Abstract
The (pp)pGpp metabolism is an important component of bacterial physiology as it is involved in various stress responses and mechanisms of cell homeostasis, e.g., the regulation of growth. However, in order to better understand the (pp)pGpp associated regulation, it is crucial to study the molecular mechanisms of (pp)pGpp metabolism. In recent years, bioinformatic analyses of the RelA/SpoT homolog (RSH) superfamily have led to the discovery of small monofunctional RSH derivatives in addition to the well-known bifunctional Rel proteins. These are also referred to as small alarmone synthetases (SASs) or small alarmone hydrolases (SAHs). In this study, the ORF cg1485 from C. glutamicum was identified as a putative SAH encoding gene, based on a high similarity of the corresponding amino acid sequence with the (pp)pGpp hydrolysis domain. The characterization of its gene product, designated as RelHCg, represents the first functional investigation of a bacterial representative of the SAH subfamily. The predicted pyrophosphohydrolase activity was demonstrated in vivo by expression in two E. coli strains, characterized by different alarmone basal levels, as well as by in vitro analysis of the purified protein. During the assay-based analysis of hydrolysis activity in relation to the three known alarmone species, both RelHCg and the bifunctional RSH enzyme RelCg were found to exhibit a pronounced substrate inhibition for alarmone concentrations of more than 0.75 mM. This characteristic of (pp)pGpp hydrolases could be an important mechanism for realizing the bistable character of the (pp)pGpp metabolism between a (pp)pGpp basal level and stress-associated alarmone production. The deletion of relHCg caused only a minor effect on growth behavior in both wild-type background and deletion mutants with deletion of (pp)pGpp synthetases. Based on this observation, the protein is probably only present or active under specific environmental conditions. The independent loss of the corresponding gene in numerous representatives of the genus Corynebacterium, which was found by bioinformatic analyses, also supports this hypothesis. Furthermore, growth analysis of all possible deletion combinations of the three active C. glutamicum RSH genes revealed interesting functional relationships which will have to be investigated in more detail in the future.
Keywords: stringent response, alarmone, (p)ppGpp, (pp)pGpp, Mesh1-L, RelH, SAH, pGpp
Introduction
The stringent response represents an important bacterial regulatory system, as it allows the organisms to respond quickly to different stress conditions. In response to unfavorable environmental conditions such as nutrient deficiency situations, the hyperphosphorylated guanosine derivatives GMP 3′diphosphate (pGpp) (Gaca et al., 2015b), guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) are produced (Cashel and Kalbacher, 1970; Haseltine et al., 1972). These messenger nucleotides, also referred to as alarmones or as (pp)pGpp, influence the expression or activity of numerous proteins and thereby orchestrate the inhibition of cell growth as well as the redistribution of cellular resources toward a state of persistence (Dalebroux and Swanson, 2012; Gaca et al., 2015a). New results also illustrate that the (pp)pGpp metabolism, which is highly conserved in bacteria and plant chloroplasts (van der Biezen et al., 2000; Masuda et al., 2008; Potrykus and Cashel, 2008), not only induces the coordinated response to various stress conditions, but also includes numerous regulatory functions for the maintenance of cell homeostasis (Potrykus and Cashel, 2008; Gaca et al., 2013) and growth rate regulation (Potrykus et al., 2011). Due to their functions in the context of stress-induced (pp)pGpp production, alarmone synthetases have long been in the focus of research. However, especially when considering the non-stress-associated functions of (pp)pGpp metabolism, the counteracting (pp)pGpp hydrolysis is likewise an important and so far little studied component of this system (Hauryliuk et al., 2015). The importance of (pp)pGpp hydrolases has already been demonstrated by the fact that the deletion of the only gene with this function in the model organism Escherichia coli, termed spoTEc, is lethal (Xiao et al., 1991). Due to the wide-ranging regulatory influences of the (pp)pGpp metabolism, a deeper understanding of this system is highly relevant for medical areas such as stress associated persistence of pathogenic species (Dalebroux et al., 2010), as well as for biotechnological applications (Michalowski et al., 2017).
The enzymes responsible for synthesis and hydrolysis of (pp)pGpp are referred to as RelA/SpoT homologs (RSHs) because of their protein sequence similarities to the corresponding E. coli proteins RelAEc and SpoTEc. However, in most organisms only one copy of this multi-domain enzyme occurs, which also contains other domains associated with regulatory functions. While the pyrophosphokinase activity has already been extensively studied due to its high relevance in the context of various stress responses, the pyrophosphohydrolase activity mediating HD domain of RSH proteins has so far been functionally characterized only for a few organisms (Aravind and Koonin, 1998). In addition to the ‘long’ multi-domain RSH enzymes which have significantly divergent activity spectra due to different regulatory mechanisms, small single-domain RSH proteins have recently been identified by bioinformatic approaches (Lemos et al., 2007; Nanamiya et al., 2008; Sun et al., 2010). These include monofunctional (pp)pGpp synthetases as well as (pp)pGpp hydrolases, also referred to as small alarmone synthetases (SASs) and small alarmone hydrolases (SAHs), respectively. A bioinformatics study of the RSH superfamily carried out by Atkinson et al. (2011) revealed the widespread occurrence of these two protein classes. By searching for stringent response-associated sequence motifs in 1,000 genomes and a subsequent phylogenetic analysis of the identified genes, a classification into 12 subgroups of SAS proteins and 7 subgroups of SAH enzymes has been proposed.
In addition to the bifunctional enzyme RelCg, the presence of the SAS enzyme RelSCg was recently reported for the industrially relevant production organism C. glutamicum (Ruwe et al., 2017). The deletion of the C. glutamicum relCg gene resulted in a growth deficit in minimal medium, probably caused by increased (pp)pGpp levels due to the synthetase activity of RelSCg and a lacking hydrolase activity by RelCg. Contrary to expectations, however, after a stationary phase lasting several hours in the low OD range, the strain began to grow again with almost normal parameters. Based on these observations the existence of a further (pp)pGpp hydrolase was assumed. A candidate protein has already been identified by bioinformatic analysis of putative C. glutamicum SAS enzymes (Ruwe et al., 2017), as well as by Atkinson et al. (2011) in the context of a global RSH classification based on amino acid sequence similarities. However, regarding this class of enzymes nearly no information is available. The only members of the SAH proteins investigated so far are eukaryotic representatives of Drosophila melanogaster and Homo sapiens (Sun et al., 2010). However, as no (pp)pGpp synthetases are known for both organisms, the functional context of these proteins is completely unknown and may deviate from those of the prokaryotic orthologs.
In addition, the production of pGpp was found for both (pp)pGpp synthetases in C. glutamicum in the course of our previous investigations. The assembly and disassembly of this substance, which has only recently been assigned to the alarmone species, could be an important link within the stringent response of C. glutamicum and related organisms. Since different biological functionalities have been identified for the three known alarmone species in recent years (Gaca et al., 2015b), differences in the activity of the hydrolytically active enzymes with regard to these substrates could represent an important and so far unexplored component of the (pp)pGpp metabolism.
The aim of the work described here is the analysis of a possible expansion of the C. glutamicum (pp)pGpp metabolism by enzymes with (pp)pGpp hydrolase activity and a further elucidation of the functional relationship between the different RSH species. By heterologous expression of a C. glutamicum SAH gene in E. coli and the development of a LC-MS based, in vitro (pp)pGpp hydrolase characterization method, we were able to confirm the predicted functionality and for the first time provide enzyme kinetics data for a bacterial monofunctional small alarmone hydrolase. Both, for the pyrophosphohydrolase activities of RelCg and the newly identified SAH enzyme, a significant substrate inhibition was found, which could be an important mechanistic component for realizing the bistable character of (pp)pGpp metabolism.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains used or constructed for this study are listed in Table 1, and plasmids are listed in Table 2.
Table 1.
Bacterial strains used in this study.
| Strain | Characteristics or genotype | Source |
|---|---|---|
| C. glutamicum | ||
| CR099 | C. glutamicum ATCC 13032, ΔCGP1, ΔCGP2, ΔCGP3, ΔISCg1, ΔISCg2 | Baumgart et al., 2013; Unthan et al., 2015 |
| CR099 Δrel | C. glutamicum CR099 Δrel | Ruwe et al., 2017 |
| CR099 ΔrelS | C. glutamicum CR099 ΔrelS | Ruwe et al., 2017 |
| CR099 ΔrelH | C. glutamicum CR099 ΔrelH | This study |
| CR099 ΔrelΔrelS | C. glutamicum CR099 ΔrelΔrelS | Ruwe et al., 2017 |
| CR099 ΔrelΔrelH | C. glutamicum CR099 ΔrelΔrelH | This study |
| CR099 ΔrelSΔrelH | C. glutamicum CR099 ΔrelSΔrelH | This study |
| CR099 ΔrelΔrelSΔrelH | C. glutamicum CR099 ΔrelΔrelSΔrelH | This study |
| E. coli | ||
| ER2566 | E. coli expression strain with a chromosomal copy of the T7 RNA polymerase gene under the control of the lac promoter | New England Biolabs |
| MG1655 | Wild-type E. coli MG1655, derived from E. coli K12 | Blattner et al., 1997 |
| MG1655 ΔrelA | MG1655 ΔrelA | Ruwe et al., 2017 |
Table 2.
