Abstract
Considered a “mixing vessel” for influenza viruses, pigs can give rise to new influenza virus reassortants that can threaten humans. During our surveillance of pigs in Guangxi, China from 2013 to 2015, we isolated 11 H1N1 and three H3N2 influenza A viruses of swine origin (IAVs-S). Out of the 14, we detected ten novel triple-reassortant viruses, which contained surface genes (hemagglutinin and neuraminidase) from Eurasian avian-like (EA) H1N1 or seasonal human-like H3N2, matrix (M) genes from H1N1/2009 pandemic or EA H1N1, nonstructural (NS) genes from classical swine, and the remaining genes from H1N1/2009 pandemic. Mouse studies indicate that these IAVs-S replicate efficiently without prior adaptation, with some isolates demonstrating lethality. Notably, the reassortant EA H1N1 viruses with EA-like M gene have been reported in human infections. Further investigations will help to assess the potential risk of these novel triple-reassortant viruses to humans.
Introduction
The segmented nature of the genome of influenza viruses is responsible for its ability to reassort its eight RNA genes when cells are co-infected with two different influenza virus strains1. Influenza A virus of swine origin (IAV-S) circulating in pigs have been known to occasionally reassort with avian and human influenza viruses to generate novel genotypes and establish new swine influenza virus lineages. Importantly, these new lineages might represent influenza pandemic threats for humans, as highlighted by the last 2009 H1N1 influenza virus pandemic2. Despite several introductions of avian influenza virus genes into swine strains by reassortment, only viruses from H1, H2, and H3 subtypes out of the 16 possible avian influenza virus subtypes have been known to circulate in pigs in the last century, mirroring the same subtypes that have been known to circulate in humans.
At present, there are H1N1, H1N2, and H3N2 major viral subtypes circulating among pigs worldwide. Among the swine H1N1 influenza lineages, the classical H1N1 swine (CS) lineage is related to the H1N1 virus that caused both the 1918 and 2009 human pandemics3,4. The Eurasian avian-like (EA) H1N1 viruses have been prevalent in European pigs since 19795 and had contributed its neuraminidase (NA) and matrix (M) segments to the H1N1/2009 pandemic virus6. In 1997–1998, double or triple-reassortant H3N2 strains emerged containing genes from classical swine H1N1, seasonal human, and avian viruses, and they became prevalent in swine in North America7–9. Following the outbreak of H1N1/2009 pandemic in humans, reassortant viruses derived from H1N1/2009 pandemic viruses and enzootic IAVs-S have been continuously detected from pigs in many regions globally10–19. Furthermore, sporadic human infection with EA H1N1 viruses and with EA-H1N1/2009 pandemic reassortant viruses has been reported20–23. Also, human infections with the swine-derived H3N2 variant (H3N2v) have been reported and they are associated with a IAV-S reassortant H3N2 strain that contains the M segment of the H1N1/2009 pandemic virus24, emphasizing the important role of pigs in the generation of reassortant influenza viruses with the potential to infect humans. Importantly, humans lack herd immunity to EA H1N1 viruses11,25, individuals aged ≤14 years show broad susceptibility against H3N2v viruses, and the seasonal human vaccine does not induce neutralizing antibodies against these IAVs-S26. The insufficient immunity in humans against H1 and H3 IAVs-S raises concerns over whether they could be the source of the next influenza pandemic.
Southern China has diverse IAV-S ecosystem, where H1N1, H1N2, and H3N2 subtypes and different lineages IAV-S are co-circulating in pigs6,11,27; however, IAV-S surveillance in this region of the world is still very limited. In this study, we performed surveillance in pigs in Guangxi. We found that novel triple-reassortant EA H1N1 and human-like (HL) H3N2 reassortants, which carried H1N1/2009 pandemic internal genes (PB2, PB1, PA, and NP) and CS H1N1 NS genes have been circulated in pigs for some time and they are predominant in the pig population in Guangxi. This is the first evidence that such novel IAV-S reassortants have been established in pigs in Guangxi, China. These viruses demonstrated variable levels of virulence in mice with some isolates being lethal in this animal model. We suggest that these novel IAV-S lineages should be closely monitored for their potential to cause human infections.
Materials and methods
Surveillance of IAV-S
From 2013 to 2015, disease outbreaks with severe respiratory signs, including fever, coughing, sneezing, nasal discharge, and low appetite occurred in ten pig farms in Guangxi. A total of 600 nasal swabs and 135 lung tissues with or without any respiratory signs were collected from farms or slaughterhouses in Nanning, Qinzhou, Chongzuo, Laibin, Liuzhou, Bobai, Guigang, Dongxing, and Baise of Guangxi (Table 1).
Table 1.
