ABSTRACT
Transcription factor-induced reprogramming of somatic cells to pluripotency is mediated via profound alterations in the epigenetic landscape. The histone variant macroH2A1 (mH2A1) is a barrier to the cellular reprogramming process. We demonstrate here that mH2A1 blocks reprogramming and contributes to the preservation of cell identity by trapping cells at the very early stages of the process, namely, at the mesenchymal-to-epithelial transition (MET). We provide a comprehensive analysis of the genomic sites occupied by the mH2A1 nucleosomes in human fibroblasts and embryonic stem (ES) cells and how they affect the reprogramming of fibroblasts to pluripotency. We have integrated chromatin immunoprecipitation sequencing (ChIP-seq) data with transcriptome sequencing (RNA-seq) data using cells containing reduced levels of mH2A1 and have inferred mH2A1-centered gene-regulatory networks that support the fibroblast and ES cell fates. We found that the exact positions of mH2A1 nucleosomes in regulatory regions of specific network genes with key regulatory roles guarantee the functional robustness of the regulatory networks. Using the reconstructed networks, we can predict and validate several components and their interactions in the establishment of stable cell types by limiting progression to alternative cell fates.
KEYWORDS: chromatin structure, gene expression, histone variants, reprogramming
INTRODUCTION
Cell identity is a reflection of cell-type-specific gene expression programs guided by unique transcription factor (TF) networks (1). Additionally, the robustness of the transcriptional state depends on the configuration and the constraints imposed by the chromatin context in which TFs operate (2, 3). Specifically, the local chromatin architecture, as defined by the positions and the density of the nucleosomes, varies among different cell types and thus contributes to differential regulation of gene expression by controlling the accessibility of TFs to regulatory DNA sequences (4–7). In addition, the presence of histone variants affects chromatin composition and complexity by creating specialized nucleosomes, which when localized on DNA-regulatory elements can have profound effects on nucleosome stability, protein accessibility to DNA, and cellular longevity (8, 9).
Direct reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) through the ectopic overexpression of the pluripotency-associated TFs OCT4, SOX2, KLF4, and c-MYC (OSKM), is the most extreme and best-studied example in which cell fate decisions can be manipulated and even reversed in vitro (10, 11). Specifically, the expression of the associated TFs can destabilize the transcriptional networks of nearly every differentiated somatic cell and induce the progressive reconstitution of embryonic stem cell (ESC) transcriptional networks, which eventually lead to the establishment of an ES-like phenotype (12, 13). Recent studies using murine somatic cells identified two major waves of gene expression changes that coincide with the repression of somatic genes at the early stage and with the activation of core pluripotency genes at the late stage (14–16). Certainly, it has been proposed that iPSC formation follows an early and a late deterministic phase, separated by a more stochastic phase (14–16). The efficiency of conversion into iPSCs remains extremely low (0.1 to 3%), and the acquisition of induced pluripotency is a remarkably slow process, especially in human cells (10, 11, 17). These observations suggest that the reprogramming factors need to overcome and/or reverse a series of epigenetic barriers that have been gradually imposed on the genome during cell differentiation to stabilize cell identity and to prevent aberrant cell fate changes. Recent studies in a variety of systems examining the mechanisms of somatic cell reprogramming revealed that mesenchymal-to-epithelial transition (MET) plays an indispensable role in the initiation of this process in many cell types (18–20). Besides cellular reprogramming, MET occurs in normal development and cancer metastasis (19, 20). Epithelial cells and mesenchymal cells are two types of cells with distinct functions in the animal body. Epithelial cells are able to build cell junctions with their neighbors, whereas mesenchymal cells are loosely connected to each other, are more motile, and lack the apicobasal polarity of epithelial cells (19, 20).