Plasmids used in this study.
| Plasmid | Characteristics | Source |
|---|---|---|
| pZMP | Constitutive E. coli – C. glutamicum shuttle expression vector | Walter et al., 2016 |
| pZMP::relCg | pZMP carrying the gene rel (cg1861) from C. glutamicum CR099 | This study |
| pZMP::relSCg | pZMP carrying the gene relS (cg2324) from C. glutamicum CR099 | This study |
| pZMP::relHCg | pZMP carrying the gene relH (cg1485) from C. glutamicum CR099 | This study |
| pK18mobsacB | Suicide vector for gene deletion by homologous recombination | Schäfer et al., 1994 |
| pK18mobsacB_relHCg | pK18mobsacB carrying the up- and downstream region (500 bp, respectively) of the gene relH from C. glutamicum CR099 | This study |
| pTXB1 | E. coli expression vector, carrying the self-cleavable Mxe intein/chitin binding domain | New England Biolabs |
| pTXB1::relCg | pTXB1 carrying the gene rel (cg1861) from C. glutamicum CR099 | Ruwe et al., 2017 |
| pTXB1::relSCg | pTXB1 carrying the gene relS (cg2324) from C. glutamicum CR099 | Ruwe et al., 2017 |
| pTXB1::relHCg | pTXB1 carrying the gene relH (cg1485) from C. glutamicum CR099 | This study |
C. glutamicum and E. coli strains were cultivated as described previously (Ruwe et al., 2017). In order to avoid the accumulation of pseudorevertants in the precultures of the minimal medium cultivation, all strains were grown in CASO broth (Carl Roth) and subsequently washed in the chemically defined CGXII main culture medium (Keilhauer et al., 1993).
Construction of C. glutamicum Deletion Mutants
The strains used in this study were constructed as described previously (Ruwe et al., 2017). All primers used in this study are listed in Supplementary Table S1 with their respective sequences and purposes. Briefly, C. glutamicum deletion mutants were generated using the pK18mobsacB suicide vector system (Schäfer et al., 1994).
Constitutive Expression of RSH Genes in E. coli MG1655 and E. coli MG1655 ΔrelA
In order to investigate the in vivo activity of putative (pp)pGpp pyrophosphohydrolases from C. glutamicum, a constitutive expression of the corresponding genes in two E. coli test strains with different (pp)pGpp basal levels was aimed at. For this purpose, the genes cg1861 (relCg), cg1485 (relHCg) and the SAS gene cg2324 (relSCg) were cloned into the vector pZMP by means of Gibson Isothermal Assembly under control of the tac promoter (Gibson et al., 2009). The resulting plasmids pZMP::relCg, pZMP::relHCg and pZMP::relSCg, as well as the empty plasmid were subsequently transformed into the strains E. coli MG1655 and E. coli MG1655 ΔrelA.
All strains were cultivated in LB medium up to an OD600 of 3, washed in PBS buffer (pH 7.4) and adjusted to an OD600 of 0.1. Subsequently, 5 μL aliquots of different dilution stages were spotted onto LB-medium plates, MOPS minimal medium plates (MM) and MOPS minimal medium plates containing each 1 mM L-serine, L-methionine, and L-glycine (MM+SMG) (Kasai, 2002). All solid media plates contained 50 μg mL-1 kanamycin and 4 g L-1 glycerol was used as C source for all minimal medium plates. Incubation was carried out at 37°C for 24 h in the case of LB solid medium and 48 h for MM and MM+SMG plates.
Protein Purification
For the tag-free purification of the putative SAH enzyme RelHCg, the IMPACT kit (New England Biolabs) was used analogously to the purification of the already available enzymes RelCg and RelSCg, which were also used in the course of this study. The corresponding gene relHCg was introduced into the vector pTXB1 and the plasmid obtained was transformed into the expression strain E. coli ER2566. The further procedure was consistent with the purification of RelCg and RelSCg already described in the previous study (Ruwe et al., 2017).
Preparation of pGpp, ppGpp, and pppGpp
The three alarmone species pGpp, ppGpp, and pppGpp were produced by RelSCg-catalyzed pyrophosphorylation of GMP, GDP or GTP and purified by chromatography. The procedure was based on a protocol for the production and purification of ppGpp and pppGpp by Mechold et al. (2013). However, it was adapted to the usage of RelSCg, optimized in terms of optimum purity, and modified for the purification of pGpp. The reaction mixtures comprising 2.5 mL contained 50 mM Tris-HCl (pH 8), 10 mM MgCl2, 12 mM ATP, 10 mM of the respective guanosine derivative and 1.5 μM purified RelSCg. After an incubation period of 16 h, phenol chloroform extraction was performed, using an equivalent volume of phenol chloroform isoamyl alcohol mixture (25:24:1). Nucleotides were precipitated by adjusting the LiCl concentration to 1 M and adding four volumes of ice-cold absolute ethanol. Following an incubation period of 30 min on ice, the suspension was centrifuged at 4°C and 7,200 × g for 10 min. The obtained pellet was washed two times with absolute ethanol, dried on ice and stored at -20°C.
For purification, the precipitated guanosine derivatives were resuspended in binding buffer [0.5 mM EDTA, 25 mM TRIS-HCl (pH 7.5)], containing 50 mM LiCl and loaded onto a 5 mL Bio-Scale Mini Macro-Prep High Q Cartridge (Bio-Rad), using an ÄKTAprime plus system. Following a washing step at a LiCl concentration of 150 mM, the elution of ppGpp and pppGpp was carried out with a linear gradient of 150–500 mM LiCl in binding buffer. In the case of pGpp, the LiCl concentration was increased in two sections with different gradient slopes from 150 mM to 350 mM and 350 to 500 mM to achieve optimal separation. The eluate was collected in 2 mL fractions. In order to allocate the obtained UV signals to the nucleotide components contained, representative fractions were investigated by means of HPLC-MS measurements using a pppGpp pyrophosphokinase assay HPLC-MS method as described previously (Ruwe et al., 2017). The fractions corresponding to the desired guanosine derivatives were combined, diluted with LiCl-free binding buffer in a ratio of 1:6 and loaded again onto the column mentioned above. This was followed by a batch elution with binding buffer containing 1 M LiCl and a precipitation as described above. The concentration of all alarmone species dissolved in water was determined by UV absorption measurement.
(pp)pGpp Pyrophosphohydrolase Assay
In order to characterize (pp)pGpp hydrolysis activities of RelCg and RelHCg, the enzymes were incubated with different alarmone species under variation of relevant process parameters. The enzyme reactions, each containing 50 μL, were stopped by a strong reduction of the pH. For this purpose, 4 μL 50% acetic acid was added, which corresponds to an optimal ratio between preferably complete inactivation and the smallest possible decay of the substrates or products. HPLC analysis of the hydrolase assays was based on the previously described method for the detection of (pp)Gpp in pyrophosphokinase assays (Ruwe et al., 2017). The concentration of the isocratic elution profile was adapted to the analyzed nucleotide species. For the evaluation of pppGpp hydrolysis 38% of 10 mM ammonium bicarbonate buffer (pH 9.3) was used. In contrast, 36% and 34% of this buffer were applied to analyze assay mixtures containing ppGpp or pGpp, respectively. The quantitative evaluation of the products GMP, GDP, or GTP was performed using the UV signal and suitable nucleotide standards, which were also incubated under identical conditions and treated with acetic acid. The reaction mixtures for the characterization of the pyrophosphohydrolase reaction with regard to divalent ions contained 50 mM HEPES-Na (pH 8.0), 200 mM NaCl, 0.5 mM ppGpp as well as 52.3 nM RelHCg or 98.2 nM RelCg, respectively. In order to avoid oxidation of Mn2+, the Mn2+ containing buffer solutions were saturated with nitrogen before the addition of substrates and enzymes to remove dissolved oxygen. Since comparatively low activities were measured using different MgCl2 values, 10 times higher enzyme concentrations were used. The incubation time was 25 min for RelHCg and 45 min for RelCg containing approaches. The pH dependence was basically determined with identical reaction components and test parameters, although different buffer systems and a defined MnCl2 concentration of 1 mM were used. Citrate, phosphate, HEPES-Na, Tris-HCL and ammonia buffer systems with a concentration of 50 mM each were applied to cover a broad pH spectrum. For the highest and lowest pH values, enzyme-free controls were carried out to verify the stability of the substrate ppGpp over the entire pH range.
For the kinetic analysis of the pyrophosphohydrolase activities of RelCg or RelHCg, assays with different concentrations of the three alarmone species were performed. Since maximum ppGpp concentrations of 4 mM were determined for E. coli, the substrate concentrations were varied from 0.1 to 4 mM. The reaction mixtures comprised 50 mM HEPES-Na (pH 8.0), 200 mM NaCl, 1 mM MnCl2 and 52.3 nM RelHCg or 98.2 nM RelCg, respectively. The conditions used were based on the characterization of ppGpp hydrolysis with respect to different reaction parameters and represent a compromise between high activity of both enzymes and physiological conditions. In order to ensure optimal evaluability, the incubation times of the different enzyme-substrate combinations were adapted to the values determined in preliminary tests and ranged from 20 to 45 min. The incubation temperature was 30°C.
Data evaluation was carried out using the program OriginPro 2018 (OriginLab). During the analysis of the enzyme kinetics with respect to different substrate concentrations, a pronounced substrate inhibition was observed. Since no converging fit of the experimental data could be realized using classical functions to describe the enzyme kinetics in the case of substrate inhibition, the kinetic parameters Km, kcat and the corresponding inhibition constants could not be determined conclusively. For this reason, the kinetic parameters Km and kcat were determined by a linearized representation according to Eadie–Hofstee using the lowest evaluable substrate concentrations (Hofstee, 1959). Only two data points could be used for two data sets in this context, so that no error analysis was possible and the determined parameters merely represent an approximation.
