G was identified in 11 out of the 36 BRCA MS sets, and COPA_I164V was the only one identified in the four different tumor datasets (Table 1). As expected, these 9 editing sites tended to have a higher editing level (i.e., the proportion of edited reads among total mapped reads at the specific site of a given sample) and reside in genes with a higher expression than RNA editing sites without detected variant peptides (Figure S2A–C). In terms of predicted functional effects, RNA editing sites with variant peptide support were not more impactful than those without variant peptide support (Figure S2D). However, 4 of the RNA editing sites with variant peptide support (COG3_I635V, COPA_I164V, FLNB_Q2327R, and IGFBP7_R78G) have been previously reported in humans and mice (Pinto et al., 2014). We next validated editing signals at these sites using two independent approaches. First, using an RNA editing fingerprint assay (Crews et al., 2015), we validated 5 of these editing sites in a breast cancer cell line, Hs578T (Figure 2A). Second, we perturbed the expression of ADAR1 and ADAR2 in this cell line. Based on RNA-seq data upon overexpression or siRNA knockdown of specific ADAR enzymes, we observed dramatic changes in editing levels in all 6 sites with sufficient coverage (Figure 2B and Figure S3). Thus, these two approaches validated 7 out of 9 RNA editing sites, with 4 sites confirmed by both approaches (COG3_I635V, COPA_I164V, FLNB_Q2327R, and IFI30_T223A). Furthermore, these perturbation experiments revealed the ADAR enzymes responsible for observed editing signals: ADAR1 was responsible for EEF1A1_T104A and SERPINB6_E337G; ADAR2 was responsible for COG3_I635V, COPA_I164V, FLNB_Q2327R and IGFBP7_R78G; and both ADAR1 and ADAR2 contributed to editing of IFI30_T223A (Figure 2A, 2B). Collectively, these results provide independent evidence for 7 RNA editing sites that contributed to the vast majority of editing-introduced variant peptides identified (i.e., 28/34 in BRCA). The corresponding editing changes at the remaining two sites (EEF1A1_I90V and HSP90AB1_K550R) were not validated in the cell line surveyed potentially due to their low editing level (e.g., < 1%).
Figure 2. Experimental validation of RNA editing events with peptide support at the RNA level.
(A) The relative expression (top) and the editing level changes (bottom) after transfection of wild-type ADAR enzymes (ADAR1/2 WT) and catalytically-inactive ADAR enzymes (ADAR1/2 MUT), GFP serves as negative control. Error bars denote ± SD. (B) ADAR-perturbed RNA-seq experiment. The editing levels of the six RNA editing sites with sufficient coverage (×10) in different perturbed conditions, and GFP, mock and RISC_free served as negative controls. See also Figure S2, Figure S3.
Given the tremendous complexity of identifying variant peptides from proteome-wide MS data (Deutsch et al., 2016; Nesvizhskii, 2014), we sought to validate the effects of RNA editing on variant peptides in independent samples using a targeted MS approach. For this purpose, we focused on two RNA editing sites (COG3_I635V and COPA_I164V) because (i) COPA_I164V is the only site consistently identified across multiple MS datasets; and (ii) we recently reported the potential functional effects of COG3_I635V on cell viability and drug sensitivity (Han et al., 2015). We enriched COG3 and COPA proteins from an ovarian cancer cell line, OVCAR-8, through immunoprecipitation, and then performed LC-MS/MS analysis with spiked-in heavy isotope labeled synthetic peptides. Importantly, endogenous and synthetic peptides corresponding to both RNA editing events appeared with the matched m/z peaks (Figure 3A, B), and they also had the same retention time as the edited peptides (Figure 3C, D). In addition, the edited peptide in COPA was previously identified in mouse liver (Wu et al., 2014). These results provide additional support for the presence of these edited proteins in tumor cells.
Figure 3. Validation of variant peptides caused by RNA editing sites through LC-MS/MS with heavy isotope labeling synthetic peptides.
(A–D) Annotated MS (A, B) and retention times (C, D) for EVSLDLKK (COG3_I635V) (A, C) and VWDVSGLR (COPA_I164V) (B, D). The results of unlabeled endogenous variant peptide are shown at the top whereas the results of the spiked, labeled synthetic peptide are shown at the bottom of each panel. The heavy isotope labeled amino acids are shown in red.
Clinically relevant patterns of RNA editing events with variant peptide support
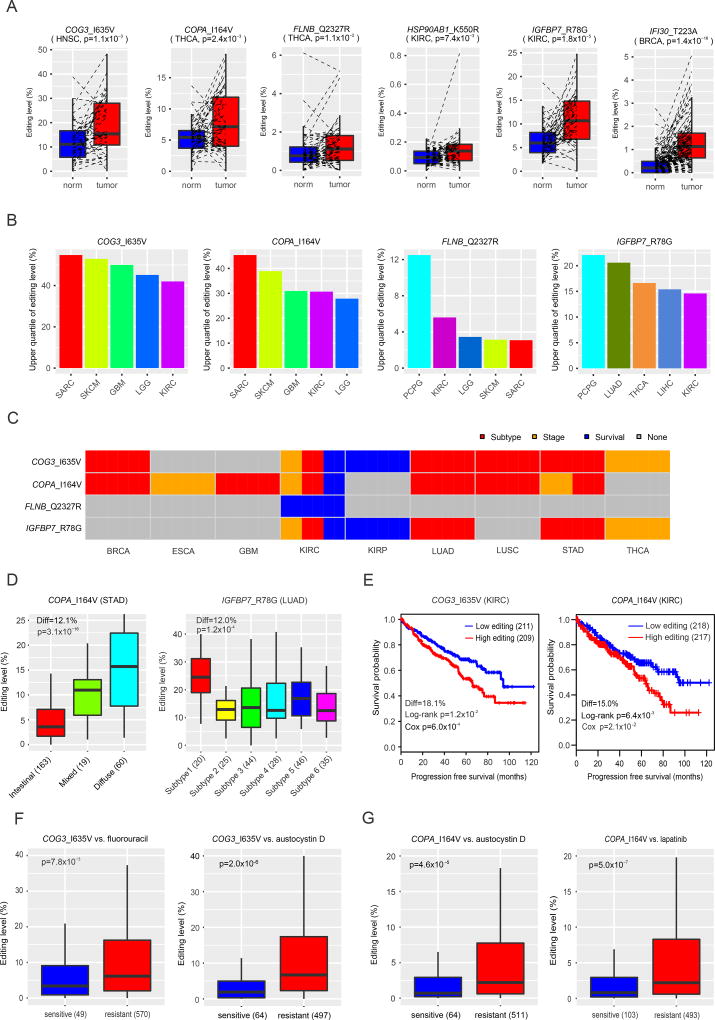
For the 8 unique RNA editing events with variant peptides detected in patient samples, one fundamental question is whether they, like “driver mutations”, can play active roles in tumor pathophysiology or simply represent “passenger” events. We carried out several analyses to address this question. First, unlike somatic mutations that are by definition cancer-specific, RNA editing usually occurs in both normal and tumor samples. To assess whether these RNA editing events are dysregulated in cancer, we compared their editing levels in tumor samples relative to the matched normal samples using TCGA RNA-seq data. Although the observed RNA editing patterns often varied in different tumor contexts, 6 out of the 8 RNA editing sites showed significant over-editing patterns in some cancer types (Figure 4A, Table S4). However, it should be noted that such tumor-normal comparisons could be misleading because tumor and normal