MacroH2A1.2 (mH2A1.2) is a vertebrate-specific histone H2A variant in which the H2A-like histone domain bears a large (~30-kDa) C-terminal globular macrodomain via a short flexible linker that protrudes from the core nucleosome structure (21, 22). mH2A1.2 has previously been reported to block cellular reprogramming of somatic cells by maintaining pluripotency loci in a repressed state (23–25). Moreover, genome-wide occupancy profiles show that in human keratinocytes, mH2A1.2 preferentially occupies genes that are expressed at low levels and are marked with the repression marker H3K27me3, including pluripotency-related genes and bivalent developmental regulators. Thus, the presence of mH2A1.2 at these genes prevents regaining of the activation marker H3K4me2 during reprogramming, imposing an additional layer of repression that preserves cell identity (24). In agreement with this, the presence of mH2A1.2 has been associated with cell resistance to efficient chromatin remodeling (7).
Despite initial observations linking mH2A1.2 to gene repression (4, 26), recent experiments suggest that mH2A1.2 nucleosomes are involved in both positive and negative regulation of transcription. For example, the knockdown of mH2A1.2 has been reported to block the induction of genes via serum starvation (26). Previous work from our laboratory has shown that singular mH2A1.2 nucleosomes occupy the transcription start sites (TSS) of subsets of both expressed and nonexpressed genes, with opposing regulatory consequences (3). Specifically, mH2A1.2 nucleosomes mask repressor binding sites in expressed genes and activator binding sites in repressed genes, thus generating distinct chromatin landscapes that limit genetic or extracellular inductive signals leading to robust gene expression programs. Therefore, the strategic positions and the stabilization of mH2A1.2 nucleosomes in specific human promoters define robust gene expression patterns. In addition, mH2A1.2 is required for the activation of muscle enhancers and the recruitment of the transcription factor PBX1 (27).
We report here that mH2A1.2 constitutes a barrier to the cellular reprogramming process (23–25). Specifically, we demonstrate that mH2A1.2 nucleosomes contribute to the preservation of cell identity by trapping cells at the very early stages of the process, namely, during MET. MET serves as the fundamental crossroads for cell fate decision during reprogramming and is characterized by the loss of key mesenchymal features, such as N-cadherin (N-CAD), and the acquisition of key epithelial characteristics, like E-cadherin (E-CAD), thus leading to loss of adhesion, transforming the elongated fibroblasts into tightly packed clusters of rounded cells (18). We have integrated chromatin immunoprecipitation sequencing (ChIP-seq) with transcriptome sequencing (RNA-seq) data derived from cells bearing reduced levels of mH2A1.2 and have inferred gene-regulatory networks (GRNs) that could identify the putative role of mH2A1.2 in controlling the activity of key regulators of MET during cellular reprogramming.
RESULTS
Role of mH2A1.2 in the reprogramming process.
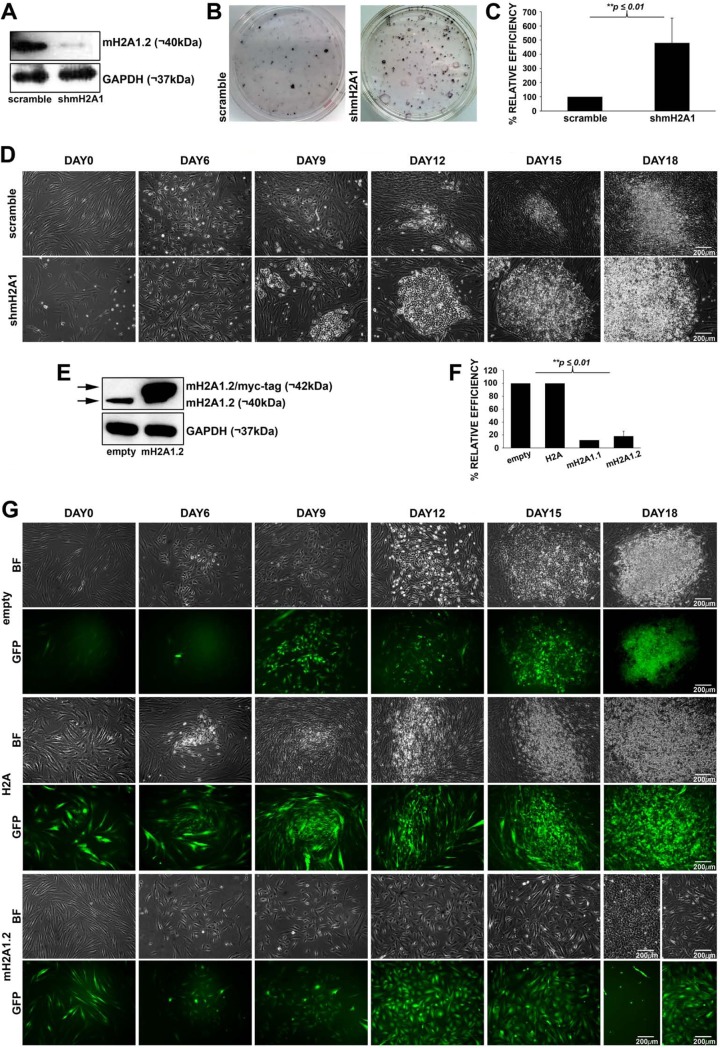
