Abstract
Wheat (Triticum aestivum L.) is a temperate cereal with an optimum temperature range of 15–22°C during the grain filling stage. Heat stress is one of the major environmental constraints for wheat production worldwide. Temperatures above 25°C during the grain filling stage significantly reduced wheat yield and quality. This reduction was reported due to the inactivation of the soluble starch synthase, a key heat-labile enzyme in starch transformation of wheat endosperm. To improve wheat productivity under heat stress, the rice soluble starch synthase I, under the control of either a constitutive promoter or an endosperm-specific promoter, was expressed in wheat and the transgenic lines were monitored for expression and the effects on yield-related traits. The results showed that the transgenic wheat events expressed rice soluble starch synthase I at a high level after four generations, and transgenic plants produced grains of greater weight during heat stress. Under heat stress conditions, the thousand kernel weight increased 21–34% in T2 and T3 transgenic plants compared to the non-transgenic control plants. In addition, the photosynthetic duration of transgenic wheat was longer than in non-transgenic controls. This study demonstrated that the engineering of a heat tolerant soluble starch synthase gene can be a potential strategy to improve wheat yield under heat stress conditions.
Keywords: Starch synthesis, Wheat transformation, Heat stress, Thermotolerance, Yield
Introduction
Wheat (Triticum aestivum L.) is one of the most important main food crops worldwide. It provides approximately 20% of the calories consumed by humans (Pfeifer et al. 2014). Given the socioeconomic importance of wheat, many efforts of breeding have focused on improving its yield, quality, and adaptability to biotic and abiotic stresses. Generally, wheat yield and quality can be affected by various environmental constraints such as high temperature, drought, or the combination of these two (Wardlaw et al. 2002; Stratonovitch and Semenov 2015). With global climate changes, the frequency of extreme high temperatures is expected to increase and negatively impact crop production, especially for cold season crops (Pradhan et al. 2012). It is reported that for every 1°C rise above the optimum temperature during the grain filling stage, wheat yield is reduced 3 to 4% (Wardlaw et al. 1989). Ever-increasing evidence in the past decades has demonstrated that elevated temperature during wheat anthesis and grain filling stages significantly reduces grain yield (Bhullar and Jenner 1986a; Keeling et al. 1993; Farooq et al. 2011; Prasad and Djanaguiraman 2014). Even short-term heat stress could cause an 11–23% reduction in grain number and 10–26% reduction in individual grain sizes (Talukder et al. 2013). In the Great Plains regions of the USA, high temperature stress is not uncommon during early reproductive development and grain filling periods of wheat when wheat is most susceptible to extreme temperatures (Assad and Paulsen 2002). Improving the heat tolerance in wheat has been considered a key goal that would result in yield stability (Kuchel et al. 2007).
A major component of wheat seeds is starch, which accounts for 55–75% of total dry grain weight (Gillies et al. 2012). Therefore, the grain yield depends largely on starch accumulation in the seed endosperm (Jenner et al. 1991). Several processes in starch deposition during the grain filling stage have been identified in response to heat stress. Starch granules in higher plants usually contain two classes of glucose polymers, amylose, and amylopectin. In wheat, amylose is a linear or less branched molecule with a low degree of polymerization comprising 25–30% of wheat grain starch, and amylopectin is a large and highly branched polymer comprising 70–75% of grain starch (Myers et al. 2000; James et al. 2003). Both amylose and amylopectin are synthesized from ADP-glucose (Myers et al. 2000). Soluble starch synthase (SSS) enzymes catalyze the extension of linear glucan chains by adding glucose units to the glucan non-reducing ends through α-1,4 linkages, and then starch branching enzymes cleave and reattach the glucan segments to form branches on the polymers (Jeon et al. 2010). In cereal endosperm, these genes have been divided into five classes, including the soluble SS I-IV (SSI, SSII, SSIII, and SSIV) and granule bound starch synthase (GBSS) (Hirose and Terao 2004).
Previous studies indicated that the adverse effects of elevated temperature on starch deposition in wheat grain endosperm are likely due to the inactivation of starch synthesis (Bhullar and Jenner 1986a, b; Rijven 1986; Jenner 1994; Zhao et al. 2008; Prakash et al. 2009) and that soluble starch synthase I was the most heat-labile enzyme limiting the starch deposition in the biosynthetic pathway in seeds (Keeling et al. 1993; Jenner 1994; Sumesh et al. 2008; Prakash et al. 2009). In the developing endosperm, the expression levels of starch biosynthesis genes have been also reported to be early induced, but significantly suppressed by heat stress (Hurkman et al. 2003). Genetic engineering of such key enzymes to increase their thermostability might be an alternative approach to improve the starch biosynthesis under heat stress conditions.
The soluble starch synthase genes from a subtropical species, rice (Oryza sativa L.), could be good candidates to be introduced into wheat by genetic engineering. Previous studies in rice have demonstrated that the predominant SSI was stable and active at high temperature up to 35°C, resulting in increased production of long chain of amylopectin in the endosperm (Jiang et al. 2003). The presence of either SSI or SSIIIa gene could sustain the starch biosynthesis in rice, and SSI may have higher activity than SSIIIa, accounting for almost 70% of the total SS activity (Fujita et al. 2006; Fujita et al. 2011). A similar phenomenon observed in maize, the purified SSI enzyme showed a higher heat stability than SSIII at an incubation temperature of 42°C (Huang et al. 2016). The rice SSI presents only one isoform in rice (Baba et al. 1993; Hirose and Terao 2004) and shares 81% amino acid similarity with wheat SSI that produces a 75-kDa protein (Li et al. 1999; McMaugh et al. 2014).
The present report evaluated the overexpression of the rice SSI gene in transgenic wheat. The transgenic plants had significantly enhanced grain yield under heat stress during the grain filling stage, likely due to higher starch deposition resulting in greater kernel weight compared to the non-transgenic wheat. These results demonstrate that engineering heat-sensitive key enzymes can be a useful strategy for improving the heat tolerance of cold season crops, and for developing food crops adapted to an increasingly changed environment.
Materials and Methods
Plant materials
A spring wheat, T. aestivum L. cv. ‘Bobwhite’, was used for transformation, as well as phenotypic comparison in this study. Plants were grown in a controlled environment with a 16-h photoperiod at 600 μmol m−2 s−1 (Sylvania 20934 T-5 fluorescent tube, Sylvania, Wilmington, MA). The day/night temperatures were set at 20/15°C. Immature embryos were separated from surface sterilized caryopses and placed the embryo axis faced down in contact with CM4 medium at 25°C for 1 wk in darkness for callus formation as described by Zhou et al. (1995). The caryopses were collected from wheat plants that were at the stage of 12–14 d post anthesis. After callus formation, the embryogenic calluses were selected and prepared for biolistic bombardment.
Thermostability prediction
Relative thermostability of the rice SSI protein, the amino acid sequences of rice (UniPort database: Q0DEC8), and wheat SSI (Genbank Accession ID: AAD54661) was performed using a novel scoring algorithm as described by Li et al. (2010).
