ABSTRACT
Insulin receptor substrate 1 (IRS-1) is a common cytosolic adaptor molecule involved in signal transduction from insulin and insulin-like growth factor I (IGF-I) receptors. IRS-1 can also be found in the nucleus. We report here a new finding of unique IRS-1 nuclear structures, which we observed initially in glioblastoma biopsy specimens and glioblastoma xenografts. These nuclear structures can be reproduced in vitro by the ectopic expression of IRS-1 cDNA cloned in frame with the nuclear localization signal (NLS–IRS-1). In these structures, IRS-1 localizes at the periphery, while the center harbors a key autophagy protein, LC3. These new nuclear structures are highly dynamic, rapidly exchange IRS-1 molecules with the surrounding nucleoplasm, disassemble during mitosis, and require a growth stimulus for their reassembly and maintenance. In tumor cells engineered to express NLS–IRS-1, the IRS-1/LC3 nuclear structures repress autophagy induced by either amino acid starvation or rapamycin treatment. In this process, IRS-1 nuclear structures sequester LC3 inside the nucleus, possibly preventing its cytosolic translocation and the formation of new autophagosomes. This novel mechanism provides a quick and reversible way of inhibiting autophagy, which could counteract autophagy-induced cancer cell death under severe stress, including anticancer therapies.
KEYWORDS: autophagy, cellular distribution, fluorescence recovery after photobleaching, glioblastoma, multiprotein complexes, nuclear suborganelle, phase transition, protein binding, signal transduction
INTRODUCTION
Insulin receptor substrate 1 (IRS-1) is a common cytosolic adaptor molecule, which transduces the signal from both insulin and insulin-like growth factor I (IGF-I) receptors (1). We previously reported that IRS-1 can be translocated to the nucleus by either the human polyomavirus JC oncoprotein, large T antigen (2), or estrogen receptors (3–5). In addition, nuclear IRS-1 (nIRS-1) has been detected in cells expressing the simian virus 40 (SV40) T antigen (6) and v-Src (7) and in breast cancer cells in association with estrogen receptor alpha (3). In other studies, nIRS-1 was found in nucleoli, where it interacted with upstream binding factor 1 (UBF1) (8), and more recently, nIRS-1 was detected in complexes with noncoding RNAs in association with Cajal bodies (9, 10).
In recent years, it has become apparent that the nucleus is highly compartmentalized (11, 12), and there are several nuclear structures, such as nucleoli, speckles, paraspeckles, Cajal bodies, PML, Polycomb, and nuclear stress bodies, that play specific roles in the function of the nucleus (13–15). Unlike typical cytoplasmic organelles, nuclear structures lack biological membranes that separate them from the surrounding nucleoplasm. Although not membrane bound, nuclear structures are temporally stable, and their stability is based on specific protein-protein and protein-RNA interactions (14). Although the lack of biological membranes allows the highly dynamic regulation of their composition and function in response to cellular signals and stress (16), it also makes biochemical analyses of these membraneless structures more challenging. In fact, most of the information is derived from structural studies using electron and confocal microscopy, and complete proteome analyses have been performed on only two nuclear structures, nucleoli (17, 18) and interchromatin granule clusters (speckles) (19). Nonetheless, the exact composition and function of most of these nuclear structures remain elusive.
Here, we report a new finding that nuclear IRS-1 can be organized in large nuclear structures, which can reach up to 2 μm in diameter. In these structures, IRS-1 is localized at the periphery, and the center harbors a typical autophagy protein, LC3; however, other autophagy proteins or biological membranes were not detected. We observed these IRS-1-containing nuclear structures for the first time in a restricted number of cells of glioblastoma biopsy specimens and glioblastoma xenografts, mostly in areas adjacent to necrotic tissue and in the infiltrating edges of the tumor. We were also able to reproduce the formation of these IRS-1-containing nuclear structures in vitro by the ectopic expression of IRS-1 cDNA cloned in frame with a nuclear localization signal (NLS–IRS-1). In living cells expressing the NLS–IRS-1–green fluorescent protein (GFP) fusion protein, IRS-1/LC3 structures are highly dynamic: they disassemble during mitosis or following prolonged serum starvation, reassemble shortly after cytokinesis in growth factor-stimulated cells, and quickly exchange IRS-1 molecules with the surrounding nucleoplasm. Importantly, tumor cells positive for the IRS-1/LC3 nuclear structures have severely impaired autophagy, which correlated with the accumulation of LC3 inside the nucleus. In summary, the IRS-1/LC3 nuclear structures provide a quick and reversible mechanism of blocking autophagy, which could play a role in tumor cell survival by counteracting the autophagy-induced death of tumor cells exposed to severe stress.
RESULTS
Detection of IRS-1 nuclear structures in human brain tumors.
We observed IRS-1-containing nuclear structures when we evaluated the possible diagnostic value of nIRS-1 in a brain tumor tissue array consisting of 64 different brain tumor clinical samples (GL803a; USBiomax, Inc.). In 25 out of a total of 64 brain tumor biopsy specimens (39.1%), IRS-1 was found in the cell nuclei (Table 1). Positive cells were grouped into clusters, predominantly near the infiltrating edges of the tumor or near necrotic areas in glioblastomas. The results in Fig. 1A show representative examples of two glioblastoma biopsy specimens, from cases C2 and A5 (Table 1), in which IRS-1 is present in either the nuclei (Fig. 1A) or the cytoplasm (Fig. 1B) of the tumor cells. Interestingly, when the same brain biopsy specimens were examined by using immunofluorescence and high-resolution confocal imaging (Fig. 1C to G), some of the tumor cells exhibited well-defined nuclear structures, which varied in size from 0.2 μm to up to 1 μm in diameter. In comparison to overall nuclear IRS-1 immunolabeling, the number of tumor cells positive for IRS-1 nuclear structures was significantly lower (0.01%) when the entire tumor biopsy specimen was analyzed. However, in some areas of the tumor, the frequency of cells positive for these structures was much higher, reaching up to 10%, an increase of several orders of magnitude (Fig. 1C). Two high-magnification images (Fig. 1F and G) demonstrate IRS-1 nuclear structures detected by either anti-IRS-1 rabbit polyclonal antibody or anti-IRS-1(pS612) mouse monoclonal antibody, respectively. We did not detect these nuclear structures in unaffected brain areas (Fig. 1E) or in tumor tissue by using either anti-IRS-1(pY) antibody (data not shown) or an irrelevant primary antibody (anti bromodeoxyuridine [anti-BrdU]) plus a secondary antibody (Fig. 1D).
TABLE 1.
IRS-1 immunohistochemistry performed on a tissue array from which 64 high-quality brain tumor biopsy specimens were selecteda
| Position | Gender | Age (yr) | Tumor type | Grade | Presence of IRS-1 |
|
|---|---|---|---|---|---|---|
| N | C | |||||
| A2 | M | 65 | Astrocytoma | 2 | + | +/− |
| A3 | M | 46 | Astrocytoma (sparse) | 1 | + | +/− |
| A4 | M | 53 | Anaplastic astrocytoma | 3 | − | + |
| A5 | M | 25 | Glioblastoma | 4 | − | ++++ |
| A6 | F | 10 | Glioblastoma | 4 | − | ++ |
| A7 | M | 33 | Glioblastoma | 4 | − | − |
| A9 | M | 34 | Astrocytoma | 2 | + | +++ |
| A10 | M | 57 | Anaplastic astrocytoma | 3 | − | ++ |
| B2 | F | 16 | Astrocytoma | 2 | + | ++ |
| B3 | M | 57 | Anaplastic astrocytoma | 3 | +++ | ++ |
| B4 | F | 60 | Anaplastic astrocytoma | 3 | + | + |
| B5 | M | 36 | Glioblastoma | 4 | ++ | + |
| B6 | F | 39 | Anaplastic astrocytoma | 3 | − | − |
| B7 | F | 66 | Astrocytoma (cortex infilb) | 2 | + | + |
| B8 | M | 52 | Anaplastic astrocytoma | 3 | + | ++ |
| B9 | M | 34 | Glioblastoma | 4 | − | +++ |
| B10 | F | 43 | Glioblastoma | 4 | − | +++ |
| C1 | F | 50 | Astrocytoma | 2 | ++ | + |
| C2 | F | 49 | Glioblastoma (cortex infil) | 4 | +++ | + |
| C3 | M | 41 | Astrocytoma | 2 | − | + |
| C4 | F | 6 | Glioblastoma | 4 | +++ | ++ |
| C6 | M | 43 | Glioblastoma | 4 | ++ | +++ |
| C7 | M | 12 | Glioblastoma (sparse) | 4 | − | + |
| C8 | M | 41 | Glioblastoma | 4 | − | + |
| C9 | F | 55 | Glioblastoma (sparse) giant cell | 4 | − | ++++ |
| C10 | M | 62 | Glioblastoma | 4 | + | ++ |
| D1 | F | 22 | Glioblastoma multiforme | 4 | − | + |
| D2 | M | 64 | Glioblastoma | 4 | − | + |
| D4 | M | 55 | Glioblastoma | 4 | +/− | − |
| D5 | M | 36 | Glioblastoma (sparse) giant cell | 4 | − | ++ |
| D6 | M | 43 | Glioblastoma | 4 | +/− | |
| D7 | M | 71 | Glioblastoma (giant cells) | 4 | − | ++ |
| D9 | F | 64 | Glioblastoma (giant cells) | 4 | − | ++ |
| D10 | F | 36 | Glioblastoma | 4 | − | +/− |
| E2 | F | 48 | Anaplastic oligodendroglioma | 3 | − | − |
| E4 | M | 59 | Glioblastoma (sparse) | 4 | − | ++ |
| E5 | M | 68 | Glioblastoma (giant cell) | 4 | − | ++ |
| E6 | F | 4 | Glioblastoma | 4 | − | ++ |
| E7 | F | 50 | Glioblastoma multiforme | 4 | +/− | − |
| E8 | M | 59 | Glioblastoma multiforme | 4 | − | + |
| E9 | M | 50 | Glioblastoma | 4 | − | − |
| E10 | M | 42 | Glioblastoma multiforme | 4 | − | +/− |
| F1 | M | 58 | Glioblastoma multiforme | 4 | − | ++ |
| F2 | F | 52 | Glioblastoma multiforme | 4 | − | + |
| F3 | F | 51 | Astrocytoma | 2 | − | − |
| F4 | M | 41 | Glioblastoma | 4 | − | + |
| F5 | M | 33 | Glioblastoma | 4 | − | ++ |
| F6 | M | 33 | Anaplastic oligodendroglioma | 3 | ++ | − |
| F7 | F | 37 | Anaplastic oligodendroglioma | 3 | − | ++ |
| F8 | M | 17 | Anaplastic oligodendroglioma | 3 | − | + |
| F9 | M | 55 | Anaplastic oligodendroglioma | 3 | − | − |
| G1 | F | 30 | Anaplastic oligodendroglioma | 3 | + | + |
| G2 | M | 39 | Anaplastic oligodendroglioma | 3 | − | − |
| G3 | F | 14 | Medulloblastoma | − | + | − |
| G4 | M | 11 | Medulloblastoma | − | − | + |
| G5 | M | 3 | Oligoastrocytoma | 2 | − | + |
| G7 | F | 46 | Oligodendroglioma | 2 | − | +/− |
| G8 | M | 52 | Anaplastic oligodendroglioma | 3 | − | − |
| G10 | M | 45 | Oligoastrocytoma | 2 | − | − |
| H1 | F | 48 | Anaplastic oligoastrocytoma | 3 | + | − |
| H2 | M | 41 | Ependymoma | − | +/− | − |
| H3 | M | 3 | Ependymoma | − | ++ | − |
| H4 | F | 15 | Astrocytoma | 1 | + | − |
| H5 | F | 14 | Pilocytic astrocytoma | 1 | +/− | − |
| H6 | F | 30 | Cancer-adjacent normal cerebral tissue | − | − | − |
| H7 | F | 49 | Cancer-adjacent normal cerebral tissue | − | − | − |
| H8 | M | 59 | Cancer-adjacent normal cerebral tissue | − | + | − |
| H9 | M | 39 | Cancer-adjacent normal cerebral tissue | − | − | − |
| H10 | M | 52 | Cancer-adjacent normal cerebral tissue | − | − | − |
| I10 | M | 58 | Malignant melanoma (marked point) | − | − | +++ |
The tissue array (GL803a; USBiomax, Inc.) includes 32 glioblastoma, 19 astrocytoma, 9 oligodendriogloma, 2 ependymoma, and 2 medulloblastoma specimens. The array also contains 5 adjacent normal brain tissues and 1 liver cancer specimen. Diagnosis of the tumor is based on World Health Organization criteria for brain tumors, and the identity and grade of each tumor of this array were additionally verified by a neuropathologist (L. Del Valle). The presence (+) or absence (−) of IRS-1 immunohistolabeling was determined by utilizing an anti-IRS-1 rabbit polyclonal antibody (Millipore), and the subcellular localization of immunolabeling, cytosolic (C) versus nuclear (N), was determined by using an Olympus BX61 bright-field light microscope with a 40× objective. Nuclear IRS-1 was detected in 25 out of a total of 64 brain tumors examined (39.1%). Two cases, cases A5 and C2, are also shown in Fig. 1A. M, male; F, female. +/−, very low level of immunolabeling.
Cortex infil, infiltrated cortex.
FIG 1.
