ABSTRACT
Eukaryotic transcription activation domains (ADs) are intrinsically disordered polypeptides that typically interact with coactivator complexes, leading to stimulation of transcription initiation, elongation, and chromatin modifications. Here we examined the properties of two strong and conserved yeast ADs: Met4 and Ino2. Both factors have tandem ADs that were identified by conserved sequence and functional studies. While the AD function of both factors depended on hydrophobic residues, Ino2 further required key conserved acidic and polar residues for optimal function. Binding studies showed that the ADs bound multiple Med15 activator-binding domains (ABDs) with similar orders of micromolar affinity and similar but distinct thermodynamic properties. Protein cross-linking data show that no unique complex was formed upon Met4-Med15 binding. Rather, we observed heterogeneous AD-ABD contacts with nearly every possible AD-ABD combination. Many of these properties are similar to those observed with yeast activator Gcn4, which forms a large heterogeneous, dynamic, and fuzzy complex with Med15. We suggest that this molecular behavior is common among eukaryotic activators.
KEYWORDS: intrinsically disordered protein, transcription activator, transcriptional regulation
INTRODUCTION
Transcription activators play essential roles in gene regulation, and regulation of activator function is often the endpoint of many signaling pathways, serving to modulate transcription in response to developmental pathways, growth, stress, and other environmental signals (1, 2). The targeting of multiple activators in different combinations to gene regulatory regions leads to diverse patterns of gene regulation. Activators can enhance RNA polymerase II transcription through binding to coactivator complexes such as Mediator, SAGA, TFIID, Swi/Snf, and NuA4, which are complexes that contact the basal transcription machinery and/or function to modify chromatin (3, 4). Most eukaryotic activators contain separate DNA-binding domains (DBDs) and transcription activation domains (ADs) (3, 5). Unlike DNA-binding domains, which are usually structurally ordered, eukaryotic ADs are intrinsically disordered, lacking a stable structure (6–10).
Many types of intrinsically disordered proteins (IDPs) bind their targets via short linear motifs, 3- to 10-residue sequences that function as recognition sites for enzymes such as kinases, acetylases, or methylases or as the substrates for peptide-binding domains such as SH2, SH3, and 14-3-3 domains (11–14). In contrast, different ADs have little primary sequence similarity, although they are often enriched for acidic, proline, and glutamine residues (15, 16). At least part of this sequence bias is due to overrepresentation of these residues in intrinsically disordered proteins (17). Known ADs range in length from an ∼5-residue sequence motif to nearly 100 residues (18–20). Mutations created within ADs have shown that their function can be remarkably resistant to mutagenesis, although hydrophobic and, sometimes, acidic residues are critical for activity (3, 21).
One of the best-characterized activators is yeast Gcn4, a transcription factor that activates a large set of genes in response to metabolic stress (22, 23). Gcn4 contains tandem acidic ADs that are unrelated with respect to sequence and interacts with the coactivators Mediator, SAGA, NuA4, TFIID, and Swi/Snf (6, 18, 19, 24–30). Binding of Gcn4 to the Mediator tail module subunit Med15 occurs via multiple heterogeneous interactions between the tandem ADs and up to 4 activator-binding domains (ABDs) on Med15 termed KIX, ABD1, ABD2, and ABD3 (ABD123) (27, 28). The measured individual binding interactions are dynamic, with half-lives on the low millisecond time scale (6). Combined biochemical and structural analyses showed that the interaction between Gcn4 ADs and Med15 is “fuzzy,” as Gcn4 binds to the Med15 activator-binding domains in multiple orientations (6, 18), and that the fuzzy nature of this complex is conserved upon interaction of the tandem ADs with full-length Med15 (20). This binding mechanism can explain the fact that many activators bind multiple unrelated targets using a variety of AD sequences. In contrast, several well-characterized activators are known from structural studies to bind their targets using a different mechanism that utilizes a higher-affinity and more specific protein-protein interface (7, 8, 31, 32).
To explore whether other activators have properties similar to Gcn4, we used molecular, genetic, and biochemical approaches to characterize two strong yeast activators, Met4 and Ino2. Both factors have tandem acidic ADs that are moderately conserved in closely related yeast species but have primary sequences that are unrelated to those of each other and to that of Gcn4. Despite these sequence differences, Met4, Ino2, and Gcn4 have been shown to have similar functions in transcription activation assays and to require Med15 for activation of Mediator tail-dependent promoters, and both ADs bind Med15 activator-binding domains with low micromolar affinities. These and other results suggest that Gcn4, Met4, and Ino2 use similar strategies to bind Mediator that involve a large, dynamic, and fuzzy protein interface.
RESULTS
Mediator tail dependence of transcription activators.
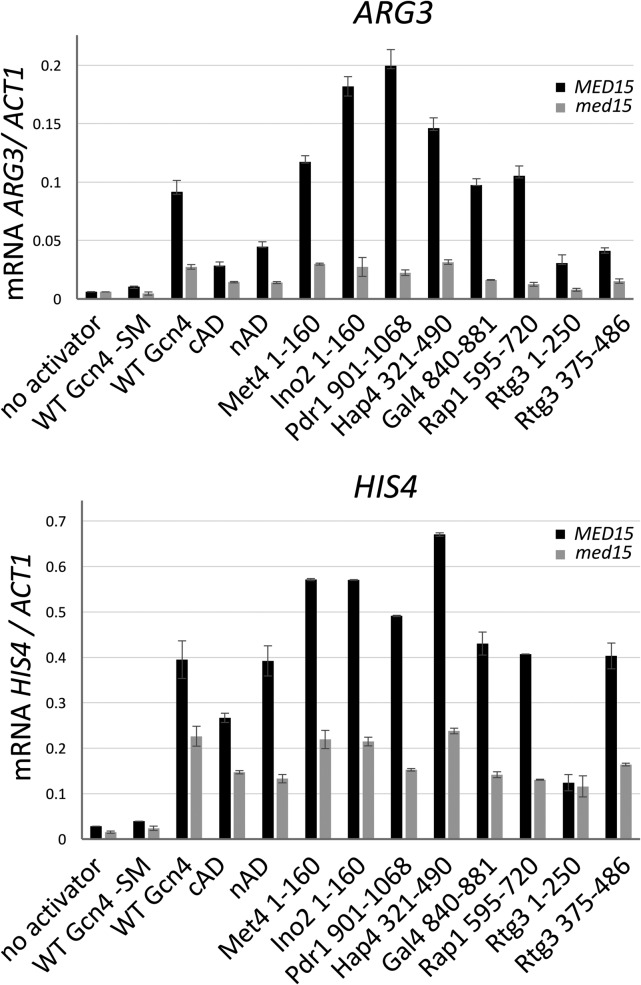
As a first step in exploring the mechanism of yeast ADs in comparison to Gcn4, we tested the activity and coactivator dependence of several previously characterized transcription factors. Segments from 7 transcription factors with published AD function were fused to the N terminus of the Gcn4 central region linker plus the Gcn4 DNA-binding domain (Gcn4 residues 124 to 281) and tested for activation of two Gcn4-dependent promoters, namely, ARG3 and HIS4 (both TATA-containing promoters; defined as TATAWAW [33]). The expression of these AD-Gcn4 derivatives was from low-copy-number autonomously replicating sequence (ARS)- and centromere sequence (CEN)-containing plasmids under the control of the Gcn4 regulatory region with ∼1 kb of DNA upstream from the Gcn4 open reading frame (ORF) containing all known Gcn4 transcription and translational regulatory regions. The regions of the factors tested for function were as follows: Met4 residues 1 to 160 (34), Ino2 residues 1 to 160 (35), Pdr1 residues 901 to 1068 (36), Hap4 residues 321 to 490 (37), Gal4 residues 840 to 881 (38, 39), and Rtg3 residues 1 to 250 and 375 to 486 (40). Fusion proteins contained a C-terminal triple-FLAG epitope tag to monitor protein expression (18). Gcn4 synthesis and activity are induced in response to amino acid starvation, so the activity of these chimeric activators was measured 90 min after addition of sulfometuron methyl (+SM), an inhibitor of Ile and Val biosynthesis, to the cell growth media (27).
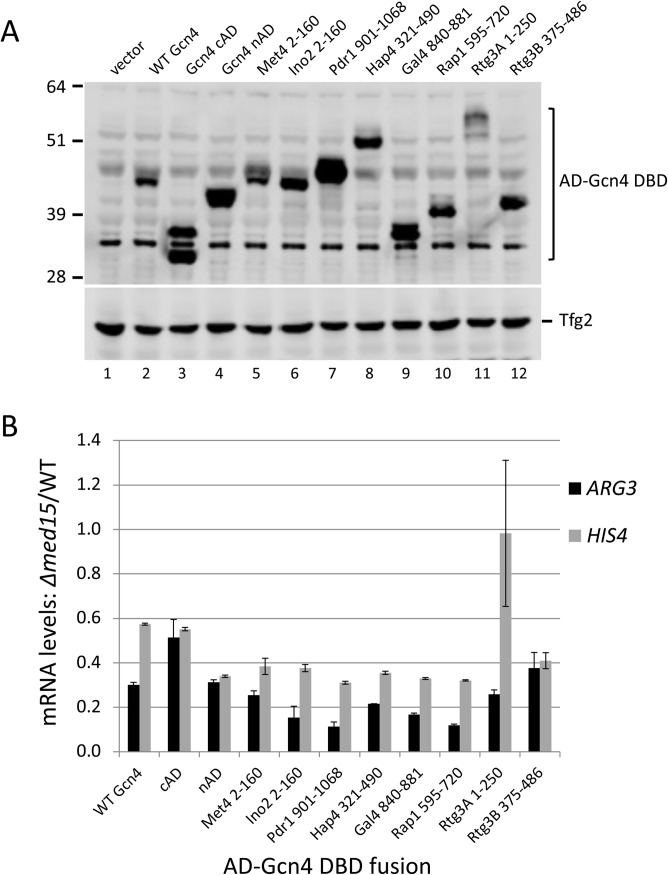
Figure 1 shows that, under conditions of fusion to the Gcn4 DBD, all these ADs function to activate transcription at ARG3 and HIS4, although their relative levels of activity depend on the presence of the specific promoter. Met4, Ino2, Pdr1, and Hap4 are strong ADs at both promoters and are comparable to or better than wild-type (WT) Gcn4 in that respect. The two Rtg3 ADs have different relative levels of activity, depending on the promoter, with the C-terminal AD (cAD) having the most activity at HIS4. Western analysis showed that all proteins were expressed and that the level of expression did not correlate with AD function (Fig. 2A).