Details of samples collected for SIVs testing in Guangxi
| City | Farm | Strains abbreviation | Sampling site | Time | Clinical signsa |
|---|---|---|---|---|---|
| Nanning | 1# | NN1994 | Farm(500 sows) | 2013.09 | Difficulty breathing; PRRSV positive |
| 2# | NNLX | Farm (900 sows) | 2014.10 | Fever, coughing, sneezing, nasal discharge | |
| 3# | NNXD | Farm(600 sows) | 2013.01 | Difficulty breathing; PRRSV and PCV positive | |
| NNXD2023 | Farm(600 sows) | 2013.09 | Coughing, sneezing, nasal discharge | ||
| 4# | JGB4 | Farm(1000 sows) | 2013.09 | Coughing and sneezing | |
| JG1 | Farm(1000 sows) | 2014.03 | Coughing and sneezing | ||
| Chongzuo | 5# | CZ7 | Farm(1000 sows) | 2014.10 | Fever, coughing, sneezing |
| Qingzhou | 6# | QZ5 | Farm(3000 sows) | 2014.08 | Fever, coughing, nasal discharge |
| Laibing | 7# | LB9 | Farm(800 sows) | 2014.10 | Fever, coughing, sneezing |
| Liuzhou | 8# | LZA3 | Farm(500 sows) | 2015.06 | Coughing and sneezing |
| Bobai | 9# | BB2 | Farm(1000 sows) | 2014.09 | Coughing and sneezing |
| Guigang | 10# | GG6 | Farm(800 sows) | 2013.05 | Coughing and sneezing |
| Dongxing | N/A | DX24 | Abattoirs | 2013.12 | N/A |
| Baise | N/A | BS30 | Abattoirs | 2014.11 | N/A |
N/A not available, PCV porcine circovirus
aPRRSV represents porcine reproductive and respiratory syndrome virus
Virus isolation and DNA Sequencing
As previously described28, RNA was directly extracted from nasal swabs or lung tissues. Reverse transcription was carried out under standard conditions with universal 12 primer. One pair of universal M gene primers was used to amplify M gene. M gene positive samples were inoculated onto monolayers of Madin–Darby canine kidney (MDCK) cells maintained in DMEM containing 1 μg/mL trypsin for 48 h at 37 °C. After three serial passages, virus isolates were identified by RT-PCR and the whole genome was amplified by eight pairs of primers and sequenced.
Phylogenetic analysis
Individual gene segments were aligned and analyzed with the Seqman and Megalign program (DNASTAR, Madison, USA). Phylogenetic trees were generated by applying the method of the Clustal IW alignment algorithm from MEGA 7.0 software (http://www.megasoftware.net/), and bootstrap values of 1000 were used.
Serological assays
A total of 1170 swine serum samples were collected from different cities of Guangxi and treated with receptor-destroying enzyme (Sigma, USA) for 18 h at 37 °C, followed by heat inactivation at 56 °C for 30 min before being tested for the presence of hemagglutination-inhibition (HI) antibodies with 1% (V/V) guinea pig erythrocytes29. HI assays were performed on all treated serum samples with two contemporary viruses, EA H1N1 virus (A/swine/Guangxi/BB2/2013) and HL H3N2 virus (A/swine/Guangxi/JGB4/2013). HI antibody titers ≥20 were determined as serological positive.
Replication and pathogenicity in mice
Groups of nine 6-week-old female BALB/c mice were anesthetized with CO2 and inoculated intranasally (i.n.) with 5 × 104 TCID50 of selected influenza viruses in a volume of 50 µL. Mock-infected mice were inoculated i.n. with 50 µL phosphate-buffered saline. Three mice were killed on days 3 and 5 post infection (p.i.), respectively. Organs (spleen, kidney, intestine, brain, and lung) were collected to assess for viral replication in MDCK cells. The remaining mice were monitored for clinical signs, body weight, and survival for 14 days. Mice were killed when the body weight loss was above 25% of their pre-challenge weight.
Results
Virus isolation and identification
A total of 735 nasal swabs or lung tissue samples were collected from pigs on farms and in slaughterhouses in Guangxi province of southern China from 2013 to 2015, containing 55 positive samples identified by RT-PCR. Fourteen strains were successfully isolated by serial passages on MDCK cells, including 11 H1N1 and three H3N2 IAVs-S. These results indicate that different subtypes of IAVs-S co-circulating in Guangxi. The nucleotide sequences of the 14 IAVs-S determined in this study will be deposited in the GenBank database under numbers KM061010 to KM061025 and MF927789 to MF927884.
Phylogenetic analysis
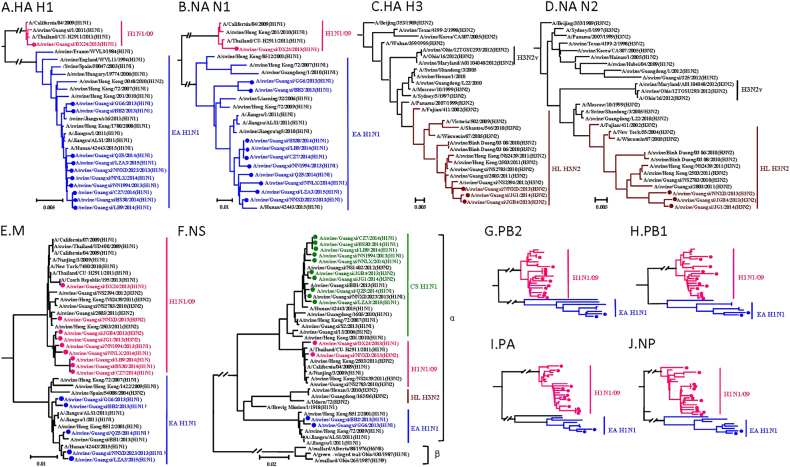
To determine the genetic characteristics of these viruses, all eight gene segments of the viruses grown in MDCKs were sequenced and phylogenetic analyses were performed. Phylogenetic analysis of the hemagglutinin (HA) reveals that the 11 H1 HA genes are separated into EA-like H1N1 lineage and H1N1/2009 pandemic lineage (Fig. 1a). All ten H1 HA genes clustered into the EA H1N1 lineage share 95.9–100% identity at the nucleotide level, whereas the interlineage homology is <74%. All the three H3 HA genes share over 98.6% of identity and are clustered into HL H3N2 lineage (Fig. 1c). Similar to the HA genes, ten N1 NA genes and one N1 NA gene are clustered into the EA H1N1 lineage and H1N1/2009 pandemic lineage, respectively (Fig. 1b). The N1 intralineage virus homology is over 94.7%, while the homology between the lineages is <89.4%. Three N2 NA genes are clustered into HL H3N2 lineage, which share over 98.7% of homology within the lineage (Fig. 1d).