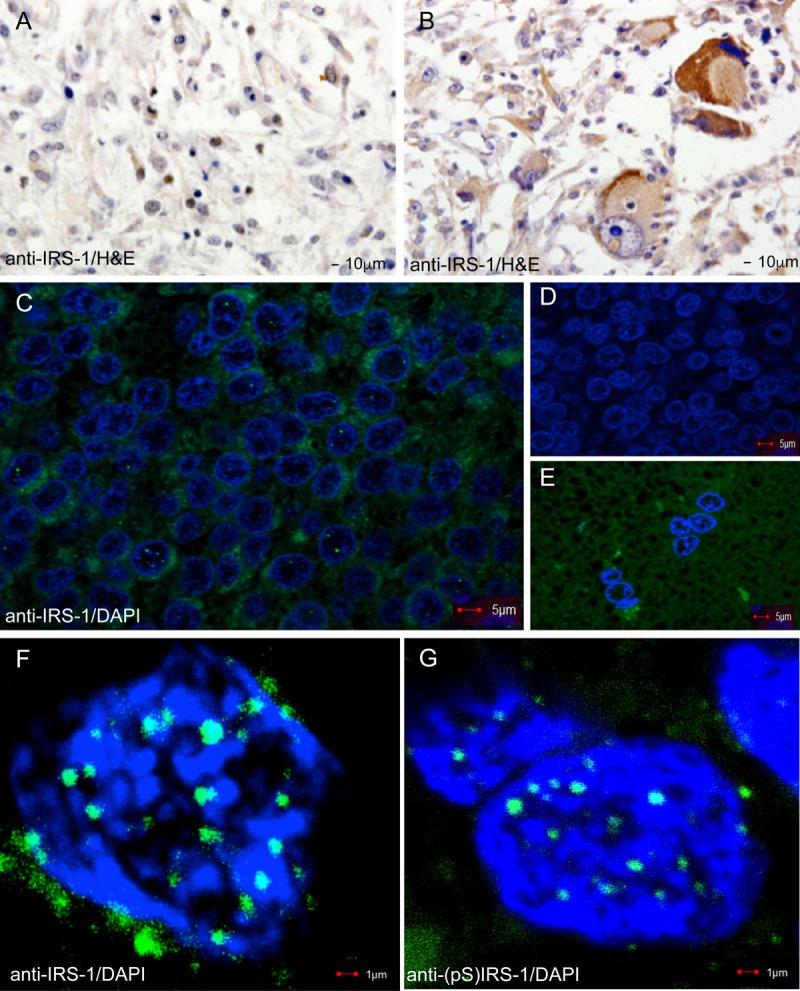
Immunohistochemical detection of IRS-1. IRS-1 was detected in formalin-fixed paraffin-embedded sections of glioblastoma tissues (GL803a tissue array; USBiomax, Inc.) by using anti-IRS-1 rabbit polyclonal (Millipore) and anti-rabbit HRP-conjugated secondary antibodies. (A and B) Two glioblastoma biopsy specimens in which IRS-1 is localized in either the nuclei of some tumor cells (case C2 from Table 1) (A) or the cytoplasm (case A5 from Table 1) (B). (C, F, and G) Examples of low-magnification (C) and high-magnification (F and G) confocal images of IRS-1 nuclear structures detected in a restricted number of cells from glioblastoma biopsy specimens. IRS-1 nuclear structures were detected with anti-IRS-1 rabbit polyclonal antibody (catalog no. 06-248; Millipore) and FITC-conjugated anti-rabbit secondary antibodies. Nuclei were counterstained with DAPI (blue fluorescence). (C) Selected area adjacent to the tumor-infiltrating margin with a high number of tumor cells positive for IRS-1 nuclear structures (15 out of a total of 121 cells show IRS-1 nuclear structures). The percentage of tumor cells positive for IRS-1 nuclear structures was evaluated by using high-magnification confocal imaging (original magnification, ×100). At least 100 randomly selected fields per biopsy specimen were examined for 10 different glioblastoma biopsy specimens. (D) The same glioblastoma multiforme biopsy specimen immunolabeled with an irrelevant antibody (anti-BrdU primary antibody and FITC-conjugated secondary antibody). (E) Unaffected brain area, adjacent to the tumor depicted in panel C, immunolabeled with anti-IRS-1 rabbit polyclonal antibody.
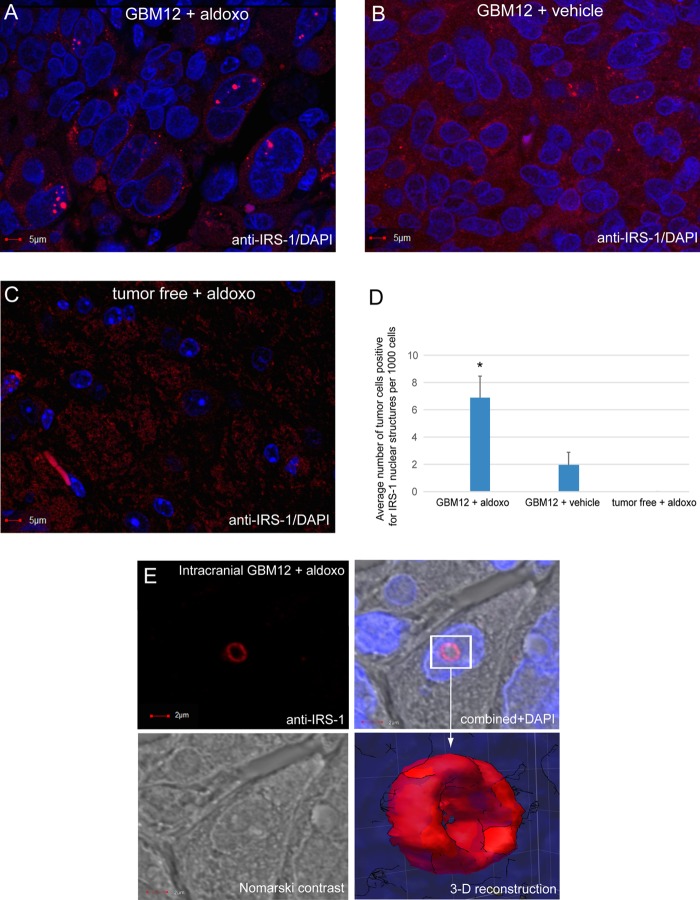
IRS-1-positive nuclear structures were also detected in glioblastoma xenografts (Fig. 2), and importantly, we observed a nearly 4-fold increase in their presence after genotoxic treatment (Fig. 2D). In this experiment, mice bearing intracranial glioblastomas formed by GBM12 native human glioblastoma cells (20–22) were treated with aldoxorubicin by tail vein injection (23). We selected aldoxorubicin (doxorubicin conjugated with an albumin affinity linker) since it has an exceptional ability to accumulate in tumor tissues (24–26) and triggered extensive genotoxicity in glioblastoma brain xenografts (23). Similar to glioblastoma biopsy specimens, we also detected areas within the xenografts in which the frequency of tumor cells positive for IRS-1 nuclear structures was much higher. Figure 2A shows an area within the xenograft in which 5 out of 65 nuclei are positive for these IRS-1-containing nuclear structures. Interestingly, by using fresh tissue explants from aldoxorubicin-treated mice, we also observed large IRS-1-containing nuclear structures, which resembled the shape of a ring (Fig. 2E).
FIG 2.
Detection of IRS-1 nuclear structures in glioblastoma multiforme xenografts. Human GBM12 cells were injected into the brains of immunodeficient mice (1 × 105 cells in 2 μl of PBS) by using a Hamilton syringe and a stereotactic frame. The cells were allowed to form brain tumors for 2 weeks, and tumor-bearing mice were subsequently treated with aldoxorubicin (aldoxo) (24 mg/kg of body weight by tail vein injection) (23). Seven days after treatment, mice were sacrificed, and brain tumors were extracted, formalin fixed, paraffin embedded, and sectioned. (A to C) Immunofluorescence detection of IRS-1 (anti-IRS-1 rabbit polyclonal antibody, catalog no. 06-248; Millipore) in brain tumor sections from aldoxorubicin-treated (A) and control vehicle-treated (B) mice and in tumor-free brain tissue from an aldoxorubicin-treated mouse (C). (D) Frequency of tumor cells positive for IRS-1 nuclear structures. The images were evaluated by using high-magnification confocal microscopy (original magnification, ×100). Three tumors per group were evaluated, in which 100 randomly selected fields per tumor were examined (n = 3). Data represent average values ± standard deviations. (E) High-magnification image of a single tumor cell from an aldoxorubicin-treated mouse in which anti-IRS-1 antibody recognized the ringlike structure. The same cell is also visualized by Nomarski contrast, and nuclei are labeled with DAPI (blue fluorescence). The rectangle indicates an IRS-1-positive nuclear structure, and the arrow points to the three-dimensional reconstruction of the IRS-1 ringlike structure. The image was acquired by using an FV1000 confocal microscope (Olympus), and the 3-D surface reconstruction was generated by using SlideBook 5 software (Intelligent Imaging Innovations).
Induction of IRS-1 nuclear structures in cell culture.
Since IRS-1 nuclear structures are relatively rare in brain tumor tissues, and therefore are difficult to study, we attempted to induce their formation in LN-229 glioblastoma cells following the ectopic expression of IRS-1 cloned in frame with a nuclear localization signal (pALS1-NLS-IRS-1/mycTag). Following immunolabeling with either anti-IRS-1 (Fig. 3A) or anti-myc tag (Fig. 3B) antibodies, some of the transfected cells contained highly organized ringlike structures, which varied in size from 0.2 μm to up to 2 μm in diameter. Interestingly, these nuclear structures can be detected without immunolabeling by using Nomarski contrast (Fig. 3C, arrows). Using the same vector, we asked if the ectopic expression of NLS–IRS-1 could trigger the formation of these nuclear structures in different cell types. The results in Fig. 3D show that the transient expression of the NLS–IRS-1–myc tag transgene also triggered the formation of the IRS-1 nuclear structures in U87MG, another glioblastoma cell line, as well as normal human astrocytes (NHAs), mouse embryo fibroblasts (MEFs), and HeLa cells (Fig. 4). These data demonstrate that the overexpression of IRS-1 in the nucleus is sufficient to trigger the formation of IRS-1 nuclear structures in five different cell types tested so far.
FIG 3.
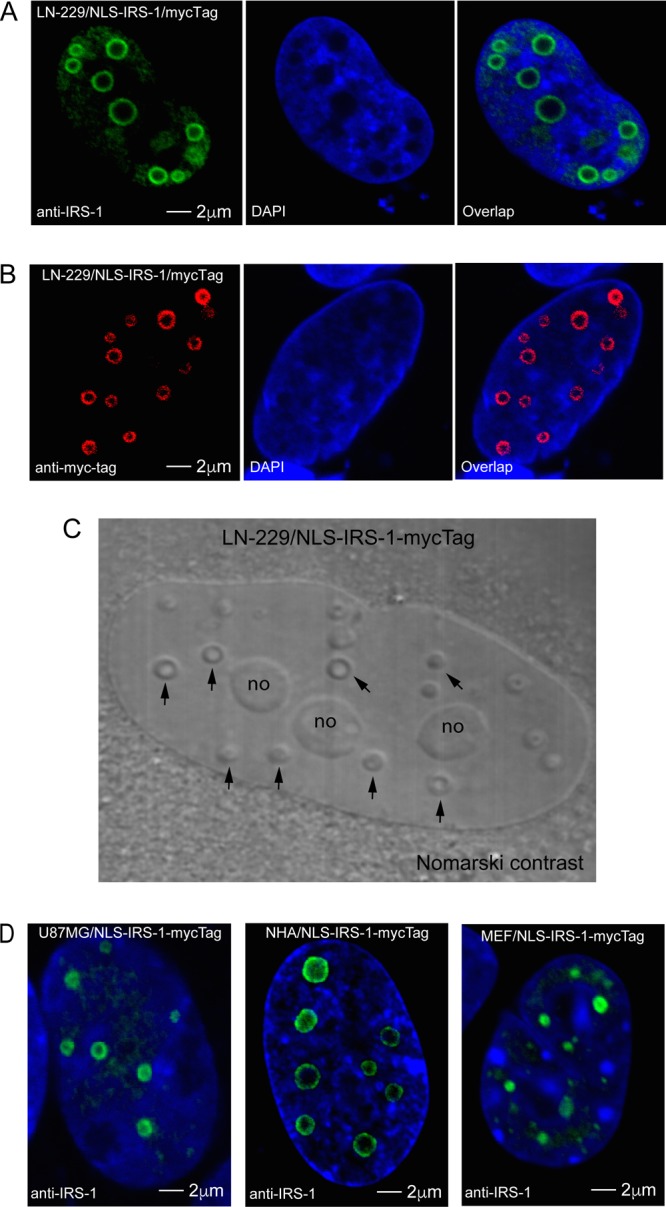
Detection of ringlike structures in nuclei of cells expressing IRS-1 fused with a nuclear localization signal (NLS). (A and B) Confocal images of LN-229 human glioblastoma cells transiently transfected with the construct in which the CMV promoter drives the expression of human IRS-1 cDNA cloned in frame with the NLS and a myc tag (pALS1-NLS-IRS-1/mycTag). The unique nuclear ringlike structures were detected by either anti-IRS-1 rabbit polyclonal antibody (green fluorescence) (A) or anti-myc tag mouse monoclonal antibody (red fluorescence) (B), and the nuclei are labeled with DAPI (blue fluorescence). (C) Detection of IRS-1 ringlike structures by Nomarski contrast (arrows) in LN-229 cells expressing the pALS1-NLS-IRS-1/mycTag construct. In the same image, nucleoli are indicated by “no.” (D) Similar ringlike structures were also observed in another human glioblastoma cell line, U87MG; in normal human astrocytes (NHA); and in mouse embryo fibroblasts (MEF), all of which were transfected (transiently) with the same pALS1-NLS-IRS-1/mycTag plasmid vector.