FIG 1.
Activity of yeast transcription factor AD-Gcn4 DBD fusions at two Gcn4 inducible genes. Previously defined AD regions were fused to Gcn4 residues 125 to 281 and expressed under the control of the Gcn4 gene regulatory region. Gcn4 induction conditions were initiated by addition of SM for 90 min, and mRNA levels from ARG3 and HIS4 were quantitated by RT-qPCR. Measurements were made in both MED15 and med15Δ strains as indicated.
FIG 2.
Expression of Gcn4 fusion proteins and Med15 dependence. (A) Western blot of whole-cell extracts from cells used as described in the Fig. 1 legend. Western was probed with anti-Gcn4 and anti-Tfg2 (TFIIF subunit) as indicated. (B). Mediator tail module dependence for the different activators at two Gcn4-responsive genes measured as the ratio of mRNA levels in the med15Δ and MED15 strains. Data are from Fig. 1.
TATA-containing Gcn4-activated genes can differ somewhat in their dependence on the Mediator tail module, a direct binding target for Gcn4. For example, ARG3 shows 5-fold to 10-fold dependence on Med15, a Mediator tail subunit, while HIS4 shows ∼2-fold dependence (27). We measured the Mediator tail dependence of these chimeric activators by comparing expression in the WT to that in a Δmed15 mutant. As previously found with Gcn4, all chimeric ADs showed the strongest Med15 dependence at ARG3 and somewhat lower dependence at HIS4 (Fig. 1 and 2B). The one outlier among these ADs is the Rtg3 N-terminal AD (nAD), which showed no Med15 dependence at HIS4. From these results, we conclude that nearly all of these ADs function similarly to Gcn4.
Met4 contains tandem conserved ADs that overlap ubiquitin (Ub)-binding domains.
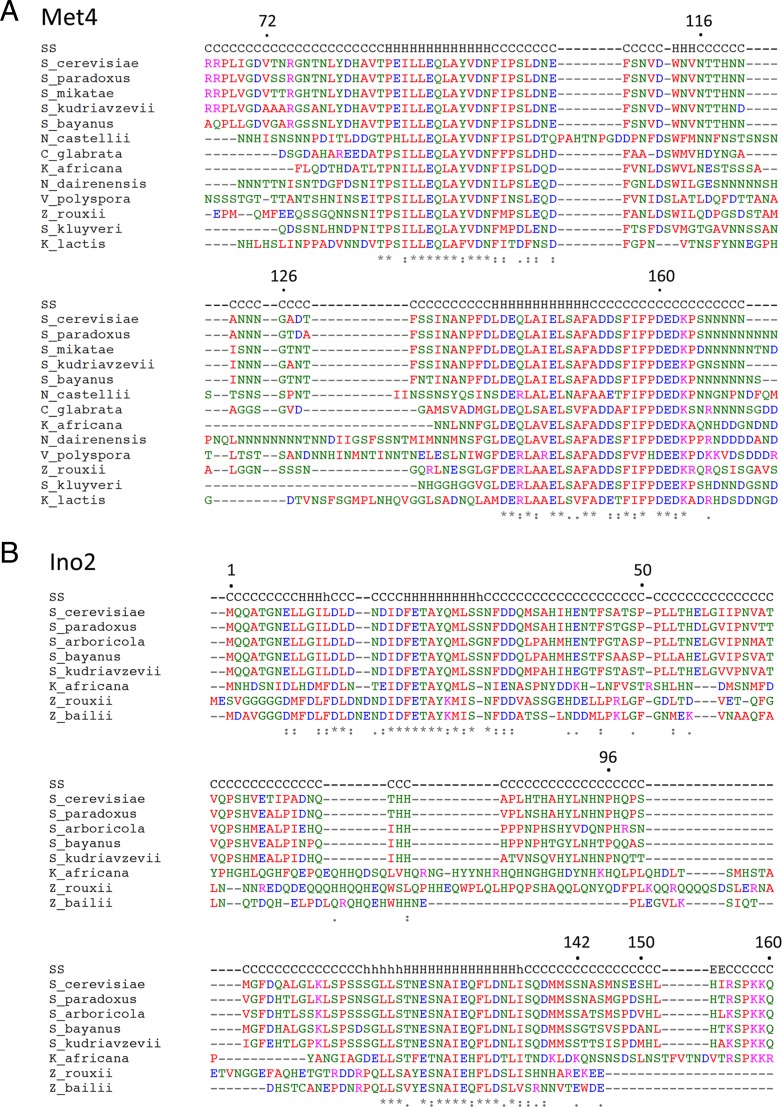
On the basis of the in vivo activity and sequence conservation and previously published work, we focused further characterization efforts on the Met4 and Ino2 ADs. Figure 3 shows that Met4 residues 65 to 170 are enriched in both hydrophobic and acidic residues and that the sequence contains tandem 22-residue-long sequence blocks that are conserved among closely related yeast species. Both of these conserved regions are predicted to have a propensity for alpha helix formation (Fig. 3A).
FIG 3.
Conservation of activation domains in closely related yeast species. (A) Met4 and (B) Ino2 sequences were aligned by Clustal Omega (58), with secondary structure predictions (SS) from Ali2D (59). S_cerevisiae, Saccharomyces cerevisiae; S_paradoxus, Saccharomyces paradoxus; S_mikatae, Saccharomyces mikatae; S_kudriavzevii, Saccharomyces kudriavzevii; S_bayanus, Saccharomyces bayanus; N_castellii, Naumovozyma castellii; C_glabrata, Candida glabrata; K_africana, Kloeckera africana; N_dairenensis, Naumovozyma dairenensis; V_polyspora, Vanderwaltozyma polyspora; Z_rouxii, Zygosaccharomyces rouxii; S_kluyveri, Saccharomyces kluyveri; K_lactis, Kluyveromyces lactis.
Yeast Met4 is a bZIP protein that activates the transcription of at least 45 genes involved in sulfur metabolism (41, 42). Met4 is recruited to regulatory regions by DNA-binding protein CBF1 and related factors Met31 and Met32 (Met31/32), while cofactor Met28 acts to stabilize these DNA-bound complexes (43, 44). Prior analysis of Met4-LexA fusions showed that the sequence consisting of Met4 residues 79 to 160 contains transcription activation function (34). Met4 activator function is known to be regulated both by ubiquitylation and by ubiquitin (Ub) binding. Met4 is modified by a relatively short poly-Ub chain at residue K163 (45), located at the C-terminal edge of the second conserved sequence block. Eliminating ubiquitylation by the K163R mutation activates Met4 similarly to growth under induction conditions but has little if any effect on protein stability. These findings suggest that Met4-Ub regulates function separately from proteolysis (46, 47). Met4 also contains tandem Ub-binding domains defined by mutations Δ85–96 and Δ135–155 (48). Inactivation of these domains leads to longer Met4 poly-Ub chains and decreased protein stability, showing that Ub binding protects Met4 from protein degradation. The Ub-binding domains are contained within the region required for transcription activation, and it has not been determined whether these activities are overlapping or independent functions.