Fig. 1.
Phylogenetic trees of the HA H1 (a), NA N1 (b), HA H3 (c), NA N2 (d), M (e), NS (f), PB2 (g), PB1 (h), PA (i), and NP (j) genes of the H1N1 and H3N2 influenza lineages. The unrooted trees were generated with the MEGA 7.0 program by using neighbor-joining analysis and reliability of the tree was assessed by bootstrap analysis with 1000 replications. Neighbor-joining bootstrap values ≥70 are shown at the major branches of the trees. The 12 trees were rooted to A/Brevig_Mission/1/18(H1N1). Viruses shown in black were downloaded from available databases. The isolates in our study were marked in different color, consistent with Fig. 2
The PB2, PB1, PA, NP, and M genes share 83–100%, 84.2–100%, 81.2–100%, 82.4–100%, and 92.5–100% identity, respectively, at the nucleotide level. The PB2, PB1, PA, and NP genes show the same clustering pattern. Specifically, 12 out of 14 viruses are clustered with the H1N1/2009 pandemic lineages, while the remaining two viruses belong to the EA H1N1 lineage (Fig. 1g–j and Figure S1). The intralineage homologies are above 97.1%, 96.7%, 95.3%, and 96.9%, respectively. However, the interlineage homologies are <84%, 84.8%, 84.7%, and 83.5%, respectively. The M gene is also clustered into two lineages: nine H1N1/2009 pandemic and five EA H1N1 (Fig. 1e). The homology of the M genes within lineages is over 96.2%, and the homology of the M genes is <95.6% between the lineages. Remarkably, the NS gene shows distinct diversity, forming three different lineages: EA H1N1 (2), H1N1/2009 pandemic (2), and CS H1N1 (10) (Fig. 1f). The NS intralineage virus homology is over 97.4%, while the interlineage homology is <91.4%. All of these NS genes belong to the alpha lineage nomenclature of NS gene, as shown in Fig. 1f.
Based on phylogenetic analyses of the gene segments, the viruses isolated and grown in MDCKs can be divided into six distinct genotypes (A, B, C, D, E, and F) (Fig. 2). The IAVs-S in genotypes A and B are prototypical EA H1N1 and H1N1/2009 pandemic, respectively. Notably, the IAVs-S in genotype C are novel triple H1N1 reassortants containing the HA and NA genes from EA H1N1, the NS gene from CS H1N1 and the rest of the gene segments are from H1N1/2009 pandemic. The genotype D IAVs-S is similar to the genotype C, except for the M gene, which is derived from EA H1N1. Genotype D IAVs-S were first reported in Tianjin, China in 201330 and caused human infection in Hunan, China in 201522, this genotype was detected continuously from 2013 to 2015 in Guangxi in our study. The three H3N2 viruses in genotypes E and F have HA and NA genes derived from swine HL H3N2 viruses. It should be noted that two viruses in genotype E are also novel triple reassortants containing five H1N1/2009 pandemic-origin internal genes and the NS gene derived from CS, similar to H1N1 viruses in genotype C. One virus in genotype F possessed six internal genes from H1N1/2009 pandemic viruses, which was soon detected after the spillovers of H1N1/2009 pandemic from human to pigs12. Our data demonstrate that there are EA H1N1 and HL H3N2 IAVs-S which have reassorted to acquire the CS H1N1 NS gene and H1N1/2009 pandemic internal genes. The similar pattern of internal gene reassortment acquired by multiple H1N1 and H3N2 IAVs-S from different times and locations in Guangxi suggest that H1N1/2009 pandemic ribonucleoprotein complex genes (PB2, PB1, PA, and NP genes) and CS H1N1 NS gene constellation is selectively advantageous and compatible with different surface genes. Importantly, IAV-S has the potential to cause epidemics in humans. The triple reassortant, A/swine/Guangxi/NNXD2023/2013(H1N1) (Sw/GX/NNXD2023/2013) in genotype D, shared 98.6–99.3% similarity at the nucleotide level with the recent human isolate A/Hunan/42443/2015 (HuN/42443/2015)22, as shown in Table 2, highlighting the potential ability of these new IAV-S reassortants to infect humans.
Fig. 2. Genotypes of H1N1 and H3N2 IAVs-S from Guangxi during 2013 to 2015.
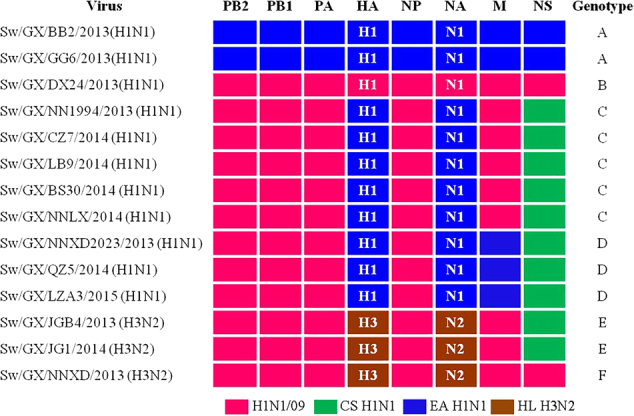
Origin of each gene segment is colored for representing the different lineages
Table 2.