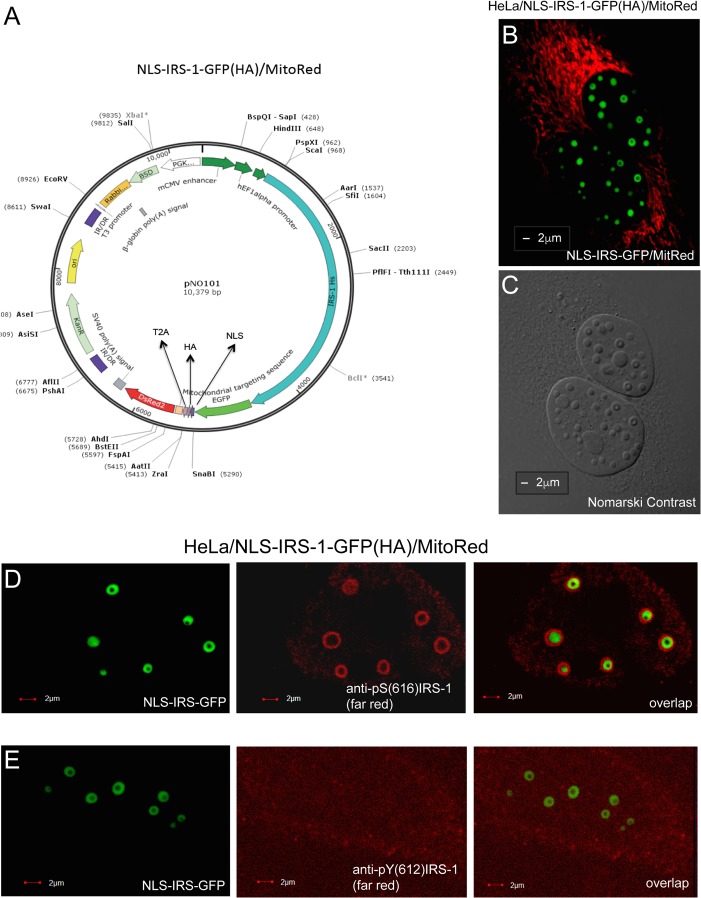
FIG 4.
Detection of IRS-1 ringlike structures in living cells. (A) Schematic illustration of the NLS–IRS-1–GFP/MitoRed expression vector in which human IRS-1 cDNA is cloned in frame with green fluorescent protein (GFP), an NLS, an HA tag, and MitoRed. Since the MitoDsRed2 sequence was inserted behind the T2A sequence, which encodes a viral self-cleaving peptide, this plasmid generates two independent proteins, NLS–IRS-1–GFP(HA) and MitoRed, from the same translation product. (B and C) Representative confocal images of live HeLa cells that stably express the NLS–IRS-1–GFP/MitoRed plasmid. IRS-1 ringlike structures were detected by either EGFP-associated green fluorescence (B) or Nomarski contrast (C). The red fluorescence in panel B indicates mitochondria (MitoRed). (D and E) HeLa clones expressing NLS–IRS-1–GFP/MitoRed were additionally immunolabeled with different anti-IRS-1 antibodies: anti-IRS-1(pS616) mouse monoclonal antibody (catalog no. 3193S; Cell Signaling Technology) (which recognizes the pS616 residue in the human IRS-1 molecule) (D) and anti-IRS-1(pY612) rabbit polyclonal antibody (catalog no. 44816G; Thermo Fisher Scientific) (which recognizes pY612 in human IRS-1) (E). The FarRed-conjugated secondary antibodies were utilized to avoid possible interference from MitoRed fluorescence. IR/DR, inverted repeat/direct repeat sequence; BSD, blasticidin S deaminase gene; PGK, phosphoglycerate kinase promoter; mCMV, murine cytomegalovirus early promoter; Hs IRS1, Homo sapiens IRS1 gene.
To enable analyses of IRS-1 nuclear structures in live cells, we constructed a new vector that enables the expression of two fluorescent proteins: NLS–IRS-1–GFP(HA) and MitoDsRed (Fig. 4A). The first protein is a fusion of IRS-1 with enhanced green fluorescent protein (EGFP), the nuclear localization signal, and a hemagglutinin (HA) epitope tag. The second protein is a fusion of red fluorescent protein (RFP) (DsRed) with a mitochondrial localization signal. Both proteins are separated by a self-cleaving T2A peptide that allows the segregation of these polypeptides during translation (27). We have selected HeLa cells stably expressing these proteins (HeLa/CL006). The confocal images in Fig. 4B and C show examples of two HeLa cells expressing the NLS–IRS-1–GFP/MitoRed vector in which IRS-1 nuclear structures are visualized by either GFP fluorescence (Fig. 4B) or Nomarski contrast (Fig. 4C). Using these cells, we also evaluated the phosphorylation status of IRS-1 molecules contained within these new nuclear structures. To achieve this, we used antibodies that recognize either serine (S616, S636, S639, and S794) or tyrosine (Y612) phosphoamino residues of IRS-1. The results in Fig. 4D demonstrate that the anti-IRS-1 antibody recognizing phosphorylated serine 616 of IRS-1 colocalizes exclusively with the outer part of the ring formed by NLS–IRS-1–GFP and does not overlap the GFP signal. This unexpected pattern of immunolabeling could indicate that multiple IRS-1 molecules forming the structure are organized in a way such that serine-containing residues are exposed outside, and the anti-IRS-1(pS616) antibody binds exclusively to the surface of the structure. Other tested phosphoserine-specific IRS-1 antibodies also bound preferentially to the outer part of the ring but with a somewhat lower affinity (not shown). In contrast, anti-IRS-1(pY612) did not recognize these structures (Fig. 4E). In summary, the presented results indicate that the pool of IRS-1 molecules that build the structures are serine phosphorylated, and serine-containing residues localize most likely at the periphery of the structures.
Dynamic properties of IRS-1 nuclear structures.
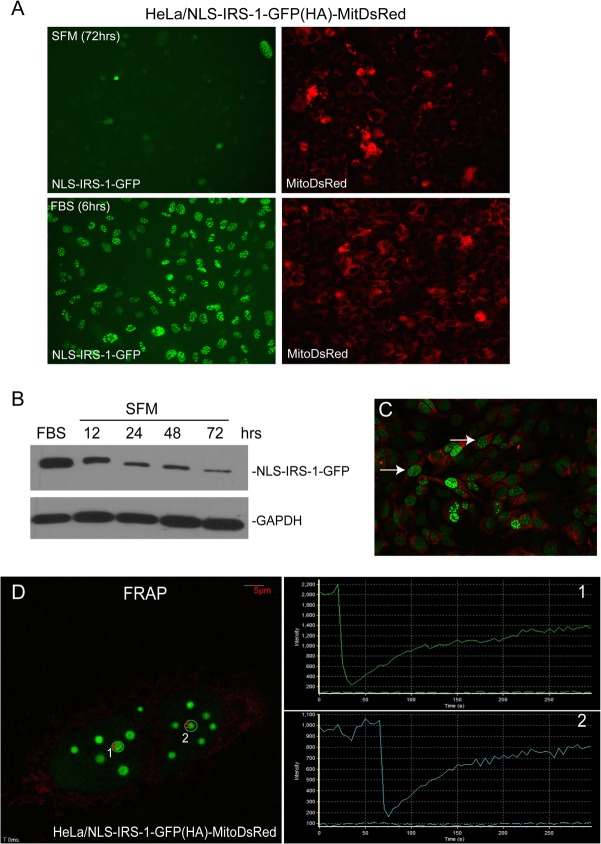
HeLa cells stably expressing the pAL-NLS-IRS-1-GFP/MitoRed transgene (Fig. 4A) allowed us to observe dynamics of IRS-1 nuclear structures in live cells (Fig. 5). Using time-lapse photography, we observed a quick (in a range of minutes) disassembly of the structures during mitosis (Fig. 5C; see also Movie S1 in the supplemental material) or following 72 h of continuous growth in serum-free medium (SFM) (Fig. 5A and Movie S2). They reassembled (in minutes) after cytokinesis (Fig. 5C and Movie S1) or when the serum-starved cells were stimulated again (Fig. 5A and Movie S2). After treatment of growth stimuli (Movie S2), the cells required between 3 and 6 h for a full reassembly of the structures after starvation. Interestingly, the cells that lost the structures in SFM were still positive for MitoDsRed fluorescence (Fig. 5A). The Western blot depicted in Fig. 5B demonstrates a significant decrease in the level of the NLS–IRS-1–GFP fusion protein in SFM over time. This decrease in the NLS–IRS-1–GFP protein level could contribute, at least partially, to the observed disassembly of the structures in SFM.
FIG 5.
Dynamics of IRS-1/LC3 nuclear structures determined in HeLa clones stably expressing NLS–IRS-1–GFP/MitoRed. (A) Cells were starved in serum-free medium (SFM) for 72 h and subsequently stimulated with 10% FBS for 6 h. Both EGFP fluorescence and MitoRed fluorescence were registered. The time-lapse images were acquired every 30 min for 6 h by using a VivaView LCV110U instrument (Olympus). The presented images show the starting point (SFM at 72 h) and the 6-h time point following FBS stimulation (FBS at 6 h) of the 24-h time-lapse experiment (see Movie S3 in the supplemental material). (B) Western blot analysis indicating IRS-1 protein levels following stimulation with 10% FBS for 6 h and following serum starvation in SFM for 12, 24, 48, and 72 h. (C) Time-lapse images were taken every 10 min for 12 h following serum stimulation (10% FBS). Two cells indicated by arrows are positive for IRS-1 ringlike structures and are undergoing mitotic division, as shown by the time-lapse experiment depicted in Movie S2. (D) Fluorescence recovery after photobleaching (FRAP). Shown are representative fluorescent images of two areas selected for photobleaching (two white circles labeled 1 and 2). The two plots on the right side of the image show increasing intensities of fluorescence after photobleaching obtained from areas 1 and 2, respectively. Time-lapse analysis of FRAP is provided in Movie S3.
To test the mobility of IRS-1 molecules within IRS-1 nuclear structures, we carried out fluorescence recovery after photobleaching (FRAP). In the FRAP experiment, a defined area of the IRS-1 nuclear structure was bleached irreversibly by a single, high-powered spot laser pulse. The recovery of the fluorescent signal in the bleached area is the consequence of the replacement of the GFP fusion protein from the surrounding nucleoplasm. The average FRAP half-time of 18.44 ± 4.4 s (n = 8) (Fig. 5D and Movie S3) indicates a rapid movement of the NLS–IRS-1–GFP molecules into and out of the nuclear structure, indicating their highly dynamic nature.
Interactions between IRS-1 nuclear structures and other nuclear suborganelles.
The IRS-1 nuclear structures share the nucleoplasm with an array of other nuclear bodies, including nucleoli, Cajal bodies, PML bodies, Polycomb bodies, speckles, and paraspeckles. Our results show that IRS-1 nuclear structures are most likely unique; do not overlap nucleoli (Fig. 6A) or other tested nuclear suborganelles, including Cajal bodies (Fig. 6B) and nuclear speckles (Fig. 6C); and do not contain large quantities of double-stranded RNA (dsRNA), as indicated by the lack of pyronin Y labeling (Fig. 6A). We also found a particular relationship between IRS-1 nuclear structures and PML bodies, in which a single PML body interacts with the surface of a single IRS-1 structure (Fig. 6D).
FIG 6.
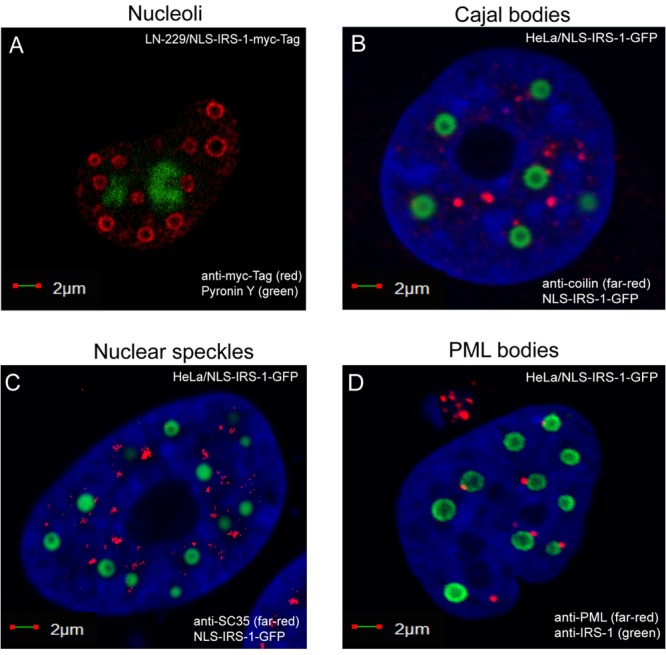
Positioning of the IRS-1 nuclear structures with respect to other nuclear suborganelles. (A) Confocal images of LN-229 cells expressing the NLS–IRS-1–myc tag construct. The cells were fixed, immunolabeled with anti-myc tag antibody (red), and labeled with pyronin Y to detect nucleolar RNA (green fluorescence). (B to D) HeLa cells expressing the NLS–IRS-1–GFP construct. The cells were additionally immunolabeled with anticoilin antibody (marker of Cajal bodies) (red fluorescence) (B), anti-SC35 antibody (marker of nuclear speckles) (red fluorescence) (C), or anti-PML antibody (marker of PML bodies) (red fluorescence) (D).