samples usually contain very different cell compositions. For example, most cells in a breast tumor are epithelial cells, whereas the epithelial proportion in normal breast tissues is typically low (e.g., a few percent). We also detected RNA editing signals in the Genotype-Tissue Expression (GTEx) RNA-seq data of normal tissues (Figure S4A) (Picardi et al., 2017). Second, somatic mutations usually occur at a high allele frequency in tumor cells (e.g., 50% for heterozyous mutations and 100% for homozyous mutations for a diploid cancer genome). To assess the editing level (equivalent to the allele frequency) of these RNA editing sites in tumor samples, we performed a pan-cancer analysis using TCGA RNA-seq data of >8,000 tumor samples of 24 cancer types (Table S5). We found that RNA editing events for these sites generally could be detected in a broad range of cancer types, but the editing level varied greatly from site to site and from cancer type to cancer type. Importantly, four RNA editing sties (COG3_I635V, COPA_I164V, FLNB_Q2327R and IGFBP7_R78G) showed relatively high editing levels in a large portion of patients (e.g. >25% of patients) of multiple cancer types (Figure 4B). Although it is under debate about what variant level (%) is required for gain-of-function activity in cancer, our results clearly showed that the functional effects for amino acid changes caused by RNA editing events cannot be simply dismissed due to low editing level. Third, to assess their clinical relevance more thoroughly, we examined the correlations of these four RNA editing events with key clinical features using TCGA pan-cancer data and identified extensive significant patterns in different cancer types (FDR < 0.05; Editing Diff > 3%, Figure 4C, Table S4). For example, the editing level of COPA_I164V was increased in stomach adenocarcinoma subtypes, from intestinal, mixed, to diffuse (p = 3.1×10−16, Editing Diff = 12.1%, Figure 4D, Table S4). RNA editing at both COG3_I635V and COPA_I164V correlated with worse progression-free patient survival time in kidney renal clear cell carcinoma (COG3, log-rank p = 1.2×10−2, Cox model p = 6.0×10−4, Editing Diff = 18.1%; COPA, log-rank p = 6.4×10−3, Cox model p = 1.9×10−2, Editing Diff = 15.0%, Figure 4E, Table S4). Notably, ADAR1 and ADAR2 expression levels did not show significant correlations with patient survival times in this disease (Figure S4B), suggesting that the signals at individual RNA editing sites contain independent prognostic information from the ADAR enzymes responsible for their generation. Finally, we examined correlations between RNA editing levels and drug sensitivity using Cancer Cell Line Encyclopedia (CCLE) cell lines and identified that editing at COG3 and COPA was significantly associated with drug sensitivity (Figure 4F, G, Table S4). For example, higher RNA editing at COG3_I635V was significantly associated with resistance to fluorouracil (Rs = 0.21, p = 4.6×10−7, FDR < 0.01; Wilcoxon rank sum test p = 7.8×10−3, Figure 4F); and higher RNA editing at COPA_I164V was significantly associated with resistance to austocystin D (Rs = 0.33, p = 2.3×10−15, FDR < 0.01; Wilcoxon rank sum test p = 4.6×10−5, Figure 4G). Thus, the identified RNA editing events may be involved in tumorigenesis and have potential clinical implications.
Figure 4. Clinical relevance of A-to-I RNA editing events with peptide support.
(A) Normal-tumor comparison of RNA editing levels. Paired t test was used to assess statistical significance. (B) The upper quartile values of RNA editing levels of four RNA editing sites. For each editing site, only the top five cancer types with the highest editing levels are shown. (C) Clinically relevant patterns of RNA editing sites with peptide evidence in different cancer types. For each cancer type, grey boxes indicate not significant, red boxes indicate significantly differential editing levels among tumor subtypes (Kruskal-Wallis or Wilcoxon rank sum test, FDR < 0.05, editing level difference > 3%), orange boxes indicate significantly differential editing levels among stage (Kruskal-Wallis or Wilcoxon rank sum test, FDR < 0.05, editing level difference > 3%), and blue boxes indicate significant associations of editing level with progression-free survival times (log-rank or Cox model test, FDR < 0.05, editing level difference >3%). (D) Differential editing level of COPA_I164V (left) and IGFBP7_R78G (right) in stomach adenocarcinoma (STAD) subtypes (left) and lung Adenocarcinoma (LUAD) subtypes (right). Kruskal-Wallis test was used to assess statistical significance. (E) Correlations of editing level in COPA_I164V (left) and IGFBP7_R78G (right) with patient progression-free survival time in kidney renal clear cell carcinoma (KIRC). Log-rank test was used to assess statistical significance. (F) The association of editing level at COG3_I635V with the drug sensitivity of fluorouracil and austocystin D. (G) The association of editing level at COPA_I164V with the sensitivity to austocystin D and lapatinib. (F) and (G) Wilcoxon rank sum test was used to assess statistical significance. In (A), (D), (F) and (G), the horizontal line in the box is the median, the bottom and top of the box are the first and third quartiles, and the whiskers extend to 1.5 IQR of the lower quartile and the upper quartile, respectively. In (D)–(G), numbers in parentheses indicate the sample numbers included in each comparison group. See also Table S4, Table S5, Figure S4.