Previous studies have indicated that mH2A1.2 prevents the reprogramming process of mouse embryonic fibroblasts (MEFs) and human keratinocytes (23–25). However, the underlying molecular mechanism by which mH2A1.2 blocks this process remains elusive. To address this issue, we reprogrammed mesoderm-derived human fibroblasts (hFBSs) isolated from skin biopsy specimens with a mixture of lentiviruses encoding the OSKM (OCT4, SOX2, KLF4, and c-MYC), along with a small hairpin RNA (shRNA) sequence against mH2A1 or a scramble sequence. Human iPSC colonies formed using our reprogramming platform were expanded in the absence of doxycycline (Dox) and verified for their pluripotency characteristics by examining the expression of key pluripotency factors (OCT4, SOX2, and NANOG) and by their ability to differentiate into other cell types by the addition of retinoic acid (data not shown). The knockdown of mH2A1 (mH2A1-KD) was confirmed by Western blot analyses indicating that its expression was reduced more than 90% relative to the control sample expressing the scramble sequence (Fig. 1A). Alkaline phosphatase (AP) staining, which marks the undifferentiated ES-like cell phenotype (28), demonstrated an ∼5-fold increase in the reprogramming efficiency of mH2A1-KD hFBS cells (Fig. 1B and C). The time course reprogramming experiment shown in Fig. 1D revealed that mH2A1-KD not only enhanced the total number of reprogrammed iPSCs that formed colonies (Fig. 1C) but also accelerated the kinetics of the process, since the first colonies appeared on day 9 as opposed to day 12 for the control cells. Additionally, the size of the iPSC colonies formed in mH2A1-KD cells is larger than in the control cells, implying that mH2A1.2 controls the cell cycle. These data are consistent with the fact that mH2A1.2 functions as a reprogramming barrier in human cells by blocking cell proliferation early in the reprogramming process.
FIG 1.
MacroH2A1.2 blocks cellular reprogramming. (A) Western blot depicting mH2A1.2 protein levels in mH2A1-KD human fibroblasts (shmH2A1) compared to the corresponding levels in a control sample (scramble). GAPDH was used as a loading control. (B) Representative AP staining of iPSC colonies formed after reprogramming of mH2A1-KD human fibroblasts (shmH2A1) or control cells (scramble). (C) Relative reprogramming efficiency of mH2A1-KD human fibroblasts (shmH2A1) compared to the control sample (scramble), which was taken as 100%. AP-positive iPSC-like colonies were counted, and their percentage was plotted relative to the scramble. The mean value and standard deviation (SD) (n = 3) are shown. **, P ≤ 0.01. (D) Phase-contrast images taken from a time course reprogramming experiment showing the morphological changes and colony formation from day 0 to day 18 of mH2A1-KD human fibroblasts (shmH2A1) (bottom) and control sample cells (scramble) (top). (E) Western blot using the anti-mH2A1.2 antibody in extracts prepared from human fibroblasts overexpressing mH2A1.2 (mH2A1.2/myc-tag) or from control cells (empty). GAPDH was used as a loading control. (F) Relative efficiencies of reprogramming of human fibroblasts overexpressing H2A (H2A), mH2A1.1 (mH2A1.1), mH2A1.2 (mH2A1.2), or control vector (empty). AP-positive colonies were counted, and their percentage was plotted in relation to the empty vector. The mean values and SD (n = 3) are shown. **, P ≤ 0.01. (G) Phase-contrast and fluorescent images taken from a time course reprogramming experiment showing the morphological changes and iPSC colony formation, from day 0 to day 18, of human fibroblasts overexpressing H2A (H2A) (middle) or mH2A1.2 (mH2A1.2) (bottom). The empty vector was used as a control (empty) (top).