Detection of LC3 inside IRS-1 nuclear structures.
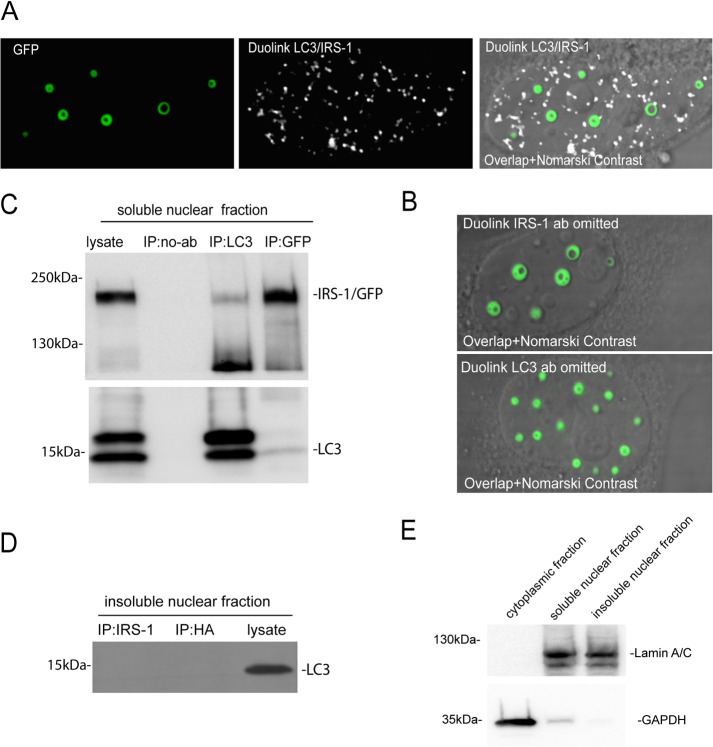
Exponentially growing HeLa clones expressing the NLS–IRS-1–GFP/MitoRed construct are very efficient in forming IRS-1 nuclear structures, which enabled the preparation of nuclear extracts enriched with these structures. Although mass spectrometry (MS) analyses based on these enriched nuclear fractions demonstrated a high degree of contamination (data not shown), we selected and tested several candidate proteins by immunocytofluorescence and immunoprecipitation. The results in Fig. 7 demonstrate that one of the tested proteins, LC3, localized within the IRS-1 nuclear structure. LC3 is a key autophagy initiator (28, 29) that has also been detected in the nucleus (30). In this experiment, LN-229 cells expressing the NLS–IRS-1–myc tag fusion protein and double immunolabeled with anti-myc tag mouse monoclonal antibody (red) and anti-LC3 rabbit polyclonal antibody (green) revealed that LC3 is localized inside the structure (Fig. 7A). We confirmed this result in HeLa cells expressing the NLS–IRS-1–GFP/MitoRed construct. In these cells, IRS-1 nuclear structures were visualized by EGFP fluorescence, and LC3 was detected by using an anti-LC3 rabbit polyclonal antibody (red) (Fig. 7B). It should be emphasized that our attempts to detect other autophagy proteins or biological membranes within the IRS-1/LC3 complexes were negative (data not shown). Importantly, we were also able to detect LC3 inside IRS-1 nuclear structures in human glioblastoma clinical samples (Fig. 7C). In these archival samples, the structures have a different appearance than in cell culture (Fig. 7A and B), which may be a result of tissue processing and/or the quality of the archival brain tumor clinical samples. In fact, we found IRS-1 ringlike nuclear structures in fresh tissue explants form mouse glioblastoma xenografts (Fig. 2E).
FIG 7.
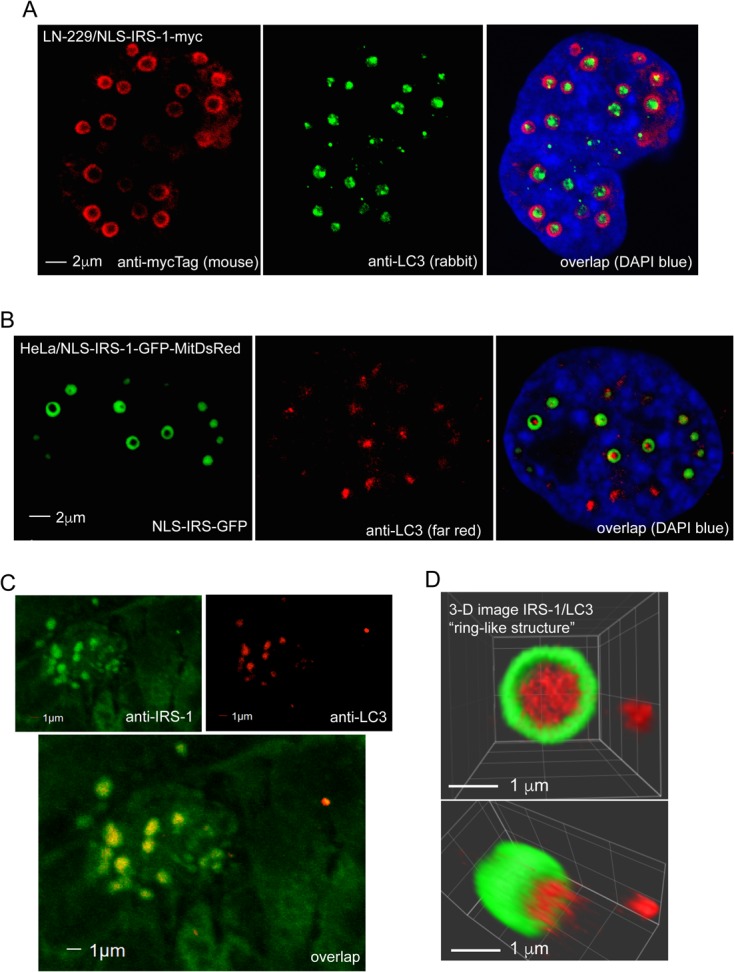
Detection of LC3 in IRS-1 ringlike structures. (A) Confocal images of two LN-229 human glioblastoma cells expressing the NLS–IRS-1–myc tag fusion protein (pALS1-NLS-IRS-1/mycTag). Double immunolabeling was performed using by anti-myc tag mouse monoclonal antibody (red) and anti-LC3 rabbit polyclonal antibody (green). DAPI labeling was used to visualize nuclei (blue). LC3 immunolabeling (green) is localized inside the ringlike structure immunolabeled with anti-myc tag antibody (red). (B) Single immunolabeling with anti-LC3 rabbit polyclonal antibody (red) was utilized to detect IRS-1/LC3 structures in HeLa cells stably expressing the NLS–IRS-1–GFP construct (see the legend to Fig. 4). To avoid potential spectral overlap (fluorescence bleedthrough), which in the case of the IRS-1–GFP nuclear rings can also excite the red channel, we used a secondary antibody conjugated with FarRed (Alexa Fluor 647). (C) Confocal images of IRS-1/LC3 nuclear structures found in glioblastoma biopsy specimens. Formalin-fixed, paraffin-embedded sections were immunolabeled with anti-IRS-1(pS612) mouse monoclonal (green) and anti-LC3 rabbit polyclonal (red) antibodies, and colocalization between IRS-1 and LC3 was evaluated by using confocal imaging in combination with SlideBook 5 Mask operation software. (D) Three-dimensional reconstruction of the IRS-1/LC3 nuclear ringlike structure from panel B. The image was acquired by using an Olympus FV1000 confocal microscope and processed by using SlideBook 5 software (3-D volume reconstruction).
Three-dimensional (3-D) confocal reconstruction of a single IRS-1/LC3 structure revealed a cylinder-like shape in which LC3 is “encapsulated” by the IRS-1 overlay (Fig. 7D). This 3-D image also suggests that IRS-1 and LC3 may not bind directly to each other, since the overlap between red fluorescence (LC3) and green fluorescence was quite low inside the structures (4.6% ± 0.3% overlap between red and green pixels). Moreover, a proximity ligation assay (PLA) (DuoLink; Sigma), which can recognize two molecules separated by about 30 nm, failed to detect IRS-1/LC3 interactions inside the structures (Fig. 8A). Instead, we detected multiple IRS-1/LC3 complexes within the nucleoplasm outside the structures (Fig. 8A, white nuclear fluorescence). These IRS-1/LC3 complexes were not detected when either anti-IRS-1 or anti-LC3 antibody was omitted (Fig. 8B) (DuoLink negative controls). We confirmed this direct interaction between IRS-1 and LC3 using reciprocal immunoprecipitation (IP)/Western blotting and soluble nuclear fractions (Fig. 8C). In contrast, IP/Western analyses performed with insoluble nuclear fractions, which are also highly enriched with the IRS-1/LC3 nuclear structures, were negative (Fig. 8D). These results support our data from the proximity ligation assay indicating a direct interaction between IRS-1 and LC3 in the nucleoplasm outside the IRS-1/LC3 structures (Fig. 8A). This binding could be mediated by the LC3-interacting region (LIR) that we identified in the IRS-1 phosphotyrosine binding (PTB) domain (amino acids [aa] 162 to 167) of human IRS-1 (31). On the other hand, the mechanism by which IRS-1 and LC3 molecules interact within the IRS-1/LC3 nuclear structures seems to be indirect. Finally, Fig. 8E shows control Western blots that demonstrate very low cross-contamination between the subcellular fractions used in this experiment.
FIG 8.
Characterization of the IRS-1/LC3 nuclear interaction. (A) Proximity ligation assay of the IRS-1/LC3 nuclear interaction. In this assay, anti-HA tag mouse monoclonal or anti-IRS-1 mouse monoclonal antibodies were used in combination with anti-LC3 rabbit polyclonal antibody in HeLa cells stably expressing the NLS–IRS-1–GFP(HA)/MitoRed construct. Subsequently, DNA-bound secondary anti-mouse and anti-rabbit antibodies were used as a substrate for DNA ligation, amplification, and in situ hybridization, all of which were performed according to the manufacturer's recommendations (DuoLink; Sigma). Green fluorescence associated with the NLS–IRS-1–GFP fusion protein indicates positions of IRS-1 nuclear structures, and white fluorescence indicates sites in which nuclear IRS-1 and LC3 are in proximity to each other (20 to 30 nm). These DuoLink-positive sites are abundant inside the nucleus but overlap the IRS-1 nuclear structures only rarely. (B) Control DuoLink reactions in which the indicated primary antibodies (ab) were omitted. (C and D) Reciprocal immunoprecipitation/Western blot analyses (IP/W) based on HeLa/NLS-IRS-I/GFP(HA)/MitoRed cells from which two nuclear fractions were prepared: a nuclear fraction enriched with IRS-1/LC3 ring structures (insoluble nuclear fraction) and the remaining nucleoplasm from which IRS-1 nuclear rings were extracted (soluble nuclear fraction). Note that reciprocal binding between IRS-1 and LC3 was detected exclusively in the soluble nuclear fraction. In this experiment, anti-GFP mouse monoclonal (catalog no. sc-9996; Santa Cruz Biotechnology) and anti-LC3 rabbit polyclonal (catalog no. M152-3; MBL) antibodies were used for immunoprecipitation, and Western blots were probed with anti-GFP mouse monoclonal (catalog no. sc-9996; Santa Cruz Biotechnology) and anti-LC3 rabbit polyclonal (catalog no. 12741S; Cell Signaling Technology) antibodies. (D) IP/Western analysis of the insoluble nuclear fraction. We used anti-IRS-1 rabbit polyclonal (catalog no. 06-248; Millipore) and anti-HA rabbit polyclonal (catalog no. sc-805; Santa Cruz Biotechnology) antibodies for immunoprecipitation, and the corresponding Western blot was developed with anti-LC3 rabbit polyclonal antibody (catalog no. 12741P; Cell Signaling Technology). Note the absence of an interaction between IRS-1 and LC3 in the insoluble nuclear fraction enriched with the IRS-1/LC3 nuclear structures. (E) Control Western blot to evaluate potential cross-contamination between subcellular fractions. We used the procedure described in Materials and Methods to obtain cytosolic as well as soluble and insoluble nuclear fractions. We used anti-GAPDH (catalog no. sc-47724; Santa Cruz Biotechnology) and anti-lamin A/C (catalog no. 2032; Cell Signaling Technology) antibodies to evaluate the purities of the cytosolic and nuclear fractions, respectively.
IRS-1/LC3 nuclear structures inhibit starvation-induced and rapamycin-induced autophagy.