Genome similarity of H1N1 novel reassortant viruses compared with the human isolated A/Hunan/42443/2015(H1N1)
| Genotype | Name of virus | Gene segment (%)a | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HA | NA | PB2 | PB1 | PA | NP | M | NS | ||
| C | A/swine/Guangxi/NN1994/2013 | 98.7 | 97.8 | 98.0 | 99.1 | 98.4 | 98.9 | 95.0 | 97.9 |
| C | A/swine/Guangxi/CZ7/2014 | 98.0 | 98.0 | 97.9 | 99.0 | 97.8 | 98.7 | 92.8 | 97.7 |
| C | A/swine/Guangxi/LB9/2014 | 98.0 | 97.9 | 97.9 | 99.0 | 97.8 | 98.7 | 94.8 | 97.6 |
| C | A/swine/Guangxi/BS30/2014 | 98.0 | 98.0 | 97.9 | 98.9 | 97.8 | 98.6 | 94.8 | 97.7 |
| C | A/swine/Guangxi /NNLX/2014 | 98.6 | 97.7 | 98.0 | 98.6 | 98.5 | 98.6 | 94.8 | 97.9 |
| D | A/swine/Guangxi NNXD2023/2013 | 99.0 | 98.9 | 98.8 | 99.3 | 98.7 | 99.1 | 99.6 | 98.6 |
| D | A/swine/Guangxi /QZ5/2014 | 98.6 | 98.1 | 97.9 | 99.0 | 98.5 | 98.6 | 97.3 | 98.0 |
| D | A/swine/Guangxi /LZA3/2015 | 98.8 | 97.9 | 98.0 | 98.5 | 98.3 | 98.7 | 99.4 | 97.6 |
aThe genome similarity was generated based on the open reading frame (ORF) sequences
Molecular features
We next analyzed if these IAVs-S possessed key molecular features associated with receptor-binding ability, increased virulence, transmission, and antiviral resistance (Table 3).
Table 3.
Amino acid substitutions in the novel H1N1 and H3N2 reassortant isolates compared with human isolates
| Lineage/subtype | Host | Virus | No. of glycosylation sites in HA | HAa | PB2 | M2 | NAc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 138 | 190 | 225 | 226 | 228 | 271 | 590 | 591 | 627 | 701 | 31 | 274 | ||||
| EA H1N1 | Human | A/Hunan/42443/2015 | 7 | A | D | E | Q | G | A | G | R | E | D | N | Y |
| EA H1N1 | Swineb | Sw/GX/NNXD2023/2013 | 6 | * | * | * | * | * | * | S | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/QZ5/2014 | 6 | * | * | * | * | * | * | S | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/LZA3/2015 | 6 | * | * | * | * | * | * | N | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/NN1994/2013 | 7 | * | * | * | * | * | * | S | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/CZ7/2014 | 6 | * | * | * | * | * | * | S | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/LB9/2014 | 6 | * | * | * | * | * | * | S | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/BS30/2014 | 6 | * | * | * | * | * | * | S | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/NNLX/2014 | 6 | * | * | * | * | * | * | S | * | * | * | * | * |
| EA H1N1 | Swineb | Sw/GX/BB2/2013 | 6 | * | * | * | * | * | T | * | Q | * | N | * | * |
| EA H1N1 | Swineb | Sw/GX/GG6/2013 | 6 | * | * | * | * | * | T | * | Q | * | N | * | * |
| Early H1N1 | Avian | A/duck/Schleswig/21/1979 | 6 | * | E | G | * | * | T | * | Q | * | * | S | * |
| Pandemic H1N1 | Swineb | Sw/GX/DX24/2013 | 6 | * | * | D | * | * | * | S | * | * | * | * | * |
| Pandemic H1N1 | Human | A/California/04/2009 | 6 | * | * | D | * | * | * | S | * | * | * | * | * |
| HL H3N2 | Swineb | Sw/GX/NNXD/2013 | 6 | S | * | D | I | S | * | S | * | * | * | * | H |
| HL H3N2 | Swineb | Sw/GX/JGB4/2013 | 6 | S | * | D | I | S | * | S | * | * | * | * | H |
| HL H3N2 | Swineb | Sw/GX/JG1/2014 | 6 | S | * | D | I | S | * | S | * | * | * | * | H |
*indicated the identical amino acids with A/Hunan/42443/2015; Sw represents swine, GX represents Guangxi
aH3 numbering
bisolates in this study
cN2 numbering
It is generally accepted that receptor-binding preference to human-type receptor is the initial key step for a novel influenza-virus-causing-pandemic. For H1 HAs, 190D and 225D/E are known to allow efficient binding to human-type α2–6 sialic acid linked receptors (H3 numbering, which is used throughout this work)31,32. All the EA H1N1 IAVs-S were highly conserved in the HA receptor-binding site, containing 138A, 190D, 225E, and 226Q, consistent with binding to α2–6 linked sialic acid. The H1N1/2009 pandemic-like swine isolate, Sw/GX/DX24/2013 contains 138A, 190D, 225D, and 226Q, also consistent with binding to α2–6 linked sialic acid. The α2–6 linked sialic acid receptor-binding specificity of H3 HAs is known to be determined by leucine and serine at positions 226 and 228. Our HL H3N2 viruses possessed characteristic residues found in human-adapted seasonal H3N2 viruses, such as 190D, 226I, and 228S32.