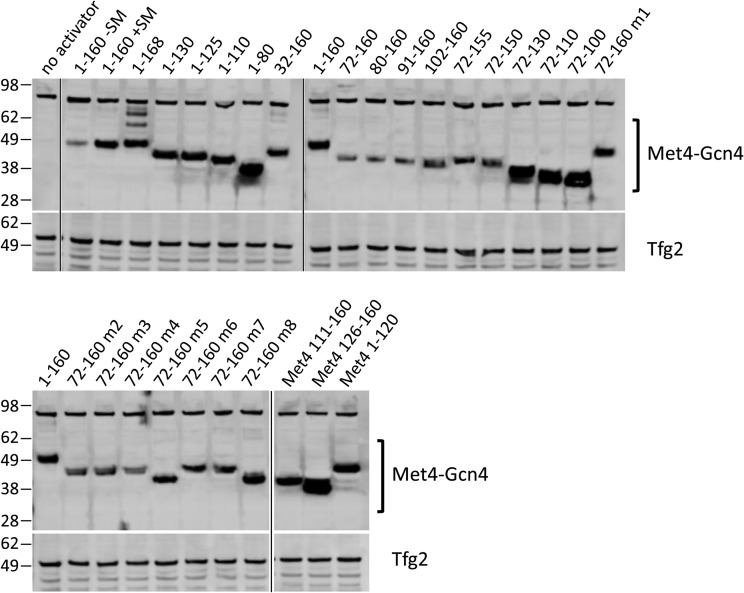
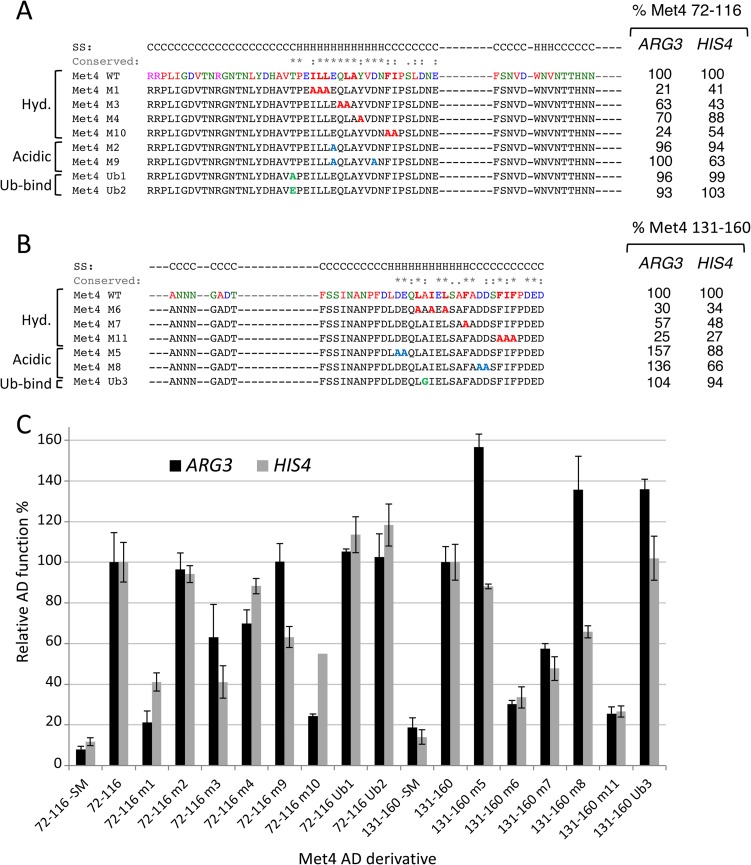
A series of deletions was constructed in the Met4-Gcn4 fusion to identify the minimal regions necessary for AD function at ARG3 and HIS4 (Fig. 4). As with the other chimeric activators described above, protein expression levels did not correlate with AD function (Fig. 5). Consistent with previous observations, Met4 residues 72 to 160 encode 85% to 92% of Met4 AD function (34). Further deletions demonstrated that Met4 contains tandem ADs, with the functional regions centered on the two conserved sequence blocks. Met4 72-116 contains 34% to 62% of Met4 AD function, depending on the target gene. Met4 126-160 contains 18% to 55% of Met4 AD function, again depending on the target gene. Because of this gene-specific activation function, the two Met4 ADs synergize at ARG3 but are approximately additive with respect to their activity at HIS4. We speculate that this may be due to different coactivator dependencies at these genes.
FIG 4.
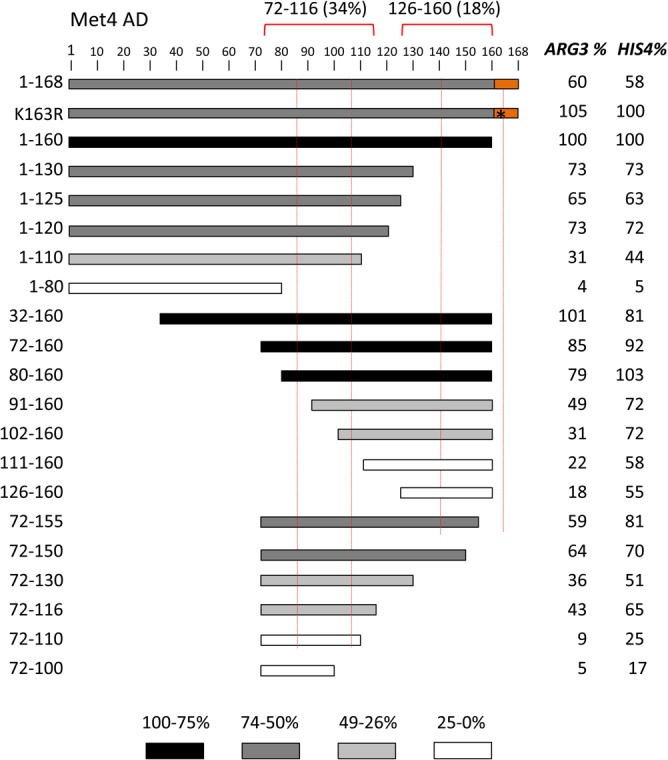
Met4 tandem activation domains. (A) Met4 derivatives fused to Gcn4 and assayed for activation of ARG3 and HIS4 as described in the Fig. 1 legend are shown. Protein segments are shaded according to the percentage of activity compared with residues 1 to 160. Red dotted lines indicate the two conserved sequence blocks shown in Fig. 3A. The orange block indicates a repressive element, and the asterisk indicates the K163R mutation that blocks protein ubiquitylation. Red brackets indicate the limits of the individual ADs at ARG3 and the percentage of activity at ARG3 compared to that of Met4 residues 1 to 160.
FIG 5.
Protein expression of Met4-Gcn4 derivatives. Western blots analyzing whole-cell extracts of cells used for the RT-qPCR assays are shown. Blots were probed with anti-FLAG or anti-Tfg2 (TFIIF subunit) as indicated.
The deletion analysis also found that Met4 residues 161 to 168 repress AD function 40% to 50%. Part of this region is conserved and contains the ubiquitinated K163 residue (46, 47). Western analysis results are consistent with modification at this residue, as this fusion protein migrated in a series of lower-mobility species in SDS-PAGE, although protein levels appeared unchanged from those seen with Met4 1-160–Gcn4 (Fig. 5). All derivatives lacking residues 161 to 168 showed no apparent modification. Mutation of K163 to R in the 1-168–Gcn4 construct eliminated both this protein modification (Fig. 5) and the repressive function (Fig. 4). Unexpectedly, blocking ubiquitination led to lower levels of the fusion protein. This again shows that there is little or no correspondence between protein levels and activation function in this system.
We next examined the importance of conserved and acidic residues within each Met4 AD for transcription of ARG3 and HIS4 (Fig. 6). For Met4 72-116, alanine substitution at three blocks of conserved hydrophobic residues showed at least a 2-fold decrease in function at one or both of the Gcn4-activated genes. In contrast, mutation of two conserved acidic residues gave no more than a 40% decrease in function. Therefore, as at Gcn4, the hydrophobic residues, and not the acidic residues, are most important for function. A similar finding was observed with the second AD, Met4 131-160, where three groups of hydrophobic residues were shown to be important for function whereas mutation of two groups of acidic residues showed no major decrease in activity.
FIG 6.
Hydrophobic (Hyd) but not acidic residues are important for Met4 AD function. (A and B) Mutations in the two Met4 ADs (residues 72 to 116 [A] and residues 131 to 160 [B]) that were targeted for alanine substitution and the resulting effects on induced expression from ARG3 and HIS4. Secondary structure predictions and sequence conservation data are from Fig. 3A. Ub-bind, ubiquitin binding mutation. (C) Quantitation of Met4-Gcn4 fusion protein activity measured by RT-qPCR. The data were used to produce the results shown in panels A and B.
As described above, Met4 contains tandem Ub-binding domains that overlap the two conserved sequence blocks in the AD region. To test whether Ub binding and AD function are separable, we tested three mutations that are known to inhibit or eliminate Ub binding (Fig. 6). Within the 72-to-116 AD, mutations T86A and T86E are both known to eliminate Ub binding (A. Tyrrell, K. Flick, and P. Kaiser, personal communication). These two mutants retained at least 96% of the wild-type AD function with no major changes in protein level (not shown). Mutation A145G in the context of residues 131 to 160, which is a mutation known to limit Ub binding (46), also caused no decrease in AD function but did significantly reduce fusion protein levels. Therefore, we conclude that the Ub binding function of Met4 is not required for activator function, although the two sequences overlap.
Ino2 contains tandem conserved ADs that require both hydrophobic and acidic residues for function.
Next, we examined residues important for Ino2 AD function in the Gcn4 chimeras. Ino2 and Ino4 are bHLH factors required for transcriptional regulation of yeast structural genes involved in phospholipid biosynthesis. Both proteins are required for sequence-specific DNA binding, but only Ino2 contains transcription activation function (35, 49). Previous analysis showed that Ino2 residues 1 to 35 and 98 to 135, when fused to the Gal4 DBD, have activator function, and they were termed AD1 and AD2 (50). Mutagenesis of AD1 showed that both hydrophobic and acidic residues are required for function. AD2 overlaps with the binding site for the Opi1 repressor (Ino2 residues 118 to 135), which targets Ino2 to repress transcription in response to high levels of inositol and choline (51). The results of mutagenesis of AD2 suggested that the residues required for Opi1-dependent repression and AD2 function overlap only partially.