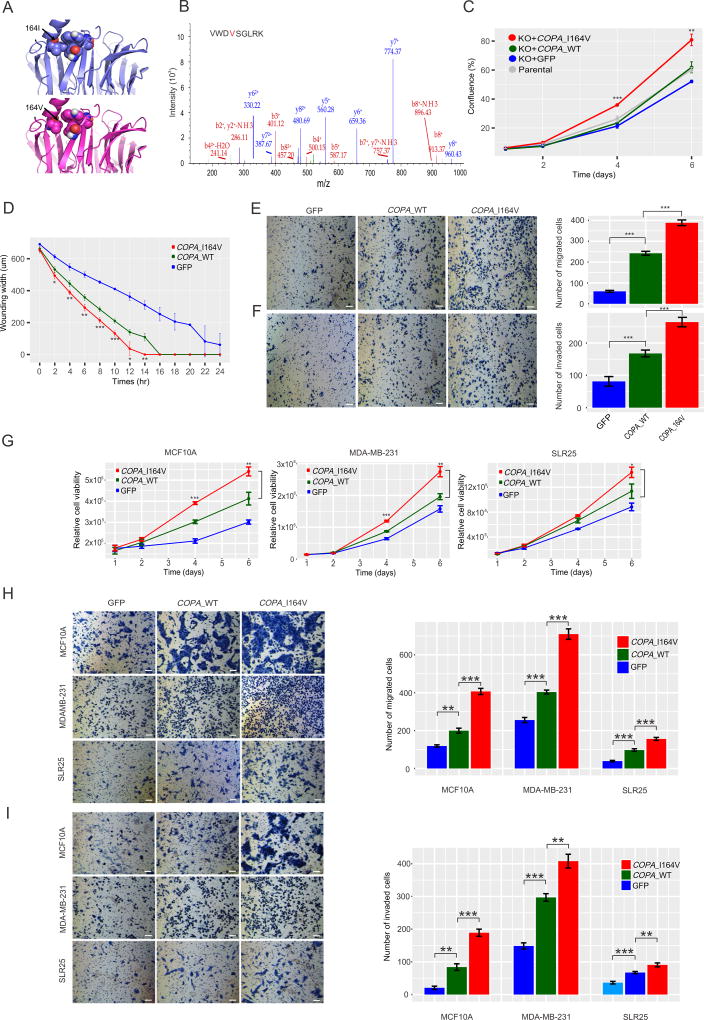
Functional effects of COPA editing on tumor cells
Since the above intriguing patterns do not necessarily imply a causal relationship, we next focused on COPA editing for experimental investigation because of its variant peptide prevalence across four MS datasets (Table 1), correlation of RNA editing level with variant peptide ion intensity (Figure S5A and B), relatively high editing level (Figure 4B), extensive clinical correlations across cancer types (Figure 4C), and a large predicted free energy change on protein conformation (Rosetta, 3.03 R.U., Figure 5A). In CRISPR/cas9 COPA knockout MDA-MB-231 (breast cancer) cells (Figure S5C), we introduced wild-type COPA and mutant COPA_I164V cDNAs, respectively, with the newly introduced COPA proteins being expressed at a level similar to that in parental cells (Figure S5D–F). The expression of edited COPA proteins was confirmed and their relative amount was assessed using LC-MS/MS (Figure 5B). Indeed, when mutant COPA was overexpressed, the proportion of edited proteins was markedly increased. We found that edited COPA_I164V significantly increased cell viability, wounding healing, migration and invasion compared with wild-type COPA (Student’s t-test, p < 0.05, Figure 5C–F). We observed similar patterns with introduction of wild-type and edited COPA into MDA-MB-231 cells where the parental RNA was depleted by shRNA targeting the 3'UTR (Figure S6, Table S6) as well as wild-type MDA-MB-231, MCF10A (a normal breast epithelial cell line) and SLR25 (a kidney cancer cell line) (Figure 5G–I). These results suggest that edited COPA can make a notable contribution to tumor development.
Figure 5. Functional effects of RNA editing at COPA_I164V.
(A) Impact of I164V on folding of COPA protein. (B) The annotated LC-MS/MS of the edited COPA peptide in the MDA-MB-231 cell line that overexpressed the edited COPA protein. (C–F) Effects of COPA editing on cell viability (C), wound healing (D), migration (E), and invasion (F) in CRISPR/cas9 COPA knockout MDA-MB-231 cells. (G–I) Effects of the edited COPA on cell viability (G) migration (H), and invasion (I) in three wild-type cell lines MCF10A, MDA-MB-231 and SLR25. All scale bars = 100 µm. All error bars denote ± SD. t test, *, p < 0.05; **, p < 0.01; ***, p < 0.001. All functional assays were performed simultaneously. See also Figure S5, Figure S6, Table S6.
Discussion
Understanding the mechanisms contributing to protein diversity in cancer cells is a fundamental issue in cancer research, since mutated proteins have been widely used as biomarkers and therapeutic targets, and more recently a major determinant for cancer immunotherapy response. This study provides large-scale direct evidence that the genetic information recoded by A-to-I RNA editing in cancer is manifest at the protein level. Although the absolute numbers of DNA mutational and RNA editing events detected at the protein level are low (likely due to the low coverage of the CPTAC MS data relative to exome-seq or RNA-seq data), our analysis provides a systematic estimation about the relative contributions of these two mechanisms to cancer protein diversity. Among the three cancer types surveyed, the contribution of RNA editing to cancer proteomic diversity is the most significant in BRCA. Even in terms of unique variants with variant peptides, the contribution of RNA editing is notable. Indeed, based on the average coverage of specific sites, the actual number of RNA editing sites that alter protein sequence could be 10~100 times that identified in this study. However, that the level of RNA editing varies from site to site, generally lower than that of somatic mutations. Several RNA editing events with variant peptide support show clinically relevant patterns; and importantly, the editing in COG3 (reported in our previous study) (Han et al., 2015) and COPA (experimentally characterized in this study) could functionally drive the growth and migration of cancer cells, in a manner similar to driver somatic mutations. Collectively, our study suggests that A-to-I RNA editing contributes to protein heterogeneity at least in some cancer types, and thus deserves more effort from the cancer research community to elucidate the molecular basis of human cancers and develop prognostic and therapeutic approaches.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Han Liang (hliang1@mdanderson.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
MDA-MB-231 and MCF10A cell lines were purchased from American Type Culture Collection (ATCC). Hs578T, OVCAR-8, SLR23 and SLR25 cells were obtained from the MD Anderson Characterized Cell Line Core Facility. All the cell lines were confirmed by short tandem repeat (STR) analysis, and mycoplasma testing was found to be negative. MCF10A cells were maintained in complete DMEM/F12 (Invitrogen) full medium with 5% horse serum (Invitrogen), 20 ng/ml EGF (Peprotech), 10 µg/ml insulin (Sigma), 100 ng/ml Cholera Toxin (Sigma), 0.5 mg/ml Hydrocortisone. MDA-MB-231, Hs578T, SLR23 and SLR25 cells were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum.
