Abstract
Mammalian innate and adaptive immune systems use the pattern recognition receptors, such as toll-like receptors, to detect conserved bacterial and viral components. Bacteria synthesize diverse D-amino acids while eukaryotes and archaea generally produce two D-amino acids, raising the possibility that many of bacterial D-amino acids are bacteria-specific metabolites. Although D-amino acids have not been identified to bind to any known pattern recognition receptors, D-amino acids are enantioselectively recognized by some other receptors and enzymes including a flavoenzyme D-amino acid oxidase (DAO) in mammals. At host–microbe interfaces in the neutrophils and intestinal mucosa, DAO catalyzes oxidation of bacterial D-amino acids, such as D-alanine, and generates H2O2, which is linked to antimicrobial activity. Intestinal DAO also modifies the composition of microbiota through modulation of growth for some bacteria that are dependent on host nutrition. Furthermore, regulation and recognition of D-amino acids in mammals have additional meanings at various host–microbe interfaces; D-phenylalanine and D-tryptophan regulate chemotaxis of neutrophils through a G-coupled protein receptor, D-serine has a bacteriostatic role in the urinary tract, D-phenylalanine and D-leucine inhibit innate immunity through the sweet taste receptor in the upper airway, and D-tryptophan modulates immune tolerance in the lower airway. This mini-review highlights recent evidence supporting the hypothesis that D-amino acids are utilized as inter-kingdom communication at host–microbe interface to modulate bacterial colonization and host defense.
Keywords: D-amino acid, D-amino acid oxidase, hydrogen peroxide, mucosal immunity, innate immunity, small intestine, neutrophil, host–microbe interaction
Introduction
Among all domains of life, bacteria have the largest capacity to produce wide variety of D-amino acids, whereas archaea and eukaryotes are thought to synthesize generally two kinds of D-amino acids, D-serine and D-aspartate. Bacteria utilize diverse D-amino acids in multiple biological processes to support their growth, to regulate spore germination, and to configure or remodel their cell wall (Cava et al., 2011). By contrast, mammals utilize D-serine in neurophysiology and D-aspartate in neurogenesis and endocrine systems (Fujii and Saito, 2004). Metabolism of D-amino acids in mammals involves two flavoenzymes: D-amino acid oxidase (DAO) and D-aspartate oxidase. DAO catalyzes stereoselective oxidative deamination of multiple neutral and basic D-amino acids, which yields alpha-keto acids, ammonium ion, and hydrogen peroxide (Figure 1A, showing the exact chemical reaction). DAO is rarely found in bacteria, but occurs widely in most eukaryotes from yeast to humans with the exception of plants (Pollegioni et al., 2007). DAO activity was first described in the porcine kidney by Krebs (1935), but its physiological role was not clear because its substrates had been regarded as “unnatural” isomers of amino acids. After discovery of D-amino acids as integral components of bacterial cell wall in 1950s, Cline and Lehrer in 1969 identified DAO activity in granule fraction of human neutrophilic leukocytes (Cline and Lehrer, 1969), which is linked to bactericidal activity of leukocytes by H2O2 produced through oxidation of bacterial D-amino acids (Eckstein et al., 1971; DeChatelet et al., 1972). DAO has received attention by neuroscientists for past few decades because DAO in the mammalian hindbrain degrades its physiological endogenous substrate D-serine, which binds to N-methyl D-aspartate (NMDA) glutamate receptors and plays crucial roles in neurophysiology and pathology (Mothet et al., 2000; Shleper et al., 2005; Sasabe et al., 2007; Basu et al., 2009; Mitchell et al., 2010; Mustafa et al., 2010; Balu et al., 2013). More recently, DAO was identified in epithelial surface of the mammalian small intestine, where interplay between mammalian DAO and bacterial D-amino acids modifies commensal bacteria and mucosal defense (Sasabe et al., 2016). Notably no mammalian genes have homology to any known bacterial genes encoding synthetic enzymes for D-amino acids, and many of bacterial D-amino acids are thought bacteria-specific metabolites. Therefore, metabolism of bacterial D-amino acids by mammalian DAO or other molecules in the host–microbial interface may serve as a type of bacterial recognition.
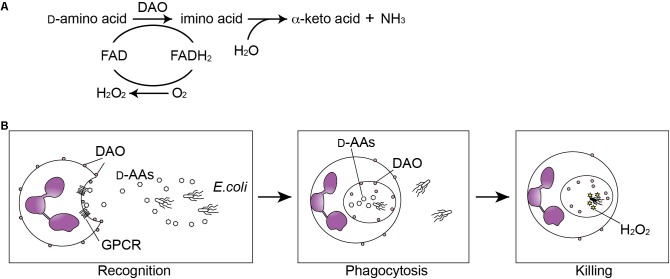
FIGURE 1.