Plasmid constructs
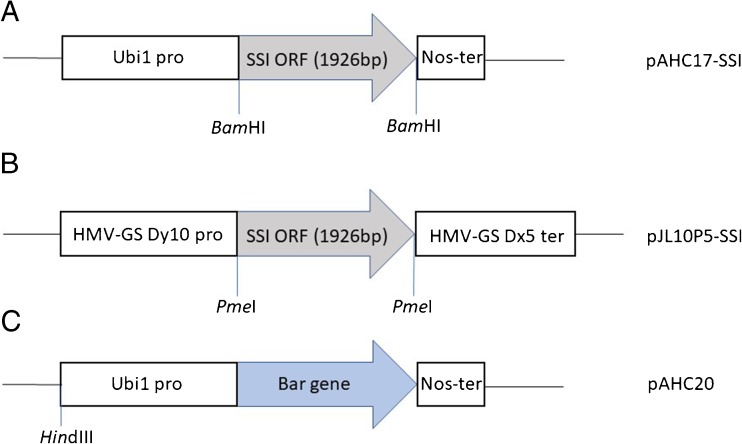
The expression cassette in the pAHC17 vector contained the maize ubiquitin 1 (Ubi1) promoter (Christensen et al. 1992) and the nopaline synthase (NOS) terminator. The vector pJL10P5 (a kind gift from Dr. Ann Blechl, USDA-ARS, Albany, CA) was driven by endosperm-specific high-molecular-weight (HMW) glutenin Dy10 promoter (Lamacchia et al. 2001) and Dx5 terminator (Fig. 1B) from T. aestivum. The 1926-bp O. sativa soluble starch synthase I (SSI) ORF (Genbank accession ID: FJ750946) was subcloned into pAHC17 using a BamHI site and pJL10P5 using PmeI restriction sites (Fig. 1A, B). The orientation within the plasmid and the integrity of the SSI gene were confirmed by PCR and sequencing. The third construct, pAHC20 contained the bar gene controlled by Ubi1 promoter and NOS terminator (Fig. 1C) which confers resistance to glufosinate-ammonium, the active ingredient of the herbicide Liberty® (AgrEvo, Wilmington, DE).
Figure 1.
Schematic representation of the constructs for SSI and bar gene expression. (A) Construction map of pAHC17-SSI expression region. It contained the maize ubiquitin 1 promoter (Ubi1), the rice SSI gene, and nopaline synthase terminator (Nos-ter). The SSI ORF was sub-cloned into pAHC17 vector using BamHI restriction site. (B) Construction map of pJL10P5-SSI expression region. It contained Dy10 high-molecular-weight glutenin promoter (HMW-GS Dy10), the rice SSI gene, and the Dx5 terminator. The SSI ORF was sub-cloned into pJL10P5 using Pme I restriction site. (C) Construction map of pAHC20 containing bar gene which confers resistance to the herbicide Liberty® (glufosinate-ammonium).
Wheat transformation
Biolistic transformation was performed using particle inflow gun as previously described by Finer et al. (1992). Plasmid DNAs (either pAHC17-SSI or pJL10P5-SSI) were mixed with pAHC20 in a 1:1 molar ratio prior to bombardment. After bombardment, all calluses were transferred to selective callus induction medium containing 5 mg L−1 glufosinate-ammonium and maintained in darkness for 2 wk. Recovery and regeneration of transgenic tissues was according to previously established protocols with slightly modification (Altpeter et al. 1996; Anand et al. 2003). The media used were described by Cheng et al. (1997). After 10–12 wk., T0 transgenic plants with developed roots and shoots were transferred into small peat pots for hardening with high humidity. These hardened plants were selected by herbicide resistance using 0.2% (v/v) Liberty® (AgrEvo) solution diluted with double-distilled water. The solution was applied using a cotton ball on adaxial leaves selected. The positive plants were transferred into Pro-Mix BX grow medium (Premier Tech, Rivière-du-Loup, Canada) and grown in the growth chamber 16 h day/8 h night, 500 μmol m−2 s−1 light intensity (Lucalux 400W High Pressure Sodium lamps, General Electric, Cleveland, OH), at 19°C.
DNA extraction, polymerase chain reaction, and Southern analysis for transgenic wheat plants
In each generation, PCR testing was performed using genomic DNA to screen transgenic wheat plants for both bar gene and the rice SSI gene. Samples of 100–150-mg leaf tissue were taken and placed in a 2-mL centrifuge tube and homogenized with liquid nitrogen. Eight hundred-microliter 2× cetyltrimethylammonium bromide (CTAB) extraction buffer containing 4 μL 2-mercaptoethanol (Sigma-Aldrich®, St. Louis, MO) was added to each sample. Genomic DNA was isolated from leaf tissue using modified CTAB method described by Rogers and Bendich (1985). The presence of bar gene from pAHC20 was determined with a primer pair UbiAB-F (5′-CTTCAGCAGGTGGGTGTAGAGCGTG) and BarAB-R (5′-CCTGCCTTCATACGCTATTTATTTGC). The rice SSI-specific primer pair, osSSI-F (5′- CCCGATCTAGAAGGTCTCACAG), and osSSI-R (5′-TATGACAAGAACACTGCGGGC) amplified a 636-bp fragment, without any amplification of wheat genomic DNA and cDNA. The amplification condition was 95°C for 5 min, 32 cycles of 95°C 30 s, 60°C 30 s, and 72°C 30 s followed a final extension step at 72°C for 10 min. Southern blot analysis was conducted using a sequence amplified by osSSI-F and osSSI-R with pAHC17-SSI plasmid DNA as template. The PCR product was purified using QIAGEN® gel purification kit (QIAGEN®, Valencia, CA) and labeled with dCTP (α-32P) using Megapriming DNA labeling system (Amersham, Piscataway, NJ) following the manufacturer’s instructions. The labeled PCR product was used as a probe in Southern blot analysis. Twenty-five-microgram genomic DNA was digested with BamHI for Dy10 transgenic plants, and HindIII for both Ub1 and Ub2 transgenic lines. Digested DNA were separated on 0.8% agarose gels and transferred to a hybridization membrane. The membrane was probed with labeled PCR product (Sambrook and Russell 2006; Southern 2006).
RNA extraction and reverse transcription polymerase chain reaction
Total RNA was isolated from both leaves and 20-d-old developing seeds of wheat. For the leaf sample, total RNA was isolated using RNeasy® Plant Mini Kit (QIAGEN®), while isolation from the seed was done by following a previously described protocol of Li and Trick (2005). Total RNA was treated with RQ1 DNase (Promega®, Madison, WI) to remove genomic DNA contaminants. One microgram of treated total RNA was used as template using a reverse transcription system kit (Catalog no. A3500, Promega®) following the product protocol. PCR was conducted using osSSI primers described above. DNA contamination was checked by performing PCR using the tubulin primers TubF:5′-ATCTGTGCCTTGACCGTATCAGG and TubR: 5′-GACATCAACATTCAGAGCACCATC. PCR conditions were the same as described above.
Western blot analysis
Total soluble proteins in flag leaves and seeds were extracted from each T2 transgenic wheat event, one non-transgenic wheat variety (BW), and one rice variety (Nipponbare) at 20 d after anthesis. Proteins were extracted by grinding and homogenizing samples in protein extraction buffer (Fu et al. 2008). Protein concentrations were estimated by using Quick Start® Bradford Protein Assay kit (Bio Rad® Inc., Hercules, CA) according to manufacturer instructions. Samples were diluted in the buffer to maintain equal concentration for equal loading. Equal loading was also maintained by checking stained gels. Protein separation was performed by loading 40 μg of each sample on 10% (w/v) sodium dodecyl sulfate (SDS) polyacrylamide gels. SDS-PAGE separated proteins were then transferred to polyvinylidene difluoride (PDVF) membranes for immunoblotting. PDVF membranes with the transferred proteins were probed for rice SSI protein using rabbit polyclonal anti-SSI antibodies (AnaSpec, Inc., Fremont, CA), and goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Heat stress bioassays and phenotyping
A total of four independent heat stress experiments were conducted following similar procedure with different transgenic generations and temperature regimes. Day length of all the experiment was maintained as 16/8 h light/dark. The transgenic generations and high temperature regimes for all the experiments were as follows: Exp. 1 (T2 generation; 31/24°C day/night), Exp. 2 (T2; 33/26°C), Exp. 3 (T3; 34/28°C), and Exp. 4 (T4; 32–33/30°C). In each experiment, phenotyping was conducted under heat stress and optimum temperature conditions (22/15°C day/night for Exp. 1–3; and 20/15°C for Exp. 4), using eight to 20 pots for each event per line. Transgenic plants from ubiquitin promoter events (Ub-1, Ub-2), a Dy10 promoter event (Dy10) and non-transgenic Bobwhite produced from tissue culture (BWTC) were grown in the greenhouse. Following germination, seedlings were PCR screened for the presence of SSI gene in the laboratory and single seedlings were transplanted to pots (14 cm height, 50 cm top perimeter, and 36 cm bottom perimeter) filled with Metro Mix® 200 potting soil (Hummert™ Intl., Topeka, KS). The greenhouse was set at day/night temperature of 22/15°C with 16-h photoperiod supplemented with 400 W high-pressure sodium light (~ 800–600 μmol m−2 s−1) for normal growth. Plants were watered daily. One or two tillers per plant were tagged at anthesis, and half the plants from each event/line were transferred to a high-temperature growth chamber at 10 d after anthesis, based on the anthesis date of the tagged tillers. The remaining plants were transferred to another growth chamber having optimum day/night temperature (22/15°C) with the same light conditions as the high temperature chamber. Thousand kernel weight (TKW) was obtained from the tagged tillers by drying to equivalent moisture content, weighing the seeds and converting to TKW using the seed number of that tagged head. The number of effective tillers was also counted during harvest. Chlorophyll content was measured as described by Uauy et al. (2006), on flag leaves every other d for 12 d, starting at 12 d after anthesis. Days required for physiological maturity were calculated from date of anthesis to date of physiological maturity (based on 50% of stems turning yellow). Non-transgenic and transgenic event means were compared using two sample t tests with unequal sample variance at α = 0.05.