METHOD DETAILS
Mass spectrum data analysis
We obtained BRCA, OV, and CRC MS datasets in mzML format from the Clinical Proteomic Tumor Analysis Consortium (CPTAC, https://cptac-data-portal.georgetown.edu/cptacPublic/). For BRCA and OV, each MS set contains the mixed data from three samples and one common internal control sample labeled for isobaric molecules; for CRC, each MS set represents the data from a single sample. For each dataset, only MS2 spectra were extracted and merged into one file in mgf format employing the msconvert from ProteoWizard. We obtained the RNA-seq BAM files of these samples from CGHub (https://cghub.ucsc.edu) and the somatic mutation data (TCGA level-3) from Firehose. We only included the samples with parallel MS, RNA-seq (BRCA and OV: UNC paired-end; CRC: UNC single-ended) and somatic mutation data for further analyses. As a result, our analysis employed 36 BRCA MS sets for 101 samples, 64 OV MS sets for 90 samples and 89 CRC MS sets for 84 samples (including 5 replicated sets). In addition, we downloaded the MS dataset of 34 NCI60 cell lines (Moghaddas Gholami et al., 2013), and the raw data (vendor format) were converted into mzML format using the command msconvert from ProteoWizard.
We downloaded the annotated RNA editing list (version 2) from RADAR (Rigorously Annotated Database of A-to-I RNA editing; http://rnaedit.com/), which included 2,576,459 entries (Ramaswami and Li, 2014). We re-annotated all the RNA editing sites and the somatic mutations using ANNOVAR based on the RefSeq annotation file (hg19_refGeneMrna.fa and hg19_refGene.txt [March 22, 2015]). Since A-to-I editing is strand-specific, we further discarded 378 sites with inconsistent strand annotation; and we also discarded the sites in 10 HLA genes due to potential mapping errors. We downloaded RNA-seq bam files (hg19) from CGhub (https://cghub.ucsc.edu). Our analysis included 1,369 missense RNA editing sites, and we screened these sites for editing signals (≥ 3 edited reads and ≥ 0.1% editing level in a given sample) based on the RNA-seq bam files.
For each MS set, the amino acid changes caused by missense mutations or RNA editing events from the related samples, together with the RefSeq mRNAs, were used to build a sample-set-specific searching database. We used the R package, sapFinder, for the searching database construction, peptide spectrum match identification and data parser (Wen et al., 2014). The tool, sapFinder, employs R version of X!Tandem and a refined FDR estimation method, which was specially designed to address the issue of high risk of false positive variant identification. In the analysis, the parent ion mass tolerance and fragment ion mass tolerance (monoisotopic mass) was set as 10 ppm and 0.1 Da, respectively. Carbamidomethylation of cysteine (57.02 Da) was considered as fixed modification, and oxidation on methioine (15.99 Da) was considered as variable modification. For BRCA and OV MS datasets, we further considered two more fixed modifications, iTRAQ 4-plex of N-terminal and lysine (144.10 Da), and two more variable modifications, acetylation of protein N-term (42.01 Da) and deamination of asparagine (0.98 Da). The specific protein cleavage site was set as “[KR]|[X]”, allowing for 2 missed cleavages. The FDR in sapFinder was set to 0.01. Only RNA editing and somatic mutations with supported uniquely mapped variant peptides were kept for further manual check by a proteomic expert. In the information flow analysis, we counted the numbers of variants at each step, and the variants recurrent within the samples of a MS set were only counted once. We also performed a simulation analysis to estimate the overall false positive rate in which the nucleotide change of A-to-C or A-to-T at the same RNA editing sites were introduced and the same analytic procedure (including the manual check) was employed. We employed x-Tracker to extract ion intensity for each peptide identified (Shadforth et al., 2005). SearchGUI and PeptideShaker were used to generate representative peptide-spectrum matches (Vaudel et al., 2011; Vaudel et al., 2015). We used xtendem-parser to view peptide-spectrum matches when necessary (Muth et al., 2010). To test correlations between the RNA editing level detected by RNA-seq data and variant peptide intensity in MS data, given three samples in each MS data set, we used the binomial test (x = 15, n = 34, p = 1/3). For peptide-spectra matches from COPA_I164V in breast cancer, we normalized the ion intensity of TCGA samples based on that of the common reference sample across MS datasets. We then classified samples into “high editing group” and “low editing group” using the upper quartile value of editing levels at the indicated site. One-tailed Wilcoxon rank sum test was used to determine the significance level between two groups.
To rule out the possibility that the detected variant peptides inferred by RNA editing in BRCA, OV and CRC were due to variants at the DNA level, we manually re-checked the allele frequency at these detected sites in whole exome sequencing (WXS) or whole-genome sequencing (WGS) data that were downloaded from Genomic Data Commons (https://portal.gdc.cancer.gov/).