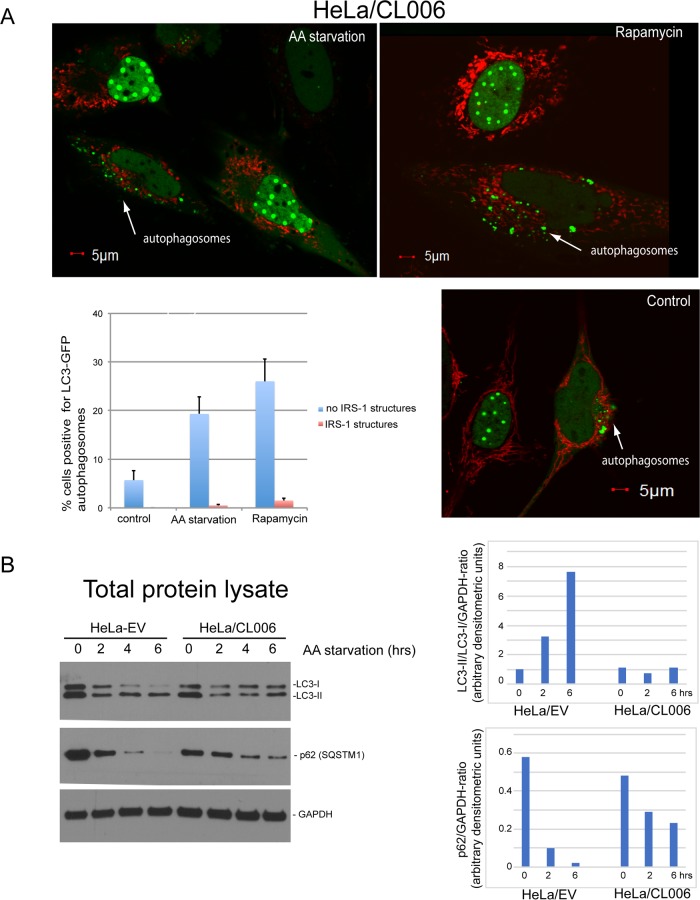
It was recently reported that nuclear LC3 is directly involved in starvation-induced autophagy (32), and our data show that LC3 can be sequestered inside the IRS-1-containing nuclear structures (Fig. 7). These data encouraged the idea that the IRS-1/LC3 nuclear structures may function as a new autophagy control mechanism. The results in Fig. 9 show that HeLa cells expressing the NLS–IRS-1–GFP(HA)/MitoRed transgene (clone 006) were capable of forming autophagosomes only when the IRS-1/LC3 nuclear structures were not assembled. In this experiment, autophagy was stimulated by either rapamycin treatment or amino acid starvation, and autophagosomes were detected by using the baculovirus-mediated expression of an LC3-GFP reporter (33). In all experiments, we used 70 to 90% confluent cultures grown in 10% fetal bovine serum (FBS) for 2 to 3 days. Under these conditions, we usually see about 60% of cells positive for IRS-1/LC3 nuclear structures and about 40% of cells with diffuse green nuclear fluorescence in the absence of the structures. This is in spite of the fact that the transgene (NLS–IRS-1–GFP) is constitutively expressed (Fig. 4A), and nearly 100% of the cells (clone 006) are still capable of assembling the structures after restimulation with fresh FBS (see Movie S2 in the supplemental material). This particular feature of the model allowed us to quantify autophagy in IRS-1/LC3-positive and IRS-1/LC3-negative cells in the same culture. The results in Fig. 9A show autophagosomes in 5.7% of the cells without the induction of autophagy (control), and all autophagosome-positive cells were negative for IRS-1/LC3 nuclear structures (arrows). In cells exposed to amino acid starvation, autophagosomes were detected in 19.3% of IRS-1/LC3-negative cells and in 0.5% of IRS-1/LC3-positive cells (Fig. 9A, histogram). Finally, cells exposed to rapamycin demonstrated autophagosomes in 26.1% of IRS-1/LC3-negative cells and in 1.5% of IRS-1/LC3-positive cells (Fig. 9A, histogram). These data indicate a strong inverse correlation between the presence of IRS-1/LC3 nuclear structures and the formation of autophagosomes.
FIG 9.
Inhibition of autophagy by IRS-1/LC3 nuclear structures. (A) Exponentially growing (10% FBS) HeLa cells stably expressing the NLS–IRS-1–GFP(HA)/MitoRed transgene (HeLa/CL006) were additionally transduced with the baculovirus-based LC3-GFP reporter (to detect autophagosomes), and autophagy was stimulated by either amino acid (AA) starvation or rapamycin treatment. Note that autophagosomes (arrows and green cytosolic vesicles) are detected almost exclusively in cells negative for IRS-1 nuclear structures (green nuclear structures). Red cytosolic fluorescence indicates mitochondria (MitoRed). The histogram demonstrates the quantification of cells positive for autophagosomes among cells that are either positive or negative for IRS-1/LC3 nuclear structures. Data represent average values with standard deviations (n = 3; at least 500 cells were counted per experiment). (B) Western blot analyses of HeLa cells transfected with the empty vector (EV) and HeLa cells expressing NLS–IRS-1–GFP(HA)/MitoRed (HeLa/CL006). The cells were exposed to amino acid starvation for 0, 2, and 6 h, and the corresponding blots were probed with anti-LC3 and anti-p62 rabbit polyclonal antibodies, respectively. Anti-GAPDH antibody was used as a loading marker. The histograms show quantifications of the corresponding blots using ImageJ. Data represent arbitrary densitometry units for LC3-II/LC3-I ratios and p62 levels normalized by the GAPDH loading control.
We also asked how the presence of the IRS-1/LC3 nuclear structures affects two molecular markers of autophagy: the LC3-II/LC3-I ratio, which is expected to increase in cells undergoing autophagy, and the stability of p62(SQSTM1), which is expected to rapidly decline during autophagy. The results in Fig. 9B demonstrate that in comparison to HeLa cells expressing an empty vector (HeLa/EV), cells with IRS-1/LC3 nuclear structures (HeLa/CL006) demonstrated a practically unchanged LC3-II/LC3-I ratio and more stable p62(SQST1) following amino acid starvation. This further confirms that cells positive for IRS-1/LC3 nuclear structures have attenuated autophagy.
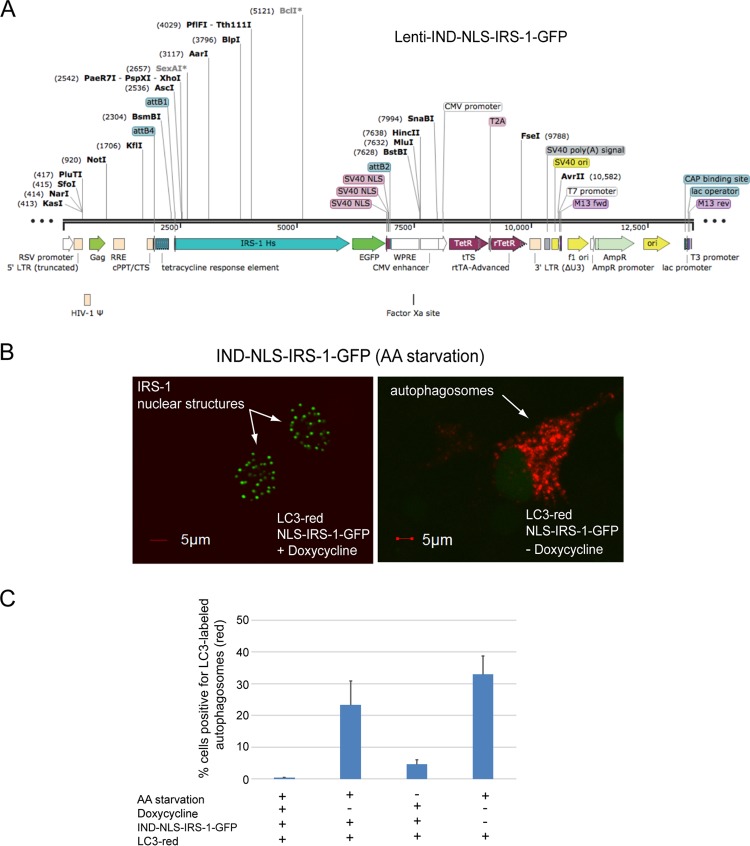
We have confirmed this result with the inducible (Tet-on) expression of nuclear IRS-1 (IND–NLS–IRS-1–GFP) using a single-vector lentiviral system (Fig. 10A). The results in Fig. 10B demonstrate that HeLa cells expressing this construct are capable of forming IRS-1 nuclear structures following doxycycline treatment (arrows and green fluorescence). In this experiment, the cells were additionally transduced with the LC3-red baculovirus system (Molecular Probes, MA) to monitor the de novo formation of autophagosomes (Fig. 10B, arrow and red fluorescence). Following doxycycline treatment (for 24 h) and amino acid starvation (for 4 h), HeLa cells transduced with the LC3-red and/or the IND–NLS–IRS-1–GFP vector were analyzed by confocal imaging. We observed red autophagosomes almost exclusively in cells negative for IRS-1 nuclear structures (Fig. 10B). Quantitatively, we detected autophagosomes in 0.41% ± 0.05% (n = 4; at least 100 cells per experiment were counted) of cells positive for IRS-1 nuclear structures (Fig. 10C, first bar) and in 23.3% ± 7.7% of cells negative for IRS-1 nuclear structures in the same culture (Fig. 10C, second bar). In the absence of amino acid starvation, we detected autophagosomes in 4.6% ± 1.4% of cells, and all cells were negative for IRS-1 nuclear structures (Fig. 10C, third bar). Finally, amino acid starvation in the absence of IRS-1 nuclear structures (Fig. 10C, fourth bar) resulted in 32.9% ± 5.8% of cells with autophagosomes. These data confirmed a reverse correlation between the presence of IRS-1 nuclear structures and the formation of autophagosomes and imply that the retention of LC3 in the nucleus may play a role in this process.
FIG 10.
Inducible expression of IRS-1 nuclear structures inhibits autophagy. (A) Linear map of the inducible (Tet-on) lentivirus single-vector system (Lenti-IND-NLS-IRS-1-GFP). LTR, long terminal repeat. (B) Confocal images demonstrating inducible expression of nuclear IRS-1 in the presence of an autophagy marker, LC3-RFP protein expressed from baculovirus (Molecular Probes). HeLa cells expressing these two constructs were cultured in the presence or absence of amino acid (AA) starvation and in the presence or absence of doxycycline treatment (1,000 nM). (C) Quantification of data from the experiment in panel B. The histogram illustrates the percentages of cells positive for LC3-labeled autophagosomes (red fluorescence) in cells that were either positive or negative for IRS-1 nuclear structures (green fluorescence). Data represent average values (± standard deviations) from 4 independent experiments in which at least 1,000 cells per experiment were counted (n = 4). RSV, Rous sarcoma virus; RRE, Rev response element; cPPT/CTS, central polypurine tract/central termination sequence; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; CAP, catabolite activator protein.
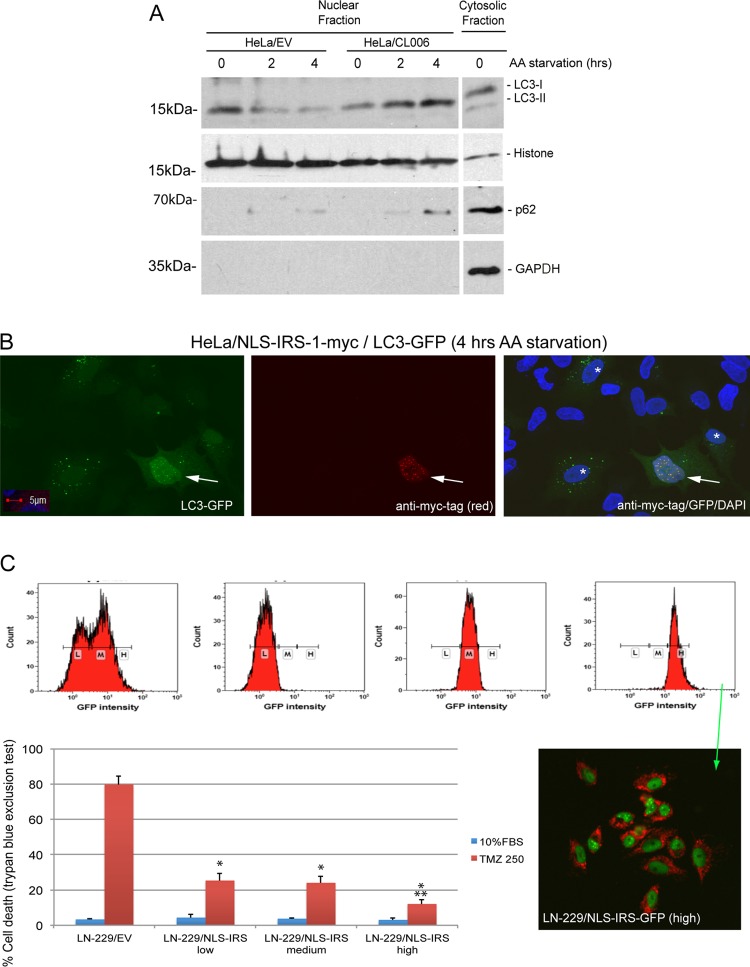
To test this possibility, we isolated nuclear extracts form HeLa/EV and HeLa/CL006 cells 0, 2, and 4 h following amino acid starvation. The Western blot analysis in Fig. 11A demonstrates an apparent presence of LC3 in the nucleus and its gradual decline following amino acid starvation. In contrast, HeLa/CL006 cells demonstrated a gradual accumulation of LC3 in the nucleus following amino acid starvation. Interestingly, we also detected p62(SQSTM1) in the nuclear fraction of HeLa/CL006 cells 4 h after starvation (Fig. 11A). This could be relevant to our study since p62(SQSTM1) was recently found in PML bodies (34), and our data demonstrate a direct interaction between IRS-1/LC3 nuclear structures and PML bodies (Fig. 6D). Finally, we also observed an apparent accumulation of nuclear LC3 in HeLa cells but only in the absence of autophagosomes (Fig. 11B, arrow). In this experiment, the cells were exposed to amino acid starvation following the coexpression of pALS-NLS-IRS-1/mycTag and LC3-GFP. This further supports our hypothesis that the retention of LC3 inside the IRS-1 nuclear structures is directly involved in the inhibition of autophagy.