Results and Discussion
Comparison between wheat and rice SSI genes
The SSI enzyme is responsible for the elongation of shorter A and B1 chains during starch biosynthesis in the soluble phase of the amylopectin (Commuri and Keeling 2001; Fujita et al. 2006) and therefore the inactivation or destruction of this gene can result in a significant impact on crystalline amylopectin matrix formation, as well as grain filling. To determine the relative thermostability of the rice SSI protein, the amino acid sequences of rice (UniPort database: Q0DEC8) and wheat SSI (Genbank Accession ID: AAD54661) were compared to each other (Fig. 2). Using a novel scoring algorithm to compare relative thermostability of proteins using an integrated statistical and machine learning approach (Li et al. 2010), it was predicted that the rice SSI protein was more thermostable than the wheat SSI protein (data not shown). This result agrees with previous reports that the wheat soluble SSI enzyme is heat-labile based on bioassays (Keeling et al. 1993; Jenner 1994; Sumesh et al. 2008; Prakash et al. 2009). To test the hypothesis that the expressed heat-stable rice SSI gene in wheat would improve the grain filling potential of wheat under moderate heat stress, the rice SSI gene was selected and introduced into the Bobwhite wheat variety.
Figure 2.
Amino acid sequence comparison for two SSI enzymes from wheat (Ta_SSI: 647aa) and rice (Os_SSI: 641aa). Two enzymes share 82% identity. X indicates amino acid mismatch, dash indicates gaps in alignment, and bullet indicates sequence alignment.
Overexpression of rice SSI gene in wheat and segregation patterns
Two constructed expression vectors, pAHC17-SSI and pJL10P5-SSI, were co-transformed into wheat with pAHC20 containing the selective marker gene bar. After transformation and herbicide selection, a total of six bar positive transgenic wheat events were identified. Five of six events contained the modified rice SSI gene as determined by PCR analysis from leaf DNA in the T0 generation (Table 1). Ultimately, three SSI-positive events were obtained for further analyses. Two transgenic events were under the control of Ubi1 promoter, assigned as Ub1 and Ub2. The third event was under the control of the wheat HMW-glutenin Dy10 promoter, assigned as Dy10, which was only expressed in seeds.
Table 1.
Presence of rice SSI gene in six Liberty® positive Triticum aestivum L. cv. ‘Bobwhite’ events and their Χ2 values
| Event no. | Promoter | PCR test in T0 plants | Number of PCR positive T1 plants | Number of PCR negative T1 plants | Χ2 value (based on 3:1 ratio) |
|---|---|---|---|---|---|
| 1286 (Ub1) | Ubiquitin | + | 37 | 6 | 2.79 |
| 1588 | Ubiquitin | + | 4 | 2 | 1.33 |
| 1700 (Ub2) | Ubiquitin | + | 7 | 2 | 0.037 |
| 2036 | DY10 | + | 46 | 20 | 0.99 |
| 2173 (Dy10) | DY10 | + | 13 | 5 | 0.074 |
| 2540 | DY10 | – | 0 | 10 | – |
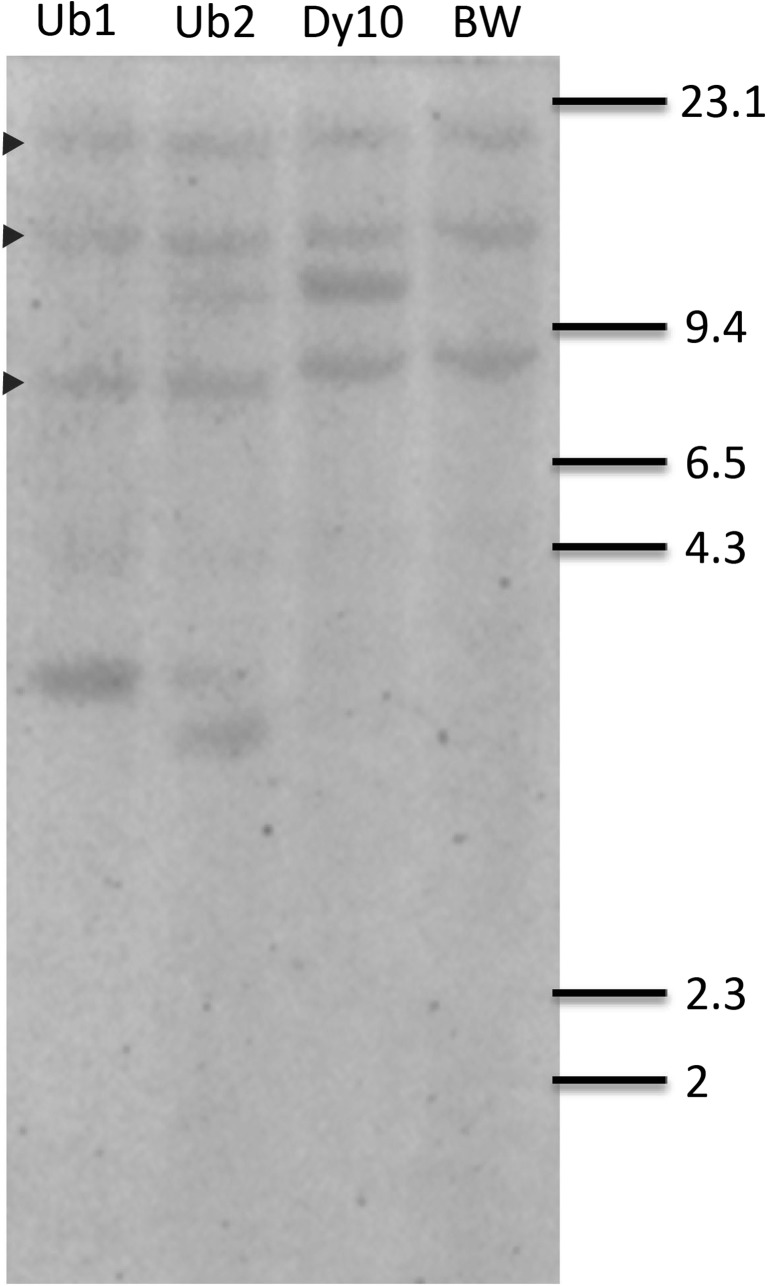
Analysis of SSI gene segregation using Χ2 test in T1 plants showed that there may be one or more transgene copies of the SSI in the events (Table 1). Southern analysis using genomic DNA from T2 leaves and the SSI gene sequence as a probe demonstrated that all three tested transgenic events and the non-transgenic BW had three bands in common (Fig. 3), indicating that the probe could be hybridized with three endogenous SSI genes, corresponding to each of the three genomes of hexaploid wheat. This observation is consistent with a previous report (Li et al. 1999) that high similarity (88%) between the rice and wheat SSI genes. Additional unique bands were presented in each transgenic line (Fig. 3), confirmed the transgene integration. Based on band numbers, Ub1 and Dy10 transgenic events appeared to harbor only a single SSI gene insertion (single copy), while Ub2 event was likely to harbor three copies of the rice SSI gene.