Bioinformatic Analyses
To assess the distribution of RelHCg within the genus Corynebacterium, an HMM of RelHCg orthologs was constructed using the program JACKHMMER (Finn et al., 2015). The amino acid sequence of Cg1485 was used as an input against the ”Reference proteomes” database with an initial significance e-value of 1e-25. After four iterations, a large increase of candidates was observed, so the third iteration was inspected. Based on the observed minimum, an e-value of 1e-37 was chosen which led to convergence after five iterations with a total of 446 sequences giving hits above the cutoff. The HMM resulting from these sequences was then used as input for HMMSEARCH to search for RelHCg in the available proteomes of Corynebacterium species strains. Hits with an e-value below 1e-37 were considered significant. The data were mapped on a phylogenetic tree of the Corynebacterium type strains, created on the basis of the respective 16S rDNA sequences according to the RDP database (Cole et al., 2014).
Results
The Genome of C. glutamicum Encodes a Putative Small Alarmone Hydrolase (SAH) Containing All Conserved (pp)pGpp Hydrolase Sequence Motifs
To identify possible monofunctional (pp)pGpp hydrolases for C. glutamicum, a BLASTP analysis of the already known bifunctional enzyme RelCg was performed with a C. glutamicum ATCC 13032 reference genome (Kalinowski et al., 2003). In the course of this investigation, a high degree of similarity was found between the RelCg hydrolase domain and the amino acid sequence of the gene product of cg1485. This result is consistent with a global ‘high throughput sensitive sequence searching’ of the RSH superfamily, where the corresponding ORF was also identified as a putative SAH (Atkinson et al., 2011). The analysis of a pooled C. glutamicum RNAseq data set, based on cultures from different cultivation- and stress conditions, revealed the full-length transcription of the ORF cg1485 (Pfeifer-Sancar et al., 2013). Based on a data set of native 5′-ends of transcripts, also created in the context of this study, a clear -10 motif was identified, which indicates a σ70 associated transcription. Hence, an expression of the SAH gene cg1485 can be assumed. However, due to the pooling of samples from different conditions for the RNAseq experiments, the exact expression conditions are not known.
Based on a phylogenetic analysis of all SAH candidates found, Atkinson et al. (2011) proposed a differentiation of seven SAH subgroups and classified cg1485 as a member of the Mesh1-L subgroup. However, since no suitable gene name has been assigned for representatives of this group, we here propose the term relH and subsequently designate the gene cg1485 as relHCg. The corresponding enzyme RelHCg is attributed to the metal-dependent phosphohydrolases due to its high level of similarity to the HD_4 domain (pfam13328) (Aravind and Koonin, 1998). In addition, the amino acid sequence of the relHCg gene product is consistent with all 6 conserved catalytic amino acid sequence motifs identified for the (pp)pGpp hydrolase domain (Figure 1) (Hogg et al., 2004; Sun et al., 2010; Steinchen and Bange, 2016). This also applies to the hydrolase domain of the bifunctional enzyme RelCg. An analysis of representatives of all other SAH subgroups showed a very strong conservation of these motifs over all previously identified (pp)pGpp hydrolases (Figure 1) (Atkinson et al., 2011). Only the motif HD1 could not be found in the analyzed representatives of the SAH subclasses pbcSpo, pbcSpo2, and divSpo.
FIGURE 1.
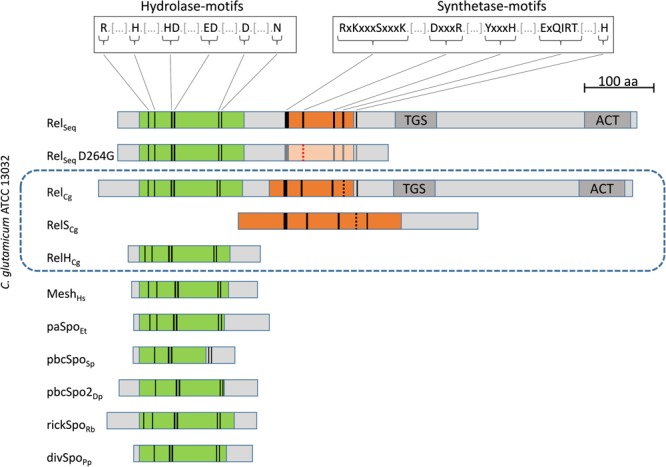
Comparative structural analysis of enzymes involved in (pp)pGpp metabolism. True-to-scale representation of domain architecture and conserved sequence motives of stringent response associated enzymes of C. glutamicum ATCC 13032 (enclosed by dashed blue lines) and further selected species. These include the native Rel enzyme of Streptococcus equisimilis RelSeq, as well as the shortened and only hydrolytically active variant RelSeq D264G (Hogg et al., 2004; Sun et al., 2010) and representatives of the six other SAH subgroups (Atkinson et al., 2011): Mesh (Homo sapiens), paSpo (Erwinia tasmaniensis), pbcSpo (S. pneumoniae), pbcSpo2 (Desulfotalea psychrophila), rickSpo (Rickettsia bellii), and divSpo (Photobacterium profundum). Hydrolase domain HD4 (pfam13328) and ppGpp synthetase catalytic domain YjbM (COG2357) are indicated in green and orange, respectively. Additional C-terminal elements of long RSH enzymes, which are likely to have regulatory functions and are classified as TGS (ThrRS, GTPase, and SpoT) and ACT (aspartate kinase, chorismate mutase and TyrA) domains (Wolf et al., 1999; Chipman and Shaanan, 2001), are also indicated. Matches with conserved synthetase and hydrolase motifs are represented by black lines (Bag et al., 2014; Steinchen and Bange, 2016). Minor deviations from the conserved sequence motifs are shown by dashed black lines. The amino acid exchange of the protein RelSeq D264G is represented by a red dashed line and the resulting non-functional YjbM domain is faded out.
The Deletion of relHCg Causes Only Minor Effects on the Growth Behavior of C. glutamicum Under Standard Growth Conditions
In order to investigate the physiological importance of RelHCg, a growth analysis of a relHCg deletion mutant was aimed at. The precise deletion was performed by homologous recombination using the vector pK18mobsacB. Since growth effects were observed for mutants with deletions in genes of the (pp)pGpp-synthesizing enzymes RelCg and RelSCg (Ruwe et al., 2017), the gene relHCg was also removed from these single deletion mutants, as well as from the double mutant strain C. glutamicum CR099 Δrel, ΔrelS. CR099 is a direct descendant of the ATCC 13032 wild-type and only the prophage regions (ΔCGP1/2/3) and all members of two families of insertion elements (ISCg1 and ISCg2) have been removed from the chromosome (Baumgart et al., 2013, 2018; Unthan et al., 2015). It is therefore considered as the wild-type (Cg) in this study. Shaking flask experiments were performed in CGXII minimal medium and the complex medium CASO broth. To avoid the generation of pseudorevertants in precultures of the growth experiment, as observed for the growth of E. coli (p)ppGpp0 strains in minimal medium (Murphy and Cashel, 2003), all precultures were grown in complex CASO broth. In the case of the growth experiment in minimal medium, the precultures were subsequently washed in CGXII main culture medium and adjusted to the desired inoculum density.
For the strain C. glutamicum CR099 ΔrelHCg no difference compared to the growth behavior of the parental strain was found in both media tested (Figure 2). While in minimal medium both strains exhibited exponential growth for the entire cultivation, the use of CASO broth resulted in a course with two characteristic growth phases. Up to an OD600 of 4–5 CR099 and CR099 ΔrelHCg grew exponentially. Subsequently, after a deceleration phase, a nearly linear growth phase occurred until the final OD600 of approximately 13 was reached. This was probably due to a change in the source of nutrients available. As expected, significant differences were observed for all strains in which the long RSH protein encoding gene relCg was deleted. In complex medium, the corresponding single deletion mutant CR099 ΔrelCg exhibited initially slightly delayed growth and also grew a little bit slower in the second growth phase from an OD of 5-6, compared to the parental strain (Figure 2A). In contrast to the unimpaired strains which attained their maximum OD600 after 12.5 h, the relCg deletion mutant reached this value only after 20 h. The additional deletion of relHCg did not affect the growth behavior compared to strain CR099 ΔrelCg. An even more pronounced growth deficit was observed for the strain with deletions of both (pp)pGpp synthetase genes, relCg and relSCg. The growth behavior of this strain was similar to that of the Δrel mutant until the second phase. However, after a much stronger deceleration phase, this strain completely stopped growing and reached only an OD600 of 8 after 35 h. Again, the additional deletion of relHCg did not result in any further alteration of growth behavior. In summary, no influence of relHCg on the growth behavior could be determined for cultivations in complex medium.
FIGURE 2.
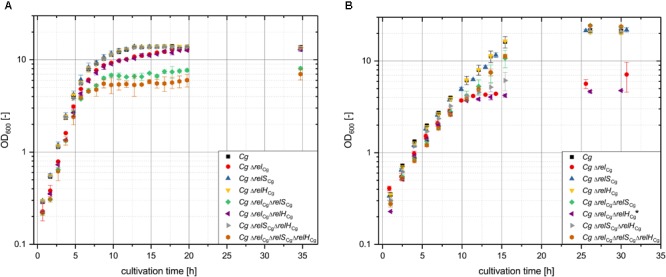
Effect of the deletion of putative (pp)pGpp metabolism associated genes on the growth of C. glutamicum CR099 in complex CASO broth (A) and minimal CGXII medium (B). To preclude possible enrichment of suppressor mutants all cultures were inoculated from precultures in complex CASO broth. An initial OD600 of 0.2 was used for the cultivation in CASO broth. For the growth analysis in CGXII minimal medium, the precultures were washed in the main culture medium and then diluted to an OD600 of 0.5. Mean values and standard deviations shown were calculated from three biological replicates. ∗In the minimal medium cultivation of the strain CR099 ΔrelSCgΔrelHCg, only two of the three replicas could be evaluated, because the third one showed a deviating growth behavior which did not correspond to the preliminary experiments and was therefore considered to be the result of a pseudoreversion.