The presence of multiple basic amino acids at the HA cleavage site is characteristic of highly pathogenic influenza virus. All of our IAVs-S possessed a single basic amino acid (PSIQSR↓G or PEKQTR↓G) in the HA cleavage site. In addition, HA protein glycosylation is known to vary during influenza virus evolution33. One of the swine H1N1 isolates, Sw/GX/NN1994/2013, has seven HA potential glycosylation sites (Asn-X-Ser/Thr), which is the same number as the EA human strain (HuN/42443/2015). The remaining 13 IAVs-S had six HA potential glycosylation sites.
Among influenza virus proteins, PB2 is considered a major viral determinant of influenza viruses in mammals. The 627K and 701N amino acid residues in PB2 are known to play an important role in mammalian fitness for influenza viruses34–38. Most of IAVs-S with EA H1N1-origin PB2 genes have 701N. For H1N1/2009 pandemic virus, it was reported that 271A in PB2 enhances the transmissibility of influenza A in ferrets and 591R is important for H1N1/2009 pandemic in mammalian adaptation38–40. In our study, the IAVs-S with H1N1/2009 pandemic PB2 contain 271A and 591R.
Antiviral compounds are the first line of defense against novel influenza viruses until vaccines become available. Currently, two classes of drugs, adamantanes and NA inhibitors are available for treatment of influenza infections. The molecular marker of amantadine resistance, 31N in the transmembrane region of the M2 protein, is observed in all the isolates (Table 3)41. Meanwhile, H274Y mutation conferring resistance to NA inhibitors is also found in all the H1N1 swine isolates. However, the H3N2 IAVs-S do not have any known resistant mutation in the N2 against NA inhibitor.
Seroprevalence of IAV-S in pigs of Guangxi
Of the 1170 serum samples collected from different cities in Guangxi, 170 (14.6%) were positive solely against EA H1, while 219 (18.8%) of the sera were positive solely for HL H3 swine virus (Table 4). Additionally, 126 (10.8%) were reactive against EA H1 and HL H3 according to the HI assay, indicating the possibility of a high frequency of coinfection, successive infection over time, and/or multiple infections by different IAV-S subtypes in the pig population. We did not test serologically for antibodies against other influenza A virus strains, our data are conservative estimates of the seroprevalence of IAV-S in the pig population.
Table 4.
Seroprevalence of antibodies against different swine influenza virus in pigs in Guangxi from 2009 to 2013
| Location | Serum samplesa | No. (%) of sera positive | ||
|---|---|---|---|---|
| EA H1N1 | HL H3N2 | EA H1N1 + HL H3N2 | ||
| Nanning | 283 | 86 (30.4) | 166 (58.7) | 45(15.9) |
| Beihai | 60 | 24 (40.0) | 29 (48.3) | 8(13.3) |
| Fangchenggang | 40 | 3 (7.5) | 5 (12.5) | 1(2.5) |
| Liuzhou | 101 | 28 (27.7) | 17 (16.6) | 8(7.9) |
| Laibin | 130 | 31 (23.8) | 4 (3.1) | 2(1.5) |
| Guilin | 135 | 33 (24.4) | 33 (24.4) | 16(11.9) |
| Hezhou | 67 | 18 (26.9) | 17 (25.4) | 9(13.4) |
| Hechi | 80 | 9 (11.2) | 3 (1.3) | 1(1.3) |
| Baise | 60 | 6 (10.0) | 1 (1.7) | 0(0) |
| Yulin | 134 | 44 (32.8) | 51 (38.1) | 25(18.7) |
| Guigang | 80 | 14 (17.5) | 19 (23.8) | 11(13.8) |
| Total | 1170 | 296 (25.3) | 345 (29.5) | 126(10.8) |
aEurasian avian-like (EA) H1N1, A/swine/Guangxi/BB2/2013(H1N1); Human-like swine (HL) H3N2, A/swine/Guangxi/JGB4/2013 (H3N2)
Replication and virulence in mice
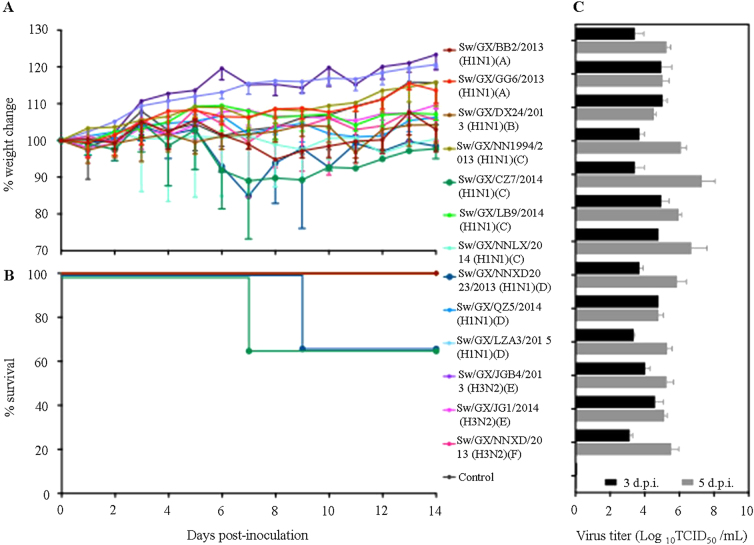
We selected ten H1N1 and three H3N2 influenza viruses, which included examples of each genotype and evaluated their replication and virulence in BALB/c mice. Groups of nine 6-week-old BABL/c mice were inoculated i.n. with 5 × 104 TCID50 of virus in a volume of 50 µL. The average body weight loss of mice caused by two H1N1 viruses (Sw/GX/CZ7/2014 (genotype C) and Sw/GX/NNXD2023/2013 (genotype D)) reached to 15.5% and 15.1%, respectively (Fig. 3a), especially Sw/GX/CZ7/2014 caused 33% of mice death (Fig. 3b). The rest did not show obvious weight lost or death during the 2-week observation. All 13 viruses replicated in lungs of mice with titers ranging from 3.1 to 5.0 log10 TCID50/mL and 4.5 to 7.3 log10 TCID50/ml at days 3 and 5 p.i., respectively (Fig. 3c), with higher titers observed by the lethal Sw/GX/CZ7/2014 virus. There was no detection in spleens, kidneys, intestines, or brains of any mice. These results indicated that novel triple-reassortant viruses could replicate well in mice lung without prior adaptation and two H1N1 IAVs-S causing sign of disease and lethality. Interestingly, although it is known that H3N2 human viruses replicate to low levels in mice without adaptation42, this was not the case for the IAV-S H3N2 viruses isolated in our surveillance.