Oxidation of D-amino acids by D-amino acid oxidase (DAO) generates anti-microbial H2O2. (A) DAO catalizes oxidative deamination of D-amino acids. The D-amino acid is oxidized to an imino acid with reduction of FAD to FADH2, which is subsequently oxidized to FAD with reduction of oxygen into hydrogen peroxide. The imino acid is then non-enzymatically hydrolyzed to the corresponding alpha-keto acid and ammonia. (B) Neutrophils recognize bacterial D-amino acids through the G protein-coupled receptor (GPCR) GPR109B and are chemoattracted by bacteria. DAO oxidizes bacterial D-amino acids during the phagocytosis and generated H2O2 kills the bacteria.
Distribution of DAO Substrates in Mammals
D-amino acid oxidase is distributed to the proximal tubules in the kidney with the highest expression, hepatocytes in the liver (exceptionally not detectable in mice), astrocytes in the hindbrain of the central nervous system, neutrophils, and epithelium of the small intestine (Pollegioni et al., 2007; Koga et al., 2017). Although DAO has oxidative activity selective to D-enantiomers of amino acids, DAO has a broad spectrum of substrates including multiple neutral and basic D-amino acids. In fact, mice lacking systemic DAO activity due to a missense mutation of G181R (DAO-null mice) show increased levels of various D-amino acids, including D-alanine, D-leucine, D-methionine, D-proline, D-phenylalanine, D-serine, and D-tyrosine, in the multiple tissues and body fluids (Koga et al., 2017). Since bacteria have the largest genetic capacity to produce wide variety of D-amino acids, most of the DAO substrates except for D-serine have been regarded to originate from bacteria. Among those D-amino acids, D-alanine and D-serine are considered major substrates of DAO in mammals because the two D-amino acids show marked increases compared to other D-amino acids in the DAO-null mice (Miyoshi et al., 2011; Koga et al., 2017).
The D-alanine and D-serine have different origins in mammals. D-Serine is produced in mammals through conversion from L-serine by an endogenous enzyme, serine racemase (SR) (Wolosker et al., 1999), expressed primarily in neurons of the central nervous system (Miya et al., 2008). The concentration of D-serine is highest (10–30% of total serine) at submillimolar level in the forebrain (Hashimoto et al., 1992; Miyoshi et al., 2009; Suzuki et al., 2017), where D-serine serves as an endogenous coagonist with L-glutamate to activate NMDA receptors (Mothet et al., 2000; Basu et al., 2009). By contrast, DAO degrades D-serine and retains it at a low level (less than 1% of total serine) in the hindbrain and spinal cord (Miyoshi et al., 2009). In the periphery, D-serine level is usually low with the exception of that in the urine (Miyoshi et al., 2009). The D-serine is over 50% of total urinary serine and is one of the most abundant amino acids in mammalian urine, where D-serine has a bacteriostatic role against uropathogenic bacteria (Roos and Klemm, 2006; Korte-Berwanger et al., 2013) by inhibiting L-serine metabolism and synthesis of pantothenate (Cosloy and McFall, 1973) and by modulating virulence gene expression (Roesch et al., 2003; Anfora and Welch, 2006; Anfora et al., 2007). It is also known that mammalian D-serine can repress efficient colonization of enterohaemorrhagic Escherichia coli (EHEC) by selectively inhibiting expression of a type III secretion system, which allows intimate attachment of EHEC to the host cells (Connolly et al., 2015). Such anti-bacterial D-serine is rarely synthesized in bacteria with exception of vancomycin-resistant Enterococci (Sieradzki and Tomasz, 1996; De Jonge et al., 2002; Reynolds and Courvalin, 2005) and therefore, bacteria have been thought to develop sensing system of host D-serine to recognize niche and colonize to favorable sites within the host (Connolly et al., 2016).