Cultivation in minimal medium basically corresponded to the growth profiles already determined in the analysis of the C. glutamicum (pp)pGpp synthetases (Ruwe et al., 2017). Again, the additional deletion of relHCg in the growth affected strains did not lead to further effects. In contrast to the cultivation in complex medium, the strain CR099 ΔrelCgΔrelHCg almost completely stopped growing from an OD600 of 4, similar to the ΔrelCg single mutant (Ruwe et al., 2017). However, unlike the ΔrelCgΔrelHCg double mutant, the strain CR099 ΔrelCg showed an increasing OD value in the cultivation period from 26 to 30 h, following a period of several hours characterized by minimal growth. It should be noted that only two of the three CR099 ΔrelCgΔrelHCg replicates were evaluated (Figure 2B). The third replicate showed no deceleration phase and exhibited exponential growth over the entire cultivation course, analogous to the parental strain (Supplementary Figure S1). A similar behavior of aberrant replicates has been observed in previous cultivations (data not shown). Since the occurrence of suppressor mutants in RSH deletion mutants is a known phenomenon (Murphy and Cashel, 2003), the two replicates complying with the strain CR099 ΔrelCg were considered correct. According to previous investigations, strains with deletion of both (pp)pGpp-synthetase genes relCg and relSCg achieved a growth rate corresponding to the parental strain after a significant growth deficit at the beginning of cultivation. Interestingly, when cultivated in CGXII minimal medium, the double deletion mutant CR099 ΔrelSCgΔrelHCg exhibited a marked growth deficit in the form of an expanded deceleration phase, whereas both single mutants showed no impairment. However, the strain reached a final OD600 corresponding to the parental strain after 26 h.
RelHCg Exhibits a (pp)pGpp Pyrophosphohydrolase Activity in Vivo
In order to test for an in vivo activity of the putative (pp)pGpp pyrophosphohydrolase RelHCg, the relHCg gene was heterologously expressed in E. coli strains characterized by different (pp)pGpp metabolism variants. In comparison to the E. coli wild-type MG1655, strain MG1655 ΔrelA has a reduced (pp)pGpp basal level (Xiao et al., 1991). Since (pp)pGpp concentration influences the expression of amino acid synthesis genes in E. coli and thereby triggers growth effects on suitable minimal medium plates, this combination of strains is well suited to analyze the influence of heterologously expressed (pp)pGpp synthetases and hydrolases (Uzan and Danchin, 1978; Kasai, 2002). In addition to the gene relHCg, the already known components of (pp)pGpp metabolism relCg and relSCg were introduced into the constitutively expressing shuttle vector plasmid pZMP and transformed into the strains E. coli MG1655 and E. coli MG1655 ΔrelA.
In line with our expectations, no significant growth differences were observed for growth on complex LB medium for all variants of both strains (Figure 3). Using minimal medium (MOPS-MM) plates, an almost complete growth inhibition was observed for the expression of relHCg in the strain MG1655 ΔrelA [(pp)pGpp basal level reduced] compared to the corresponding empty plasmid control. In the wild type strain, however, the expression of the SAH gene only led to minimal reduction in growth. The growth inhibition of the ΔrelA strain is most likely due to a further reduction of the (pp)pGpp level. Since both strains differ only in the deletion of the monofunctional pyrophosphokinase gene relA, no difference in growth behavior would be expected for the wild-type strain if other relH associated growth-limiting reasons were responsible for the observed phenotype. Thus, the different effects of relHCg expression in the E. coli test strains, in combination with the empty plasmid controls, clearly indicate that RelHCg exhibits the expected pyrophosphohydrolase activity. Using MOPS-MM plates supplemented with L-serine, L-methionine and L-glycine (SMG-MM) (Kasai, 2002), which induces an even higher (pp)pGpp requirement and thereby inhibits growth of both RelHCg expressing E. coli strains, a significant growth reduction was also triggered by the expression of RelCg. In analogy to the RelHCg associated effects, this could be due to a weak constitutive pyrophosphohydrolase activity of RelCg in vivo. The control strain E. coli MG1655 ΔrelA could only grow under these conditions in the presence of the SAS enzyme RelSCg, whereas the constitutive expression of relCg had no complementing effect. This is in line with the (pp)pGpp synthesizing activity of RelSCg (Ruwe et al., 2017).
FIGURE 3.
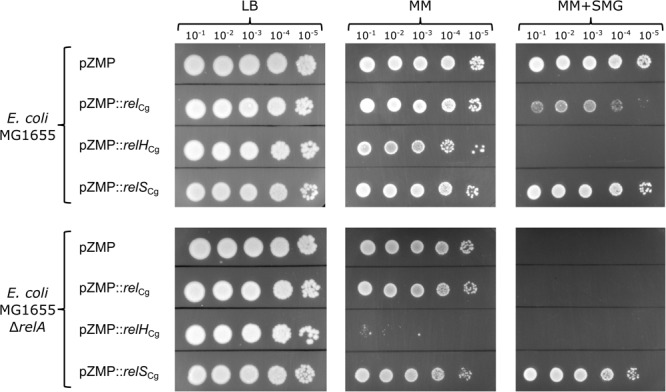
Growth characterization of E. coli MG1655 and E. coli MG1655 ΔrelA, each heterologously expressing the C. glutamicum genes relCg, relHCg or relSCg, on different solid medium plates. The empty vector pZMP was used as control. All strains were cultivated using initial OD600 values of 0.05 in LB-medium for 3 h. The cells were washed and serially diluted in PBS-buffer. 5 μL aliquots of every strain and dilution stage were spotted on LB medium plates (LB), minimal medium plates (MM) and minimal medium plates containing each 1 mM L-serine, L-methionine, and L-glycine (MM+SMG). All solid media plates contained 50 μg mL-1 kanamycin and 4 g L-1 glycerol was used as C source for all minimal medium plates. Incubation was carried out at 37°C for 24 h in the case of LB solid medium and 48 h for MM and MM+SMG plates.
RelHCg Possesses a Mn2+ Dependent Pyrophosphohydrolase Activity in Basic pH Range in Vitro
In order to analyze the hydrolysis of different alarmone species by RelHCg and the bifunctional enzyme RelCg in vitro, both enzymes were expressed heterologously in E. coli, purified by affinity chromatography and an in vitro pyrophosphohydrolase assay was established. The guanosine derivatives applied as substrate were prepared using the SAS enzyme RelSCg and purified by precipitation and chromatography steps. Since a strong manganese dependence of the pyrophosphohydrolase reaction was found for the already investigated eukaryotic SAH enzymes as well as bacterial bifunctional RSH enzymes (Aravind and Koonin, 1998; Hogg et al., 2004; Sun et al., 2010), first the dependence of the analyzed enzymes on divalent cations was investigated. In addition, the pH dependence was determined in order to find suitable parameters for further analyses.
The suspected Mn2+ dependence of the ppGpp hydrolysis was confirmed for both enzymes, as no enzyme activity could be detected without the presence of manganese ions (Figure 4). It was found that the maximum activities for the conversion of ppGpp to GDP for RelCg and RelHCg were achieved at manganese concentrations of 1 and 1.75 mM, respectively. Higher Mn2+ concentrations led to a significant reduction in pyrophosphohydrolase activity. For RelHCg, the ppGpp hydrolase activity was also detected in the presence of magnesium ions, whereby the maximum activity was achieved at a significantly higher concentration of 7.5 mM. Furthermore, the maximal specific activity of 0.059 kat mol-1 was only about 20% of the value of 0.32 kat mol-1, which was achieved at 1.75 mM Mn2+. For RelCg no ppGpp hydrolase activity could be detected in the presence of Mg2+ Ions (data not shown). The analysis of pH dependence of both (pp)pGpp hydrolases revealed quite similar characteristics. While both enzymes are inactive in the acid pH range, the specific activity increases continuously with increasing pH from a value of 7. RelHCg seems to be dependent on the buffer components used, since a remarkably low specific pyrophosphohydrolase activity deviating from the basic trend was measured for a pH value of 8.5 in TRIS buffer.
FIGURE 4.
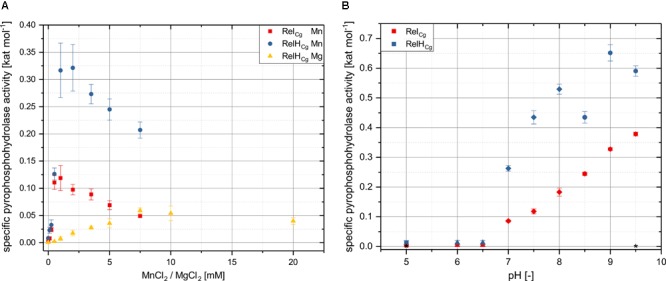
In vitro characterization of RelCg and RelHCg with respect to divalent ion concentration and pH. The specific pyrophosphohydrolase activities of the purified enzymes were determined by (pp)pGpp pyrophosphohydrolase assays and subsequent HPLC analysis (see Material and Methods) under variation of MgCl2/MnCl2 concentration (A) and pH-value (B). The reaction mixture for determining the dependence on divalent ions contained 50 mM HEPES-Na (pH 8.0), 200 mM NaCl, 0.5 mM ppGpp as well as 52.3 nM RelHCg or 98.2 nM RelCg respectively, and was incubated at 30°C. In order to avoid oxidation of Mn2+, the corresponding solutions were saturated with nitrogen before the addition of substrates and enzyme to remove dissolved oxygen. The incubation time was 25 min for RelHCg and 45 min for RelCg containing approaches. Since comparatively low activities were measured using magnesium, 10 times higher enzyme concentrations were used. The pH dependency was determined with identical reaction components and experimental parameters, whereby different buffer systems and a defined MnCl2 concentration of 1 mM were used. The various buffer systems (50 mM each) are illustrated by different icons: square: citrate buffer; triangle: phosphate buffer; rhombus: HEPES-buffer; circle: TRIS buffer; hexagon: ammonia buffer. Mean values and standard deviations shown were calculated from three replicates. ∗ Enzyme free controls were carried out at the highest and lowest pH values of 5 and 9.5 and did not yield any unspecific production of GDP or other components, so that the substrate ppGpp is considered to be stable at the pH conditions used.