Pan-cancer clinical relevance analysis of the RNA editing sites
We downloaded the RNA-seq bam files of 8,223 samples from TCGA 24 cancer types from CGHub, and only paired-end RNA-seq data were used in the analysis. To reliably estimate the RNA editing level at a site of interest, we only considered the samples where the site was covered by at least 10 high-quality reads (base quality ≥ 20 and mapping quality ≥ 20), and the editing level was defined as the fraction of edited reads among all the reads covering that position as previously described (Han et al., 2015). We obtained the clinical information of patient samples, including subtypes, clinical stages, and patient progression-free survival time from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). We employed the Kruskal-Wallis test or Wilcoxon rank sum test or detect RNA editing sites with a differential editing frequency among different tumor stages or subtype and considered FDR < 0.05 as statistically significant. We used the log-rank test and the univariate Cox proportional hazard model test to assess whether the RNA editing level was significantly correlated with progression-free survival (PFS) and considered FDR < 0.05 per cancer type in either test as statistically significant. We only reported the sites with a median editing-level difference between comparison of >3%. We downloaded the RNA editing matrix from REDIportal (http://srv00.recas.ba.infn.it/atlas/), which is based on RNA-seq data in normal tissues from the GTEx project, and calculated RNA editing levels of the sites of interest.
We obtained 946 RNA-seq BAM files of CCLE cell lines (Barretina et al., 2012) from CGHub and the drug sensitivity data from Cancer Therapeutics Response Portal (http://www.broadinstitute.org/ctrp/) (Seashore-Ludlow et al., 2015), which included the drug sensitivity data of 481 compounds across 664 cell lines. For each RNA editing site, we first calculated the Spearman rank correlation between the RNA editing level (|Rs| > 0.2) and considered FDR < 0.01 as statistically significant. To confirm the patterns of significant hits, we implemented the waterfall method to categorize sensitive and resistant cell lines as previously described (Barretina et al., 2012), and applied Wilcoxon rank sum test to assess the editing level difference between the two cell line groups and considered p < 0.05 as statistically significant.
We compared the expression level and editing level difference between the RNA editing sites with and without peptide evidence. For the 8 RNA editing with peptide evidence detected in BRCA, we compared them with the remaining missense RNA editing events that could be theoretically detectable. We also assessed the functional effects of amino acid changes introduced by RNA editing events, by polyPhen-2 score (http://genetics.bwh.harvard.edu/pph2/bgi.shtml).
Modeling of protein folding energy changes
To understand the impact of the amino acid changes introduced by COPA_I164V at the atomic level, we first used I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) to generate protein models (Roy et al., 2010; Yang et al., 2015; Zhang, 2008), and selected the representative protein models of the largest cluster in the subsequent analysis. Next, we estimated the protein stability upon the amino acid change by applying Rosetta ddg protocol to calculate free energy changes (ΔΔGs) based on the RNA edited sequence. In the simulation, we generated 50 models for both wild-type and edited proteins, allowing all side-chains to be repacked, followed by backbone and side-chain minimization (Kellogg et al., 2011). The predicted ΔΔG is the energy difference between wild-type and edited sequence based on mean values of the three models with the lowest energy. To further confirm prediction accuracy, we incorporated I-Mutant2.0 (Capriotti et al., 2005) and estimated ΔΔGs for the proteins using structural information.
Generation of stable cell lines
The mutant open reading frames (ORFs) corresponding to the RNA editing sites in COPA or mutant ADAR1 and ADAR2 were made by site-directed mutagenesis and confirmed by Sanger sequencing as previously described (Han et al., 2015). ADAR1-E912A and ADAR2-E396A contain an E-to-A amino acid change that abolishes ADAR editase activity (Macbeth et al., 2005). Virus were produced by transfecting HEK293PA cells with the GFP control vectors, pHAGE-V5-puromycin expression vectors (carrying COPA-WT or COPA-I164V; ADAR1-WT or ADAR1-E912A; ADAR2-WT or ADAR2-E396A), pZIP-hEF1a-Blast-Zsgreen/non-targeting shRNA constructs or pZIP-hEF1a-Blast-Zsgreen/COPA shRNA constructs (Transomic technologies, Table S6) and the Lentiviral Packaging Mix (psPAX2 and pMD2.G). For RNA editing fingerprint assay, Hs578T cells were transfected by the virus followed by selection with puromycin (1 µg/ml). For COPA functional assays, MDA-MB-231, MCF10A and SLR25 cells were transduced by the virus followed by selection with puromycin (MDA-MB-231 1 µg/ml, MCF10A 0.75 µg/ml, and SLR25 1 µg/ml), or Blasticidin (MDA-MB-231 10 µg/ml); and after 7 days of antibiotic selection, expression of the constructs was verified by Western blots.
RNA isolation and quantitative real-time RT-PCR
RNAs were isolated using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). RNAs were transcribed into cDNAs using the High-Capacity cDNA Reverse Transcription Kit (Life technologies, CA, USA). Quantitative real-time PCR (qPCR) was performed by Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems, Darmstadt, Germany). Expression levels were normalized to β-actin. Reactions were done in duplicate using TaqMan® Fast Universal PCR Master Mix (2X), no AmpErase® UNG (Life technologies). The relative expression was calculated by the 2(−ΔΔCt) method. The primers from ThermoFisher were as follows: ADAR1 primer (Hs00241666_m1), ADAR2 primer (Hs00953724_m1), COPA primer (Hs00189232_m1), and β-actin primer (Hs99999903_m1).
RNA editing fingerprint assay
Lentivirus-transduced cells were harvested and total RNA was isolated using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) with TURBO DNA-free™ Kit (ThermoFisher Scientific) incubation step to digest any trace genomic DNA present. RNAs were transcribed into cDNAs using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). We designed RNA editing site-specific primers that were compatible with SYBR green qRT-PCR protocols according to previous studies of the RNA editing fingerprint assay (Crews et al., 2015). We performed qRT-PCR in triplicate using cDNA on an Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems, Darmstadt, Germany) and SYBR® Select Master Mix (ThermoFisher Scientific). Melting curve analysis was performed on each plate according to the manufacturer’s instructions. The relative expression was calculated by the 2(−ΔΔCt) method, and the expression levels were normalized to GAPDH. The relative RNA editing level (edit/WT RNA ratio) were calculated as 2(CtEdit − CtWT).