To further examine the role of mH2A1.2 in the reprogramming of hFBSs to pluripotency, we tested the effect of mH2A1.2 overexpression. hFBSs undergoing reprogramming were infected with a lentivirus coexpressing mH2A1.2 and green fluorescent protein (GFP), and their reprogramming efficiency was compared to that of control reprogrammable cells infected with a lentivirus expressing GFP only (empty vector). Western blots confirmed that mH2A1.2 was expressed at ∼20-fold higher levels than the endogenous protein (Fig. 1E). As predicted, the number of AP-positive colonies formed by cells overexpressing mH2A1.2 or mH2A1.1 was ∼5-fold lower than that of control cells (Fig. 1F). Importantly, Fig. 1G shows that cells overexpressing mH2A1.2 did not produce iPSC colonies, since GFP-positive cells maintained their initial elongated shape. In contrast, all iPSC colonies formed did not express GFP, indicating that they had not been transduced with the mH2A1.2 coexpressing GFP lentiviral vectors. As a control, we showed that lentivirus-dependent overexpression of the canonical H2A did not affect the efficiency of cellular reprogramming (Fig. 1F and G). Taken together, our data suggest that mH2A1.2 plays a critical role in blocking cellular reprogramming, probably by intervening in early binary cell fate decisions.
The role of mH2A1.2 at the early stages of the reprogramming process.
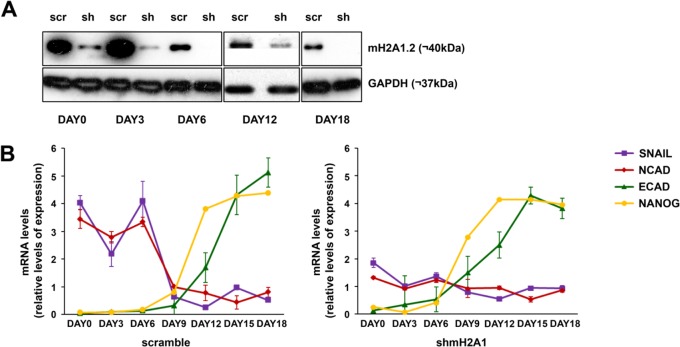
We have shown that hFBSs overexpressing mH2A1.2 become resistant to reprogramming and that KD of mH2A1 accelerates reprogramming. Therefore, we asked whether mH2A1.2 prevents reprogramming by interfering with the MET process shown to be required for reprogramming (18). MET is characterized by the so-called “cadherin switch,” which involves downregulation of N-CAD and the simultaneous upregulation of E-CAD (18, 19). The transcription factor SNAI1 plays a critical role in the cadherin switch by directly repressing the expression of E-CAD (29, 30). To test this possibility, we quantitated the relative expression levels of N-CAD, E-CAD, SNAI1, and NANOG in control and mH2A1-KD hFBSs during a time course reprogramming experiment. We found that in mH2A1-KD cells, expression of both SNAI1 and N-CAD is significantly reduced, whereas in contrast, the E-CAD gene expression program is shifted earlier, since its expression begins on day 3 compared to control cells, where E-CAD starts to be expressed on day 12 (Fig. 2). Similarly, NANOG expression is shifted earlier, from day 12 in wild-type (WT) cells to day 9 in mH2A1-KD cells (Fig. 2). Thus, a decrease of the endogenous levels of mH2A1.2 enhances cellular reprogramming by facilitating the N-CAD-to-E-CAD transcriptional switch, which is required for MET, presumably due to the reduced SNAI1 expression levels.