Another major substrate of DAO in mammals is bacterial D-alanine, one of most common D-amino acids present in the bacterial cell wall. While mammals are not capable of synthesizing D-alanine, most bacteria encode two different PLP-dependent alanine racemases, DadX and Alr, which are established drug targets for antibiotics. Both racemases catalyze the same reaction, but are components of distinct molecular pathways. In contrast to association of DadX with L-alanine catabolism (D-alanine is subsequently converted into pyruvate), Alr synthesizes the D-alanine that is utilized for peptidoglycan synthesis (Walsh, 1989; Watanabe et al., 2002). The D-alanine is incorporated into the peptides to cross-linking repeated disaccharide and provides chemical resistance to most known proteases (Nagata et al., 1998). It has become clear that intestinal D-alanine as well as other D-amino acids including D-proline, and D-glutamate, is produced exclusively by intestinal microbiota (Sasabe et al., 2016). Furthermore, Konno et al. (1990, 1993) have shown using antibiotic-treated and germ-free (GF) mice that vast majority of D-alanine in the serum and urine is of bacterial origin. Therefore, D-alanine produced by intestinal microbiota is, at least in part, uptaken in the intestine, circulated, and excreted into the urine. Mechanisms regulating in vivo D-alanine kinetics, such as D-alanine transport and metabolism, still remain largely unclear.
Role of DAO in the Innate Defense by Leukocytes
D-amino acid oxidase, conserved widely in eukaryotes but not in bacteria (Pollegioni et al., 2007), is able to generate H2O2 through catabolism of D-amino acids (usually of bacterial origin), and therefore, DAO has classically been considered a potential component of the innate defense in mammals. Cline and Lehrer (1969) first identified DAO activity in granule fraction of guinea pig and human neutrophilic leukocytes, which is linked to bactericidal activity of leukocytes (Eckstein et al., 1971; DeChatelet et al., 1972). They have also shown in vitro that oxidation of bacterial metabolites such as D-amino acids by DAO generates H2O2 and subsequently activates chloride ions together with myeloperoxidase to kill E. coli. A later study using electron microscope showed that DAO is localized to the neutrophilic surface, internalized during phagocytosis, and is able to produce H2O2 within the phagosome (Robinson et al., 1978). On the other hand, in vivo bactericidal effect of neutrophilic DAO remains controversial. Neutrophils obtained from patients with chronic granulomatous disease (CGD), which is a primary immunodeficiency that interferes production of reactive oxygen species by phagocytes (i.e., neutrophils and macrophages) and leads to recurrent or persistent intracellular bacterial and fungal infections, have comparable DAO activity to those from control patients with bacterial infections (Eckstein et al., 1971). On the basis of the observation, it seems unlikely that oxidation of D-amino acids is the primary source of H2O2 generation during phagocytosis, where NADPH oxidase plays a major role (Nguyen et al., 2017). However, patients with CGD are rarely infected with catalase-negative organisms (Winkelstein et al., 2000), suggesting that a source of H2O2 stress independent of NADPH oxidase, such as DAO, may be selective for certain infections in these patients. It had remained unclear for more than 40 years if DAO plays a role in vivo against bacterial infection until the study using the DAO-null mice by Nakamura et al. (2012). The DAO-null mice were injected intravenously with Staphylococcus aureus, and showed increased number of the bacteria in the kidney and reduced survival rate compared to wild-type controls. However, they did not find ex vivo bactericidal effect of DAO in neutrophils derived from peritoneal cavity against S. aureus, warranting further examination using conditional knockout to unveil the physiological role of neutrophilic DAO. Another in vivo study has shown that neutrophil DAO functions to exert bactericidal activity on intraperitoneally injected Salmonella typhimurium at early stages of infection (Tuinema et al., 2014), when neutrophils are the major cell type infected by Salmonella (Loetscher et al., 2012). Interestingly, S. typhimurium limits exposure to oxidative damage elicited by DAO through importing D-alanine by dalS, an ABC importer specific for D-alanine. Indeed, dalS mutants of S. typhimurium are exposed to greater H2O2 stress than the wild type in vivo, which is attenuated by the presence of a chemical DAO inhibitor, 6-chloro-1,2-benzisoxazol-3(2H)-one (CBIO) (Tuinema et al., 2014). On the other hand, aromatic D-amino acids, such as D-phenylalanine and D-tryptophan, act as chemoattractant factors for human leukocytes through a G protein-coupled receptor, GPR109B (Irukayama-Tomobe et al., 2009). Collectively, these studies imply a mechanistic insight into a host–pathogen interaction; neutrophils are chemo-attracted by bacterial D-amino acids and kill bacteria through oxidation of the amino acids in phagosome (Figure 1B), while bacteria evade bactericidal activity of DAO through actively importing its substrates.