FIG 11.
Accumulation of LC3 in the nucleus in cells positive for IRS-1 nuclear structures is associated with inhibition of autophagy. (A) Western blot analysis of nuclear extracts from HeLa/EV and HeLa/CL006 cells 0, 2, and 4 h following amino acid starvation. The last lane in the blot represents the cytosolic fraction from HeLa/CL006 cells without amino acid starvation (control). The blots were probed with anti-LC3 (Cell Signaling Technology) and anti-p62(SQSTM1) (Santa Cruz). Anti-histone H3 (Cell Signaling Technology) and anti-GAPDH (Santa Cruz Biotechnology) were used as nuclear and cytosolic markers, respectively. (B) Confocal images of HeLa cells cotransfected with the pALS-NLS-IRS-1/mycTag plasmid and the LC3-GFP baculovirus vector. IRS-1 nuclear structures (red fluorescence) were immunolabeled with mouse monoclonal anti-myc tag antibody (catalog number sc-40; Santa Cruz Biotechnology). Both nuclear LC3 and LC3 within autophagosomes are labeled with GFP. Colocalization between nuclear IRS-1 and nuclear LC3 is indicated by an arrow, and cells with autophagosomes are indicated by a white asterisk. Nuclei were counterstained with DAPI (blue fluorescence). (C) Drug resistance detected in tumor cells positive for IRS-1/LC3 nuclear structures. Temozolomide (TMZ)-sensitive LN-229 human glioblastoma cells were transfected with NLS–IRS-1–GFP(HA)/MitoRed and sorted to obtain three mixed populations, expressing low (L), medium (M), and high (H) levels of the transgenes (NLS–IRS-1–GFP and MitoRed). Resistance to temozolomide was tested in monolayer cultures in the presence of 10% FBS with or without 250 μM TMZ. LN-229 cells transfected with an empty vector were used as a control. In both panels, data represent average values ± standard deviations (n = 3). * indicates statistically significant differences between HeLa/EV and HeLa/CL006 cells or between LN-229/EV and LN229/NLS-IRS-1 cells; ** indicates statistically significant differences between LN-229/NLS-IRS-1(low) and LN-229/NLS-IRS-1(high) cells. The inset shows cell sorting and confocal analyses of LN-229 cells expressing the NLS–IRS-1–GFP(HA)/MitoRed transgene. Cells with low, medium, and high levels of the transgene (NLS–IRS-1–GFP) were selected and used for the TMZ resistance assay.
Drug resistance of glioblastoma cells engineered to form IRS-1 nuclear structures.
Drug resistance represents one of the major reasons for the failure of antiglioblastoma therapies, and autophagy may play a role in this process. Therefore, we evaluated whether glioblastoma cells engineered to form IRS-1 nuclear structures become resistant to temozolomide (TMZ), the standard of care for glioblastoma patients (35, 36). The results in Fig. 11C demonstrate that LN-229 human glioblastoma cells, which are TMZ sensitive (37), acquired partial resistance to this anticancer drug after the ectopic expression of NLS–IRS-1. In this experiment, we sorted LN-229 cells transfected with the NLS–IRS-1–GFP/MitoRed construct into three cell subpopulations, expressing low (L), medium (M), or high (H) levels of the transgene (Fig. 11C, inset). Importantly, all LN-229 cells expressing the NLS–IRS-1–GFP transgene demonstrated a survival advantage in the presence of 250 μM TMZ, and the population characterized by the highest level of GFP fluorescence associated with IRS-1 nuclear structures was the most resistant to TMZ (Fig. 11C).
DISCUSSION
In this study, we report a new finding of nuclear structures that consist of at least two protein components, IRS-1, which is a typical signaling molecule (1), and LC3, which is a typical autophagy protein (38). These new nuclear structures were first detected in a limited number of tumor cells from glioblastoma biopsy specimens (Fig. 1) and in glioblastoma xenografts (Fig. 2). We were also able to reproduce them in cell culture by the ectopic expression of IRS-1 cloned in frame with a nuclear localization signal (NLS–IRS-1) (Fig. 3 and 4). This in vitro approach allowed us to conduct a further evaluation of their properties, function, and structure. Using confocal imaging, we demonstrated that cells expressing the NLS–IRS-1 transgene form nuclear structures, which vary in size and can reach up to 2 μm in diameter. They often develop a ringlike appearance in which IRS-1 localizes at the periphery and the center harbors LC3 (Fig. 7). Once the IRS-1/LC3 complexes are formed, they are highly dynamic, as evidenced by the fact that they disassemble during mitosis, reappear shortly after cytokinesis (Fig. 5C; see also Movie S1 in the supplemental material) or when serum-starved cells are stimulated to grow (Fig. 5A and Movie S2), and rapidly exchange IRS-1 molecules with the surrounding nucleoplasm (Fig. 5D and Movie S3). These dynamic properties indicate that the structures could have a specific nuclear function and refute the argument that they are simply aggregates of misfolded/overexpressed proteins destined for degradation.
The fact that we observed these new IRS-1-containing nuclear structures only in a small population of glioblastoma tumor cells suggests that they could be produced by tumor stem cells. In fact, we observed a significant increase in their appearance in glioblastoma-bearing mice following genotoxic treatment (Fig. 2D), conditions under which tumor stem cells usually acquire a growth advantage. However, our preliminary searches indicated that neither CD133-positive nor CD44-positive glioma-initiating cells were positive for IRS-1-containing nuclear structures. We found instead that glioblastoma cells positive for the IRS-1 nuclear structures have elevated levels of Bmi1-containing Polycomb bodies (data not shown). This could be relevant since the repression of the Ink4a-Arf locus by Polycomb repressor complex 1 was closely associated with stemness (39–42). Further experiments are required to verify if the formation of IRS-1/LC3 nuclear structures is indeed associated with a stemlike phenotype.
Another interesting observation is that all IRS-1-containing nuclear structures have a circular shape in the same plane, which indicates that their 3-D images should resemble the shape of a sphere (a sphere is the only 3-D object that can generate a perfect circle in all planes). However, our 3-D confocal image reconstructions generated oval-like or barrel-like images of the IRS-1/LC3 structures (Fig. 7D). This discrepancy could be explained by a well-known phenomenon, the distortion of confocal/fluorescent images along the z axis, which is defined by the point spread function (PSF) (43). Considering this interpretation, the IRS-1/LC3 nuclear structures most likely have the shape of a sphere.
It is well accepted that the membraneless nuclear suborganelles play a fundamental role in nuclear function. However, the molecular mechanism(s) controlling their formation and stability is much less well understood. It was recently proposed that these nuclear suborganelles can be assembled through the phase separation of their specific molecular components, and importantly, they are capable of forming multiple functional compartments within the structure. Recent work by Feric et al. (44) demonstrates that the mixing of specific purified nuclear proteins results in their phase separation into droplets containing noncoalescing liquid phases, which under certain conditions can form multicompartmental sphere-like structures morphologically resembling native nuclear suborganelles even as complex as nucleoli. Considering these new findings, the assembly and stability of our IRS-1-containing nuclear structures could also involve liquid-phase separation based on the immiscibility of their components. In our experimental setting, the formation of the nuclear structures happens within the native nuclear environment and is initiated by the ectopic overexpression of IRS-1 in the nucleus (Fig. 3 and 4). In addition, these IRS-1-containing nuclear structures recruit at least one additional component, LC3, and assemble it in a way such that the outer part consists of serine-phosphorylated IRS-1 molecules and the center harbors endogenous LC3 (Fig. 7). Also, in comparison to ex vivo data (44), our experimental model involves additional intrinsic control mechanisms, which are provided by the native nuclear environment. These mechanisms are responsible for the observed disassembly of the IRS-1/LC3 nuclear structures during mitosis (in minutes) and following prolonged serum starvation (in days) and for their reassembly after cytokinesis is completed (in minutes), and the cells are stimulated to grow (Movies S1 and S2). All these regulatory events happen in spite of the fact that the main stimulus, the accumulation of the NLS–IRS-1 fusion protein, is constitutively delivered (Fig. 4). Therefore, in addition to the specific physical traits associated with liquid-phase separation and the availability of substrates (nuclear IRS-1 and nuclear LC3), there are also intrinsic mechanisms in the nucleus that affect/control the assembly and stability of these new nuclear suborganelles. In addition, negative data from pyronin Y labeling (Fig. 6A) suggest that either RNA is not present or its content is low in IRS-1/LC3 structures.
Highly organized IRS-1/LC3 ringlike structures are formed in vitro following the ectopic overexpression of NLS–IRS-1. However, they have a different appearance in glioblastoma biopsy specimens. In archival tumor samples, the IRS-1/LC3 structures are often less regular, are smaller (up to 1 μm), and show a high degree of colocalization between LC3 and IRS-1 (compare Fig. 7A and B with C and with Fig. 1F and G). These morphological differences may suggest that the structures formed in vitro could be different from those found in archival tissues. However, they may also indicate that these differences could be an artifact of tissue processing, such as formalin fixation, tissue dehydration, paraffin embedding, and subsequent rehydration, which may contribute to the shrinkage and distortion of these delicate membraneless structures. However, on rare occasions, we observed ringlike structures in fresh tumor explants (Fig. 2E).
One obvious clue in determining how the IRS-1-containing nuclear structures may function is the presence of LC3 detected in the core of the structure. LC3, in addition to being a key component of the autophagosome, has also been detected in the nucleus (29, 30), yet autophagy inside the nucleus has never been reported. Indeed, when the IRS-1/LC3 structures were tested for the presence of other autophagy proteins or biological membranes, the results were negative (data not shown). This challenged our initial thought that the IRS-1/LC3 structures may act as putative nuclear autophagosomes. Instead, our data indicate that the IRS-1/LC3 nuclear structures are involved in the regulation of autophagy in the cytoplasm, most likely by controlling a nuclear pool of LC3. It was recently proposed that nuclear LC3 is both directly involved in and indispensable for starvation-induced autophagy (32). Our data support this mechanism and provide an additional, so-far-unknown level of autophagy control.
In summary, this is the first demonstration of nuclear structures that contain two common cytoplasmic molecules, IRS-1 and LC3. They are highly dynamic and may play a critical role in controlling autophagy. Further studies are necessary to determine their complete molecular composition and structure and to explain how the temporal appearance and disappearance of these nuclear structures could link autophagy with the resistance of tumor cells to stress, including the resistance of glioblastoma cells to temozolomide (Fig. 11C).
MATERIALS AND METHODS
Human brain tumor biopsy specimens.
A commercially available brain tumor tissue microarray was purchased from US Biomax, Inc. (Rockville, MD). The slide (GL803a) consisted of 80 cores from 80 different cases, which included low-grade diffuse astrocytomas (10 cases), anaplastic astrocytomas (9 cases), glioblastomas (33 cases), oligodendrogliomas (6 cases), anaplastic oligodendrogliomas (4 cases), mixed oligoastrocytomas (3 cases), medulloblastomas (3 cases), ependymomas (2 cases), anaplastic ependymomas (2 cases), normal brain tissue (5 cases), and melanoma (1 case). The tissues were previously fixed in formalin and embedded in paraffin. Following hematoxylin-and-eosin (H&E) staining the tumors were reevaluated by a neuropathologist (L. Del Valle) for accurate diagnosis and quality of the tissue. Using this additional evaluation, we selected 64 brain tumor tissues and evaluated them for IRS-1 subcellular localization (Table 1 and Fig. 1).
Cell culture.
The cell culture and animal experiments were based on a human glioblastoma cell line, LN-229 (ATCC CRL-2611), and two glioblastoma isolates, GBM12 ARG (TMZ sensitive) and GBM12 TMZ (TMZ resistant). GBM12 cells were kindly provided by Jann Sarkaria (Mayo Clinic, Rochester, MN) and are routinely propagated in subcutaneous tissue of immunodeficient mice. These cells were used for both intracranial xenografts and short-term cell culture experiments (20). In addition, HeLa cells (cervical adenocarcinoma; ATCC CCL-2) were used for the large-scale production and purification of the IRS-1 nuclear structures and for analyses of IRS-1/LC3 nuclear structures in living cells. We also employed another human glioblastoma cell line, U87MG (ATCC HTB-14); MEFs (45); and NHAs (Lonza/Clonetics, Walkersville, MD).
Intracranial tumor growth.
All described procedures were performed according to an approved LSU Health Sciences Center (LSUHSC) IACUC protocol (IACUC protocol no. 3444). Foxn1nu immunodeficient mice (Charles River) were anesthetized with 4% isoflurane, and the depth of anesthesia was monitored by toe and tail pinch reflexes. The temperature was maintained at 36.6°C to 37.5°C with a homeothermic blanket throughout the procedure. Mice were placed into a stereotactic frame (Stoeliting), and a 2- to-3 mm incision was made on the head just to the right of the midline. A small burr hole (0.45 mm) was made in the skull by using a small dental drill. Mice were inoculated with 1 × 105 GBM12 glioblastoma cells in 2 μl phosphate-buffered saline (PBS) at a rate of 0.8 μl/min by using a microprocessor-controlled injector (Harvard Apparatus) with the following coordinates: 2 mm posterior to the bregma and 1.5 mm lateral to the sagittal suture, at a depth of 3.4 mm into the brain parenchyma. After inoculation, the skin was sealed with 3M Vetbond tissue-adhesive glue. The mice were kept warm until they recovered from anesthesia. During the procedure and recovery, mice were continuously monitored for any signs of distress. At the endpoint of each experiment (20% weight loss), or when mice exhibited signs of pain or distress, according to IACUC tumor endpoint policy, mice were euthanized by CO2 inhalation.