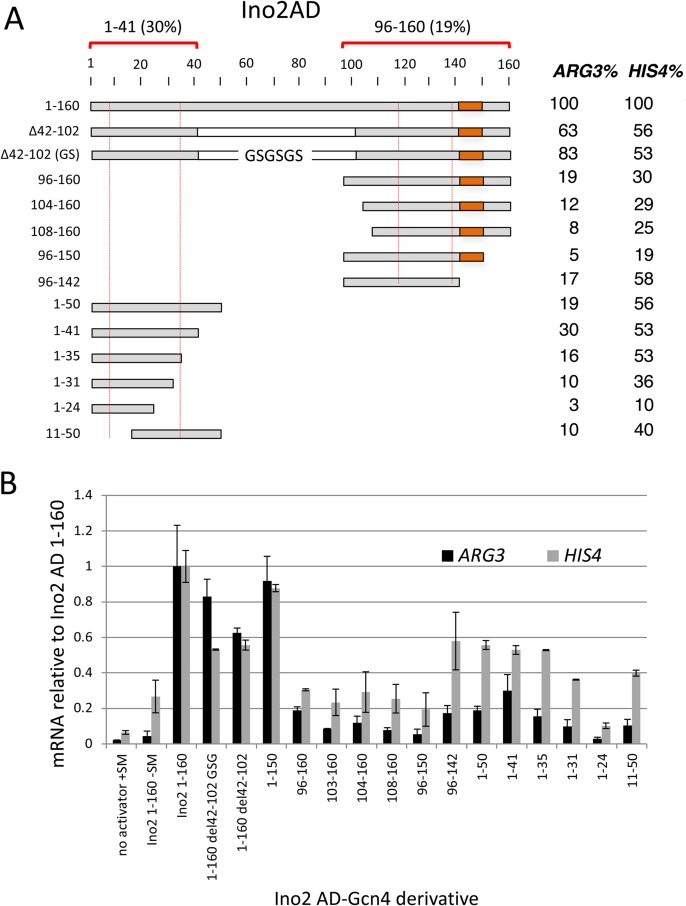
Like Met4, the Ino2 AD region contains two blocks of conserved sequence enriched for hydrophobic and acidic residues within 29 and 21 residues that overlap Ino2 AD1 and AD2 (Fig. 3B). Both of these conserved regions are predicted to have a propensity for alpha helix formation. In agreement with earlier work, we found that each of the two Ino2 ADs, when fused to the Gcn4 DBD, activated ARG3 and HIS4 (Fig. 7). In our system, the minimal segments necessary for AD function are Ino2 residues 1 to 41 and 96 to 160. The intervening region between these two regions can be deleted with less than a ∼2-fold decrease in function. Unexpectedly, we found that the C-terminal AD contained a region that repressed function. Deletion of residues 143 to 150 increased activator function 2-fold to 3-fold, depending on the promoter assayed (Fig. 7; orange rectangle). There were no obvious features in the sequence of this region that explained this repressive activity. Although the Ino2 C-terminal AD is reportedly targeted by the Opi1 repressor (51), we found that the addition of inositol and choline did not repress the activity of either AD (not shown). This is consistent with a previous report that chimeric Ino2-LexA constructs lacking the Ino2 DBD are refractory to Opi1 repression (52).
FIG 7.
Activity of the Ino2 tandem activation domains. (A) Ino2 derivatives fused to Gcn4 and assayed for activation of ARG3 and HIS4 as described in the Fig. 1 legend are shown. Red dotted lines indicate the two conserved sequence blocks shown in Fig. 3B. The orange block indicates an inhibitory element. Red brackets indicate the limits of the two ADs at ARG3 and the percentage of activity on ARG3 compared to Ino2 residues 1 to 160. (B) Quantitation of Ino2-Gcn4 fusion protein activity measured by RT-qPCR.
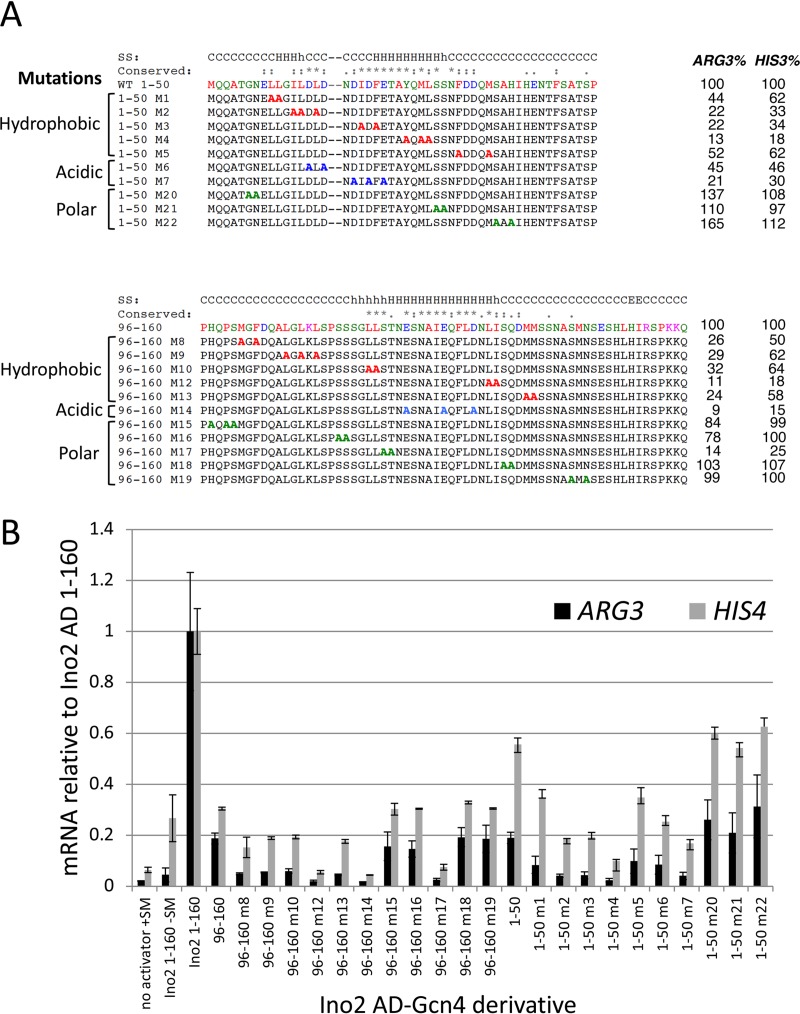
To explore residues important for function of the Ino2 ADs beyond those found in previous work, we mutagenized the individual ADs by double or triple substitution of Ala for hydrophobic, acidic, and polar residues. Function was monitored at ARG3 and HIS3 under induction conditions that included +SM. Residues required for the Ino2 N-terminal AD were distributed over 29 amino acids, almost all of which were in the conserved sequence block (Fig. 8). We found that 5 sets of hydrophobic mutations reduced activity ∼50% or more for at least one Gcn4-dependent gene. In addition, a triple mutation of conserved acidic residues was as detrimental as most mutations of hydrophobic residues. For the C-terminal AD, we found that mutations reducing function were located within a 40-residue segment that was much larger than the conserved sequence block (Fig. 8). Ala substitutions that reduced function by at least 60% for one or both of the Gcn4-dependent genes included 5 sets of hydrophobic residues and one triple mutation of three conserved acidic residues. We found that, in a manner unique to this AD, mutation of conserved residues S120 and T121 to Ala reduced activity by at least 4-fold. Two mutations of other polar residues did not affect function. In summary, residues important for the Ino2 ADs are distributed over 29 to 40 residues and include hydrophobic, acidic, and polar side chains.
FIG 8.
Hydrophobic, acidic, and polar residues are important for Ino2 AD function. (A) Mutations in the two Ino2 ADs (residues 1 to 50 [A] and residues 96 to 160 [B]) that were targeted for alanine substitution and the resulting effects on induced expression from ARG3 and HIS4. Residues are color-coded by amino acid type. Secondary structure predictions and sequence conservation data are from Fig. 3. (B) Quantitation of Ino2-Gcn4 fusion protein activity measured by RT-qPCR.
Met4 and Ino2 bind multiple Med15 activator-binding domains.
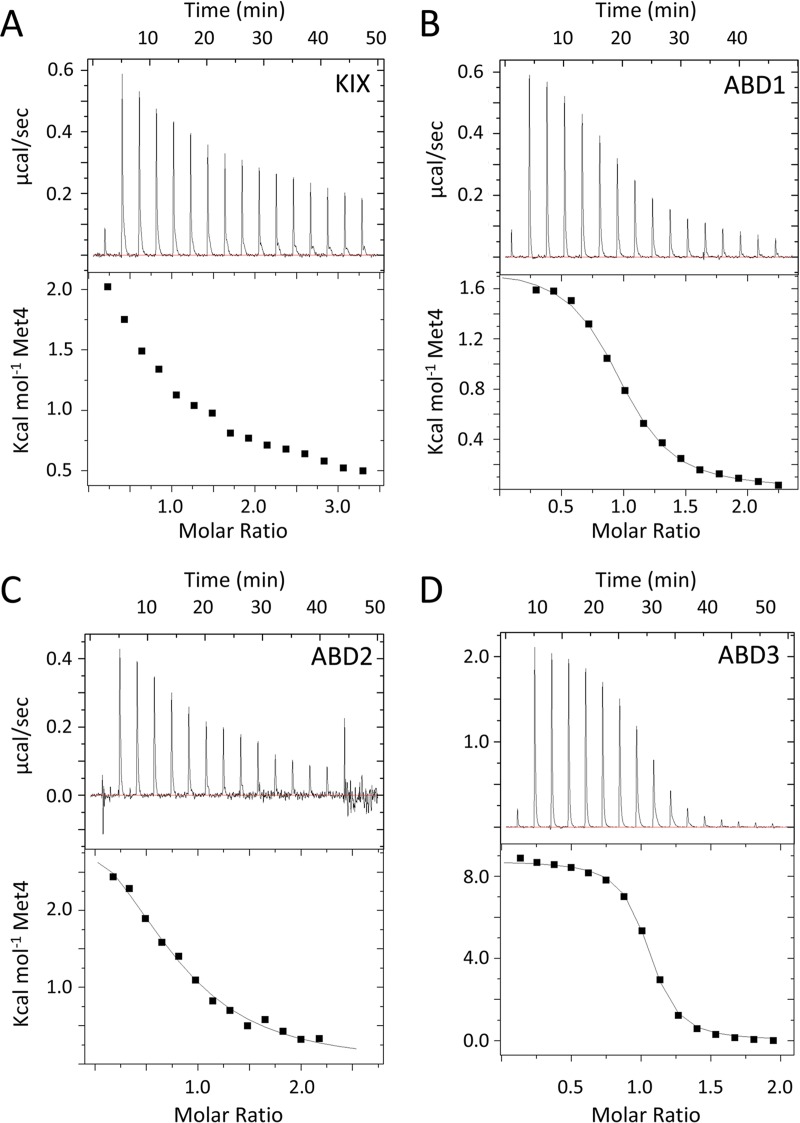
To explore the interactions between Med15 and the Ino2 and Met4 tandem ADs, we used purified proteins in combination with isothermal titration calorimetry (ITC) and/or fluorescence polarization (FP) to measure the affinities and thermodynamic properties of these interactions (Fig. 9, 10, and 11; summarized in Table 1). Binding between the ADs and the individual Med15 activator-binding domains was monitored using ITC. We were unable to use ITC to monitor binding to the longer Med15 polypeptides (KIX plus ABD1, -2, and -3 and ABD1, -2, and -3), so FP was employed to quantitate binding of N-terminal fluorescently labeled AD peptides to Med15. We used FP and ITC to monitor AD binding to ABD3 for comparisons of the two methods, and the results were similar. Met4 affinities monitored were within 20% by either approach, and Ino2 affinities were within ∼3-fold. For the discussion below, the affinities were compared using the ITC values where available.