The Pyrophosphohydrolase Activities of RelHCg and RelCg Show Substrate Inhibition for Various Alarmone Species
In order to analyze the physiological significance of the two alarmone hydrolases in the context of the (pp)pGpp metabolism of C. glutamicum, the enzyme kinetics for the hydrolysis of the three known alarmone species pGpp, ppGpp, and pppGpp were determined. The analysis of the respective activity and specificity parameters for the different substrates is especially interesting to explore the background of different biological functions of the alarmone species (Mechold et al., 2013). In addition, equivalent activity differences were already determined in the course of previous studies of (pp)pGpp synthetases (Steinchen et al., 2015; Ruwe et al., 2017).
In the course of studying the specific pyrophosphohydrolase activities as a function of the substrate concentration used, fundamentally similar activity levels were found for both RelCg and RelHCg. Interestingly, the turnover numbers as well as the corresponding kinetic profiles differed significantly with respect to individual alarmone species (Figure 5). Furthermore, a significant reduction in hydrolase activity at high substrate concentrations was observed for both enzymes and all substrates. Therefore, the enzyme parameters Km and kcat could only be determined by a linearized representation according to Eadie–Hofstee (Hofstee, 1959), using the lowest substrate concentrations (Supplementary Figures S2, S3). For this reason, the values given in Table 3 are approximate values and would have to be verified using further low substrate concentrations in the uninhibited range of the enzyme kinetics. RelHCg achieved the highest specific pyrophosphohydrolase activity for the conversion of ppGpp to GDP. The determined turnover number of 0.43 s-1 was significantly higher than the values of 0.19 s-1 and 0.08 s-1 which were calculated for the hydrolysis of pGpp and pppGpp, respectively (Table 3). The maximum specific RelHCg activity was achieved for all substrates already at very low concentrations of 0.25–0.75 mM. Higher concentrations lead to a drastic decrease in activity. At a substrate concentration of 1.75 mM, the specific activities reached only between 10% and 27% of the respective maximum values (Figure 5). In contrast, for the hydrolysis activity of the bifunctional enzyme RelCg, the highest values were obtained using the substrate pGpp. The maximum activity was 0.51 kat mol-1 at a pGpp concentration of 1.75 mM. ppGpp and pppGpp were hydrolyzed by RelCg with significantly lower maximum hydrolase activities of 0.17 kat mol-1 each. In contrast to the kinetics of RelCg for pGpp, however, its activity for the hydrolysis of ppGpp and pppGpp collapsed at substrate concentrations of more than 0.75 mM, analogous to the kinetics determined for RelHCg.
FIGURE 5.
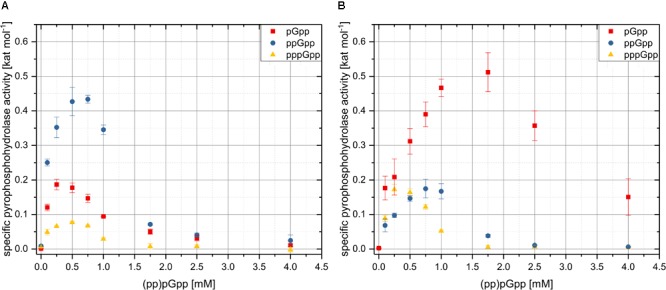
Kinetics of RelHCg (A) and RelCg (B) using pGpp, ppGpp and pppGpp as substrates. Specific activities were determined for different substrate concentrations by in vitro analysis and given in kat per mol of the respective enzyme. The reaction mixtures contained 50 mM HEPES-Na (pH 8.0), 200 mM NaCl, 1 mM MnCl2 as well as 52.3 nM RelHCg or 98.2 nM RelCg, respectively. The incubation times of the different enzyme-substrate combinations were adapted to the values determined in preliminary tests and ranged from 20 to 45 min. The incubation temperature was 30°C. Mean values and standard deviations shown were calculated from three replicates.
Table 3.
Specific enzyme parameters determined for the hydrolysis of pGpp, ppGpp, and pppGpp by RelCg and RelHCg, respectively.
| RelCg |
RelHCg |
|||||
|---|---|---|---|---|---|---|
| Substrate | Km [mM] | kcat [s-1] | kcat/Km [M-1 s-1] | Km [mM] | kcat [s-1] | kcat/Km [M-1 s-1] |
| pGpp | 0.734 ± 0.127 | 0.794 ± 0.071 | 1.08 × 103 | 0.144∗ | 0.294∗ | 2.04 × 103 |
| ppGpp | 0.511 ± 0.01 | 0.296 ± 0.004 | 5.79 × 102 | 0.101 ± 0.007 | 0.502 ± 0.018 | 4.97 × 103 |
| pppGpp | 0.422∗ | 0.563∗ | 1.34 × 103 | 0.09 ± 0.011 | 0.09 ± 0.003 | 1.01 × 103 |
As a result of significant substrate inhibition, Km and kcat were calculated by linearized representation according to Eadie–Hofstee (Hofstee, 1959), using the lowest substrate concentrations (Supplementary Figures S2, S3). ∗Due to the substrate inhibition, the parameters shown were only generated on the basis of two data points, which means that no error indication of the regression is possible.
The Distribution of the relH Homolog in the Genus Corynebacterium Indicates a Frequent Gene Loss During Evolution
The RelHCg enzyme appears to constitute an active functional part of the C. glutamicum (pp)pGpp metabolism. Since Atkinson et al. (2011) identified representatives of the Mesh-1L subgroup in numerous bacterial groups and in an archaeon during the analysis of the RSH superfamily, studying the relHCg phylogeny represents an interesting approach to generate information on a possible association with specific habitats or lifestyles. An initial BLASTP study revealed that many corynebacteria, in contrast to the highly conserved rel and relS genes, contain no ORF with a significant similarity to the amino acid sequence of RelHCg besides the hydrolase domain of the respective rel gene. Based on this observation we assessed the presence of this gene in the whole genus of which more than 110 species were described (Oliveira et al., 2017). These species were isolated from the environment but also from humans and animals, where in some cases, a pathogenic potential has been documented. The information obtained was added to a phylogenetic tree based on the 16S rRNA, supplemented additionally with information on the isolation site and the associated biosafety level (Figure 6). It was found that species closely related to C. glutamicum such as C. efficiens and C. deserti also possess a relH gene. These species represent environmental isolates without known pathogenic association. Interestingly, the more distantly related species of the genus Corynebacterium showed a rather heterogeneous occurrence of the analyzed gene. Within the identified phylogenetic subgroups, several representatives lack a relH homolog, indicating the occurrence of several independent loss events during evolution. The loss occurs in species isolated from different habitats and both potentially pathogenic and apathogenic species. Since no clear patterns could be observed, there is no apparent association with specific environmental conditions based on the evaluated data. An independent uptake of relH orthologs is unlikely due to the high sequence similarities of the existing genes and the resulting delineation within the Mesh-1L subgroup (data not shown).
FIGURE 6.
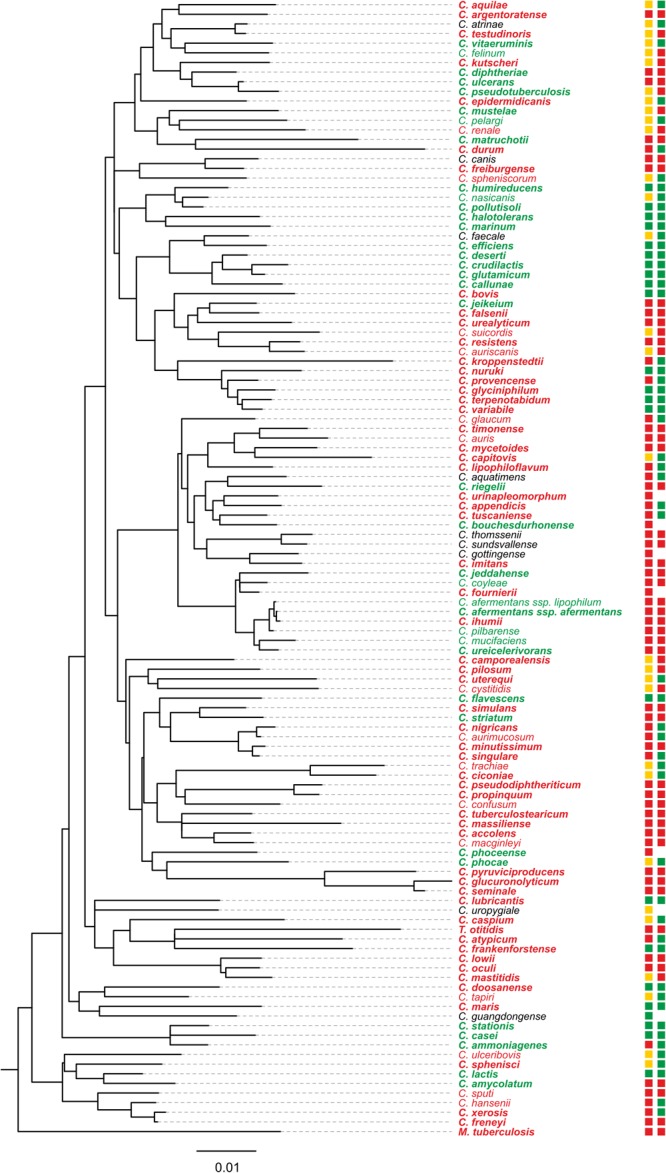
Phylogenetic tree of the genus Corynebacterium with additional species-specific information about the occurrence of a relHCg ortholog, the respective isolation site and the biosafety level. The phylogenetic tree was generated on the basis of the respective 16S rDNA sequences using the RDP database (Cole et al., 2014). The analysis of available genomes for the presence of a relHCg ortholog was based on a Hidden Markov model (see Materials and Methods). Species with a respective gene are shown in green, species without relH gene in red and species without an available genome in black. The genomes of species printed in bolt type are available in databases, whereas the other genomes are our unpublished data. The first box after the species name represents the isolation site, where green illustrates environmental isolates, yellow those isolated from animals, and red human isolates. The second box corresponds to the biosafety classification (according to German law); green: level one; red: level two; no box: no classification available.