RNA-seq based ADAR perturbation experiments
To validate the RNA-editing sites, we chose Hs578T, SLR23 and SLR25 for perturbation studies. To knockdown ADAR enzymes, the cells were transfected by ADAR1-siRNA (Catalog#:4390824, siRNA ID: s1007, ThermoFisher Scientific), ADAR2-siRNA (Catalog#:4392420, siRNA ID: s1010, ThermoFisher Scientific), RISC-free control (Catalog#:AM4611, ThermoFisher Scientific) with 50 nM, or MOCK only transfection reagent Lipofectamine® RNAiMAX (ThermoFisher Scientific). To overexpress ADAR enzymes, virus was produced by transfecting the cells with the GFP control vectors, or pHAGE expression vectors (carrying ADAR1-WT or ADAR2-WT). Total RNA of 96 h post-transfection cells was subjected to mRNA paired-end sequencing (the sequencing platform was HiSeq2000) at the MD Anderson Sequencing and Microarray Core Facility. We mapped FASTQ raw reads with Tophat2 (Kim et al., 2013) and performed the RNA editing analysis in the same way for TCGA RNA-seq BAM files. The related FASTQ files have been deposited to NCBI SRA (SRP082419).
CRISPR/Cas9 knockout experiments
To generate a clean background to assess the effect of COPA RNA editing, the CRISPR/Cas9 experiment was performed according to previous studies (Ran et al., 2013). Briefly, gRNA targeting COPA exon 2 was cloned into pSpCas9(BB)-2A-GFP (PX458). PX458 was a gift from Feng Zhang (Addgene, #48138). The plasmid (10 µg) was transfected into 2 million MDA-MB-231 cells in 10-cm-diameter tissue culture dish by Lipofectamine 3000 reagent (Life Technology, L3000015). Two days after transfection, GFP positive cells were sorted by Moflo Astrios Cell Sorter in MD Anderson Cancer Center Flow Cytometry Core Facility. Individual GFP positive cell were seeded in wells of a 96-well plate. Four weeks after sorting, potential COPA knockout clones were verified by Western blot using COPA antibody (Sigma-Aldrich, HPA028024). To further confirm the results, we used PCR to amplify the region around the gRNA targeting sequence and cloned the PCR product into TA cloning vector. After transformation, 10 bacterial clones were sequenced by M13 primers. Table S5 lists the gRNA sequence, targeting sequence and PCR primers.
Immunoblotting
Whole-cell lysates for western blotting were extracted with RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, protease, and phosphatase inhibitor cocktail). Protein concentrations were determined using bicinchoninic acid (Pierce, Rockford, IL, USA) assays according to the manufacturer’s instruction. Cell lysates (30 µg) were loaded onto 8% or 12% SDS-PAGE and transferred to a polyvinylidene fluoride membrane and protein expression was depicted with an enhanced chemiluminescence Western blot detection kit (Amersham Biosciences, Little Chalfont, UK). The following antibodies were used: COPA (1:1000, SIGMA, HPA028024), V5 (1:5000, life technologies, R960-25), and ERK2 (1:3000, Santa Cruz Biotechnology, sc-154), a-Tubulin (1:1000, Cell Signaling Technology, CST-2144), GAPDH (1:3000, Santa Cruz Biotechnology, sc-25778).
Immunofluorescence
Cells were cultured in chamber slides overnight and fixed with ice-cold methanol for 10 min at room temperature. Cells were then blocked for non-specific binding with 8% bovine serum albumin (BSA) serum in PBS for 1 hr at room temperature, and incubated with the anti-V5 antibody (1:200, Life Technologies, R960-25) overnight at 4°C, followed by incubation with Alexa Fluor 568 goat anti-mouse IgG (1:500, Invitrogen, A11004) for 2 hr at room temperature. Cover slips were mounted on slides using Prolong gold antifade mountant with DAPI (ThermoFisher Scientific, P36935). Immunofluorescence images were acquired on a fluorescence microscope.
Cell viability assay
The MDA-MB-231, MCF10A, and SLR25 stable cell lines were seeded into 96-well plates, and the assays were performed at day 1, 2, 4, and 6 time points. CellTiter-Glo 2.0 (Promega, Madison, WI, USA) was added to assess cell viability according to the manufacturer's instructions. MDA-MB-231 parental cells, and COPA knock-out cells with GFP, or COPA expression vectors were seeded into 24-well plates and viability was accessed by measuring cell confluence (%) using IncuCyte Zoom live imaging. The significance of the differences was analyzed with Student’s t-test, and p < 0.05 was considered statistically significant.
Wound healing assay
MDA-MB-231 cells (6×104) were seeded into 96-well ImageLock plates for 24 hr in RPMI-1640 medium included with 5% fetal bovine serum. Automated 96-well cell migration (scratch wound) on IncuCyte was analyzed by IncuCyte™ Cell Migration Kit (Essen BioScience, Ann Arbor, Michigan, USA), which comprises of a 96-pin woundmaking tool (WoundMaker™), Cell Migration Analysis software module and 96-well ImageLock Plates.
In vitro migration and invasion assay
For transwell migration assays, 2.5×104 to 1×105 cells were plated in the top chamber with a non-coated membrane (Corning BioCoat Control Insert; 8.0 µm; 24-well; 24/CS 354578). For invasion assays, 2.5×104 to 1×105 cells were plated in the top chamber with Matrigel-coated membrane (Corning BioCoat Matrigel Invasion Chamber; 24-well; 24/CS 354483). In both assays, cells were plated in medium without serum or growth factors, and medium supplemented with growth factors and serum (for MCF10A) or serum (for MDA-MB-231 and SLR25) was used as a chemoattractant in the lower chamber. The cells were incubated for 18 hr or 30 hr and cells that did not migrate or invade through the pores were removed with a cotton swab. Cells on the lower surface of the membrane were fixed with ethanol, and then stained with Coomassie brilliant blue and counted in 10 different low-power (100×) microscopic fields.