Figure 3.
Southern blot analysis of three T2 generation transgenic Triticum aestivum L. cv. ‘Bobwhite’ events and a non-transgenic Bobwhite (BW) control. Twenty-five micrograms of leaf DNA was digested with BamHI for DY10, or HindIII for Ub1 and Ub2 events, fractioned on 0.8% agarose gel, transferred to a nylon membrane, and hybridized with 32p-labeled PCR product of rice SSI gene. Ub1, event number 1286 with ubiquitin promoter; Ub2, event number 1700 with ubiquitin promoter; Dy10, event number 2173 with Dy10 promoter; BW, non-transgenic Bobwhite variety. Arrows indicated the endogenous SSI genes in hexaploid wheat.
Molecular analysis of transgenic wheat plants
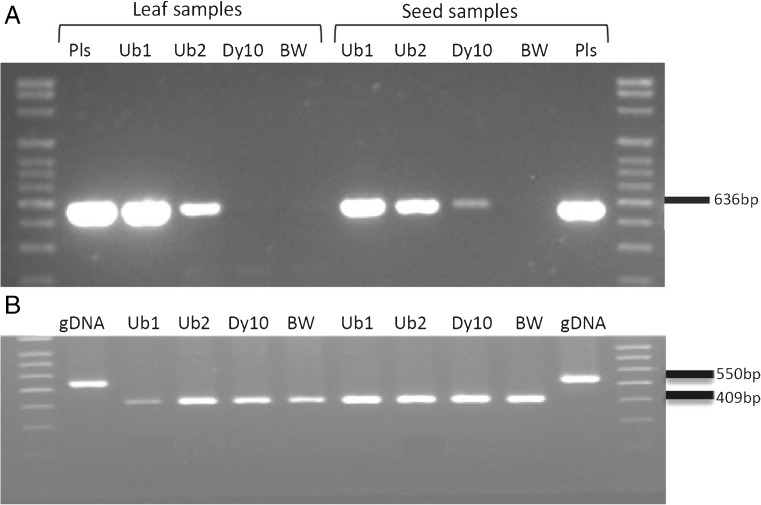
To confirm the transgene expression in wheat plants, RT-PCR was conducted using total RNA isolated from both leaf and seed samples of the three transgenic lines and non-transgenic Bobwhite. The leaf samples from Ub1 and Ub2 produced a band matching the expected size from the rice SSI gene, while the Dy10 event and the non-transgenic BW did not show any bands (Fig. 4A). The failure of the Dy10 event to produce a band using cDNA from the leaf sample was due to the tissue specificity of the Dy10 promoter as expected. The seeds of non-transgenic BW did not show any bands, while the seed samples of all three independent events showed expected rice SSI bands (Fig. 4A). It confirmed the expression of the rice SSI gene driven by both Ubi1 and Dy10 promoters in the transgenic wheat. Single tubulin bands with different sizes between genomic DNA samples and RNA samples demonstrated that RNA samples were not contaminated with DNA (Fig. 4B).
Figure 4.
RT-PCR for leaf samples (left) and seed sample (right) of Triticum aestivum L. cv. ‘Bobwhite’. (A) Total RNA was isolated from both leaf and seed samples, and was used to carry out RT-reaction, following PCR was done with SSI-specific primers and visualized on 1% agarose gel. Pls plasmid DNA having SSI gene, Ub1 event number 1286 with Ubi1 promoter, Ub2 event number 1700 with Ubi1 promoter, Dy10 event number 2173 with Dy10 promoter, BW non-transgenic Triticum aestivum L. cv. ‘Bobwhite’. (B) RT-PCR with corresponding cDNA samples using tubulin gene primers for leaf samples (left) and seed sample (right). The tubulin primers amplified a 409-bp band for cDNA, and 550-bp band for genomic DNA, separately. gDNA genomic DNA of BW.
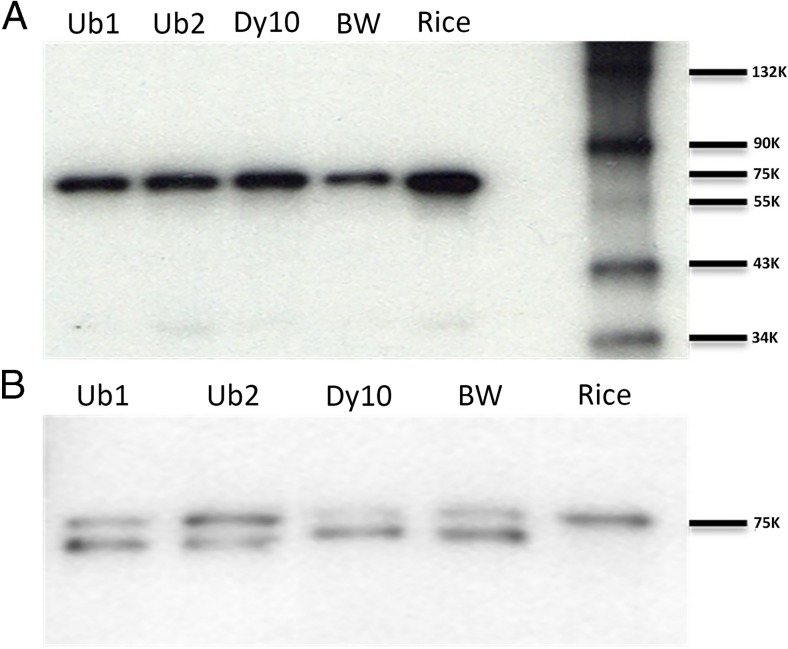
Western blot analysis was conducted to detect the presence of SSI proteins in wheat. A rice leaf sample was included as a control. In western blot analysis, the differentiation between rice SSI from wheat SSI was not successful as the antibody bound to both rice and wheat proteins (Fig. 5). This cross-reactivity was likely due to the high similarity of the amino acid sequences and length between the two proteins (Fig. 2). In seeds, only one SSI protein band was detected in the three transgenic events, non-transgenic BW, and rice. The SSI proteins showed a similar molecular weight (71 kDa) in both wheat and rice with stronger intensities in the transgenic wheat events than in non-transgenic wheat (Fig. 5A), indicating that the rice SSI protein was probably produced in the endosperm of all three transgenic wheat events. A previous study showed that the rice SSI protein produced a 57-kDa protein band (Baba et al. 1993). However, in the present study, an approximate 71-kDa band was detect by the western blot, which was similar to the 71-kDa wheat SSI band. The cDNA sequence of the rice SSI was used to query the UniProt database (www. Uniport.org), and the expected size of the protein (accession no. Q0DEC8) of 70.95 kDa was predicted, which was supportive of this current finding.
Figure 5.
Western blot analysis of seed and leaf samples from T2 generation of Triticum aestivum L. cv. ‘Bobwhite’. Three T2 generation transgenic events, one non-transgenic T. aestivum L. cv. ‘Bobwhite’ and one rice sample was used. Total proteins extracted from both leaf and seed samples were fractionated on 10% sodium dodecyl sulfate-polyacrylamide gel (SDS page gel) and transferred to PDVF membrane, probed with polyclonal antibody raised against rice SSI. Equal amount of total protein (40 μg) was loaded in each lane. In seed samples (A), all transgenic events along with rice showed stronger signal than non-transgenic wheat. The antibody bound with both wheat endogenous SSI and rice SSI protein. In leaf sample (B), antibody bound with two SSI isoforms of wheat. Comparing the signal strength of the second isoforms of SSI, Ub1, Ub2, and rice leaf sample showed enhanced signal for SSI.