DAO as an Anti-Microbial Factor in the Mucosal Innate Defense
In addition to the bactericidal function of neutrophilic DAO, regulation of D-amino acids by DAO has recently been associated with mucosal homeostasis (Sasabe et al., 2016). DAO activity was reported in luminal epithelium of the small intestine in fish (Sarower et al., 2003a,b), chickens (Brachet and Puigserver, 1992), mice and humans (Sasabe et al., 2016). Intestinal DAO was described in common carp as a metabolizing agent for D-alanine because free D-alanine is abundant exceptionally in aquatic invertebrates, besides bacteria, such as crustaceans and bivalve mollusks (D’Aniello and Giuditta, 1980; Matsushima et al., 1984), which are potential food sources of these fish. Of note, feeding carps with D-alanine increases intestinal DAO activity by eightfold, suggesting that inducible nature of fish DAO is associated with its role in metabolizing exogenous D-alanine (Sarower et al., 2003b). Distribution of intestinal DAO in chickens shows similar pattern as that in mice. In chicken and mice, DAO activity is detected in the mucosa of small intestine and higher in the proximal part compared to the distal part within small intestine (Brachet and Puigserver, 1992; Sasabe et al., 2016). Importantly, intestinal DAO in mice and humans localizes to Muc2-positive secretory vesicles of goblet cells as well as to enterocytes, which is inducible by the presence of vancomycin-sensitive intestinal microbiota, and oxidizes DAO substrates such as D-alanine in the mucosa and epithelium (Sasabe et al., 2016). Therefore, host DAO is induced and released in response to certain intestinal microbiota to react to microbial metabolites. Such microbe–host interaction can affect intestinal microbiota presumably in two ways (Sasabe et al., 2016) (Figure 2A). The first is the similar way as described in the leukocytes that oxidation of bacterial D-amino acids generates bactericidal H2O2, which limits colonization of pathogenic bacteria including Vibrio cholerae in the small intestine. V. cholerae and V. parahaemolyticus are more sensitive to DAO than Listeria monocytogenes, EHEC, or Salmonella enterica (Sasabe et al., 2016), in part, because Vibrio produces a good substrate of DAO D-methionine by expressing a broad spectrum amino acid racemase in Vibrio (bsrV) (Lam et al., 2009). On the other hand, commensal bacteria Lactobacilli are resistant to H2O2 (Serata et al., 2012) and not killed by DAO. Therefore, bactericidal activity of DAO may be defined by bacterial production/release of DAO substrates and resistance to H2O2 stress. The second is associated with nutritional niche for certain bacteria that are dependent on host nutrients and receive benefit from D-amino acids for their growth, which modifies the composition of the commensal microbiota. For example, loss of DAO results in increase of Lactobacillus johnsonii (Sasabe et al., 2016), which completely lacks genes encoding biosynthetic pathways for amino acids (Pridmore et al., 2004) and obtains growth support by D-alanine (van der Kaaij et al., 2004). Furthermore, of note, loss of such mucosal homeostasis by DAO increases the level of secretory IgA in the feces (Sasabe et al., 2016). Thus, these findings identify previously unrecognized physiological role of metabolizing D-amino acid by intestinal DAO in mucosal immunity.
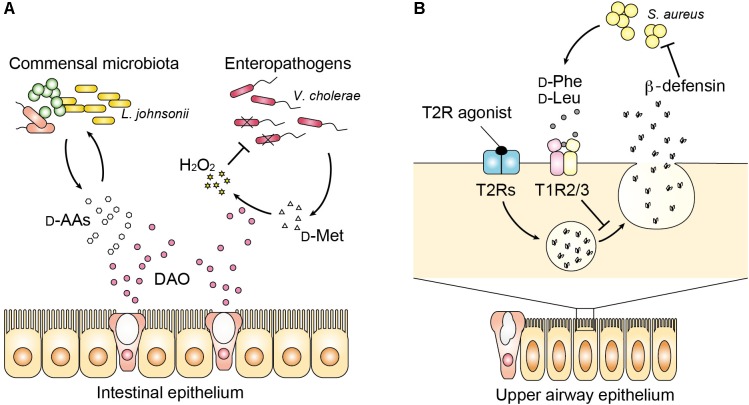
FIGURE 2.
Host–microbe communication with D-amino acids in the mucosa. (A) DAO influences luminal bacteria in the intestinal mucosa with bimodal functions. DAO modifies the composition of commensal microbiota partly by modulating availability of D-amino acids for bacterial growth. On the other hand, DAO limits colonization of enteropathogens such as Vibrio cholerae by generation of H2O2 through oxidation of bacterial D-amino acids. (B) In the upper airway, bacterial D-phenylalanine and D-leucine bind to the sweet taste receptor (T1R2/3). Release of antimicrobial peptides including beta-defensin by activation of the bitter taste receptor (T2Rs) is suppressed by signaling from the sweet taste receptor.