Protein sample preparation and LC-MS/MS analysis
We enriched the two proteins, COG3 and COPA, through immunoprecipitation from an ovarian cancer cell line, OVCAR-8. We also collected the protein, COPA, from the COPA knock-out clone transfected with the plasmids containing GFP, wild-type and edited COPA ORFs, respectively. We used the following antibodies: COPA, Sigma-Aldrich, HPA028024; COG3, Proteintech, 11130-1-AP. After one-day growth, cells were lysed in ice-cold lysis buffer (50 mM Tris HCl PH7.5, 150 mM NaCl, 2% CHAPS, 1mM EDTA) supplemented with halt protease and phosphatase inhibitor cocktail (Thermo Scientific, Catalog#78440). Protein extracts were quantified with a BCA protein assay kit (Thermo Scientific, Catalog#23225). Two µg of antibodies were incubated with SureBeads protein G Magnetic Beads (Biorad, Catalog#161-4023) for 10 min at room temperature. Then, 2–6 mg protein extracts were added to the beads and incubate 12 hr at 4 °C. After the beads were washed and boiled, the immunoprecipitated protein complexes were eluted and resolved on 4–12% Criterion XT Precast Gels (Biorad, Catalog#161-0789) and visualized by silver staining (Thermo Scientific, Catalog#24600). The corresponding bands were excised and were further subjected to LC-MS/MS analysis at the MDACC Proteomics and Metabolomics Core.
Synthetic peptides were prepared using standard FMOC chemistry, the carboxy-terminal residues were introduced using FMOC-13C6-15N2-Lysine-, or FMOC-13C6-15N4-Arginine-resin (Cambridge Isotope labs). In the experiments, these “heavy” (stable-isotope labeled) peptides were spiked into digests to validate extracted digests for the presence of the expressed (WT or edited) peptide sequences by chromatographic co-elution.
Silver-stained gel pieces were diced, washed, destained using reagents from the Pierce silver-stain kit, and digested in-gel with 200 ng modified trypsin (sequencing grade, Promega) and Rapigest (TM, Waters Corp.) for 18 hr at 37°C. Resulting peptides were extracted and analyzed by high-sensitivity LC-MS/MS on an Orbitrap Fusion mass spectrometer (Thermo Scientific, Waltham MA). Both wild-type and RNA-edited peptides were targeted specifically. Proteins were identified by database searching of the fragment spectra against the SwissProt (EBI) protein database using Mascot (v2.5, Matrix Science, London, UK) or Sequest HT and Proteome Discoverer (v1.4, Thermo Scientific), or a custom database included the expected edited sequence. Typical search settings were as follows: mass tolerances, 10 ppm precursor, 0.8 Da fragments; variable modifications, methionine sulfoxide, pyro-glutamate formation; enzyme, trypsin, up to 2 missed cleavages. In some cases, 13C6-15N2-Lys and 13C6-15N4-Arg were also included as variable modifications. Peptides were subject to 1% FDR using reverse-database searching.
QUANTIFICATION AND STATISTICAL ANALYSIS
We used paired Student’s t-test to assess the editing level differences of RNA editing sites with peptide evidence between tumor and matched normal samples. We used the log-rank test and the univariate Cox proportional hazard model test to assess whether the RNA editing level at individual sites and ADAR1/2 mRNA expression were significantly correlated with progression free survival (PFS). We used the Kruskal-Wallis test (three groups or more in comparison) or Wilcoxon rank sum test (two groups in comparison) to detect RNA editing sites with a differential editing level among different tumor stages or subtypes. We used Wilcoxon rank sum test to determine differential editing level between drug sensitive and resistant cancer cell lines. We used Wilcoxon rank sum test to compare the expression level, editing level, edited expression amount and predicted function effect between sites with peptide evidence and those without peptide evidence. We used Wilcoxon rank sum test to determine the relative ion intensity difference between COPA_I164V high and low editing groups. Cell viability, wounding length, migration and invasion data were analyzed using Student’s t-test and the graphs show mean ± SD We also described the detailed information on p values for the statistical significance in the figure legends and Methods Details.
DATA AND SOFTWARE AVAILABILITY
The RNA-seq data from ADAR1/2 perturbation experiments have been deposited in Sequence Read Archive at NCBI with the accession number, SRP082419.
Supplementary Material
Table S2. Information about RNA editing events with variant peptide support, Related to Figure 1.
Table S3. Information about peptide spectrum matches of variant peptides caused by RNA editing and somatic mutations, Related to Figure 1.
Table S4. The numeric data matrix and statistical method used in this study, Related to Figure 4.
Significance.
Understanding the molecular mechanisms contributing to protein variation and diversity is a fundamental question in biology and has significant clinical implications in cancer treatment. Through an integrated analysis of TCGA genomic data and CPTAC proteomic data, our study provides large-scale direct evidence that A-to-I RNA editing is a source of proteomic diversity in cancer cells. Thus, RNA editing represents an exciting paradigm for understanding the molecular basis of human cancer and developing the strategies for precision cancer medicine.
Highlights.
Direct assessment of A-to-I RNA editing to proteomic diversity in cancer specimens
A rigorous computational strategy to detect variant peptides caused by RNA editing
Independent experimental evidence of RNA-editing-induced variant peptides in tumors
Effects of COPA editing on proliferation, migration, and invasion of cancer cells
Acknowledgments
This study was supported by the National Institutes of Health (CA168394, CA098258 and CA143883 to G.B.M., CA175486 to H.L., CA20985 to H.L. and G.B.M., and CCSG grant CA016672); grants from the Cancer Prevention and Research Institute of Texas (RP140462 to H.L.; RR150085 to L.H.); a University of Texas System STARS award (to H.L.); the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine; Natural Scientific Foundation of China (No. 81572777 to X.X.); the Adelson Medical Research Foundation (to G.B.M.). The UT MD Anderson Proteomics and Metabolomics Facility would like to thank the MD Anderson Cancer Center, NIH High-End Instrumentation program grant 1S10OD012304-01, and CPRIT Core Facility Grant RP130397 for generous support. We thank the MD Anderson high-performance computing core facility for computing, and LeeAnn Chastain for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.L. supervised the whole project. H.L. and G.B.M. conceived of and designed the research. X.P., Y.W., D.H.H., L.H., M.Z., P.H., H.L. and G.B.M. contributed to the data analysis and helpful discussion. X.X., D.H.H., S.Y., Z.Z., K.M., K.J., M.L., Y.T., Y.L., and S.L.K. performed the experiments. X.P., X.X., H.L. and G.B.M. wrote the manuscript, with input from all other authors.