In leaves, two SSI protein bands (isoforms 60 and 71 kDa) were detected in three transgenic events and non-transgenic BW. The 71-kDa SSI isoform showed stronger band intensity in the Ub2 event than in the Ub1 event, Dy10, and non-transgenic BW. However, the 50-kDa SSI isoform showed weaker band intensity in the Ub2 even than in the Ub1 event, Dy10, and non-transgenic BW (Fig. 5B). The results indicated that there were probably two isoforms of SSI in wheat leaf tissue, but only one isoform existed in seeds. This observation is consistent with a previous report that two SSI isoforms were expressed in wheat leaf tissues, and only one isoform in seeds (Denyer et al. 1995). Only the 71-kDa isoform was detected in rice leaf tissue (Fig. 5B).
The effects of elevated temperature on transgenic wheat plant
Moderately high temperatures reduce the SSI activity of wheat at both the transcription (Hurkman et al. 2003) and enzyme activity levels (Rijven 1986), while the reduction of transcription and enzyme activity in rice is mild (Yamakawa et al. 2007). Accordingly, the transgenic wheat plants in the present study were placed under the elevated temperature to determine if the thermostable rice SSI could compensate for the low activity of wheat SSI during heat stress. Successful compensation would be expected to increase the individual grain weight by sustaining the enzyme activity. Based on previous molecular analysis above, all three events (Ub1, Ub2, and Dy10) positive for transgene (SSI) were used for heat stress bioassays. The PCR negative plants from different transgenic events (BWTC) and Bobwhite plants not derived from tissue culture (BWNTC) were used as controls.
Three independent experiments were conducted for phenotyping under increasing temperature ranges (31/24, 33/26, and 34/28°C with 16/8 h day/night) compared to optimal temperature conditions (22/15°C). In Exp. 1, T2 plants from Ub1, Dy10, and BWTC were exposed to heat stress at 31/24°C (day/night) from 10 d of anthesis until harvest. Significant differences were found for TKW for Ub1 (3.22%) and Dy10 (6.71%) events compared to BWTC (Table 2). Effective tiller number per plant and days required for physiological maturity showed no significant difference, but seed number per selected head was significantly lower for the Dy10 event (Table 2). Under optimum temperature conditions, there was no significant difference between transgenic and non-transgenic events for any studied traits (Fig. 6).
Table 2.
Comparison of agronomic traits between transgenic plants and non-transgenic Triticum aestivum L. cv. ‘Bobwhite’ in heat stress Exp. 1
| Temp. | Event name† | TKW (g) | Seed number per selected head | ||
|---|---|---|---|---|---|
| Mean ± STD | T stat | Mean ± STD | T stat | ||
| 31/24°C | BWTC | 29.5 ± 1.21 | 54.33 ± 11.58 | ||
| Ub1 | 30.45 ± 1.32 | 1.85* | 54.2 ± 14.24 | 0.02 | |
| Dy10 | 31.48 ± 0.83 | 3.47** | 41.25 ± 4.03 | 2.54* | |
| 22/15°C | BWTC | 35.62 ± 1.5 | 59.33 ± 11.5 | ||
| Ub1 | 37.71 ± 1.96 | 1.92 | 58.55 ± 11.1 | 0.1 | |
| Dy10 | 37.53 ± 2.89 | 1.13 | 45.75 ± 12.44 | 1.49 | |
TKW thousand kernel weight, BWTC non-transgenic Bobwhite but established through tissue culture, Ub1 event 1286 having ubiquitin promoter, Dy10 event 2173 having Dy10 promoter, STD standard deviation
†Number of plants studied in this experiment is accompanied by event name in parenthesis
*, **, and *** denote P < 0.05, 0.01, and 0.001 respectively
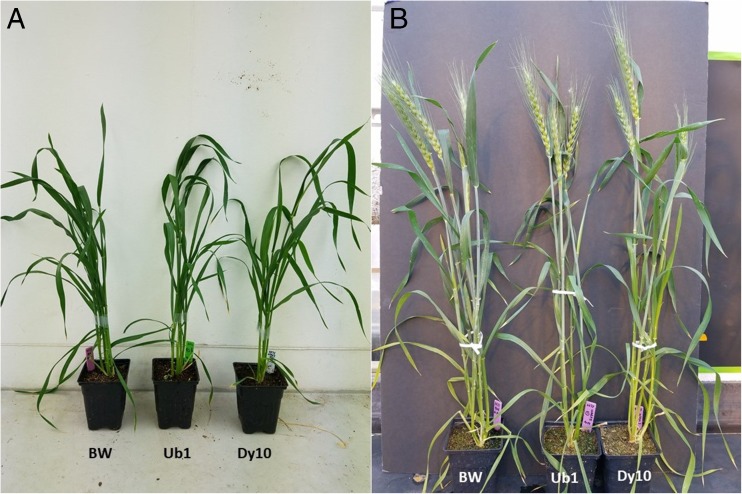
Figure 6.
Phenotype comparison of transgenic lines and non-transgenic control lines. Plants were growing under optimum temperature. The transgenic lines showed similar phenotypes as non-transgenic control. Photos were taken at 40 d after germination (A), and 50 d after germination during Triticum aestivum L. cv. ‘Bobwhite’ heading stage (B).
In Exp. 2, an increased temperature (33/26°C) was used for all three (Ub1, Ub2, and Dy10) transgenic events. Both BWTC and BWNTC were pooled as controls since there were no significant differences observed for the traits evaluated under heat and normal growing conditions. Transgenic events with the Ub1 (10.70%) and Dy10 (25.67%) promoters showed significant increases in TKW compared to control plants (Table 3). Individually, all events with the transgene had higher TKW than non-transgenic lines. The Dy10 event produced significantly lower number of seeds than non-transgenic control (BW) in both heat stress (33/26°C) and optimum temperature (22/15°C) conditions (Table 3). This result could be indicative of an interaction between the Dy10 promoter and seed setting capability, or simply the effect of somaclonal variation, which might have been fixed with advancing the generation. There was no evidence in the previous literatures suggesting that the Dy10 promoter would compromise seed setting capability. Additional events with the Dy10 promoter may be necessary to draw a more definite conclusion. Under optimum temperature conditions, tiller number and days required for physiological maturity were not significantly different (Table 3). Moreover, the Dy10 event took significantly longer to reach physiological maturity (32.66 d) under heat stress than non-transgenic events. It has been reported that pulling sugar from the leaf to seed helps to reduce feedback inhibition of leaf sugar on photosynthesis (Smidansky et al. 2002). It is possible that altered source-sink relationships resulted in a longer grain fill period, although additional events would need to be examined to draw a firm conclusion. In Exp. 3, an even higher temperature regime (34/28°C) was used in the bioassays with T3 plants. Under heat stress condition, transgenic events with both Ubi1 (22.94%) and Dy10 (34.59%) promoters produced seeds having significantly higher TKW (Tables 3 and 4) and visibly larger kernels than non-transgenic events under heat stress condition (Fig. 7). Comparably, the Dy10 event in T3 generation still produced lower number of seeds per selected head, while Ub1 and Ub2 events showed no difference compared to controls (Table 3). Tiller number per plant and chlorophyll content was not significantly different between transgenic and non-transgenic events (data not shown). Days required for physiological maturity differed significantly between transgenic and non-transgenic events, with transgenic events averaging 1.28 additional days of grain filling duration than non-transgenic plants. This observation infers the photosynthetic duration of the heat-stressed plants were longer than non-transgenic control plants.
Table 3.