FIG 9.
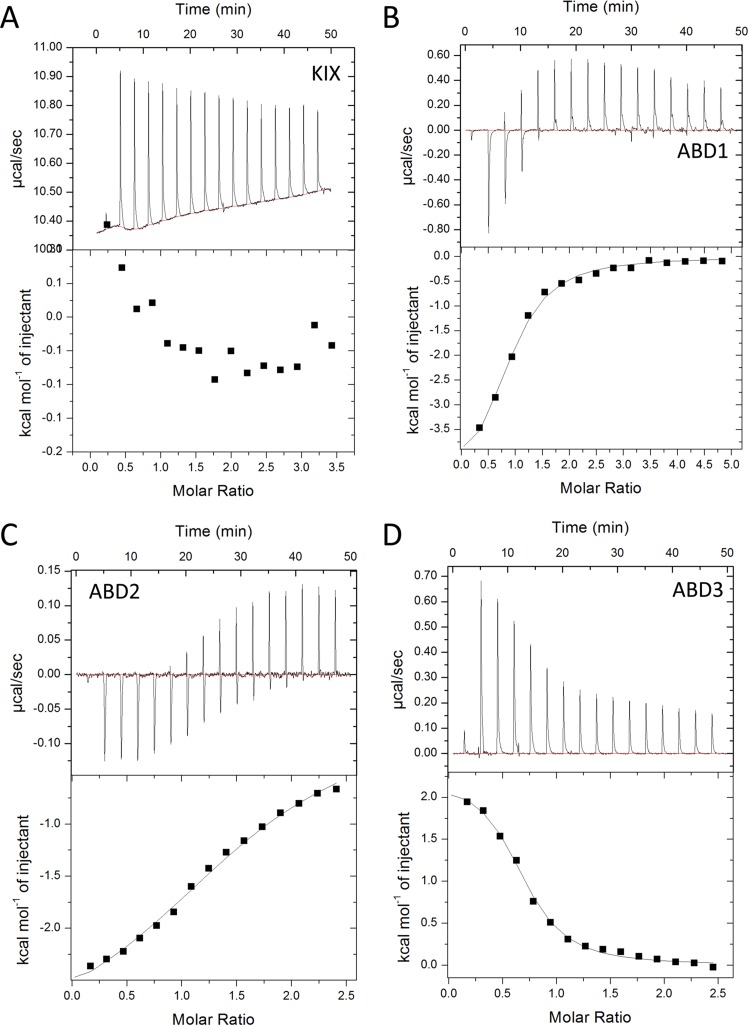
Measurement of Met4-Med15 binding by isothermal titration calorimetry. ITC was used to determine the affinity and thermodynamic parameters of interactions of Met4 residues 72 to 160 with the Med15 KIX domain (A), Med15 ABD1 (B), Med15 ABD2 (C), and Med15 ABD3 (D). All assays were performed and curves fitted as described in Materials and Methods.
FIG 10.
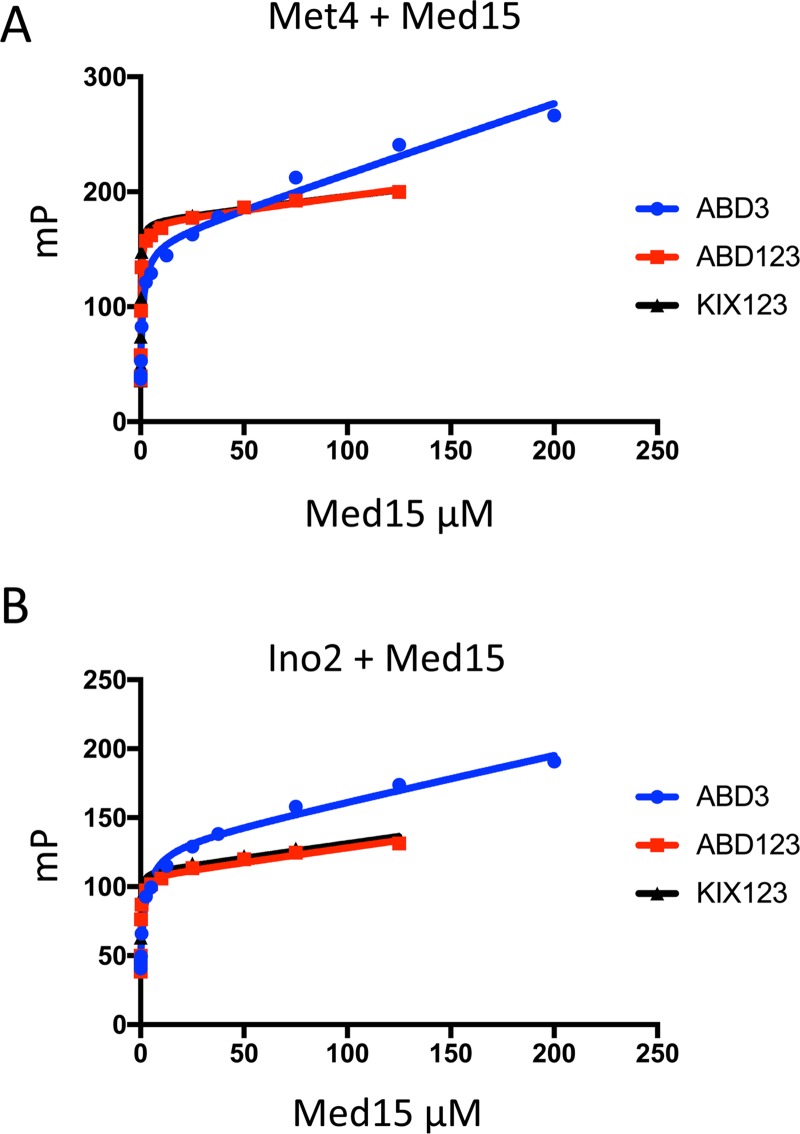
Measurement of activator-Med15 binding by fluorescence polarization. FP was used to assay binding of Oregon green-labeled Met4 72-160 (A) or Ino2 1-41-(GS)3-96-160 (B) to Med15 ABD3, Med15 ABD123, and Med15 KIX plus ABD1, -2, and -3. All assays were performed in triplicate, and curves were fitted as described in Materials and Methods.
FIG 11.
Measurement of Ino2-Med15 binding. ITC was used to determine the affinity and thermodynamic parameters of Ino2 1-41-(GS)3-96-160 interactions with the Med15 KIX domain (A), Med15 ABD1 (B), Med15 ABD2 (C), and Med15 ABD3 (D). All assays were performed and curves fitted as described in Materials and Methods.
TABLE 1.
Affinity of binding of Met4 and Ino2 ADs to Med15 derivativesa
| Med15 derivativeb | Kd (μM) | ΔH | ΔS | N | Method |
|---|---|---|---|---|---|
| Met4-Med15 interactions | |||||
| KIX | NM | N/A | N/A | N/A | ITC |
| ABD1 | 5.9 ± 0.5 | 1,780 ± 26 | 30 | 0.85 | ITC |
| ABD2 | 20.4 ± 3.4 | 3,789 ± 335 | 34.3 | 0.80 | ITC |
| ABD3 | 1.36 ± 0.1 | 8,920 ± 46 | 57 | 1.01 | ITC |
| ABD3 | 1.11 ± 0.23 | N/A | N/A | N/A | FP |
| ABD123 | 0.283 ± 0.027 | N/A | N/A | N/A | FP |
| KIX + ABD123 | 0.196 ± 0.015 | N/A | N/A | N/A | FP |
| Ino2-Med15 interactions | |||||
| KIX | NM | N/A | N/A | N/A | ITC |
| ABD1 | 25.3 ± 3.4 | −4,916 ± 286 | 4.4 | 0.85 | ITC |
| ABD2 | 33.8 ± 3.6 | −2,440 ± 73 | 12.2 | 1.48 | ITC |
| ABD3 | 7.75 ± 0.9 | 2,250 ± 61 | 31 | 0.66 | ITC |
| ABD3 | 2.33 ± 0.4 | N/A | N/A | N/A | FP |
| ABD123 | 0.314 ± 0.051 | N/A | N/A | N/A | FP |
| KIX + ABD123 | 0.213 ± 0.019 | N/A | N/A | N/A | FP |
ITC, isothermal titration calorimetry; FP, fluorescence polarization. For ITC measurements, calculated values of ΔH cal/mole (enthalpy), ΔS cal/mole/degree (entropy), and N (molar ratio) are given. NM, not measurable; N/A, not applicable.