Declaration of Interests
G.B.M. has sponsored research support from AstraZeneca, Critical Outcomes Technology, Karus, Illumina, Immunomet, Nanostring, Tarveda and Immunomet and is on the Scientific Advisory Board for AstraZeneca, Critical Outcomes Technology, ImmunoMet, Ionis, Nuevolution, Symphogen, and Tarveda. H.L. is a shareholder and scientific advisor of Precision Scientific Ltd., and Eagle Nebula Inc.
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity (vol 483, pg 603, 2012) Nature. 2012;492:290–290. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. RNA Editing by Adenosine Deaminases That Act on RNA. Annual Review of Biochemistry. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, Levanon EY. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome research. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic acids research. 2005;33:W306–310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan THM, Qamra A, Tan KT, Guo J, Yang H, Qi L, Lin JS, Ng VHE, Song Y, Hong H, et al. ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer. Gastroenterology. 2016;151:637–650.e610. doi: 10.1053/j.gastro.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Li Y, Lin CH, Chan THM, Chow RKK, Song YY, Liu M, Yuan YF, Fu L, Kong KL, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nature Medicine. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Liao XY, Zhang JB, Wang F, Qin HD, Zhang L, Shugart YY, Zeng YX, Jia WH. ADAR2 functions as a tumor suppressor via editing IGFBP7 in esophageal squamous cell carcinoma. Int J Oncol. 2017;50:622–630. doi: 10.3892/ijo.2016.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews LA, Jiang Q, Zipeto MA, Lazzari E, Court AC, Ali S, Barrett CL, Frazer KA, Jamieson CH. An RNA editing fingerprint of cancer stem cell reprogramming. Journal of translational medicine. 2015;13:52. doi: 10.1186/s12967-014-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Overall CM, Van Eyk JE, Baker MS, Paik YK, Weintraub ST, Lane L, Martens L, Vandenbrouck Y, Kusebauch U, et al. Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 2.1. Journal of proteome research. 2016;15:3961–3970. doi: 10.1021/acs.jproteome.6b00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Qin YR, Ming XY, Zuo XB, Diao YW, Zhang LY, Ai J, Liu BL, Huang TX, Cao TT, et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E4631–E4640. doi: 10.1073/pnas.1703178114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli D, Gacquer D, Rothe F, Lefort A, Libert F, Brown D, Kheddoumi N, Shlien A, Konopka T, Salgado R, et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell Rep. 2015;13:277–289. doi: 10.1016/j.celrep.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano F, Rossetti C, Tomaselli S, Cifaldi L, Lezzerini M, Pezzullo M, Boldrini R, Massimi L, Di Rocco CM, Locatelli F, Gallo A. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene. 2013;32:998–1009. doi: 10.1038/onc.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Kossenkov AV, Sakurai M, Yan J, Li Y, Xu H, Wang J, Zhang PJ, Zhang L, et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nature communications. 2016;7:10715. doi: 10.1038/ncomms10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HM, Eterovic AK, Yuan Y, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SW, Kim HP, Shin JY, Jeong EG, Lee WC, Kim KY, Park SY, Lee DW, Won JK, Jeong SY, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. The Journal of experimental medicine. 2014;211:613–621. doi: 10.1084/jem.20132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddas Gholami A, Hahne H, Wu Z, Auer FJ, Meng C, Wilhelm M, Kuster B. Global proteome analysis of the NCI-60 cell line panel. Cell reports. 2013;4:609–620. doi: 10.1016/j.celrep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Muth T, Vaudel M, Barsnes H, Martens L, Sickmann A. XTandem Parser: an open-source library to parse and analyse X!Tandem MS/MS search results. Proteomics. 2010;10:1522–1524. doi: 10.1002/pmic.200900759. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nature methods. 2014;11:1114–1125. doi: 10.1038/nmeth.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N, Bazak L, Buchumenski I, Porath HT, Danan-Gotthold M, Knisbacher BA, Eisenberg E, Levanon EY. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015;13:267–276. doi: 10.1016/j.celrep.2015.08.080. [DOI] [PubMed] [Google Scholar]
- Peng ZY, Cheng YB, Tan BCM, Kang L, Tian ZJ, Zhu YK, Zhang WW, Liang Y, Hu XD, Tan XM, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nature biotechnology. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- Picardi E, D'Erchia AM, Lo Giudice C, Pesole G. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Research. 2017;45:D750–D757. doi: 10.1093/nar/gkw767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto Y, Cohen HY, Levanon EY. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome biology. 2014;15:R5. doi: 10.1186/gb-2014-15-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic acids research. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nature methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O'Connell MA, Li JB. Identifying RNA editing sites using RNA sequencing data alone. Nature methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, Jones V, Bodycombe NE, Soule CK, Gould J, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov. 2015;5:1210–1223. doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadforth IP, Dunkley TPJ, Lilley KS, Bessant C. i-Tracker: For quantitative proteomics using iTRAQ™. BMC Genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. Journal of proteome research. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Netowrk. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012a;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012b;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudel M, Barsnes H, Berven FS, Sickmann A, Martens L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 2011;11:996–999. doi: 10.1002/pmic.201000595. [DOI] [PubMed] [Google Scholar]
- Vaudel M, Burkhart JM, Zahedi RP, Oveland E, Berven FS, Sickmann A, Martens L, Barsnes H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nature Biotechnology. 2015;33:22. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- Wen B, Xu S, Sheynkman GM, Feng Q, Lin L, Wang Q, Xu X, Wang J, Liu S. sapFinder: an R/Bioconductor package for detection of variant peptides in shotgun proteomics experiments. Bioinformatics. 2014;30:3136–3138. doi: 10.1093/bioinformatics/btu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Zhang H, Lin W, Hao Y, Ren L, Zhang C, Li N, Wei H, Jiang Y, He F. Discovery of novel genes and gene isoforms by integrating transcriptomic and proteomic profiling from mouse liver. Journal of proteome research. 2014;13:2409–2419. doi: 10.1021/pr4012206. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell. 2016 doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Information about RNA editing events with variant peptide support, Related to Figure 1.
Table S3. Information about peptide spectrum matches of variant peptides caused by RNA editing and somatic mutations, Related to Figure 1.
Table S4. The numeric data matrix and statistical method used in this study, Related to Figure 4.