Expression vectors.
We constructed three different expression vectors for this study. The initial expression vector pALS1-NLS-IRS-1/mycTag is based on the pCMV/myc/nuc plasmid (catalog no. 35-0838; Invitrogen), which contains a human cytomegalovirus (CMV) promoter, three nuclear localization signals (3× DPKKKRKV), a myc tag, an SV40 poly(A) signal, and neomycin resistance. The fragment between NcoI and NotI of this vector was replaced with the PshAI site by using an annealed 5′-phosphorylated pair of oligonucleotides (/5Phos/CATGGGACTAGCGTC and /5Phos/GGCCGACGCTAGTCC). Ultimately, we blunt-end cloned the human IRS-1 open reading frame (ORF) (RefSeq accession no. NM_005544.2), amplified without the stop codon, using a total cDNA library prepared from RNA isolated from HeLa cells.
The pAL-NLS-IRS-1-GFP(HA)/MitoRed vector was constructed from the pKT2/PGK-Bsd:GFP_CLP-Luc plasmid (46). In the first step, Klenow-mediated cloning was used to assemble two fragments amplified from the pKT2/PGK-Bsd:GFP_CLP-Luc vector (first pair of oligonucleotides, CAGAGGGAAAAAGAATTCTTAGCCCTCCCACACATAACCAGAG and CCAGCTAGCTACGTAGGTGGAAGCTACTGTACACCAACC; second pair of oligonucleotides, CTTCCACCTACGTAGCTAGCTGGCCAGACATGATAAGA and GGAGGGCTAAGAATTCTTTTTCCCTCTGCCAAAAATTATGGGGAC). In the next step, a self-cleaving E2A peptide (27) and the HA epitope tag were blunt-end cloned from a pair of annealed 5′-phosphorylated oligonucleotides (forward [F] oligonucleotide GTATACCCATACGACGTACCAGATTACGCTGGCAGTGGAGAGGGCAGAGGAAGTCTGCTAACATGCGGTGACGTCGAGGAGAATCCTGGCCCAACGCGTTGA and reverse [R] oligonucleotide TCAACGCGTTGGGCCAGGATTCTCCTCGACGTCACCGCATGTTAGCAGACTTCCTCTGCCCTCTCCACTGCCAGCGTAATCTGGTACGTCGTATGGGTATAC) into the SnaBI site generated in the previous step of cloning. In the following step, EGFP was Klenow cloned into the SnaBI site from a fragment amplified from pEGFP-N1 (Clontech Laboratories) (F oligonucleotide GGTTGGTGTACAGTAGCTTCCACCATGCCCGGGGAGCTCGTGAGCAAGGGCGAGGAGC and R oligonucleotide GCGTAATCTGGTACGTCGTATGGGTATACGTAGAGCTCCTTGTACAGCTCGTCCATGCCG). In the next step, the dsRed2 fluorescent protein targeted to a mitochondrion sequence (MitoRed), amplified from the pDsRed2-Mito vector (Clontech Laboratories) with a pair of oligonucleotides (F oligonucleotide GAATCCTGGCCCAACGCGTATGTCCGTCCTGACGCCG and R oligonucleotide CCAGCTAGCTACTCAACGCGTCTACAGGAACAGGTGGTGGCG), was introduced by Klenow cloning into the MluI site of the previously generated vector. To target EGFP to the nucleus, a Xenopus laevis nucleoplasmin nuclear localization signal (KRPAATKKAGQAKKKK) (GenBank accession no. BC072778) was prepared from an annealed and 5′-phosphorylated pair of oligonucleotides (F oligonucleotide GTAAAGAGGCCTGCGGCTACCAAAAAAGCAGGCCAGGCAAAGAAGAAGAAA and R oligonucleotide TTTCTTCTTCTTTGCCTGGCCTGCTTTTTTGGTAGCCGCAGGCCTCTTTAC) and cloned into the SnaBI site of the previously cloned vector.
Ultimately, human IRS-1 cDNA (RefSeq accession no. NM_005544.2) was cloned in frame with all cloned coding sequences amplified with a pair of oligonucleotides (F oligonucleotide ACAGTAGCTTCCACCATGCCCGCGAGCCCTCCGGAGAGC and R oligonucleotide CCTTGCTCACGAGCTCCCCCTGACGGTCCTCTGGCTGC) from a cDNA library prepared from total RNA isolated from HeLa cells and cloned into the SmaI site of the vector resulting from the previous cloning step (Fig. 4A). The expression of this vector generates a single translation product that brakes at the T2A sequence to create two separate peptides: the first one is targeted to the nucleus fusion protein between the IRS-1 and EGFP proteins, and second one is tagged with the mitochondrial DsRed fluorescent protein.
The custom-cloned Lenti-IND-NLS-IRS-1-GFP construct (Cyagen Biosciences, CA) is a lentiviral vector encoding all Tet-on-inducible system elements in a single plasmid, which drives the doxycycline-induced expression of the NLS–IRS-1–GFP fusion protein. Besides lentiviral packaging components, it includes a reverse-tetracycline-controlled transactivator (rtTA-Advanced) driven by a CMV promoter and the human IRS-1 ORF fused in frame with EGFP and three SV40 nuclear localization signals driven by an rtTA-Advanced-regulated tetracycline response element (TRE) promoter (Fig. 9A).
Immunohistochemistry.
The avidin-biotin-peroxidase (ABC) methodology was used according to the manufacturer's instructions (Vector Laboratories, Burlingame, CA). Briefly, our modified protocol includes deparaffinization in xylene, rehydration through descending grades of alcohol up to water, nonenzymatic antigen retrieval in citrate buffer (pH 6.0) for 30 min at 95°C, and endogenous peroxidase quenching with H2O2 in methanol for 20 min. After rinsing with PBS, the samples were blocked with 5% normal goat serum (for rabbit polyclonal primary antibodies) or normal horse serum (for mouse monoclonal primary antibodies) in 0.1% PBS–bovine serum albumin (BSA). The following primary antibodies were used: anti-IRS-1 rabbit polyclonal antibody (catalog no. 06-248; Millipore) (1:500 dilution) and anti-IRS-1(pS612) mouse monoclonal antibody (catalog no. 3193S; Cell Signaling) (1:100 dilution). After washing with PBS, sections were incubated with biotinylated anti-rabbit or anti-mouse secondary IgG for 20 min and then rinsed, incubated with ABC complexes, and developed with diaminobenzidine (Sigma). Finally, the sections were counterstained with hematoxylin, dehydrated in alcohol, cleared in xylene, and mounted with Permount (Fisher).
Immunofluorescence.
The cells were fixed and permeabilized with buffer containing 0.02% Triton X-100 and 4% formaldehyde in PBS, followed by washing (three times in PBS) and blocking in 5% BSA for 30 min. The subcellular distribution of IRS-1 was evaluated by utilizing an anti-IRS-1 rabbit polyclonal antibody (06-248; Millipore) followed by a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Inc., Eugene, OR), and the nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI). Other tested anti-IRS-1 antibodies included IRS-1(pY612) (catalog no. 44816G; Thermo Fisher Scientific), IRS-1(pS612) (catalog no. 3193S; Cell Signaling Technology), IRS-1(pS612) (catalog no. 3203S; Cell Signaling Technology); IRS-1(pS636/639) (catalog no. 2388S; Cell Signaling Technology), and IRS-1(pS789) (catalog no. 2389S; Cell Signaling Technology) antibodies. In cases where the NLS–IRS-1–myc tag vector was used, IRS-1 nuclear structures were also detected with an anti-myc tag mouse monoclonal antibody (catalog no. sc-40; Santa Cruz). Other primary antibodies employed in this study included anti-LC3 rabbit polyclonal antibody (catalog no. PD014; MBL), anti-PML mouse monoclonal antibody (catalog no. sc-966; Santa Cruz), anticoilin mouse monoclonal antibody (catalog no. ab11822; Abcam), anti-BMI1 rabbit polyclonal antibody (catalog no. 5856S; Cell Signaling Technology), and anti-SC35 mouse monoclonal antibody (catalog no. 556363; BD Pharmingen), and nucleoli were detected by using pyronin Y staining (18). Epifluorescent images were captured by using a FluoView FV1000 confocal microscope (Olympus of America, Center Valley, PA).
Confocal microscopy and live-cell imaging.
All fluorescent and Nomarski contrast images of fixed cells and tissues were acquired by using a FluoView FV1000 confocal laser scanning microscope equipped with a multiline argon laser (458 nm, 488 nm, and 515 nm) and diode lasers (405 nm, 559 nm, and 635 nm) (Olympus of America, Center Valley, PA). For live-cell imaging, cells expressing the NLS–IRS-1–GFP/MitoRed construct were plated onto 35-mm dishes with a no. 1.5 thick glass bottom (MatTek, MA). To ensure the formation of large quantities of the IRS-1/LC3 nuclear structures, the attached cells were first starved with SFM (Dulbecco's modified Eagle's medium [DMEM] plus 0.1% BSA) for 72 h and subsequently stimulated with 10% FBS for 6 h. Once the nuclear structures were formed, live-cell time-lapse imaging was performed by using either a FluoView FV1000 confocal laser scanning microscope equipped with a live-cell stage-top incubation system (Pathology Devices, Inc., MD) for high-magnification, high-resolution, single-cell imaging or a VivaView LCV110U incubator microscope (Olympus) for low-magnification time-lapse imaging of a large number of cells.
For quantitative FRAP analysis, the cells were imaged with a UPlan 100×/1.3 oil objective and a 488-nm argon laser set to a 0.5% nominal output to minimize fluorescence loss during the imaging phase. The dimension of the imaged area was 256 by 256 pixels (0.49 μm per pixel) at a ×20 optical zoom. Each pixel was imaged for 2 ms. For photobleaching, the selected circular region of the specimen (1 μm/21 pixels in diameter) was irradiated by a 405-nm blue diode laser set to 100% for 50 ms in tornado mode. For each experiment, 20 images were acquired before and 600 images were acquired following photobleaching for a total time of ∼2 min. Before the FRAP curve was generated, each time point was corrected for background (Fb) and photofading due to imaging. The Fb value was generated from a cell-free area of the image measured under the same conditions as those under which FRAP was performed. To correct for photofading, a time series, Ff(t), was collected. Each time point of raw data was corrected according to the formula FC(t) = [Fraw(t) − Fb]/[Ff(t) − Fb]. Finally, each time point was normalized with the corrected initial fluorescence, FC(0), averaged over 20 time points acquired before photobleaching: FN(t) = FC(t)/FC(0). To compute τ1/2 from the FRAP data, the linear interpolation method was used with GraphPad Prism.
For longer time-lapse imaging of a large number of cells, a VivaView LCV110U system (Olympus) equipped with a motorized inverted fluorescence microscope was used. For each experiment, three separate fields were imaged every 10 to 30 min during a 24-h image acquisition session. For each field and time point, three separate images were acquired: differential interference contrast (DIC), GFP fluorescence, and RFP fluorescence images. An advanced binary autofocus was used to compensate for minor sample movements. Finally, the sets of images acquired during time-lapse experiments were converted into videos by using VivaView 4.19 software.
Cell sorting.
A mixed population of LN-229 human glioblastoma cells expressing the NLS–IRS-1–GFP transgene was sorted by using a BD FACSAria II cell sorter and BD FACSDiva v8.0.1 analysis software. Prior to sorting, the cells were suspended in PBS at 5 × 106 cells/ml. Cell sorting was performed by using a 100-μm nozzle and pressure at 20 lb/in2 with a speed of 2,000 events/s. Three distinct cell populations, with low, medium, and high GFP fluorescence, were selected and used for drug testing (Fig. 6).
Immunoprecipitation, Western blots, and subcellular fractionation.
Total protein extracts and nuclear and cytosolic extracts were isolated from LN-229 and HeLa cells expressing NLS–IRS-1 constructs [pALS1-NLS-IRS-1/mycTag or pAL-NLS-IRS-1-GFP(HA)/MitoRed]. Sample preparation, immunoprecipitation, and Western blotting were performed according to standard procedures described in our previous studies (9, 25). For Western blotting, total protein extracts (50 mg) were separated on a 4-to-15% gradient SDS-PAGE gel (Bio-Rad, Hercules, CA) and transferred by using the Trans-Blot Turbo system (Bio-Rad, Hercules, CA). The resulting blots were probed with the following primary antibodies: anti-GFP mouse monoclonal antibody (catalog no. sc-9996; Santa Cruz Biotechnology), anti-IRS-1 rabbit polyclonal antibody (catalog no. 06-248; Millipore), anti-LC3 rabbit polyclonal antibody (catalog no. 12741P; Cell Signaling Technology), and anti-p62 mouse monoclonal antibody (catalog no. sc-28359; Santa Cruz Biotechnology). We used anti-Grb-2 mouse monoclonal antibody (BD Bioscience, San Jose, CA), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mouse monoclonal antibody (catalog no. sc-47724; Santa Cruz Biotechnology), and histone H3 (catalog no. 4499T; Cell Signaling Technology) as loading and subcellular fraction markers, respectively.
Subcellular fractionation.