Comparison of agronomic traits between transgenic and non-transgenic Triticum aestivum L. cv. ‘Bobwhite’ in Exp. 2 and Exp. 3 at different temperature ranges
| Temp. | Event name | TKW | Seed numbers per selected head | Physiological maturity (days) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2nd experiment (high temp 33/26°C) | 3rd experiment (high temp 34/28°C) | 2nd experiment (high temp 33/26°C) | 3rd experiment (high temp 34/28°C) | 2nd experiment (high temp 33/26°C) | 3rd experiment (high temp 34/28°C) | ||||||||
| Mean ± STD | T stat | Mean ± STD | T stat | Mean ± STD | T stat | Mean ± STD | T stat | Mean ± STD | T stat | Mean ± STD | T stat | ||
| High temperature | BWTC | 22.87 ± 1.94 | 0.301 | 20.23 ± 2.65 | 0.134 | 47.40 ± 4.56 | 0.48 | 46.6 ± 11.74 | 0.32 | 26.8 ± 3.27 | 0.25 | 22.8 ± 1.30 | 0.63 |
| BWNTC | 22.51 ± 1.67 | 19.94 ± 3.89 | 46.00 ± 4.24 | 48.8 ± 9.73 | 27.25 ± 2.22 | 22.4 ± 0.54 | |||||||
| BW | 22.71 ± 1.73 | – | 20.09 ± 3.14 | – | 46.78 ± 4.20 | – | 47.7 ± 10.23 | – | 27 ± 2.69 | – | 22.6 ± 0.97 | – | |
| Ub1 | 24.45 ± 1.44 | 1.89* | 24.35 ± 4.31 | 1.96* | 46.00 ± 4.08 | 0.31 | 50.4 ± 12.80 | 0.41 | 29 ± 3.92 | 0.93 | 23.4 ± 0.89 | 1.59 | |
| Ub2 | 25.17 ± 2.35 | 1.88* | 25.03 ± 2.46 | 3.50** | 49.50 ± 13.47 | 0.40 | 48.33 ± 4.13 | 0.17 | 28 ± 2.45 | 0.66 | 23.66 ± 0.82 | 2.36* | |
| Ub | 24.81 ± 1.84 | 2.41* | 24.70 ± 3.24 | 3.30** | 47.75 ± 9.41 | 0.27 | 49.27 ± 8.67 | 0.38 | 28.5 ± 3.07 | 1.06 | 23.54 ± 0.82 | 2.41* | |
| Dy10 | 28.54 ± 1.85 | 6.14*** | 27.04 ± 3.12 | 3.75** | 36.83 ± 9.22 | 2.48* | 25.25 ± 5.85 | 5.14** | 29.66 ± 2.50 | 1.96* | 24.25 ± 0.96 | 2.90* | |
| Optimum temperature 22/15°C | BWTC | 40.95 ± 2.27 | 0.144 | 40.91 ± 2.68 | 0.932 | 39.0 ± 8.18 | 0 | 50.2 ± 11.26 | 0.989 | 40.8 ± 1.79 | 1.31 | 38.8 ± 1.30 | 0 |
| BWNTC | 40.69 ± 3.00 | 39.45 ± 2.25 | 39.0 ± 10.61 | 56.4 ± 8.35 | 43 ± 2.94 | 38.8 ± 4.82 | |||||||
| BW | 40.84 ± 2.41 | – | 40.18 ± 2.46 | – | 39.0 ± 8.70 | – | 53.3 ± 9.9 | – | 41.78 ± 2.49 | – | 38.8 ± 3.33 | – | |
| Ub1 | 42.11 ± 3.37 | 0.68 | 43.57 ± 4.28 | 1.77 | 37.5 ± 9.15 | 0.276 | 53.83 ± 10.06 | 0.103 | 42 ± 3.46 | 0.115 | 36.5 ± 4.42 | 1.10 | |
| Ub2 | 40.27 ± 1.36 | 1.81 | 43.17 ± 2.16 | 2.55* | 49.5 ± 11.15 | 1.67 | 53.84 ± 9.54 | 0.106 | 41 ± 3.37 | 0.414 | 37.17 ± 5.19 | 0.69 | |
| Ub | 41.20 ± 2.57 | 0.292 | 43.38 ± 3.25 | 2.63** | 43.5 ± 20.06 | 0.587 | 53.83 ± 9.35 | 0.129 | 41.5 ± 3.21 | 0.20 | 36.83 ± 4.61 | 1.16 | |
| Dy10 | 40.53 ± 1.76 | 0.276 | 44.68 ± 4.54 | 1.87 | 30.0 ± 7.11 | 1.96* | 36.75 ± 16.52 | 1.87 | 41.2 ± 1.64 | 0.52 | 40.25 ± 4.03 | 0.64 | |
The Ub observation combines the Ub1 and Ub2 observations
TKW thousand kernel weight, BWTC non-transgenic Bobwhite established through tissue culture, BWNTC non-transgenic Bobwhite not regenerated through tissue culture, Ub1 event 1286 having ubiquitin promoter, Ub2 event 1700 having ubiquitin promoter, Dy10 event 2173 having Dy10 promoter, STD standard deviation
*, **, and *** denote P < 0.05, 0.01, and 0.001 respectively
Table 4.
Comparison of Triticum aestivum L. cv. ‘Bobwhite’ thousand kernel weight (TKW), TKW percent change, and mean seed number at high temperature and optimum temperature conditions
| Experiment | Event | TKW | % TKW increment | Seed number per selected head | ||
|---|---|---|---|---|---|---|
| Mean ± STD | T stat | Mean ± STD | T stat | |||
| Heat stress (32–33/30°C) | BW | 23.55 ± 4.88 | 47.17 ± 8.7 | |||
| Ub1 | 26.75 ± 2.55 | 1.39* | 13.58 | 52.6 ± 1.95 | 1.48 | |
| Ub2 | 26.39 ± 3.89 | 1.11 | 12.06 | 51.17 ± 6.55 | 0.90 | |
| Dy10 | 30.50 ± 3.38 | 2.94** | 29.51 | 46.57 ± 3.78 | 0.16 | |
| Optimum temperature (20/15°C) | BW | 34.27 ± 2.10 | 47.0 ± 12.0 | |||
| Ub1 | 34.72 ± 1.00 | 0.43 | 1.31 | 50.4 ± 10.41 | 0.47 | |
| Ub2 | 35.21 ± 0.78 | 0.95 | 2.74 | 49.83 ± 9.26 | 0.43 | |
| Dy10 | 37.59 ± 1.21 | 2.97 | 9.67 | 49 ± 10.1 | 0.27 | |
STD standard deviation
* and ** denote P < 0.05 and 0.01 respectively
Figure 7.
Triticum aestivum L. cv. ‘Bobwhite’ seed phenotype comparison under heat stress and normal temperature. (A) Harvested seeds were produced by plants grown under elevated temperature (34/28°C) after anthesis. The seeds of non-transgenic plants (BW) were visually smaller than seeds of transgenic lines (Ub1 and Dy10). (B) Harvested seeds were produced by plants grew under optimal temperature condition (22/15°C). There was no significant difference among seed size of transgenic seeds (Ub1 and Dy10) and non-transgenic controls (BW). The scale bars represent 1 cm.
In Exp. 4, the heat stress bioassay with a higher nighttime temperature and lower daytime temperature (32/33°C) was also performed. Similar to previous results, Ub1 (13.58%), Ub2 (12.06%), and Dy10 (29.51%) showed considerable increase in TKW than non-transgenic controls under high temperature (Table 4). There was high variability in kernel weight within the events; as a result, even though there was a remarkable increment in Ub1 and Ub2, they were not significantly different. None of the transgenic events showed significant variation for seed number per head in any temperature condition, indicating seed set was not affected. This result indicates such differences are probably due to starch deposition rather than compensation for reduced seed set.