Discussion
In this study, the putative SAH gene relHCg (cg1485) from C. glutamicum was investigated in in vivo and in vitro analyses with regard to the basic activity of its gene product and its functions within the (pp)pGpp metabolism. Both, the heterologous expression of the relHCg gene in two E. coli strains and the hydrolase assay-based study of the purified enzyme confirmed the proposed (pp)pGpp pyrophosphohydrolase activity in vivo and in vitro. The in vitro analysis of the hydrolase activities of RelHCg and of the bifunctional enzyme RelCg with regard to the different alarmone species revealed interesting enzyme characteristics as well as significant differences between both enzymes. The pyrophosphokinase activities of RelCg and RelHCg are strongly dependent on the presence of Mn2+ ions. This property fits expectations, as both enzymes exhibit a conserved His-Asp-box motive for Mn2+ binding that is characteristic for the HD domain of RSH type hydrolases (Aravind and Koonin, 1998; Mittenhuber, 2001). The corresponding Mn2+ dependency of (pp)pGpp hydrolases has already been demonstrated for other organisms (Hogg et al., 2004; Sun et al., 2010). Furthermore, the almost identical pH-dependencies of both (pp)pGpp hydrolases indicate similar reaction mechanisms and comparable enzyme properties. However, while RelCg has the highest activity for the substrate pGpp with a comparatively high Km value of 0.734 mM, the SAH enzyme RelHCg prefers the substrate ppGpp with a significantly lower Km value of 0.1 mM. The high hydrolysis activity of RelCg with respect to pGpp underscores the potential importance of this guanosine derivative which has recently been assigned to the alarmone species (Gaca et al., 2015b). The potential of C. glutamicum to synthesize pGpp has already been proven for the SAS enzyme RelSCg from C. glutamicum (Ruwe et al., 2017). The Km value of 0.51 mM determined for the hydrolysis of ppGpp by RelCg fits well to the data for the Rel ortholog from Streptococcus equisimilis, for which a Km value of 0.58 mM was determined (Sun et al., 2010). This result illustrates the fundamental comparability of both investigations. In contrast, two analyzed eukaryotic SAH enzymes show significantly higher Km values of approximately 3 mM. Furthermore, the maximum activities determined in this study for both RelCg and RelHCg are significantly lower than those identified for eukaryotic SAH enzymes. While RelHCg possesses a kcat value of 0.502 s-1 for ppGpp, corresponding measurements for the human SAH enzyme Mesh1 showed a turnover number of 36.6 s-1 (Sun et al., 2010). The significant Km and kcat differences, together with the unknown function of these SAH enzymes in eukaryotic systems, suggest considerable differences. Since no RSH enzyme could be identified so far as (pp)pGpp source for the corresponding organisms, there may even be completely different hyperphosphorylated substrates present. Also, the enzyme RelSeq from S. equisimilis exhibits a more than 10 times higher kcat value of 6 s-1. In contrast, the catalytic efficiency for ppGpp hydrolysis by RelSeq, with a value of 10.34 × 103 M-1 s-1 (Sun et al., 2010), is only about twice as high as the efficiency of 4.97 × 103 M-1 s-1 determined for RelHCg. However, the RelSeq variant analyzed is a truncated version of the enzyme with an additional base exchange to eliminate its synthetase activity, so that a direct comparability with RelCg and RelHCg is uncertain. Nevertheless, the small turnover numbers of the two (pp)pGpp hydrolases of C. glutamicum correspond to the turnover values determined for the (pp)pGpp synthetase activity of the SAS enzyme RelSCg in a physiological substrate concentration range. At a GDP concentration of 3.125 mM, RelSCg achieved a specific activity of 0.327 kat mol-1 (Ruwe et al., 2017). Therefore, a functional interaction of both monofunctional enzymes in the context of the C. glutamicum (pp)pGpp metabolism is realistic on the basis of the determined activity and specificity values.
One of the most interesting observations in this study is the reduction of the pyrophosphohydrolase activity of RelCg and RelHCg at high substrate concentrations. In particular, the activity of RelHCg decreases significantly at (pp)pGpp concentrations above 0.75 mM. Such substrate inhibition effects may not have been observed so far as insufficient substrate concentrations of maximally 400 μM were used in other studies. The use of these low concentrations is puzzling as the maximum ppGpp concentrations in E. coli were found to be in the range of 0.9–4 mM (Sy, 1980; Traxler et al., 2008). However, using classical equations to describe substrate inhibition kinetics based on the formation of an ESS complex, no convergent fit of the experimental data could be obtained when applying realistically dimensioned parameters (Ramsay and Tipton, 2017). The observed behavior therefore is probably not associated with the binding of the substrate to an alternative binding site. Furthermore, the analysis of the inhibition mechanism is complicated by the fact that (pp)pGpp hydrolysis is a reaction with only one substrate. As a result of this, the type of substrate inhibition cannot be verified by varying a non-inhibitory substrate. For the hydrolysis reaction, two further inhibition mechanisms are conceivable. On the one hand, parts of two substrate molecules could bind to the active center of the enzyme and thus form a dead-end ES2 complex (Lopina, 2017). The conformational flexibility of the 3′- and 5′-phosphate moieties observed for the alarmone species, which allows interaction with a broad spectrum of intracellular targets (Steinchen and Bange, 2016), could facilitate this process. If an ordered release of the products takes place, which is common for many hydrolytic enzymes in a Ping-Pong type reaction with water as the second substrate, the interaction of the substrate with the enzyme-substrate complex could also take place after the release of the first product (Cleland, 1979). The interaction with this intermediate complex, also known as EQ, in dead-end fashion could also explain the strong decrease in enzyme activity at high substrate concentrations. However, in order to elucidate the molecular background of enzyme behavior, more detailed knowledge of the so far unknown hydrolysis mechanism, in the form of crystal structure analyses or the use of hydrogen deuterium exchange mass spectrometry, is indispensable. During initial analyses of the hydrolysis reaction of long RSH enzymes, for example, the cyclic guanosine intermediate ppG2′:3′p (GPX) was found (Hogg et al., 2004), the function of which has not yet been understood (Steinchen and Bange, 2016). Furthermore, the formation of multimeric enzyme states could make the inhibition mechanism even more complex. A regulatory influence of the oligomerization is already known for long RSH enzymes and SAS representatives (Avarbock et al., 2005; Steinchen et al., 2015; Irving and Corrigan, 2018). The discovery of substrate inhibition in the long RSH representative RelCg as well as in the SAH enzyme RelHCg suggests that this enzymatic property of (pp)pGpp hydrolases may be not limited to C. glutamicum. In the context of the stringent response, this effect could be an important part of realizing a bistable system: In response to numerous stressful situations, this global and highly conserved regulatory system activates (pp)pGpp synthetases. The resulting sudden increase in (pp)pGpp levels leads to the activation of a persistence state as the hyperphosphorylated guanosine derivatives influence the biosynthesis or activity of various enzymes. Under these circumstances, the observed substrate inhibition reduces the activity of (pp)pGpp hydrolases which would quickly reduce (pp)pGpp levels back to the basal level otherwise. The substrate inhibition allows the persistence of the alarmone signal while synthesis is turned up, realizing the bistable switch. Indeed, in addition to the genetic network, enzyme properties such as substrate inhibition are known to be elementary components of bistable responses, which allow living cells a specific biochemical response to external stimuli (Tiwari et al., 2011). Moreover, the observed enzyme property may contribute to the realization of specialized (pp)pGpp metabolism-associated functionalities such as phenotypic heterogeneity, which is an important component of the lifestyle of some pathogenic bacteria like Mycobacterium tuberculosis (Ghosh et al., 2011). Last but not least, substrate inhibition would avoid an ATP-consuming futile cycle which, given the already limited resources under deficiency conditions, would be detrimental for the cells. Mechanisms that counteract (pp)pGpp futile cycling have already been described for bifunctional long RSH enzymes based on ‘intramolecular regulation of the opposing (p)ppGpp catalytic activities’ (Mechold et al., 2002).
The growth analysis of various RSH mutants in two different media, combined with the results of the in vitro analysis of the involved components, provides first insights into the functional relationships of (pp)pGpp metabolism in C. glutamicum. For the strains CR099 ΔrelCgΔrelSCg and CR099 ΔrelCgΔrelSCgΔrelHCg, growth characteristics similar to that of the parental strain were observed in minimal medium after an initially slightly delayed growth. This is a very interesting result as, according to the present state of knowledge, these strains are (pp)pGpp0 strains due to the removal of all (pp)pGpp synthetases, and (pp)pGpp0 strains of the model organisms E. coli and Bacillus subtilis are not capable of growth in minimal medium (Xiao et al., 1991; Kriel et al., 2014). C. glutamicum therefore does not seem to require a (pp)pGpp basal level for continuous growth in minimal medium, which illustrates the wide range of (pp)pGpp metabolism associated physiology. A (pp)pGpp0 strain which is not auxotrophic for amino acids has already been proven for Pseudomonas putida (Bernardo et al., 2009). In contrast to the presumed C. glutamicum ppGpp0 strains, the single ΔrelCg mutant shows a drastic decline in growth above an OD of 4. This is most likely due to increased (pp)pGpp levels resulting from a pyrophosphokinase activity of RelSCg in this growth phase combined with the lack of (pp)pGpp hydrolysis activity of RelCg (Nanamiya et al., 2008; Ruwe et al., 2017). The additional deletion of RelHCg causes an additional slightly negative effect on the growth behavior only in the final phase of cultivation from 25 h onward. Therefore, the hydrolase activity of RelCg is likely to be the key activity in the degradation of RelSCg products under these conditions. On the other hand, RelHCg does not appear to be actively present under these conditions or to participate only insignificantly in the hydrolysis of the RelSCg products.