Fig. 3.
Weight variation (a), survival rates (b), and replication (c) of novel reassortant IAVs-S in mice. Mice in each group were infected intranasally with 5 × 104 TCID50 of virus in a volume of 50 µL. The body weight and survival rates of mice were measured over 14 days. Virus titers of lung on 3 and 5 days post infection (d.p.i.) were shown as the mean titers of three mice
Discussion
Here our surveillance study reveals that EA H1N1, H1N1/2009 pandemic, and HL H3N2 IAVs-S have been co-circulating in pigs in Guangxi, China from 2013 to 2015 (Table 1). Serological data suggested that multiple subtype exposure occurred (Table 4). Co-circulation of parent viruses in pigs could facilitate reassortment events. After the repeated introduction of pandemic H1N1 virus into pig population, novel reassortant viruses with H1N1/2009 pandemic-origin genes have been isolated frequently from pigs globally19,43–45. Notably, in this study, the predominant strains are novel triple EA H1N1 and HL H3N2 reassortants, which contains the CS H1N1 NS genes and the remaining five or four genes originating from H1N1/2009 pandemic. Swine triple-reassortant viruses with H1N1/2009 pandemic internal genes and CS H1N1 NS gene may have then become established in Southern China.
The transition from avian-type to human-type receptor-binding preference is a crucial step for influenza viruses to replicate efficiently in humans46,47. However, the specific amino acids that determine receptor-binding specificity vary among the different HA subtypes. For the H1 virus subtype, substitutions at E190D and G225D/E are important for the change in preference from avian to human receptors31,48. Yang et al.25 showed that pandemic H1N1 and EA H1N1 IAVs-S preferentially bind to human-type receptor. For the H3 virus subtype, substitutions Q226L and G228S are important for the change in preference from avian to human receptors31,48. Swine H3N2 displayed binding preference for α2, 6-SA receptors, if they possess the 190D, 226V/I/L, and 228S32. Our H1N1/2009 pandemic and all EA H1N1 IAVs-S possess 190D and 225D/E, and HL H3N2 IAVs-S contain 190D, 226I, and 228S, which indicates that these isolates possibly bind to human-type receptors with high affinity.
In recent years, concerns about low-pathogenic influenza has been increased, since it may be able to replicate well and possibly cause disease in humans, as observed during the 2009 H1N1 pandemic4,49,50. Mutations in the polymerase complex are critical for influenza viruses to infect and adapt mammals; notable examples of such mutations are 271A, 627K, 701N, and 591R36–39,51,52. Our prototypical EA H1N1 IAVs-S have 701N. For H1N1/2009 pandemic PB2, it is reported that 271T and 591R confer efficient replication and transmission in mammals38,39. All of our strains with H1N1/2009 pandemic PB2 bear 271A and 591R. These indicate that 14 IAVs-S in our study have mammalian-adapting mutations in their PB2 genes (Table 3). Importantly, IAVs-S containing these particular marker combinations are transmissible in ferrets, as previously described25.
A key prerequisite for influenza pandemic is that the virus becomes highly transmissible in humans. Numerous studies have reported that the internal genes from H1N1/2009 pandemic are a critical factor to promote aerosol transmissibility for reassortant viruses38,39,50,52–56. In this study, we identified novel swine triple H1N1 and H3N2 reassortants with similar internal genetic composition (genotypes C, D, and E). This provides direct evidence that pandemic H1N1 internal genes with or without the CS H1N1 NS gene complex have been successfully incorporated by reassortment and fixed into EA H1N1 and HL H3N2 IAVs-S at least during our surveillance period of time.