FIG 2.
mH2A1.2 blocks the cadherin switch required for cellular reprogramming. (A) Western blot analysis of hFBS extracts prepared from mH2A1.2-KD (sh) or control (scr) cells undergoing reprogramming at the indicated time points using an antibody against mH2A1.2. GAPDH was used as a loading control. (B) Normalized levels of N-CAD, E-CAD, SNAI1, and NANOG mRNA at the indicated time points during the reprogramming process of control hFBSs (scramble) or mH2A1-KD cells (shmH2A1) as quantified by qPCR. The data were normalized to the levels of GAPDH and plotted in relation to the median expression value. The error bars indicate SD.
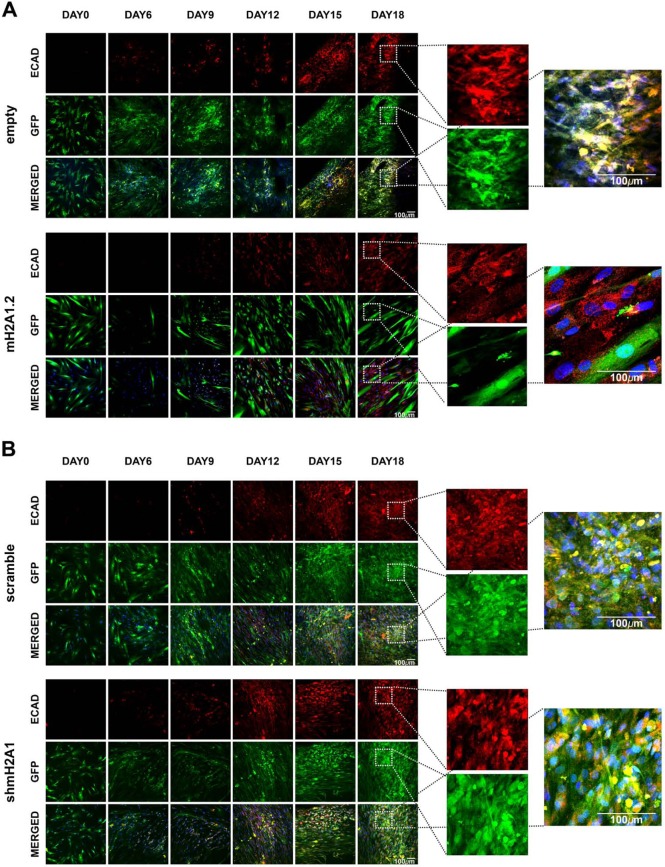
The data presented above are consistent with the idea that, ordinarily, mH2A1.2 controls MET by delaying and/or inhibiting the process. To further test this hypothesis, we carried out immunofluorescence staining for E-CAD in either mH2A1-KD or mH2A1.2-overexpressing hFBSs undergoing reprogramming compared to control cells. We discovered that there was no detectable expression of E-CAD in cells overexpressing mH2A1.2 (Fig. 3A, bottom). As a result, these cells maintained their initial elongated shape, being refractory to reprogramming. In contrast, empty-GFP-vector-expressing cells expressed E-CAD and formed GFP-positive colonies up to day 18 (Fig. 3A, top). On the other hand, in mH2A1-KD cells engineered to express GFP, E-CAD-expressing iPSC colonies were easily formed (Fig. 3B, bottom). Notably, E-CAD–GFP-positive colonies derived from mH2A1-KD–GFP cells were formed earlier (day 9) than those derived from the control cells (Fig. 3B, compare top 3 rows with bottom 3 rows, scramble-GFP, day 12). In summary, these data are consistent with the notion that mH2A1.2 prevents cellular reprogramming by trapping cells during MET.