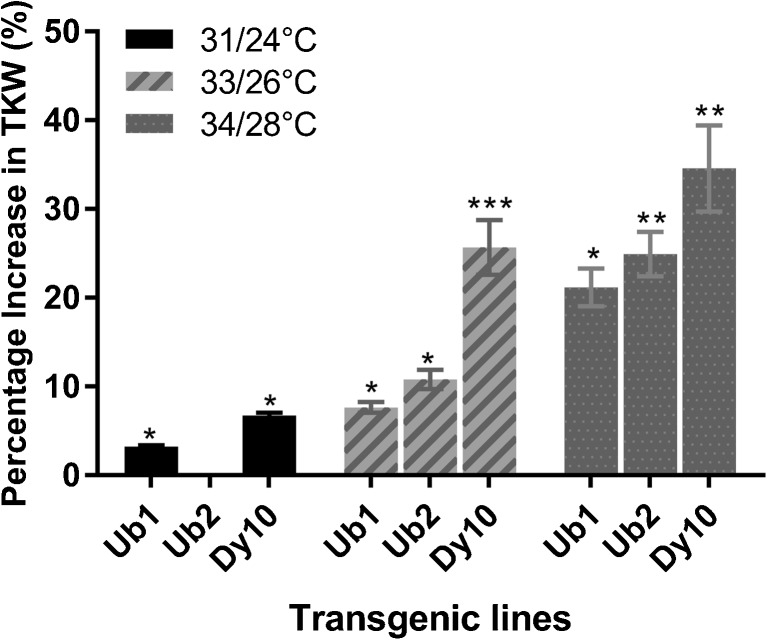
Taken together, three transgenic lines produced significantly heavier seeds compared to non-transgenic lines under heat stress in three independent experiments. This data suggests that starch synthase activity was maintained under high temperature and likely allowed continuous starch accumulation. The increased grain fill duration observed at optimum temperature may be due to reduction of feedback inhibition of photosynthesis by reducing the accumulation of sugars. Under optimum temperature, some transgenic events produced slightly heavier seeds (but not significantly) compared to controls. This observation may be due to the additive effects with the rice SSI transgene, or, more likely, may indicate other wheat starch synthase genes are affected by temperature fluctuations within the growth chambers. Nonetheless, these experiments demonstrate SSI is likely the most important enzyme for sustaining starch synthesis under heat stress. There was no significant variation among transgenic and non-transgenic events in optimum temperature conditions. It was clear that the TKW differences between the transgenic and non-transgenic lines increased (Fig. 8) as the temperatures were increased. These results support the hypothesis that wheat SSI enzyme is less stable than rice SSI at high temperature, and the additional stable rice SSI expressed in wheat is able to reduce the affect in grain yield by heat stress. It also supports previous research regarding the importance of SSI activity related to grain filling (Prakash et al. 2009). No significant variation in chlorophyll content was observed between transgenic and non-transgenic events, indicating that differences in starch deposition were not due to the differences in photosynthetic capacity. This result is in line with a previous report that reduced grain filling at high temperature was not likely due to the supply of the assimilate from photosynthesis (Sofield et al. 1977).
Figure 8.
Percent change in thousand kernel weight (TKW) compared to control for three independent experiments using Triticum aestivum L. cv. ‘Bobwhite’. The Ub2 was absent for the 31/24°C treatment. *, **, and *** denote P < 0.05, 0.01 and 0.001 respectively found in the comparison.
Conclusion
With global warming, abiotic stresses, especially heat and drought, could be a critical limiting factor for crop production. These results indicate that introduction of thermostable SSS in wheat could be a useful strategy for improving heat tolerance. This approach might be beneficial for other cool season species that are grown under conditions where heat stress may limit production. In addition, it is useful to explore the heat stability of SSS from other species typically grown under high-temperature conditions to identify the most heat-stable sources of the enzyme. It is also very useful to explore genetic variability within the wheat gene pool for the ability to maintain greenness under heat stress (Ristic et al. 2008). Research on heat tolerance in wheat has also focused on the ability to retain green leaf area under stress. This ability to retain photosynthetic capacity under stress is important. It may be necessary to identify and utilize heat-stable starch synthase enzymes to be able to fully take advantage of genetic mechanism that retain green leaf area and preserve photosynthetic function. The observation of increased grain fill of transgenic plants under the highest stress levels further indicates the connection between these traits needs to be explored more fully. New biotechnology, coupled with knowledge on starch metabolism, will help provide new strategies for improving the heat stress tolerance in wheat grains.
Acknowledgements
The authors would like to thank Julie Essig and Sheila Stevens for their technical assistance
Funding information
This work was supported by funding from the Kansas Wheat Commission. This article is contribution on 18-031-J from the Kansas Agricultural Experiment Station, Kansas State University, Manhattan, KS.
References
- Altpeter F, Vasil V, Srivastava V, Stöger E, Vasil IK. Accelerated production of transgenic wheat (Triticum aestivum L.) plants. Plant Cell Rep. 1996;16:12–17. doi: 10.1007/BF01275440. [DOI] [PubMed] [Google Scholar]
- Anand A, Zhou T, Trick HN, Gill BS, Bockus WW, Muthukrishnan S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot. 2003;54:1101–1111. doi: 10.1093/jxb/erg110. [DOI] [PubMed] [Google Scholar]
- Assad MT, Paulsen GM. Genetic changes in resistance to environmental stresses by U.S. Great Plains wheat cultivars. Euphytica. 2002;128:85–96. doi: 10.1023/A:1020688606174. [DOI] [Google Scholar]
- Baba T, Nishihara M, Mizuno K, Kawasaki T, Shimada H, Kobayashi E, Ohnishi S, Tanaka K, Arai Y. Identification, cDNA cloning, and gene expression of soluble starch synthase in rice (Oryza sativa L.) immature seeds. Plant Physiol. 1993;103:565–573. doi: 10.1104/pp.103.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar S, Jenner C. Effects of a brief episode of elevated temperature on grain filling in wheat ears cultured on solutions of sucrose. Aust J Plant Physiol. 1986;13:617–626. doi: 10.1071/PP9860617. [DOI] [Google Scholar]
- Bhullar S, Jenner C. Effects of temperature on the conversion of sucrose to starch in the developing wheat endosperm. Aust J Plant Physiol. 1986;13:605–615. doi: 10.1071/PP9860605. [DOI] [Google Scholar]
- Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 1997;115:971–980. doi: 10.1104/pp.115.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Commuri PD, Keeling PL. Chain-length specificities of maize starch synthase I enzyme: studies of glucan affinity and catalytic properties. Plant J. 2001;25:475–486. doi: 10.1046/j.1365-313x.2001.00955.x. [DOI] [PubMed] [Google Scholar]
- Denyer K, Hylton CM, Jenner CF, Smith AM. Identification of multiple isoforms of soluble and granule-bound starch synthase in developing wheat endosperm. Planta. 1995;196:256–265. doi: 10.1007/BF00201382. [DOI] [Google Scholar]
- Farooq M, Bramley H, Palta JA, Siddique KHM. Heat stress in wheat during reproductive and grain-filling phases. Crit Rev Plant Sci. 2011;30:491–507. doi: 10.1080/07352689.2011.615687. [DOI] [Google Scholar]
- Finer JJ, Vain P, Jones MW, McMullen MD. Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep. 1992;11:323–328. doi: 10.1007/BF00233358. [DOI] [PubMed] [Google Scholar]
- Fu J, Momčilović I, Clemente TE, Nersesian N, Trick HN, Ristic Z. Heterologous expression of a plastid EF-Tu reduces protein thermal aggregation and enhances CO2 fixation in wheat (Triticum aestivum) following heat stress. Plant Mol Biol. 2008;68:277–288. doi: 10.1007/s11103-008-9369-6. [DOI] [PubMed] [Google Scholar]
- Fujita N, Satoh R, Hayashi A, Kodama M, Itoh R, Aihara S, Nakamura Y. Starch biosynthesis in rice endosperm requires the presence of either starch synthase I or IIIa. J Exp Bot. 2011;62:4819–4831. doi: 10.1093/jxb/err125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006;140:1070–1084. doi: 10.1104/pp.105.071845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies SA, Futardo A, Henry RJ. Gene expression in the developing aleurone and starchy endosperm of wheat. Plant Biotechnol J. 2012;10:668–679. doi: 10.1111/j.1467-7652.2012.00705.x. [DOI] [PubMed] [Google Scholar]
- Hirose T, Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.) Planta. 2004;220:9–16. doi: 10.1007/s00425-004-1314-6. [DOI] [PubMed] [Google Scholar]