The proteins used were as follows: Met4, residues 72 to 160 (72-160); Ino2, 1-41-(GS)3-96-160; KIX, Med15 6-90; ABD1, Med15 158-238; ABD2, Med15 277-368; ABD3, Med15 484-651; ABD123, Med15 158-651, Δ239-272, and Δ373-483; KIX + ABD123, Med15 1-651, Δ239-272, and Δ373-483.
We were unable to detect binding to the Med15 KIX domain for either AD (Fig. 9A and 11A). This behavior is identical to that of the Gcn4 ADs (27). In contrast, both ADs bound to ABD1, -2, and -3 (Fig. 9B to D and 11B to D). For Met4, the order of highest to lowest binding was ABD3>ABD1>ABD2, with affinities ranging from 1 to 20 μM. The relative order of Ino2 interactions was the same, but all of the individual interactions were weaker than those seen with Met4, ranging from 8 to 34 μM. The highest-affinity interactions were seen with Med15 polypeptides containing all ABDs: KIX plus ABD123 and ABD123 (Fig. 10). For both activators, constructs containing the KIX domain had the highest affinity for the ADs even though binding to the isolated KIX domain was undetectable in our assays. This is consistent with results found for Gcn4, where the KIX domains seemed as functionally important as any of the Med15 ABDs (27) and where KIX plus ABD123 had the highest affinity for the tandem Gcn4 ADs (20). Combined, our results show that Med15 polypeptides with multiple ABDs have much higher affinity for Met4 and Ino2. For example, Met4 binds KIX plus ABD123 with ∼7-fold-higher affinity than ABD3 (Kd [dissociation constant] of 0.196 versus 1.36 μM) and Ino2 binds KIX plus ABD123 with 36-fold-higher affinity than ABD3 (Kd of 0.21 versus 7.8 μM). Finally, despite our finding that Met4 had higher affinity than Ino2 for the individual Med15 ABDs, the affinities of Ino2 and Met4 for the longer Med15 polypeptides were remarkably similar (Kd of ∼0.2 μM for KIX plus ABD123 and ∼0.3 μM for ABD123).
Our previous work showed differing thermodynamic behavior in the mechanism of Med15 binding to the two Gcn4 ADs (20). For example, the Gcn4 central activation domain binding to ABD1 is exothermic, with a favorable change in enthalpy and a small but positive entropy change. In contrast, the binding of the Gcn4 nAD to the individual Med15 ABD1, -2, and -3 domains is endothermic, with unfavorable changes in enthalpy counteracted by large positive changes in entropy. Binding of Met4 and Ino2 ADs also showed surprising and varied thermodynamic behavior depending on the combination of activator and Med15 ABD (Fig. 9 and 11) (Table 1). For example, binding of Gcn4 nAD, Met4 AD, and Ino2 AD to ABD3 was consistently endothermic. In contrast, binding to ABD1 and ABD2 was found to be endothermic or exothermic depending on the activator.
Cross-linking (CL) reveals heterogeneous AD-ABD interactions within the Met4-Med15 complex.
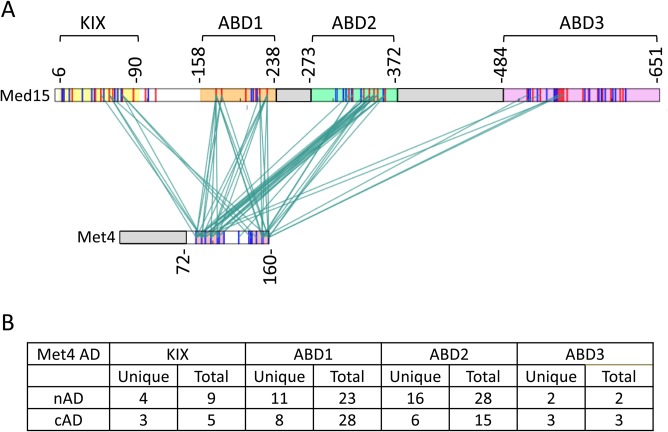
The individual binding measurements described above showed that Met4 and Ino2 interact with both the individual ABDs and the longer Med15 polypeptides, but such experiments cannot show whether the relative levels of affinity or ABD specificity change in the larger complexes. For example, these studies showed that the KIX domain contributes to overall affinity, but the data do not resolve the issue of whether there is direct contact between the AD and KIX. To examine the binding mechanism of the Met4 tandem ADs with the full-length Med15 activator-binding regions, we used the cross-linker EDC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide], which cross-links acidic side chains to lysine (Fig. 12) (see Table S3 in the supplemental material). EDC is a zero-length cross-linker, linking only closely positioned residues and leaving no linker in the cross-linked product. Analysis of the cross-linked products by mass spectrometry (MS) identified cross-links between the individual Met4 ADs and Med15 KIX, ABD1, ABD2, and ABD3. All of the intermolecular cross-links were between acidic residues in Met4 and lysine residues in Med15. Surprisingly, fewer cross-links were detected with ABD3, which individually had the highest affinity for Met4 among the ABDs. Our combined results show that Met4 made direct contacts with KIX and that there was no unique protein complex formed upon binding of Met4 to Med15. Rather, our results are consistent with the tandem ADs rapidly sampling the Med15 ABDs in a large dynamic fuzzy complex as previously proposed (6, 20).
FIG 12.
Met4 ADs interact via a heterogeneous complex with the three ABDs of Med15. (A) Mass spectrometry cross-linking experiments showed that cross-links were formed between regions throughout Met4 AD and each of the Med15 ABD regions and to KIX. Cross-links between Met4 72-160 and Med15 KIX123 are shown in the context of Met4 1-160 and Med15 1-651. Deleted regions are indicated by the gray boxes. Red bars indicate lysine residues. Blue bars indicate aspartic acid and glutamic acid residues. Conserved regions of the Met4 tandem ADs (shaded pink) are indicated as follows: N-terminal AD (nAD) residues 82 to 112 and C-terminal AD (cAD) residues 139 to 160. Regions of Med15 containing the ABDs are colored as follows: KIX (amino acids [aa] 6 to 90), yellow; ABD1 (aa 158 to 238), orange; ABD2 (aa 272 to 372), green; ABD3 (aa 484 to 651), purple. (B) Summary of the number of unique cross-links and frequency observed (Total) between the two Met4 ADs and the Med15 activator-binding domains. A cross-link outside the Met4 AD conserved region (Met4 128-Med15 333) is not listed. Data are from Table S3. Met4 N-terminal AD (nAD) and C-terminal AD (cAD) correspond to residues 72 to 116 and 126 to 160, respectively.
DISCUSSION
Compared with most protein-protein interactions, interactions of transcription activators with their targets are unusual. The primary sequence of ADs is not obviously conserved among different activators; the factors are intrinsically disordered, and they interact with multiple distinct targets having no obvious similarity. However, these properties undoubtedly allow many activators to function through a variety of coactivators and to modulate transcription at various promoters with different coactivator requirements. Here, we have focused our investigations on characterizing two strong yeast activators, Met4 and Ino2, to identify common and distinct features of yeast ADs. Examining Met4, Ino2, and seven other strong yeast activation domains, we found that all but one has similar levels of dependence on Mediator tail module subunit Med15 for activation of two TATA-containing reporter genes.
Like Gcn4, both Met4 and Ino2 have tandem ADs that are enriched for acidic and hydrophobic residues. Tandem ADs may be another feature common to strong activators in eukaryotes, as mammalian viral and human activators such as VP16 and p53 also have tandem ADs. For Met4 and Ino2, these individual ADs were identified both functionally and as blocks of moderately conserved sequences with helical propensity imbedded in nonconserved flanking sequences. This, along with previous work, shows that, although ADs do not have a common primary sequence motif, there are specific sequence requirements that constitute a functional AD.
Both individual ADs of Met4 are of intermediate length; the conserved sequence blocks are 22 residues long for both, and mutagenesis of conserved residues showed that conserved sequences of 13 and 15 residues are required for most of the AD function. Mutagenesis of the ADs showed that only hydrophobic residues were critical for normal function—identical to the finding of critical hydrophobic but not acidic residues in the Gcn4 ADs (18, 19, 24). The individual Ino2 ADs are larger than the Met4 ADs, with 29- and 21-residue conserved sequence blocks. Mutagenesis of the N-terminal AD found that a stretch of 29 residues was required for maximum function that almost precisely matched the conserved sequence block. However, the C-terminal AD was larger, with functionally important amino acids distributed over a span of 40 residues. We also found that both Ino2 ADs contained functionally important hydrophobic and acidic residues. The acidic residues may function through nonspecific electrostatic interactions with the coactivator targets or, alternatively, may make direct and specific contacts.
Monitoring the binding of Ino2 and Met4 to Med15 showed that they behaved in many respects like Gcn4. All bound Med15 ABD1, ABD2, and ABD3 with micromolar affinity, and binding to the Med15 KIX domain was undetectable in our assays. Binding of the tandem ADs to larger Med15 polypeptides showed that all had much higher affinity than the individual ABDs and that the KIX domain contributed to overall affinity under these conditions. These biochemical findings are consistent with those of our earlier study that showed that the normal in vivo response to Gcn4 activation requires multiple Med15 ABDs and the KIX domain. It seems likely that, since these individual binding interactions are weak, multiple binding sites are required to increase the affinity and specificity to reach a biologically meaningful range (53, 54).