Currently, influenza A H1N1 and H3N2 viruses are the circulating seasonal influenza A viruses subtypes in human. The H1N1/2009 pandemic became the current seasonal H1N1 virus. Our EA H1N1 HAs share <73.7 and 78.1% similarity with the H1N1/2009 pandemic vaccine strain (A/Michigan/45/2015 H1N1), at nucleotide level and amino acid level, respectively. Our H3N2 IAVs-S share <94.1 and 91.5% similarity with the H3N2 vaccine strain (A/Hong Kong/4801/2014 H3N2), at nucleotide level and amino acid level, respectively. Studies have reported that seasonal trivalent inactivated influenza vaccine induce poor cross-reactive antibodies to EA H1N1 virus23 and does not protect against swine H3N257. Importantly, according to the risk assessment tool, which is developed by the Centers for Disease Control and Prevention in the United States to evaluate the pandemic potential of different influenza strains58, we found that the EA H1N1 and swine H3N2 viruses are among the animal viruses with the highest risk score in Yang’s analysis26. Besides, at least one human infection with a similar reassortant IAV-S has been reported22. We suggest that intensive surveillance of IAV-S and of swine-to-human infections with the IAV-S described in our study should be a priority for future research.
Electronic supplementary material
Figure S1 Phylogenetic analysis of the PB2 (A), PB1 (B), PA (C) and NP (D) of the 14 IAVs-S. The unrooted trees were based on nucleotides sequences of PB2, PB1, PA and NP and were generated with the MEGA 7.0 program by using neighbor-joining analysis and reliability of the tree was assessed by bootstrap analysis with 1000 replications. Neighbor-joining bootstrap values ≥70 are shown at the major branches of the trees
Acknowledgements
We thank Gene S. Tan for editing the manuscript. We also thank Richard Cadagan for excellent technical support. The study was supported by the Guangxi Natural Science Foundation under Grant No. 2015GXNSFCA139002 and 2016GXNSFBA380219, Inovation-Driven Development Foundation of Guangxi under Grant No. Guike AA17204057 and Doctor Startup Funds of Guangxi University under Grant No. XBZ160123. This study was also partially supported by CRIP (Center for Research on Influenza Pathogenesis), an NIAID funded Center of Excellence for Influenza Surveillance and Research (CEIRS, contract HHSN272201400008C).
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Adolfo García-Sastre, Email: Adolfo.Garcia-Sastre@mssm.edu.
Ying Chen, Email: yingchen@gxu.edu.cn.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0088-z).
References
- 1.McDonald SM, et al. Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat. Rev. Microbiol. 2016;14:448–460. doi: 10.1038/nrmicro.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mena I, et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. eLife. 2016;5:e16777. doi: 10.7554/eLife.16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger JK, et al. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 4.Novel Swine-Origin Influenza AVIT. Dawood FS, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 5.Pensaert M, et al. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull. World Health Organ. 1981;59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 6.Smith GJ, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 7.Zhou NN, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karasin AI, et al. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000;68:71–85. doi: 10.1016/S0168-1702(00)00154-4. [DOI] [PubMed] [Google Scholar]
- 9.Webby RJ, et al. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 2000;74:8243–8251. doi: 10.1128/JVI.74.18.8243-8251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tremblay D, et al. Emergence of a new swine H3N2 and pandemic (H1N1) 2009 influenza A virus reassortant in two Canadian animal populations, mink and swine. J. Clin. Microbiol. 2011;49:4386–4390. doi: 10.1128/JCM.05676-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijaykrishna D, et al. Reassortment of pandemic H1N1/09 influenza A virus in swine. Science. 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan X, et al. Emergence and dissemination of a swine H3N2 reassortant influenza virus with 2009 pandemic H1N1 genes in pigs in China. J. Virol. 2012;86:2375–2378. doi: 10.1128/JVI.06824-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiromoto Y, et al. Isolation of the pandemic (H1N1) 2009 virus and its reassortant with an H3N2 swine influenza virus from healthy weaning pigs in Thailand in 2011. Virus Res. 2012;169:175–181. doi: 10.1016/j.virusres.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, et al. Emergence of novel reassortant H3N2 swine influenza viruses with the 2009 pandemic H1N1 genes in the United States. Arch. Virol. 2012;157:555–562. doi: 10.1007/s00705-011-1203-9. [DOI] [PubMed] [Google Scholar]
- 15.Lange J, et al. Reassortants of the pandemic (H1N1) 2009 virus and establishment of a novel porcine H1N2 influenza virus, lineage in Germany. Vet. Microbiol. 2013;167:345–356. doi: 10.1016/j.vetmic.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 16.Pascua PN, et al. Emergence of H3N2pM-like and novel reassortant H3N1 swine viruses possessing segments derived from the A (H1N1) pdm09 influenza virus, Korea. Influenza Other Respir. Viruses. 2013;7:1283–1291. doi: 10.1111/irv.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehira K, et al. Reassortant swine influenza viruses isolated in Japan contain genes from pandemic A(H1N1) 2009. Microbiol. Immunol. 2014;58:327–341. doi: 10.1111/1348-0421.12152. [DOI] [PubMed] [Google Scholar]
- 18.Baudon E, et al. Detection of novel reassortant influenza A (H3N2) and H1N1 2009 pandemic viruses in Swine in Hanoi, Vietnam. Zoonoses Public. Health. 2015;62:429–434. doi: 10.1111/zph.12164. [DOI] [PubMed] [Google Scholar]