- Huang B, Keeling PL, Hennen-Bierwagen TA, Myers AM. Comparative in vitro analyses of recombinant maize starch synthases SSI, SSIIa, and SSIII reveal direct regulatory interactions and thermosensitivity. Arc Biochem Biophys. 2016;596:63–72. doi: 10.1016/j.abb.2016.02.032. [DOI] [PubMed] [Google Scholar]
- Hurkman WJ, McCue KF, Altenbach SB, Korn A, Tanaka CK, Kothari KM, Johnson EL, Bechtel DB, Wilson JD, Anderson OD, DuPont FM. Effect of temperature on expression of genes encoding enzymes for starch biosynthesis in developing wheat endosperm. Plant Sci. 2003;164:873–881. doi: 10.1016/S0168-9452(03)00076-1. [DOI] [Google Scholar]
- James MG, Denyer K, Myers AM. Starch synthesis in the cereal endosperm. Curr Opin Plant Biol. 2003;6:215–222. doi: 10.1016/S1369-5266(03)00042-6. [DOI] [PubMed] [Google Scholar]
- Jenner C. Starch synthesis in the kernel of wheat under high temperature conditions. Aust J Plant Physiol. 1994;21:791–806. doi: 10.1071/PP9940791. [DOI] [Google Scholar]
- Jenner C, Ugalde T, Aspinall D. The physiology of starch and protein deposition in the endosperm of wheat. Aust J Plant Physiol. 1991;18:211–226. doi: 10.1071/PP9910211. [DOI] [Google Scholar]
- Jeon J-S, Ryoo N, Hahn T-R, Walia H, Nakamura Y. Starch biosynthesis in cereal endosperm. Plant Physiol Biochem. 2010;48:383–392. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Jiang H, Dian W, Wu P. Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry. 2003;63:53–59. doi: 10.1016/S0031-9422(03)00005-0. [DOI] [PubMed] [Google Scholar]
- Keeling PL, Bacon PJ, Holt DC. Elevated temperature reduces starch deposition in wheat endosperm by reducing the activity of soluble starch synthase. Planta. 1993;191:342–348. doi: 10.1007/BF00195691. [DOI] [Google Scholar]
- Kuchel H, Williams K, Langridge P, Eagles HA, Jefferies SP. Genetic dissection of grain yield in bread wheat. II. QTL-by-environment interaction. Theor Appl Genet. 2007;115:1015–1027. doi: 10.1007/s00122-007-0628-8. [DOI] [PubMed] [Google Scholar]
- Lamacchia C, Shewry PR, Di Fonzo N, Forsyth JL, Harris N, Lazzeri PA, Napier JA, Halford NG, Barcelo P. Endosperm-specific activity of a storage protein gene promoter in transgenic wheat seed. J Exp Bot. 2001;52:243–250. doi: 10.1093/jexbot/52.355.243. [DOI] [PubMed] [Google Scholar]
- Li Y, Middaugh CR, Fang J. A novel scoring function for discriminating hyperthermophilic and mesophilic proteins with application to predicting relative thermostability of protein mutants. BMC Bioinformatics. 2010;11:62. doi: 10.1186/1471-2105-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Rahman S, Kosar-Hashemi B, Mouille G, Appels R, Morell MK. Cloning and characterization of a gene encoding wheat starch synthase I. Theor Appl Genet. 1999;98:1208–1216. doi: 10.1007/s001220051186. [DOI] [Google Scholar]
- Li ZW, Trick HN. Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. BioTechniques. 2005;38:872–876. doi: 10.2144/05386BM05. [DOI] [PubMed] [Google Scholar]
- McMaugh SJ, Thistleton JL, Anschaw E, Luo J, Konik-Rose C, Wang H, Huang M, Larroque O, Regina A, Jobling SA, Morell MK, Li Z. Suppression of starch synthase I expression affects the granule morphology and granule size and fine structure of starch in wheat endosperm. J Exp Bot. 2014;65:2189–2201. doi: 10.1093/jxb/eru095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–998. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, Mayer KFX, Olsen O-A. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014;345:1250091. doi: 10.1126/science.1250091. [DOI] [PubMed] [Google Scholar]
- Pradhan GP, Prasad PVV, Fritz AK, Kirkham MB, Gill BS. Effects of drought and high temperature stress on synthetic hexaploid wheat. Funct Plant Biol. 2012;39:190–198. doi: 10.1071/FP11245. [DOI] [PubMed] [Google Scholar]
- Prakash P, Kumari A, Singh DV, Pandey R, Sharma-Natu P, Ghildiyal MC. Starch synthase activity and grain growth in wheat cultivars under elevated temperature: a comparison of responses. Indian J Plant Physiol. 2009;14:364–369. [Google Scholar]
- Prasad PVV, Djanaguiraman M. Response of floret fertility and individual grain weight of wheat to high temperature stress: sensitive stages and thresholds for temperature and duration. Funct Plant Biol. 2014;41:1261–1269. doi: 10.1071/FP14061. [DOI] [PubMed] [Google Scholar]
- Rijven AHGC. Heat inactivation of starch synthase in wheat endosperm tissue. Plant Physiol. 1986;81:448–453. doi: 10.1104/pp.81.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic Z, Bukovnik U, Momčilović I, Fu J, Vara Prasad PV. Heat-induced accumulation of chloroplast protein synthesis elongation factor, EF-Tu, in winter wheat. J Plant Physiol. 2008;165:192–202. doi: 10.1016/j.jplph.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol. 1985;5:69–76. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Southern blotting: capillary transfer of DNA to membranes. Cold Spring Harb Protoc. 2006;2006:pdb.prot4040. doi: 10.1101/pdb.prot4040. [DOI] [PubMed] [Google Scholar]
- Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci U S A. 2002;99:1724–1729. doi: 10.1073/pnas.022635299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofield I, Evans L, Cook M, Wardlaw I. Factors influencing the rate and duration of grain filling in wheat. Aust J Plant Physiol. 1977;4:785–797. doi: 10.1071/PP9770785. [DOI] [Google Scholar]
- Southern E. Southern blotting. Nat Protocols. 2006;1:518–525. doi: 10.1038/nprot.2006.73. [DOI] [PubMed] [Google Scholar]
- Stratonovitch P, Semenov MA. Heat tolerance around flowering in wheat identified as a key trait for increased yield potential in Europe under climate change. J Exp Bot. 2015;66:3599–3609. doi: 10.1093/jxb/erv070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumesh KV, Sharma-Natu P, Ghildiyal MC. Starch synthase activity and heat shock protein in relation to thermal tolerance of developing wheat grains. Biol Plant. 2008;52:749–753. doi: 10.1007/s10535-008-0145-x. [DOI] [Google Scholar]
- Talukder ASMHM, McDonald GK, Gill GS. Effect of short-term heat stress prior to flowering and at early grain set on the utilization of water-soluble carbohydrate by wheat genotypes. Field Crops Res. 2013;147:1–11. doi: 10.1016/j.fcr.2013.03.013. [DOI] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw I, Dawson I, Munibi P. The tolerance of wheat to high temperatures during reproductive growth. 2. Grain development. Aust J Agric Res. 1989;40:15–24. doi: 10.1071/AR9890015. [DOI] [Google Scholar]
- Wardlaw IF, Blumenthal C, Larroque O, Wrigley CW. Contrasting effects of chronic heat stress and heat shock on kernel weight and flour quality in wheat. Funct Plant Biol. 2002;29:25–34. doi: 10.1071/PP00147. [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Hirose T, Kuroda M, Yamaguchi T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007;144:258–277. doi: 10.1104/pp.107.098665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Dai T, Jiang D, Cao W. Effects of high temperature on key enzymes involved in starch and protein formation in grains of two wheat cultivars. J Agron Crop Sci. 2008;194:47–54. doi: 10.1111/j.1439-037X.2007.00283.x. [DOI] [Google Scholar]
- Zhou H, Arrowsmith JW, Fromm ME, Hironaka CM, Taylor ML, Rodriguez D, Pajeau ME, Brown SM, Santino CG, Fry JE. Glyphosate-tolerant CP4 and GOX genes as a selectable marker in wheat transformation. Plant Cell Rep. 1995;15:159–163. doi: 10.1007/BF00193711. [DOI] [PubMed] [Google Scholar]