Modification of Mucosal Immunity by Bacterial D-Amino Acids Beyond DAO
In the upper respiratory airway, it has become clear that some D-amino acids modify innate immunity although involvement of DAO has not been studied yet. Human saliva contains substantial amount of free D-amino acids such as D-alanine (∼30% of total alanine), D-proline (∼20% of total proline), and D-aspartate (∼10% of total aspartate) (Battistone and Burnett, 1961; Syrjanen et al., 1990; Nagata et al., 2006). The whole chiral profile of salivary amino acid remains uncertain, but more kinds of D-amino acids may exist considering their diverse origins including food and oral commensal microbiota. Whereas most L-amino acids are known to taste bitter, D-amino acids taste usually sweet presumably because of stereoselectivity of sweet taste receptors (T1R2/3), which preferentially bind the D-amino acids including D-tryptophan, D-phenylalanine, D-leucine, and D-histidine (Bassoli et al., 2014). Interestingly, sweet and bitter taste receptors present in the upper airway are known to influence antimicrobial innate immune responses. Activation of bitter taste receptors (T2Rs) stimulates surrounding epithelial cells to release antimicrobial peptides (Lee et al., 2014), but the sweet taste receptor (T1R) inhibits this response (Margolskee, 2002). Two D-amino acids (D-leucine and D-phenylalanine), found in respiratory isolates of Staphylococcus species inhibit the release of antimicrobial peptides by activating T1R2/3, and increase cell death of human sinonasal epithelial culture in response to infection with methicillin-resistant S. aureus (Lee et al., 2017) (Figure 2B). Thus, D-amino acids produced by nasal microbiota can inhibit innate immune response through sweet taste receptors and may shape the microbial community of the upper airways. Kepert et al. (2017) have shown further evidence to support the role of bacterial D-amino acids in the mucosal immunity both in the lower airway and intestine. They identified D-tryptophan, screened from supernatants of probiotic bacteria, to reduce secretion of chemokine ligand 17 (CCL17) in a human Hodgkin lymphoma T-cell line and to induce IL-10 and decrease LPS-induced IFN-gamma, IL-12, and IL-5 in human monocyte-derived dendritic cells. Oral supplementation of D-tryptophan in mice alters diversity of gut microbiota, increases numbers of regulatory T cells in the lung and colon, decreases lung Th2 responses, and ameliorates allergic airway inflammation and hyperresponsiveness (Kepert et al., 2017). Although further mechanisms underlying connection between innate and acquired immunity modified by bacterial D-amino acids remain largely unknown, recognition of bacterial D-amino acids by the mammalian enzyme or receptors may play a significant role in the mucosal immunity and homeostasis.
Conclusion
In mammals, intrinsic D-serine and D-aspartate have received great attention for their neuromodulatory roles in the central nervous system. In this mini-review, we have shed light on previously less-focused D-amino acids originated from commensal or pathogenic bacteria. As bacteria produce and release a largely distinct set of D-amino acids from mammals, accumulating evidence show that bacterial D-amino acids serve as inter-kingdom signals linked to innate defense in mammals. At the host–microbe interfaces, host reacts to bacteria through enantioselective recognition of D-amino acids by DAO or sweet taste receptors, and provides direct toxic response or indirect actions through modulating antimicrobial peptides. Furthermore, such recognition of bacterial D-amino acids by host may further mediate signals to modulate adaptive immunity.
Author Contributions
JS and MS contributed to the planning and writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank M. Yasui for indispensable support.
Footnotes
Funding. JS was funded by JSPS KAKENHI Grant Number 16K09327, Moritani Scholarship Foundation, and Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research. MS was supported by JSPS KAKENHI Grant Number 18K07181 and Grant-in-Aid for JSPS Research Fellow (Grant Number 17J10213).