The cells were placed on ice, scraped, suspended in PBS, and centrifuged. The resulting cellular pellet was suspended in TNMT (100 mM Tris-HCl [pH 7.5], 1,000 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100) supplemented with protease inhibitors. The cell suspension was passed five times through a 23-gauge needle and centrifuged for 5 min at 193 × g at 4°C. The collected supernatant represented the cytoplasmic fraction. The remaining pellet of cell nuclei was suspended in TNMT with protease inhibitors and sonicated six times for 3 s at 50% power. The homogenate was subsequently centrifuged for 10 min at 14,000 rpm at 4°C, and the resulting supernatant was considered the soluble nuclear fraction. The remaining pellet was suspended in TNMT with protease inhibitors and sonicated 10 times for 3 s at 50% power, and the resulting homogenate was considered the insoluble nuclear fraction. Immunoprecipitations were carried out with anti-IRS-1 rabbit polyclonal antibody (catalog no. 06-248; Millipore), anti-HA rabbit polyclonal antibody (catalog no. sc-805; Santa Cruz Biotechnology), and anti-LC3 rabbit polyclonal antibody (catalog no. L7543; Sigma), and the corresponding blots were developed with anti-LC3 rabbit polyclonal antibody (catalog no. PD014; MBL) and anti-GFP mouse monoclonal antibody (catalog no. sc-9996; Santa Cruz Biotechnology).
Autophagy.
Detection and quantification of autophagy were performed with exponentially growing (DMEM plus 10% FBS) HeLa cells stably expressing the NLS–IRS-1–GFP/MitoRed transgene (clone 006). The cells were plated onto glass-bottom 35-mm dishes (MatTek Corporation) at 2 × 104 cells per dish. Following attachment, the cells were infected with the BacMam 2.0 autophagy reporter at a multiplicity of infection (MOI) of 30. To induce autophagy, infected cells were treated with 100 nM rapamycin or amino acid starvation medium (1% BSA, 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose, and 20 mM HEPES [pH 7.4]) for 1.5 to 5 h. The formation of autophagosomes was monitored by using live-cell time-lapse imaging with an Olympus FV1000 confocal laser scanning microscope equipped with a live-cell stage-top incubation system (Pathology Devices, Inc., MD). The percentage of cells positive for autophagosomes was determined in cells that were either positive or negative for the IRS-1/LC3 nuclear structures within the same culture. The cell was counted as autophagy positive if it had at least five green fluorescent vesicles in the cytoplasm, and the cell was determined to be IRS-1/LC3 positive when at least one IRS-1 nuclear structure was detected within the entire volume of the nucleus (Fig. 9 and 10).
Proximity ligation assay.
We employed a proximity ligation assay (PLA) (DuoLink; Sigma) to analyze IRS-1/LC3 protein-protein interactions, according to the manufacturer's recommendations. Briefly, IRS-1 and LC3 proteins were probed with mouse monoclonal (catalog no. sc-8038; Santa Cruz) (1:200) and rabbit polyclonal (catalog no. PD014; MBL) (1:200) antibodies, respectively. The primary antibodies were probed with a pair of DuoLink in situ PLA secondary antibodies, anti-mouse (minus) and anti-rabbit (plus). After ligation and amplification, the PCR products were detected with FarRed DuoLink in situ detection reagent. Images from stained cells were acquired with a FluoView FV1000 confocal laser scanning microscope (Olympus) (Fig. 8).
Statistical analysis.
The data were analyzed with homoscedastic Student's t test. Differences between the control and experimental groups were considered significant at P values of <0.05 (marked with asterisks in the figures).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Susan Theodosiou for her editorial efforts.
This work was initially supported by grant R01-CA095518 (K.R.) and later supported by grants P20-GM103501 (A.O. [principal investigator {PI}] and K.R. [project leader]) and P20-GM121288-01 (K.R. [PI]), LCRC startup funds (K.R.), and a donation from the VFW Auxiliary for Glioblastoma Project (K.R.). In addition, A.O. is supported by grants 2R01CA107974-06A1 and 1R01AI112402-01, and F.P. is supported by LCRC startup funds and grant 1U54 GM104940 from the Louisiana Clinical and Translational Science Center. All confocal images were prepared in collaboration with the LSUHSC/LCRC Molecular Histopathology and Analytical Microscopy Core (MHAM) (partially supported by grant P30GM106392). All cell sorting and flow cytometry data were prepared in collaboration with the LSUHSC/LCRC Cellular Immunology Metabolism Core (CIMC) (supported by grant P20-GM103501), and statistical evaluations were prepared in collaboration with the LSUHSC Biostatistics Bioinformatics Core (BBC) (partially supported by grant P20-GM121288-01). Glioblastoma clinical samples were collected by the LSUHSC/LCRC HIV Clinical Tumor Biorepository Core (HCTB) (partially supported by grant P20-GM121288-01).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00608-17.
REFERENCES
- 1.White MF. 1994. The IRS-1 signaling system. Curr Opin Genet Dev 4:47–54. doi: 10.1016/0959-437X(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 2.Lassak A, Del Valle L, Peruzzi F, Wang JY, Enam S, Croul S, Khalili K, Reiss K. 2002. Insulin receptor substrate 1 translocation to the nucleus by the human JC virus T-antigen. J Biol Chem 277:17231–17238. doi: 10.1074/jbc.M110885200. [DOI] [PubMed] [Google Scholar]
- 3.Morelli C, Garofalo C, Sisci D, del Rincon S, Cascio S, Tu X, Vecchione A, Sauter ER, Miller WH Jr, Surmacz E. 2004. Nuclear insulin receptor substrate 1 interacts with estrogen receptor alpha at ERE promoters. Oncogene 23:7517–7526. doi: 10.1038/sj.onc.1208014. [DOI] [PubMed] [Google Scholar]
- 4.Urbanska K, Pannizzo P, Lassak A, Gualco E, Surmacz E, Croul S, Del Valle L, Khalili K, Reiss K. 2009. Estrogen receptor beta-mediated nuclear interaction between IRS-1 and Rad51 inhibits homologous recombination directed DNA repair in medulloblastoma. J Cell Physiol 219:392–401. doi: 10.1002/jcp.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilk A, Waligorska A, Waligorski P, Ochoa A, Reiss K. 2012. Inhibition of ERbeta induces resistance to cisplatin by enhancing Rad51-mediated DNA repair in human medulloblastoma cell lines. PLoS One 7:e33867. doi: 10.1371/journal.pone.0033867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prisco M, Santini F, Baffa R, Liu M, Drakas R, Wu A, Baserga R. 2002. Nuclear translocation of insulin receptor substrate-1 by the simian virus 40 T antigen and the activated type 1 insulin-like growth factor receptor. J Biol Chem 277:32078–32085. doi: 10.1074/jbc.M204658200. [DOI] [PubMed] [Google Scholar]
- 7.Tu X, Batta P, Innocent N, Prisco M, Casaburi I, Belletti B, Baserga R. 2002. Nuclear translocation of insulin receptor substrate-1 by oncogenes and Igf-I. Effect on ribosomal RNA synthesis. J Biol Chem 277:44357–44365. doi: 10.1074/jbc.M208001200. [DOI] [PubMed] [Google Scholar]
- 8.Wu A, Tu X, Prisco M, Baserga R. 2005. Regulation of upstream binding factor 1 activity by insulin-like growth factor I receptor signaling. J Biol Chem 280:2863–2872. doi: 10.1074/jbc.M406138200. [DOI] [PubMed] [Google Scholar]
- 9.Ozoe A, Sone M, Fukushima T, Kataoka N, Chida K, Asano T, Hakuno F, Takahashi S. 2014. Insulin receptor substrate-1 associates with small nucleolar RNA which contributes to ribosome biogenesis. Front Endocrinol (Lausanne) 5:24. doi: 10.3389/fendo.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozoe A, Sone M, Fukushima T, Kataoka N, Arai T, Chida K, Asano T, Hakuno F, Takahashi S. 2013. Insulin receptor substrate-1 (IRS-1) forms a ribonucleoprotein complex associated with polysomes. FEBS Lett 587:2319–2324. doi: 10.1016/j.febslet.2013.05.066. [DOI] [PubMed] [Google Scholar]
- 11.Cremer T, Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 12.Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell 128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Lamond AI, Spector DL. 2003. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 14.Mao YS, Zhang B, Spector DL. 2011. Biogenesis and function of nuclear bodies. Trends Genet 27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dundr M, Misteli T. 2010. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phair RD, Misteli T. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 17.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. 2005. Nucleolar proteome dynamics. Nature 433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 18.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. 2002. Directed proteomic analysis of the human nucleolus. Curr Biol 12:1–11. doi: 10.1016/S0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 19.Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. 2004. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell 15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. 2011. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol Chapter 14:Unit 14.16. doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cen L, Carlson BL, Pokorny JL, Mladek AC, Grogan PT, Schroeder MA, Decker PA, Anderson SK, Giannini C, Wu W, Ballman KV, Kitange GJ, Sarkaria JN. 2013. Efficacy of protracted temozolomide dosing is limited in MGMT unmethylated GBM xenograft models. Neuro Oncol 15:735–746. doi: 10.1093/neuonc/not010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tivnan A, Zakaria Z, O'Leary C, Kogel D, Pokorny JL, Sarkaria JN, Prehn JH. 2015. Inhibition of multidrug resistance protein 1 (MRP1) improves chemotherapy drug response in primary and recurrent glioblastoma multiforme. Front Neurosci 9:218. doi: 10.3389/fnins.2015.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marrero L, Wyczechowska D, Musto AE, Wilk A, Vashistha H, Zapata A, Walker C, Velasco-Gonzalez C, Parsons C, Wieland S, Levitt D, Reiss K, Prakash O. 2014. Therapeutic efficacy of aldoxorubicin in an intracranial xenograft mouse model of human glioblastoma. Neoplasia 16:874–882. doi: 10.1016/j.neo.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kratz F. 2008. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release 132:171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Kratz F. 2014. A clinical update of using albumin as a drug vehicle—a commentary. J Control Release 190:331–336. doi: 10.1016/j.jconrel.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Lebrecht D, Geist A, Ketelsen UP, Haberstroh J, Setzer B, Kratz F, Walker UA. 2007. The 6-maleimidocaproyl hydrazone derivative of doxorubicin (DOXO-EMCH) is superior to free doxorubicin with respect to cardiotoxicity and mitochondrial damage. Int J Cancer 120:927–934. doi: 10.1002/ijc.22409. [DOI] [PubMed] [Google Scholar]
- 27.Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N, Yoshimori T. 2007. How to interpret LC3 immunoblotting. Autophagy 3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 29.Wild P, McEwan DG, Dikic I. 2014. The LC3 interactome at a glance. J Cell Sci 127:3–9. doi: 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 30.Drake KR, Kang M, Kenworthy AK. 2010. Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3. PLoS One 5:e9806. doi: 10.1371/journal.pone.0009806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacomin AC, Samavedam S, Promponas V, Nezis IP. 2016. iLIR database: a Web resource for LIR motif-containing proteins in eukaryotes. Autophagy 12:1945–1953. doi: 10.1080/15548627.2016.1207016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, Liu W. 2015. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell 57:456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Wilk A, Wyczechowska D, Zapata A, Dean M, Mullinax J, Marrero L, Parsons C, Peruzzi F, Culicchia F, Ochoa A, Grabacka M, Reiss K. 20 October 2014. Molecular mechanisms of fenofibrate-induced metabolic catastrophe and glioblastoma cell death. Mol Cell Biol doi: 10.1128/MCB.00562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Souquere S, Weil D, Pierron G. 2015. Comparative ultrastructure of CRM1-nucleolar bodies (CNoBs), intranucleolar bodies (INBs) and hybrid PML/p62 bodies uncovers new facets of nuclear body dynamic and diversity. Nucleus 6:326–338. doi: 10.1080/19491034.2015.1082695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hottinger AF, Stupp R, Homicsko K. 2014. Standards of care and novel approaches in the management of glioblastoma multiforme. Chin J Cancer 33:32–39. doi: 10.5732/cjc.013.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitange GJ, Carlson BL, Schroeder MA, Grogan PT, Lamont JD, Decker PA, Wu W, James CD, Sarkaria JN. 2009. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol 11:281–291. doi: 10.1215/15228517-2008-090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, Sharp SY, Vassal G, Pearson AD, Reis RM, Hargrave D, Workman P, Jones C. 2010. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res 70:9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, et al. . 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. 2009. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci 29:8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Facchino S, Abdouh M, Chatoo W, Bernier G. 2010. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci 30:10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. 2005. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev 19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith KS, Chanda SK, Lingbeek M, Ross DT, Botstein D, van Lohuizen M, Cleary ML. 2003. Bmi-1 regulation of INK4A-ARF is a downstream requirement for transformation of hematopoietic progenitors by E2a-Pbx1. Mol Cell 12:393–400. doi: 10.1016/S1097-2765(03)00277-6. [DOI] [PubMed] [Google Scholar]
- 43.Krishnamurthi V, Liu YH, Bhattacharyya S, Turner JN, Holmes TJ. 1995. Blind deconvolution of fluorescence micrographs by maximum-likelihood estimation. Appl Opt 34:6633–6647. doi: 10.1364/AO.34.006633. [DOI] [PubMed] [Google Scholar]
- 44.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiss K, Valentinis B, Tu X, Xu SQ, Baserga R. 1998. Molecular markers of IGF-I-mediated mitogenesis. Exp Cell Res 242:361–372. doi: 10.1006/excr.1998.4113. [DOI] [PubMed] [Google Scholar]
- 46.Ohlfest JR, Demorest ZL, Motooka Y, Vengco I, Oh S, Chen E, Scappaticci FA, Saplis RJ, Ekker SC, Low WC, Freese AB, Largaespada DA. 2005. Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the sleeping beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol Ther 12:778–788. doi: 10.1016/j.ymthe.2005.07.689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.