An unexpected observation with Gcn4, Met4, and Ino2 binding to Med15 was that interactions with the individual ABDs could be either exo- or endothermic. The endothermic interactions all had large unfavorable changes in enthalpy and were driven by large positive changes in entropy. This behavior is opposite that expected because of the entropic penalty paid upon binding of a disordered protein. However, it has been proposed that, even in the bound state, IDPs can retain conformational entropy due to fuzzy protein interfaces and conformational flexibility of the protein region not in direct contact with the binding partner (55). However, these mechanisms do not seem to fully explain the large, positive entropy changes observed. At this time, we do not understand the mechanism for the large increase in entropy that occurs upon binding, but it seems to be ABD and activator specific and is likely to result at least partially from release of solvent during binding. As an example of thermodynamic specificity, binding of ABD3 to Gcn4, Met4, and Ino2 was endothermic whereas the thermodynamics of binding to ABD1 and ABD2 was activator specific. Understanding the mechanism of endothermic binding will be important for understanding not only activator mechanisms and specificity but, more generally, its characteristics as a mechanism likely used for molecular recognition by other disordered proteins.
Finally, the Met4-Med15 cross-linking experiments allowed us to probe larger and more physiologically relevant complexes. Upon mixing the Met4 tandem ADs with KIX plus ABD123, cross-linking revealed that the individual Met4 ADs directly interacted with each of the Med15 structured domains. This shows that there is no unique Met4-Med15 protein complex and is consistent with the model that multiple Gcn4 ADs rapidly sample individual Med15 ABDs in a large dynamic fuzzy complex (20). Since this cross-linking behavior is identical to that observed with Gcn4, and because Met4 and Ino2 have generally similar properties, we think it likely that all three activators function by similar mechanisms. In the future, it will be important to understand more about both the biochemical properties of these interactions and how frequently eukaryotic activators use this mode of protein-protein interaction.
MATERIALS AND METHODS
Strains and plasmids.
All yeast strains and primary plasmids used in this work are listed in Table S1 in the supplemental material.
Cell growth assays and measurement of steady-state mRNA levels.
Yeast strains were grown in duplicate to an optical density at 600 nm (OD600) of 0.5 to 0.8 in 2% (wt/vol) dextrose synthetic complete Ile-Val-Leu medium at 30°C. Cells were induced with 0.5 μg/ml SM for 90 min to induce amino acid starvation (27), RNA was extracted and assayed in triplicate by reverse transcription-quantitative PCR (RT-qPCR), and the results were analyzed as described previously (27). Data are listed in Table S2 as mRNA ratios of ARG3 to ACT1 or HIS4 to ACT1.
Quantitation of in vivo AD-Gcn4 levels.
Cells (1.5 ml) from the cultures used for the mRNA analysis described above were pelleted and incubated in 0.1 M NaOH for 5 min at room temperature. Cells were then pelleted and resuspended in 1× lithium dodecyl sulfate sample buffer (Life Technologies) containing 50 mM dithiothreitol (DTT) and were treated and analyzed as previously described (18).
Protein purification.
All proteins were expressed in BL21(DE3) RIL Escherichia coli. Med15 constructs were expressed and purified as described by Tuttle et al. (20). Ino2 1-41-(GS)3-96-160 [(GS)3 is the linker; GSGSGS] and Met4 72-160 constructs were expressed as N-terminal His6-SUMO-tagged proteins (Invitrogen). Cells were lysed in a mixture containing 50 mM HEPES (pH 7.0), 500 mM NaCl, 40 mM imidazole, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 5 mM DTT and purified using nickel-Sepharose High Performance resin (GE Healthcare). Proteins were eluted in a mixture containing 50 mM HEPES (pH 7.0), 500 mM NaCl, 500 mM imidazole, 10% glycerol, 1 mM PMSF, and 1 mM DTT. Purified SUMO-tagged proteins were concentrated using 10K-molecular-weight-cutoff (MWCO) centrifugal filters (Millipore), diluted 10× in a mixture containing 50 mM HEPES (pH 7.0), 500 mM NaCl, 40 mM imidazole, 10% glycerol, 1 mM All proteins, and 5 mM DTT, and digested with SUMO protease for 3 to 5 h at room temperature using an ∼1:800 protease/protein ratio. Cleaved His6-Sumo tag was removed using nickel-Sepharose. Peptides were further purified by chromatography on a Source 15Q ion exchanger (GE Healthcare) using a 50 to 350 mM NaCl gradient. To remove residual SUMO tag present in the sample due to coelution on the Source 15Q ion exchanger, Ino2 peptides were purified over SUMO-1(CR) resin (Nectagen) and collected in the flowthrough. All proteins were further purified using size exclusion chromatography and Superdex 75 10/30 (GE Healthcare). Proteins used in fluorescence polarization and isothermal titration calorimetry were eluted in 20 mM KH2PO4 (pH 7.5)–200 mM KCl. Proteins used in cross-linking–mass spectrometry (CL-MS) were eluted in phosphate-buffered saline (PBS) (pH 7.2). The concentration of the purified proteins was determined by UV light/visible light (UV/Vis) spectroscopy with extinction coefficients calculated with ProtParam (56).
FP and ITC binding experiments.
The Ino2 1-41-(GS)3-96-160 and Met4 72-160 used in fluorescence polarization were labeled with Oregon green 488 dye (Invitrogen) as described in reference 27. FP measurements were conducted using a Beacon 2000 instrument as described in reference 27, with concentrations of Med15 spanning 0 to 200 μM (ABD3) or 0 to 125 μM (ABD1, ABD2, and ABD3 and KIX plus ABD123). FP data were analyzed using Prism 7 (Graphpad Software, Inc.) to perform nonlinear regression analysis using the one-site total binding model Y = Bmax · X/(Kd + X) + NS · X + background, where Bmax represents maximum binding, NS represents the number of binding sites, Y represents arbitrary polarization units, and X represents the Med15 concentration.
ITC titrations were performed using a Microcal ITC200 microcalorimeter and 20 mM KH2PO4 (pH 7.5)–200 mM KCl as described in reference 6. The following proteins (concentrations) were used: Med15 6-90 (79.7 μM) versus Ino2 1-41-(GS)3-96-160 (1.32 mM); Med15 6-90 (79.7 μM) versus Met4 72-160 (1.27 mM); Med15 158-238 (111 μM) versus Ino2 1-41-(GS)3-96-160 (2.59 mM); Med15 158-238 (117 μM) versus Met4 72-160 (1.27 mM); Med15 277-368 (113 μM) versus Ino2 1-41-(GS)3-96-160 (1.32 mM); Med15 277-368 (59.7 μM) versus Met4 72-160 (732 μM); Med15 484-651 (111 μM) versus Ino2 1-41-(GS)3-96-160 (1.32 mM); Med15 484-651 (119 μM) versus Met4 72-160 (1.12 mM). The following parameters were the same for all runs: cell temperature, 22°C; reference power, 11 μcal/s; initial delay, 120 s; stir speed, 1,000 rpm; injection spacing, 180 s; filter period, 5 s; injection rate, 0.5 μl/s. Activator was added over 16 injections (injection 1, 0.4 μl; injections 2 to 16, 2.55 μl). Calorimetric data were plotted and fitted with a single binding site model using Origin 7.0 software (Microcal).
EDC cross-linking and MS sample preparation.
A 50-μg volume of Med15 1-651 Δ239-272 and Δ373-483 (KIX + ABD123) was mixed with a 3× molar excess of Ino2 1-41-(GS)3-96-160 or Met4 72-160. Samples were incubated with 15 mM (Met4) or 10 mM (Ino2) EDC and 2 mM Sulfo-NHS (Thermo Scientific) in a 50-μl total volume with PBS at pH 7.2 (Met4) or a 150-μl total volume with PBS at pH 6.5 for 2 h at room temperature. Proteins were processed for MS analysis in a manner similar to that described by Tuttle et al. (20). Protein samples were reduced with 50 mM TCEP [tris(2-carboxyethyl)phosphine] and denatured with 8 M urea at 37°C for 15 min. The samples were then alkylated in the dark at 37°C with 15 mM iodoacetamide for 1 h. The samples were then diluted 10-fold with 100 mM ammonium bicarbonate and digested with Glu-C (20:1 [wt/wt]) overnight at 37°C. Samples were then digested with trypsin (1:15 [wt/wt]) overnight at 37°C. Digested samples were purified by C18 chromatography (Waters), eluted in 80% acetonitrile–0.15 trifluoroacetic acid, and dried in a SpeedVac.
MS and data analysis.
EDC-cross-linked peptides were analyzed on a Thermo Scientific Orbitrap Elite mass spectrometer at the proteomics facility at the Fred Hutchinson Cancer Research Center, and data were analyzed as described in reference 57. Spectra were manually evaluated using a Comet/Lorikeet spectrum viewer (Trans-Proteomic Pipeline) as described in reference 57.
Supplementary Material
ACKNOWLEDGMENTS
We thank Johannes Soeding and Eckhardt Guthöhrlein (Max Plank, Göttingen, and the Gene Center, Munich) for many insightful discussions and analysis of the Ino2 and Met4 AD sequences, Peter Kaiser (University of California, Irvine [UC Irvine]) for discussions and permission to cite unpublished work, Laurent Kuras (CEA, CNRS, Université Paris Sud) for discussions, members of the laboratory of S.H. for assistance and comments during the course of this work, and Lisa Tuttle for comments on the manuscript.
The work described here was funded by a grant from the University of Washington (UW) School of Medicine to H.R. and M.B., grants 2P50GM076547 and 5R01GM110064 to J.R., and NIH grant 5RO1GM075114 to S.H. H.R. and M.B. were affiliated with the University of Washington Medical Student Research Training Program.
D.P., L.W., M.B., H.R., and S.H. performed all experimental work. D.P. purified proteins, measured binding affinities, and along with M.B., performed and analyzed cross-linking reactions. L.W., H.R., and S.H. created fusion proteins and derivatives, and L.W. and H.R. analyzed activator function and protein expression. J.L. and J.R. performed cross-linking–MS analysis. D.P., L.W., and S.H. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00038-18.