References
- Anfora A. T., Haugen B. J., Roesch P., Redford P., Welch R. A. (2007). Roles of serine accumulation and catabolism in the colonization of the murine urinary tract by Escherichia coli CFT073. 75 5298–5304. 10.1128/IAI.00652-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfora A. T., Welch R. A. (2006). DsdX is the second D-serine transporter in uropathogenic Escherichia coli clinical isolate CFT073. 188 6622–6628. 10.1128/JB.00634-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu D. T., Li Y., Puhl M. D., Benneyworth M. A., Basu A. C., Takagi S., et al. (2013). Multiple risk pathways for schizophrenia converge in serine racemase knockout mice, a mouse model of NMDA receptor hypofunction. 110 E2400–E2409. 10.1073/pnas.1304308110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassoli A., Borgonovo G., Caremoli F., Mancuso G. (2014). The taste of D- and L-amino acids: In vitro binding assays with cloned human bitter (TAS2Rs) and sweet (TAS1R2/TAS1R3) receptors. 150 27–33. 10.1016/j.foodchem.2013.10.106 [DOI] [PubMed] [Google Scholar]
- Basu A. C., Tsai G. E., Ma C. L., Ehmsen J. T., Mustafa A. K., Han L., et al. (2009). Targeted disruption of serine racemase affects glutamatergic neurotransmission and behavior. 14 719–727. 10.1038/mp.2008.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistone G. C., Burnett G. W. (1961). The free amino acid composition of human saliva. 3 161–170. 10.1016/0003-9969(61)90133-9 [DOI] [PubMed] [Google Scholar]
- Brachet P., Puigserver A. (1992). Regional differences for the D-amino acid oxidase-catalysed oxidation of D-methionine in chicken small intestine. 101 509–511. 10.1016/0305-0491(92)90329-P [DOI] [PubMed] [Google Scholar]
- Cava F., Lam H., de Pedro M. A., Waldor M. K. (2011). Emerging knowledge of regulatory roles of D-amino acids in bacteria. 68 817–831. 10.1007/s00018-010-0571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. J., Lehrer R. I. (1969). D-amino acid oxidase in leukocytes: a possible D-amino-acid-linked antimicrobial system. 62 756–763. 10.1073/pnas.62.3.756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J. P., Gabrielsen M., Goldstone R. J., Grinter R., Wang D., Cogdell R. J., et al. (2016). A highly conserved bacterial D-serine uptake system links host metabolism and virulence. 12:e1005359. 10.1371/journal.ppat.1005359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J. P., Goldstone R. J., Burgess K., Cogdell R. J., Beatson S. A., Vollmer W., et al. (2015). The host metabolite D-serine contributes to bacterial niche specificity through gene selection. 9 1039–1051. 10.1038/ismej.2014.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosloy S. D., McFall E. (1973). Metabolism of D-serine in Escherichia coli K-12: mechanism of growth inhibition. 114 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aniello A., Giuditta A. (1980). Presence of D-alanine in crustacean muscle and hepatopancreas. 66 319–322. 10.1016/0305-0491(80)90071-1 [DOI] [Google Scholar]
- De Jonge B. L., Gage D., Xu N. (2002). The carboxyl terminus of peptidoglycan stem peptides is a determinant for methicillin resistance in Staphylococcus aureus. 46 3151–3155. 10.1128/AAC.46.10.3151-3155.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChatelet L. R., McCall C. E., Cooper M. R. (1972). Amino acid oxidase in leukocytes: evidence against a major role in phagocytosis. 5 632–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein M. R., Baehner R. L., Nathan D. G. (1971). Amino acid oxidase of leukocytes in relation to H2O2 -mediated bacterial killing. 50 1985–1991. 10.1172/JCI106690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N., Saito T. (2004). Homochirality and life. 4 267–278. 10.1002/tcr.20020 [DOI] [PubMed] [Google Scholar]
- Hashimoto A., Nishikawa T., Hayashi T., Fujii N., Harada K., Oka T., et al. (1992). The presence of free D-serine in rat brain. 296 33–36. 10.1016/0014-5793(92)80397-Y [DOI] [PubMed] [Google Scholar]
- Irukayama-Tomobe Y., Tanaka H., Yokomizo T., Hashidate-Yoshida T., Yanagisawa M., Sakurai T. (2009). Aromatic D-amino acids act as chemoattractant factors for human leukocytes through a G protein-coupled receptor, GPR109B. 106 3930–3934. 10.1073/pnas.0811844106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepert I., Fonseca J., Muller C., Milger K., Hochwind K., Kostric M., et al. (2017). D-tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. 139 1525–1535. 10.1016/j.jaci.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Koga R., Miyoshi Y., Sakaue H., Hamase K., Konno R. (2017). Mouse D-amino-acid oxidase: distribution and physiological substrates. 4:82. 10.3389/fmolb.2017.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno R., Niwa A., Yasumura Y. (1990). Intestinal bacterial origin of D-alanine in urine of mutant mice lacking D-amino-acid oxidase. 268 263–265. 10.1042/bj2680263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno R., Oowada T., Ozaki A., Iida T., Niwa A., Yasumura Y., et al. (1993). Origin of D-alanine present in urine of mutant mice lacking D-amino-acid oxidase activity. 265(4 Pt 1) G699–G703. 10.1152/ajpgi.1993.265.4.G699 [DOI] [PubMed] [Google Scholar]