The analysis of the available corynebacterial genomes with regard to possible RelHCg orthologs suggests the function as a complementary (pp)pGpp hydrolase under certain conditions. Numerous species across the genus have apparently lost the corresponding gene in independent events. Therefore, an essential role in the global regulation of cell homeostasis is unlikely. An inadequate hydrolysis of the RelSCg products by RelHCg is supported by the unexpected growth deficit of the strain CR099 ΔrelSCgΔrelHCg, whereas both single mutants are not impaired under these conditions. One possible scenario for explaining this issue could be a negative activity coupling of RelCg and RelSCg. In the absence of RelSCg, RelCg appears to provide the required (pp)pGpp synthesis activity. The negative effect of the additional relHCg deletion suggests that the RelHCg hydrolase activity represents the counterpart for the synthetically active RelCg, resulting in a balanced (pp)pGpp level. The lack of RelHCg would therefore increase the (pp)pGpp levels, analogous to the ΔrelCg situation and thus trigger a growth effect. A study of RelSeq from S. equisimilis supports the previously established hypothesis, as it has shown that long RSH proteins are most likely not active synthetically and hydrolytically at the same time due to a conformation based regulation of their catalytic activities (Hogg et al., 2004).
Synthetase and hydrolase kinetics of the enzymes involved, determined by in vitro assays, also support this hypothesis. pGpp, which appears to be produced almost exclusively by RelSCg (Ruwe et al., 2017), is preferably degraded by RelCg. This could explain the minimal effects of the additional relHCg gene deletion in the strain CR099 ΔrelCgΔrelHCg, as pGpp which is produced by RelSCg may not be sufficiently degraded. However, when interpreting the in vitro enzyme characterization, it must be noted that the assays performed represent strongly reduced systems. In particular, the activities of RelCg will most likely be significantly influenced by other cellular components. Previous studies have shown that a 12 bp deletion in the large ribosomal subunit protein L11 impairs C. glutamicum in (pp)pGpp accumulation upon amino acid starvation (Wehmeier et al., 2001). The amino acid starvation-associated activation of stringent response is therefore probably carried out by a complex of RelCg, ribosomes and uncharged tRNAs analogous to numerous other organisms (Haseltine and Block, 1973; Avarbock et al., 2000). Furthermore, the activity of the single domain RSH proteins could also be influenced by various cell internal mechanisms. For example, the SAS enzyme RelQBs from B. subtilis is allosterically activated in its active tetrameric form by its own product pppGpp (Steinchen et al., 2015). This SAS feature also indirectly complies with the substrate inhibition observed for (pp)pGpp hydrolases in this study and may also contribute to the bistable nature of (pp)pGpp metabolism.
In comparison to the cultivation in minimal medium, in which a metabolically quite constant state is present due to the degradation of glucose as the sole carbon source, the growth analysis in amino acid containing complex medium provides further aspects of the (pp)pGpp metabolism in C. glutamicum. The cultivation can be divided into two sections. After a period of exponential growth, the wild-type and similarly growing strains enter a second phase with a more linear character. This is most likely due to the consumption of ingredients, which must subsequently be synthesized by C. glutamicum. Since the (pp)pGpp0 strains have a significant growth deficit in the second cultivation section, the presence of (pp)pGpp seems to be highly relevant under these conditions. Because of the described regulatory effects in other organisms like E. coli (Mechold et al., 2013), the alarmone species appear to be crucial for a correct cellular adaptation to new nutritional conditions. For example, an influence of (pp)pGpp on the transcriptional activation of amino acid synthesis clusters due to the consumption of components originally contained in the complex CASO broth is conceivable. Such an influence of the alarmone species on the transcription of amino acid synthesis clusters has already been observed in previous studies as a reaction to the induction of an amino acid starvation in C. glutamicum (Brockmann-Gretza and Kalinowski, 2006). The strains CR099 ΔrelCg and CR099 ΔrelCgΔrelHCg also exhibit delayed growth in the second cultivation phase, but unlike the presumed (pp)pGpp0 strains, they do not completely stop growth and reach the maximum OD with some delay. This can possibly be traced back to increased (pp)pGpp levels due to the lack of RelCg hydrolase activity, similar to cultivation in minimal medium. However, insufficient (pp)pGpp levels are also possible, provided that the pyrophosphokinase activities of both RelCg and RelSCg are required under these conditions.
It is important to note that future measurements of the internal (pp)pGpp pools of various mutants are essential to verify the hypotheses based on growth data and in vitro enzyme characterization. This is a major challenge as the highly relevant alarmone basal levels are very low and cannot be exactly quantified even with sensitive 32P measurements. Using recently established HPLC-based methods for the quantification of messenger nucleotides it is possible to analyze the basal level of ppGpp in Staphylococcus aureus, Ralstonia eutropha, and E. coli (Kästle et al., 2015; Juengert et al., 2017; Varik et al., 2017). However, the other alarmone species are still below the limit of detection in a non-stress induced state. Moreover, it should be noted that 32P-based analyses for C. glutamicum have shown that the (pp)pGpp concentrations in this organism are significantly lower than the values determined for E. coli (Wehmeier et al., 1998; Tauch et al., 2001). The quantitative analysis of the alarmone species at different growth conditions as well as an gene expression analysis of the functional components could provide important information on the roles of the three known alarmone species, the mechanistic processes during stringent response and the functions of the (pp)pGpp basal level. This could also help to understand the role of (pp)pGpp metabolism in the natural habitat and thus possibly provide valuable information on economically interesting mechanisms such as the growth rate regulation or bacterial persistence and virulence mechanisms.
Author Contributions
MR, JK, and MP designed, analyzed, and interpreted the performed experiments. JK and MP supervised the research. MR performed the wet lab experiments. CR performed the taxonomic analyses. The manuscript was written by MR and revised by MP, CR, and JK.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the German Ministry of Education and Research for financial funding in the frame of the e:Bio initiative (contract no. 031A302F), Vera Ortseifen (CeBiTec) and Julia Voss (CeBiTec) for the help with MALDI-TOF measurements, and the working group Fermentation Technology (Bielefeld University) for providing the ÄKTA system. Furthermore, we acknowledge support for the Article Processing Charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00916/full#supplementary-material
References
- Aravind L., Koonin E. V. (1998). The HD domain defines a new superfamily of metal-dependent phosphohydrolases. 23 469–472. 10.1016/S0968-0004(98)01293-6 [DOI] [PubMed] [Google Scholar]
- Atkinson G. C., Tenson T., Hauryliuk V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. 6:e23479. 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarbock A., Avarbock D., Teh J.-S., Buckstein M., Wang Z.-M., Rubin H. (2005). Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. 44 9913–9923. 10.1021/bi0505316 [DOI] [PubMed] [Google Scholar]
- Avarbock D., Avarbock A., Rubin H. (2000). Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. 39 11640–11648. 10.1021/bi001256k [DOI] [PubMed] [Google Scholar]
- Bag S., Das B., Dasgupta S., Bhadra R. K. (2014). Mutational analysis of the (p)ppGpp synthetase activity of the Rel enzyme of Mycobacterium tuberculosis. 196 575–588. 10.1007/s00203-014-0996-9 [DOI] [PubMed] [Google Scholar]
- Baumgart M., Unthan S., Kloß R., Radek A., Polen T., Tenhaef N., et al. (2018). Corynebacterium glutamicum chassis C1∗: building and testing a novel platform host for synthetic biology and industrial biotechnology. 7 132–144. 10.1021/acssynbio.7b00261 [DOI] [PubMed] [Google Scholar]
- Baumgart M., Unthan S., Rückert C., Sivalingam J., Grünberger A., Kalinowski J., et al. (2013). Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. 79 6006–6015. 10.1128/AEM.01634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo L. M. D., Johansson L. U. M., Skärfstad E., Shingler V. (2009). sigma54-promoter discrimination and regulation by ppGpp and DksA. 284 828–838. 10.1074/jbc.M807707200 [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Plunkett G., III, Bloch C. A., Perna N. T., Burland V., Riley M., et al. (1997). The complete genome sequence of Escherichia coli K-12. 277 1453–1462. 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- Brockmann-Gretza O., Kalinowski J. (2006). Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase. 7:230. 10.1186/1471-2164-7-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M., Kalbacher B. (1970). The control of ribonucleic acid synthesis in Escherichia coli. V. Characterization of a nucleotide associated with the stringent response. 245 2309–2318. [PubMed] [Google Scholar]
- Chipman D. M., Shaanan B. (2001). The ACT domain family. 11 694–700. 10.1016/S0959-440X(01)00272-X [DOI] [PubMed] [Google Scholar]
- Cleland W. W. (1979). “[20] Substrate inhibition,” in ed. Purich D. L. (New York, NY: Elsevier; ) 500–513. [Google Scholar]
- Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. 42 D633–D642. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D., Svensson S. L., Gaynor E. C., Swanson M. S. (2010). ppGpp conjures bacterial virulence. 74 171–199. 10.1128/MMBR.00046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D., Swanson M. S. (2012). ppGpp: magic beyond RNA polymerase. 10 203–212. 10.1038/nrmicro2720 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., et al. (2015). HMMER web server: 2015 update. 43 W30–W38. 10.1093/nar/gkv397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O., Colomer-Winter C., Lemos J. A. (2015a). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. 197 1146–1156. 10.1128/JB.02577-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O., Kajfasz J. K., Miller J. H., Liu K., Wang J. D., Abranches J., et al. (2013). Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. 4:e00646-13. 10.1128/mBio.00646-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O., Kudrin P., Colomer-Winter C., Beljantseva J., Liu K., Anderson B., et al. (2015b). From (p)ppGpp to (pp)pGpp: characterization of regulatory effects of pGpp synthesized by the small alarmone synthetase of Enterococcus faecalis. 197 2908–2919. 10.1128/JB.00324-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Sureka K., Ghosh B., Bose I., Basu J., Kundu M. (2011). Phenotypic heterogeneity in mycobacterial stringent response. 5:18. 10.1186/1752-0509-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., III, Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. 6 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Block R. (1973). Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. 70 1564–1568. 10.1073/pnas.70.5.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A., Block R., Gilbert W., Weber K. (1972). MSI and MSII made on ribosome in idling step of protein synthesis. 238 381–384. 10.1038/238381a0 [DOI] [PubMed] [Google Scholar]
- Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T., Gerdes K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. 13 298–309. 10.1038/nrmicro3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstee B. H. J. (1959). Non-inverted versus inverted plots in enzyme kinetics. 184 1296–1298. 10.1038/1841296b0 [DOI] [PubMed] [Google Scholar]
- Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R. (2004). Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. 117 57–68. 10.1016/S0092-8674(04)00260-0 [DOI] [PubMed] [Google Scholar]
- Irving S. E., Corrigan R. M. (2018). Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. 164 268–276. 10.1099/mic.0.000621 [DOI] [PubMed] [Google Scholar]
- Juengert J. R., Borisova M., Mayer C., Wolz C., Brigham C. J., Sinskey A. J., et al. (2017). Absence of ppGpp leads to increased mobilization of intermediately accumulated Poly(3-Hydroxybutyrate) in Ralstonia eutropha H16. 83:e00755-17. 10.1128/AEM.00755-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., et al. (2003). The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. 104 5–25. 10.1016/S0168-1656(03)00154-8 [DOI] [PubMed] [Google Scholar]
- Kasai K. (2002). A RelA-SpoT homolog (Cr-RSH) identified in Chlamydomonas reinhardtii generates stringent factor in vivo and localizes to chloroplasts in vitro. 30 4985–4992. 10.1093/nar/gkf628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kästle B., Geiger T., Gratani F. L., Reisinger R., Goerke C., Borisova M., et al. (2015). rRNA regulation during growth and under stringent conditions in Staphylococcus aureus. 17 4394–4405. 10.1111/1462-2920.12867 [DOI] [PubMed] [Google Scholar]
- Keilhauer C., Eggeling L., Sahm H. (1993). Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. 175 5595–5603. 10.1128/jb.175.17.5595-5603.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel A., Brinsmade S. R., Tse J. L., Tehranchi A. K., Bittner A. N., Sonenshein A. L., et al. (2014). GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. 196 189–201. 10.1128/JB.00918-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos J. A., Lin V. K., Nascimento M. M., Abranches J., Burne R. A. (2007). Three gene products govern (p)ppGpp production by Streptococcus mutans. 65 1568–1581. 10.1111/j.1365-2958.2007.05897.x [DOI] [PubMed] [Google Scholar]
- Lopina O. D. (2017). “Enzyme inhibitors and activators,” in ed. Senturk M. (Rijeka: InTech; ). [Google Scholar]
- Masuda S., Mizusawa K., Narisawa T., Tozawa Y., Ohta H., Takamiya K.-I. (2008). The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. 49 135–141. 10.1093/pcp/pcm177 [DOI] [PubMed] [Google Scholar]
- Mechold U., Murphy H., Brown L., Cashel M. (2002). Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from Streptococcus equisimilis. 184 2878–2888. 10.1128/JB.184.11.2878-2888.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U., Potrykus K., Murphy H., Murakami K. S., Cashel M. (2013). Differential regulation by ppGpp versus pppGpp in Escherichia coli. 41 6175–6189. 10.1093/nar/gkt302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski A., Siemann-Herzberg M., Takors R. (2017). Escherichia coli HGT: engineered for high glucose throughput even under slowly growing or resting conditions. 40 93–103. 10.1016/j.ymben.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Mittenhuber G. (2001). Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel. RelA and SpoT proteins). 3 585–600. [PubMed] [Google Scholar]
- Murphy H., Cashel M. (2003). Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. 371 596–601. 10.1016/S0076-6879(03)71044-1 [DOI] [PubMed] [Google Scholar]
- Nanamiya H., Kasai K., Nozawa A., Yun C.-S., Narisawa T., Murakami K., et al. (2008). Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. 67 291–304. 10.1111/j.1365-2958.2007.06018.x [DOI] [PubMed] [Google Scholar]
- Oliveira A., Oliveira L. C., Aburjaile F., Benevides L., Tiwari S., Jamal S. B., et al. (2017). Insight of genus Corynebacterium: ascertaining the role of pathogenic and non-pathogenic species. 8:1937. 10.3389/fmicb.2017.01937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer-Sancar K., Mentz A., Rückert C., Kalinowski J. (2013). Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. 14:888. 10.1186/1471-2164-14-888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K., Cashel M. (2008). (p)ppGpp: still magical? 62 35–51. 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- Potrykus K., Murphy H., Philippe N., Cashel M. (2011). ppGpp is the major source of growth rate control in E. coli. 13 563–575. 10.1111/j.1462-2920.2010.02357.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay R. R., Tipton K. F. (2017). Assessment of enzyme inhibition: a review with examples from the development of monoamine oxidase and cholinesterase inhibitory drugs. 22:E1192. 10.3390/molecules22071192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe M., Kalinowski J., Persicke M. (2017). Identification and functional characterization of small alarmone synthetases in Corynebacterium glutamicum. 8:1601. 10.3389/fmicb.2017.01601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. 145 69–73. 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- Steinchen W., Bange G. (2016). The magic dance of the alarmones (p)ppGpp. 101 531–544. 10.1111/mmi.13412 [DOI] [PubMed] [Google Scholar]
- Steinchen W., Schuhmacher J. S., Altegoer F., Fage C. D., Srinivasan V., Linne U., et al. (2015). Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. 112 13348–13353. 10.1073/pnas.1505271112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Lee G., Lee J. H., Kim H.-Y., Rhee H.-W., Park S.-Y., et al. (2010). A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. 17 1188–1194. 10.1038/nsmb.1906 [DOI] [PubMed] [Google Scholar]
- Sy J. (1980). “ppGpp, a signal molecule,” in eds Chapeville F., Haenni A.-L. (Berlin: Springer; ) 197–204. 10.1007/978-3-642-81503-4_16 [DOI] [Google Scholar]
- Tauch A., Wehmeier L., Götker S., Pühler A., Kalinowski J. (2001). Relaxed rrn expression and amino acid requirement of a Corynebacterium glutamicum rel mutant defective in (p)ppGpp metabolism. 201 53–58. 10.1111/j.1574-6968.2001.tb10732.x [DOI] [PubMed] [Google Scholar]
- Tiwari A., Ray J. C. J., Narula J., Igoshin O. A. (2011). Bistable responses in bacterial genetic networks: designs and dynamical consequences. 231 76–89. 10.1016/j.mbs.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler M. F., Summers S. M., Nguyen H.-T., Zacharia V. M., Hightower G. A., Smith J. T., et al. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. 68 1128–1148. 10.1111/j.1365-2958.2008.06229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unthan S., Baumgart M., Radek A., Herbst M., Siebert D., Bruhl N., et al. (2015). Chassis organism from Corynebacterium glutamicum–a top-down approach to identify and delete irrelevant gene clusters. 10 290–301. 10.1002/biot.201400041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzan M., Danchin A. (1978). Correlation between the serine sensitivity and the derepressibility of the ilv genes in Escherichia coli relA- mutants. 165 21–30. 10.1007/BF00270372 [DOI] [PubMed] [Google Scholar]
- van der Biezen E. A., Sun J., Coleman M. J., Bibb M. J., Jones J. D. (2000). Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. 97 3747–3752. 10.1073/pnas.060392397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varik V., Oliveira S. R. A., Hauryliuk V., Tenson T. (2017). HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. 7:11022. 10.1038/s41598-017-10988-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter F., Grenz S., Ortseifen V., Persicke M., Kalinowski J. (2016). Corynebacterium glutamicum ggtB encodes a functional γ-glutamyl transpeptidase with γ-glutamyl dipeptide synthetic and hydrolytic activity. 232 99–109. 10.1016/j.jbiotec.2015.10.019 [DOI] [PubMed] [Google Scholar]
- Wehmeier L., Brockmann-Gretza O., Pisabarro A., Tauch A., Pühler A., Martin J. F., et al. (2001). A Corynebacterium glutamicum mutant with a defined deletion within the rplK gene is impaired in (p)ppGpp accumulation upon amino acid starvation. 147 691–700. 10.1099/00221287-147-3-691 [DOI] [PubMed] [Google Scholar]
- Wehmeier L., Schafer A., Burkovski A., Kramer R., Mechold U., Malke H., et al. (1998). The role of the Corynebacterium glutamicum rel gene in (p)ppGpp metabolism. 144 1853–1862. 10.1099/00221287-144-7-1853 [DOI] [PubMed] [Google Scholar]
- Wolf Y. I., Aravind L., Grishin N. V., Koonin E. V. (1999). Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. 9 689–710. [PubMed] [Google Scholar]
- Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. (1991). Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. 266 5980–5990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