REFERENCES
- 1.Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 2.Levine M, Cattoglio C, Tjian R. 2014. Looping back to leap forward: transcription enters a new era. Cell 157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hahn S, Young ET. 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weake VM, Workman JL. 2010. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet 11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 5.Ptashne M, Gann AA. 1990. Activators and targets. Nature 346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 6.Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, Hahn S. 2011. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell 44:942–953. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, Jenkins LMM, Durell SR, Hayashi R, Mazur SJ, Cherry S, Tropea JE, Miller M, Wlodawer A, Appella E, Bai Y. 2009. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure 17:202–210. doi: 10.1016/j.str.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 9.Sigler PB. 1988. Transcriptional activation. Acid blobs and negative noodles. Nature 333:210–212. [DOI] [PubMed] [Google Scholar]
- 10.Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. 1997. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 11.Stein A, Pache RA, Bernadó P, Pons M, Aloy P. 2009. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J 276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen Ba AN, Yeh BJ, van Dyk D, Davidson AR, Andrews BJ, Weiss EL, Moses AM. 2012. Proteome-wide discovery of evolutionary conserved sequences in disordered regions. Science Signaling 5:rs1. doi: 10.1126/scisignal.2002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das RK, Mao AH, Pappu RV. 2012. Unmasking functional motifs within disordered regions of proteins. Science Signaling 5:pe17. doi: 10.1126/scisignal.2003091. [DOI] [PubMed] [Google Scholar]
- 14.Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, Seiler M, Davey NE, Haslam N, Weatheritt RJ, Budd A, Hughes T, Pas J, Rychlewski L, Trave G, Aasland R, Helmer-Citterich M, Linding R, Gibson TJ. 2010. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res 38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell PJ, Tjian R. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 16.Ptashne M, Gann A. 1997. Transcriptional activation by recruitment. Nature 386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 17.Tompa P. 2002. Intrinsically unstructured proteins. Trends Biochem Sci 27:527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 18.Warfield L, Tuttle LM, Pacheco D, Klevit RE, Hahn S. 2014. A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc Natl Acad Sci U S A 111:E3506-13. doi: 10.1073/pnas.1412088111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drysdale CM, Dueñas E, Jackson BM, Reusser U, Braus GH, Hinnebusch AG. 1995. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol 15:1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuttle LM, Pacheco D, Warfield L, Luo J, Ranish J, Hahn S, Klevit RE. 2018. Gcn4-Mediator specificity is mediated by a large and dynamic fuzzy protein-protein complex. Cell Rep 22:3251–3264. doi: 10.1016/j.celrep.2018.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erkina TY, Erkine AM. 2016. Nucleosome distortion as a possible mechanism of transcription activation domain function. Epigenetics Chromatin 9:40. doi: 10.1186/s13072-016-0092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu H, Chereji RV, Hu C, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG. 2016. Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res 26:211–225. doi: 10.1101/gr.196337.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittal N, Guimaraes JC, Gross T, Schmidt A, Vina-Vilaseca A, Nedialkova DD, Aeschimann F, Leidel SA, Spang A, Zavolan M. 2017. The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat Commun 8:457. doi: 10.1038/s41467-017-00539-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson BM, Drysdale CM, Natarajan K, Hinnebusch AG. 1996. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol 16:5557–5571. doi: 10.1128/MCB.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 26.Fishburn J, Mohibullah N, Hahn S. 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell 18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, Moorefield B, Pacheco D, Hahn S. 2010. Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol Cell Biol 30:2376–2390. doi: 10.1128/MCB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP, Hinnebusch AG. 2010. Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit Mediator to target genes in vivo. J Biol Chem 285:2438–2455. doi: 10.1074/jbc.M109.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim S-J, Natarajan K, Yoon S, Hinnebusch AG. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol 23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon S, Qiu H, Swanson MJ, Hinnebusch AG. 2003. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol Cell Biol 23:8829–8845. doi: 10.1128/MCB.23.23.8829-9945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Lello P, Jenkins LMM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, Dikeakos JD, Appella E, Legault P, Omichinski JG. 2006. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell 22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Langlois C, Mas C, Di Lello P, Jenkins LMM, Legault P, Omichinski JG. 2008. NMR structure of the complex between the Tfb1 subunit of TFIIH and the activation domain of VP16: structural similarities between VP16 and p53. J Am Chem Soc 130:10596–10604. doi: 10.1021/ja800975h. [DOI] [PubMed] [Google Scholar]
- 33.Donczew R, Hahn S. 13 December 2018. Mechanistic differences in transcription initiation at TATA-less and TATA-containing promoters. Mol Cell Biol doi: 10.1128/MCB.00448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuras L, Thomas D. 1995. Functional analysis of Met4, a yeast transcriptional activator responsive to S-adenosylmethionine. Mol Cell Biol 15:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schüller HJ. 1995. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res 23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolaczkowska A, Kolaczkowski M, Delahodde A, Goffeau A. 2002. Functional dissection of Pdr1p, a regulator of multidrug resistance in Saccharomyces cerevisiae. Mol Genet Genomics 267:96–106. doi: 10.1007/s00438-002-0642-0. [DOI] [PubMed] [Google Scholar]
- 37.Stebbins JL, Triezenberg SJ. 2004. Identification, mutational analysis, and coactivator requirements of two distinct transcriptional activation domains of the Saccharomyces cerevisiae Hap4 protein. Eukaryot Cell 3:339–347. doi: 10.1128/EC.3.2.339-347.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, Ptashne M. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847–853. doi: 10.1016/0092-8674(87)90081-X. [DOI] [PubMed] [Google Scholar]
- 39.Leuther KK, Salmeron JM, Johnston SA. 1993. Genetic evidence that an activation domain of GAL4 does not require acidity and may form a beta sheet. Cell 72:575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 40.Rothermel BA, Thornton JL, Butow RA. 1997. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J Biol Chem 272:19801–19807. doi: 10.1074/jbc.272.32.19801. [DOI] [PubMed] [Google Scholar]
- 41.Thomas D, Jacquemin I, Surdin-Kerjan Y. 1992. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol Cell Biol 12:1719–1727. doi: 10.1128/MCB.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TA, Jorgensen P, Bognar AL, Peyraud C, Thomas D, Tyers M. 2010. Dissection of combinatorial control by the Met4 transcriptional complex. Mol Biol Cell 21:456–469. doi: 10.1091/mbc.E09-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. 1996. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J 15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 44.Blaiseau PL, Thomas D. 1998. Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J 17:6327–6336. doi: 10.1093/emboj/17.21.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flick K, Ouni I, Wohlschlegel JA, Capati C, McDonald WH, Yates JR, Kaiser P. 2004. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat Cell Biol 6:634–641. doi: 10.1038/ncb1143. [DOI] [PubMed] [Google Scholar]
- 46.Flick K, Raasi S, Zhang H, Yen JL, Kaiser P. 2006. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nat Cell Biol 8:509–515. doi: 10.1038/ncb1402. [DOI] [PubMed] [Google Scholar]
- 47.Ouni I, Flick K, Kaiser P. 2010. A transcriptional activator is part of an SCF ubiquitin ligase to control degradation of its cofactors. Mol Cell 40:954–964. doi: 10.1016/j.molcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyrrell A, Flick K, Kleiger G, Zhang H, Deshaies RJ, Kaiser P. 2010. Physiologically relevant and portable tandem ubiquitin-binding domain stabilizes polyubiquitylated proteins. Proc Natl Acad Sci U S A 107:19796–19801. doi: 10.1073/pnas.1010648107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ambroziak J, Henry SA. 1994. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem 269:15344–15349. [PubMed] [Google Scholar]
- 50.Dietz M, Heyken W-T, Hoppen J, Geburtig S, Schüller H-J. 2003. TFIIB and subunits of the SAGA complex are involved in transcriptional activation of phospholipid biosynthetic genes by the regulatory protein Ino2 in the yeast Saccharomyces cerevisiae. Mol Microbiol 48:1119–1130. doi: 10.1046/j.1365-2958.2003.03501.x. [DOI] [PubMed] [Google Scholar]
- 51.Heyken W-T, Repenning A, Kumme J, Schüller H-J. 2005. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol Microbiol 56:696–707. doi: 10.1111/j.1365-2958.2004.04499.x. [DOI] [PubMed] [Google Scholar]
- 52.Kumme J, Dietz M, Wagner C, Schüller H-J. 2008. Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor. Curr Genet 54:35–45. doi: 10.1007/s00294-008-0197-7. [DOI] [PubMed] [Google Scholar]
- 53.Klein P, Pawson T, Tyers M. 30 September 2003. Mathematical modeling suggests cooperative interactions between a disordered polyvalent ligand and a single receptor site. Curr Biol doi: 10.1016/j.cub.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 54.Olsen JG, Teilum K, Kragelund BB. 2017. Behaviour of intrinsically disordered proteins in protein-protein complexes with an emphasis on fuzziness. Cell Mol Life Sci 12:269–269. doi: 10.1007/s00018-017-2560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flock T, Weatheritt RJ, Latysheva NS, Babu MM. 2014. Controlling entropy to tune the functions of intrinsically disordered regions. Curr Opin Struct Biol 26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 57.Knutson BA, Luo J, Ranish J, Hahn S. 17 August 2014. Architecture of the Saccharomyces cerevisiae RNA polymerase I core factor complex. Nat Struct Mol Biol doi: 10.1038/nsmb.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alva V, Nam S-Z, Söding J, Lupas AN. 2016. The MPI bioinformatics toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.