- Korte-Berwanger M., Sakinc T., Kline K., Nielsen H. V., Hultgren S., Gatermann S. G. (2013). Significance of the D-serine-deaminase and D-serine metabolism of Staphylococcus saprophyticus for virulence. 81 4525–4533. 10.1128/IAI.00599-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs H. A. (1935). Metabolism of amino-acids: deamination of amino-acids. 29 1620–1644. 10.1042/bj0291620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H., Oh D. C., Cava F., Takacs C. N., Clardy J., de Pedro M. A., et al. (2009). D-amino acids govern stationary phase cell wall remodeling in bacteria. 325 1552–1555. 10.1126/science.1178123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. J., Hariri B. M., McMahon D. B., Chen B., Doghramji L., Adappa N. D., et al. (2017). Bacterial d-amino acids suppress sinonasal innate immunity through sweet taste receptors in solitary chemosensory cells. 10:eaam7703. 10.1126/scisignal.aam7703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. J., Kofonow J. M., Rosen P. L., Siebert A. P., Chen B., Doghramji L., et al. (2014). Bitter and sweet taste receptors regulate human upper respiratory innate immunity. 124 1393–1405. 10.1172/JCI72094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loetscher Y., Wieser A., Lengefeld J., Kaiser P., Schubert S., Heikenwalder M., et al. (2012). Salmonella transiently reside in luminal neutrophils in the inflamed gut. 7:e34812. 10.1371/journal.pone.0034812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolskee R. F. (2002). Molecular mechanisms of bitter and sweet taste transduction. 277 1–4. 10.1074/jbc.R100054200 [DOI] [PubMed] [Google Scholar]
- Matsushima O., Katayama H., Yamada K., Kado Y. (1984). Occurrence of free D-alanine and alanine racemase activity in bivalve molluscs with special reference to intracellular osmoregulation. 5 217–225. [Google Scholar]
- Mitchell J., Paul P., Chen H. J., Morris A., Payling M., Falchi M., et al. (2010). Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. 107 7556–7561. 10.1073/pnas.0914128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya K., Inoue R., Takata Y., Abe M., Natsume R., Sakimura K., et al. (2008). Serine racemase is predominantly localized in neurons in mouse brain. 510 641–654. 10.1002/cne.21822 [DOI] [PubMed] [Google Scholar]
- Miyoshi Y., Hamase K., Okamura T., Konno R., Kasai N., Tojo Y., et al. (2011). Simultaneous two-dimensional HPLC determination of free D-serine and D-alanine in the brain and periphery of mutant rats lacking D-amino-acid oxidase. 879 3184–3189. 10.1016/j.jchromb.2010.08.024 [DOI] [PubMed] [Google Scholar]
- Miyoshi Y., Hamase K., Tojo Y., Mita M., Konno R., Zaitsu K. (2009). Determination of D-serine and D-alanine in the tissues and physiological fluids of mice with various D-amino-acid oxidase activities using two-dimensional high-performance liquid chromatography with fluorescence detection. 877 2506–2512. 10.1016/j.jchromb.2009.06.028 [DOI] [PubMed] [Google Scholar]
- Mothet J. P., Parent A. T., Wolosker H., Brady R.O, Jr, Linden D. J., Ferris C. D., et al. (2000). D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. 97 4926–4931. 10.1073/pnas.97.9.4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa A. K., Ahmad A. S., Zeynalov E., Gazi S. K., Sikka G., Ehmsen J. T., et al. (2010). Serine racemase deletion protects against cerebral ischemia and excitotoxicity. 30 1413–1416. 10.1523/JNEUROSCI.4297-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y., Fujiwara T., Kawaguchi-Nagata K., Fukumori Y., Yamanaka T. (1998). Occurrence of peptidyl D-amino acids in soluble fractions of several eubacteria, archaea and eukaryotes. 1379 76–82. 10.1016/S0304-4165(97)00084-6 [DOI] [PubMed] [Google Scholar]