- 19.Watson SJ, et al. Molecular epidemiology and evolution of influenza viruses circulating within european Swine between 2009 and 2013. J. Virol. 2015;89:9920–9931. doi: 10.1128/JVI.00840-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Jong JC, et al. Isolation of swine-like influenza A(H1N1) viruses from man in Switzerland and The Netherlands. Ann. Inst. Pasteur Virol. 1988;139:429–437. doi: 10.1016/S0769-2617(88)80078-9. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, et al. Human infection from avian-like influenza A (H1N1) viruses in pigs, China. Emerg. Infect. Dis. 2012;18:1144–1146. doi: 10.3201/eid1807.120009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W, et al. Reassortant Eurasian avian-like influenza A(H1N1) virus from a severely Ill child, Hunan Province, China, 2015. Emerg. Infect. Dis. 2016;22:1930–1936. doi: 10.3201/eid2211.160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang DY, et al. Human infection with Eurasian avian-like influenza A(H1N1) virus, China. Emerg. Infect. Dis. 2013;19:1709–1711. doi: 10.3201/eid1910.130420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jhung MA, et al. Outbreak of variant influenza A(H3N2) virus in the United States. Clin. Infect. Dis. 2013;57:1703–1712. doi: 10.1093/cid/cit649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, et al. Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses. Proc. Natl Acad. Sci. USA. 2016;113:392–397. doi: 10.1073/pnas.1522643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skowronski DM, et al. Cross-reactive and vaccine-induced antibody to an emerging swine-origin variant of influenza A virus subtype H3N2 (H3N2v) J. Infect. Dis. 2012;206:1852–1861. doi: 10.1093/infdis/jis500. [DOI] [PubMed] [Google Scholar]
- 27.Lam TT, et al. Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic. J. Virol. 2011;85:10279–10285. doi: 10.1128/JVI.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, et al. Emergence of human-like H3N2 influenza viruses in pet dogs in Guangxi, China. Virol. J. 2015;12:10. doi: 10.1186/s12985-015-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization. WHO manual on animal influenza diagnosis and surveillance. WHO/CDS/CSR/NCS/2002. http://apps.who.int/iris/bitstream/10665/68026/1/WHO_CDS_CSR_NCS_2002.5.pdf (2002).
- 30.Sun YF, et al. Novel triple-reassortant H1N1 swine influenza viruses in pigs in Tianjin, Northern China. Vet. Microbiol. 2016;183:85–91. doi: 10.1016/j.vetmic.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Glaser L, et al. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearce MB, et al. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc. Natl Acad. Sci. USA. 2012;109:39 44–3949. doi: 10.1073/pnas.1119945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao D, et al. Glycosylation of the hemagglutinin protein of H5N1 influenza virus increases its virulence in mice by exacerbating the host immune response. J. Virol. 2017;91:e02215–e02216. doi: 10.1128/JVI.02215-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatta M, et al. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 36.Bussey KA, et al. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J. Virol. 2010;84:4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res. 2017;27:1409–1421. doi: 10.1038/cr.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J. Virol. 2012;86:9666–9674. doi: 10.1128/JVI.00958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl Acad. Sci. USA. 2009;106:21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada S, et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6:e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holsinger LJ, et al. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pica N, et al. The DBA.2 mouse is susceptible to disease following infection with a broad, but limited, range of influenza A and B viruses. J. Virol. 2011;85:12825–12829. doi: 10.1128/JVI.05930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson MI, et al. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009-2011. J. Virol. 2012;86:8872–8878. doi: 10.1128/JVI.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang H, et al. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J. Virol. 2014;88:10864–10874. doi: 10.1128/JVI.01327-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong W, et al. Novel reassortant influenza viruses between pandemic (H1N1) 2009 and other influenza viruses pose a risk to public health. Microb. Pathog. 2015;89:62–72. doi: 10.1016/j.micpath.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Schrauwen EJ, Fouchier RA. Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microbes Infect. 2014;3:e9. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neumann G, Kawaoka Y. Transmission of influenza A viruses. Virology. 2015;479-480:234–246. doi: 10.1016/j.virol.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matrosovich M, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 2000;74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G, et al. H6 influenza viruses pose a potential threat to human health. J. Virol. 2014;88:3953–39564. doi: 10.1128/JVI.03292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 51.Li Z, et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steel J, et al. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimble JB, et al. Compatibility of H9N2 avian influenza surface genes and 2009 pandemic H1N1 internal genes for transmission in the ferret model. Proc. Natl Acad. Sci. USA. 2011;108:12084–12088. doi: 10.1073/pnas.1108058108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lakdawala SS, et al. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog. 2011;7:e1002443. doi: 10.1371/journal.ppat.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell PJ, et al. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J. Virol. 2014;88:3802–3814. doi: 10.1128/JVI.03607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abente EJ, et al. A highly pathogenic avian-derived influenza virus H5N1 with 2009 pandemic H1N1 internal genes demonstrates increased replication and transmission in pigs. J. Gen. Virol. 2017;98:18–30. doi: 10.1099/jgv.0.000678. [DOI] [PubMed] [Google Scholar]
- 57.Houser KV, Katz JM, Tumpey TM. Seasonal trivalent inactivated influenza vaccine does not protect against newly emerging variants of influenza A (H3N2v) virus in ferrets. J. Virol. 2013;87:1261–1263. doi: 10.1128/JVI.02625-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trock SC, Burke SA, Cox NJ. Development of framework for assessing influenza virus pandemic risk. Emerg. Infect. Dis. 2015;21:1372–1378. doi: 10.3201/eid2108.141086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic analysis of the PB2 (A), PB1 (B), PA (C) and NP (D) of the 14 IAVs-S. The unrooted trees were based on nucleotides sequences of PB2, PB1, PA and NP and were generated with the MEGA 7.0 program by using neighbor-joining analysis and reliability of the tree was assessed by bootstrap analysis with 1000 replications. Neighbor-joining bootstrap values ≥70 are shown at the major branches of the trees