- Nagata Y., Higashi M., Ishii Y., Sano H., Tanigawa M., Nagata K., et al. (2006). The presence of high concentrations of free D-amino acids in human saliva. 78 1677–1681. 10.1016/j.lfs.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Nakamura H., Fang J., Maeda H. (2012). Protective role of D-amino acid oxidase against Staphylococcus aureus infection. 80 1546–1553. 10.1128/IAI.06214-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen G. T., Green E. R., Mecsas J. (2017). Neutrophils to the ROScue: mechanisms of NADPH oxidase activation and bacterial resistance. 7:373. 10.3389/fcimb.2017.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollegioni L., Piubelli L., Sacchi S., Pilone M. S., Molla G. (2007). Physiological functions of D-amino acid oxidases: from yeast to humans. 64 1373–1394. 10.1007/s00018-007-6558-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridmore R. D., Berger B., Desiere F., Vilanova D., Barretto C., Pittet A. C., et al. (2004). The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. 101 2512–2517. 10.1073/pnas.0307327101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. E., Courvalin P. (2005). Vancomycin resistance in enterococci due to synthesis of precursors terminating in D-alanyl-D-serine. 49 21–25. 10.1128/AAC.49.1.21-25.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. M., Briggs R. T., Karnovsky M. J. (1978). Localization of D-amino acid oxidase on the cell surface of human polymorphonuclear leukocytes. 77 59–71. 10.1083/jcb.77.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch P. L., Redford P., Batchelet S., Moritz R. L., Pellett S., Haugen B. J., et al. (2003). Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. 49 55–67. 10.1046/j.1365-2958.2003.03543.x [DOI] [PubMed] [Google Scholar]
- Roos V., Klemm P. (2006). Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. 74 3565–3575. 10.1128/IAI.01959-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarower M. G., Matsui T., Abe H. (2003a). Distribution and characteristics of D-amino acid and D-aspartate oxidases in fish tissues. 295 151–159. 10.1002/jez.a.10217 [DOI] [PubMed] [Google Scholar]
- Sarower M. G., Okada S., Abe H. (2003b). Molecular characterization of D-amino acid oxidase from common carp Cyprinus carpio and its induction with exogenous free D-alanine. 420 121–129. [DOI] [PubMed] [Google Scholar]
- Sasabe J., Chiba T., Yamada M., Okamoto K., Nishimoto I., Matsuoka M., et al. (2007). D-serine is a key determinant of glutamate toxicity in amyotrophic lateral sclerosis. 26 4149–4159. 10.1038/sj.emboj.7601840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe J., Miyoshi Y., Rakoff-Nahoum S., Zhang T., Mita M., Davis B. M., et al. (2016). Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. 1:16125. 10.1038/nmicrobiol.2016.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serata M., Iino T., Yasuda E., Sako T. (2012). Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. 158(Pt 4) 953–962. 10.1099/mic.0.053942-0 [DOI] [PubMed] [Google Scholar]
- Shleper M., Kartvelishvily E., Wolosker H. (2005). D-serine is the dominant endogenous coagonist for NMDA receptor neurotoxicity in organotypic hippocampal slices. 25 9413–9417. 10.1523/JNEUROSCI.3190-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieradzki K., Tomasz A. (1996). A highly vancomycin-resistant laboratory mutant of Staphylococcus aureus. 142 161–166. 10.1111/j.1574-6968.1996.tb08424.x [DOI] [PubMed] [Google Scholar]
- Suzuki M., Imanishi N., Mita M., Hamase K., Aiso S., Sasabe J. (2017). Heterogeneity of D-serine distribution in the human central nervous system. 9:1759091417713905. 10.1177/1759091417713905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrjanen S. M., Alakuijala L., Alakuijala P., Markkanen S. O., Markkanen H. (1990). Free amino acid levels in oral fluids of normal subjects and patients with periodontal disease. 35 189–193. 10.1016/0003-9969(90)90054-E [DOI] [PubMed] [Google Scholar]
- Tuinema B. R., Reid-Yu S. A., Coombes B. K. (2014). Salmonella evades D-amino acid oxidase to promote infection in neutrophils. 5:e01886-14. 10.1128/mBio.01886-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kaaij H., Desiere F., Mollet B., Germond J. E. (2004). L-alanine auxotrophy of Lactobacillus johnsonii as demonstrated by physiological, genomic, and gene complementation approaches. 70 1869–1873. 10.1128/AEM.70.3.1869-1873.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T. (1989). Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. 264 2393–2396. [PubMed] [Google Scholar]
- Watanabe A., Yoshimura T., Mikami B., Hayashi H., Kagamiyama H., Esaki N. (2002). Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5’-phosphopyridoxyl)alanine. 277 19166–19172. 10.1074/jbc.M201615200 [DOI] [PubMed] [Google Scholar]
- Winkelstein J. A., Marino M. C., Johnston R.B, Jr, Boyle J., Curnutte J., Gallin J. I., et al. (2000). Chronic granulomatous disease. Report on a national registry of 368 patients. 79 155–169. 10.1097/00005792-200005000-00003 [DOI] [PubMed] [Google Scholar]
- Wolosker H., Sheth K. N., Takahashi M., Mothet J. P., Brady R.O, Jr, Ferris C. D., et al. (1999). Purification of serine racemase: biosynthesis of the neuromodulator D-serine. 96 721–725. 10.1073/pnas.96.2.721 [DOI] [PMC free article] [PubMed] [Google Scholar]