FIG 3.
mH2A1.2 blocks MET transition. (A) Confocal images, taken from a time course reprogramming experiment, of human fibroblasts overexpressing mH2A1.2 or control cells (empty). The cells were fixed at the indicated time points and stained for E-CAD (red), and nuclei were stained with DAPI (blue). GFP was coexpressed with mH2A1.2/myc-tag or the empty vector. The insets on day 18 depict human fibroblasts overexpressing mH2A1.2-GFP, which were not stained for E-CAD. However, colocalization of GFP with E-CAD was observed in empty transduced human fibroblasts, indicating that the presence of mH2A1.2 prevents the reprogramming process at the stage of MET. (B) Same as panel A except that the cells were transduced with a vector expressing shmH2A1. The insets for day 18 indicate the colocalization of GFP and E-CAD in cells expressing either scramble or shmH2A1.
Cell-type-specific distribution of mH2A1.2 nucleosomes in the hESC and hFBS genomes.
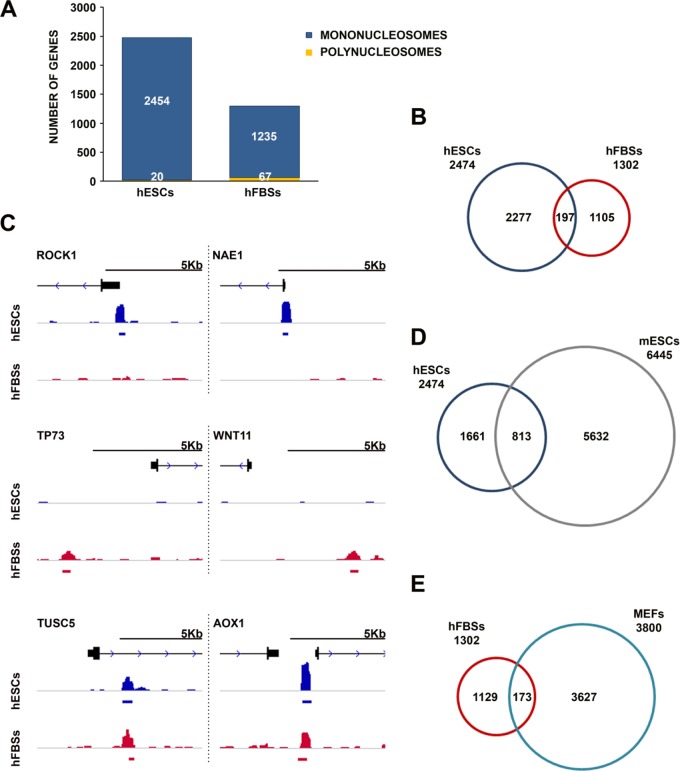
To investigate how mH2A1.2 nucleosomes could inhibit cellular reprogramming, we determined their precise genome-wide localization in hFBSs and in human ESCs (hESCs) by combining micrococcal nuclease digestion of native chromatin with immunoprecipitation (native ChIP [N-ChIP]), followed by deep DNA sequencing (3). After aligning the sequences with the human genome, model-based analysis of ChIP-seq (MACS) and SICER algorithms were used in order to detect single nucleosomes and polynucleosomes (2 or more), respectively (31, 32). Figure 4A shows that mH2A1.2 exists mainly as a component of singular nucleosomes in both cell types. A comparison of the genomic sites occupied by mH2A1.2 nucleosomes between hFBSs and hESCs revealed only a small overlap (8% of hESC sites), suggesting that mH2A1.2-containing nucleosomes are distributed across the human genome in a cell-type-specific manner (Fig. 4B). For example, Fig. 4C compares Integrative Genomics Viewer (IGV) snapshots of the same genomic region (promoter or enhancer) of six different genes between hFBSs and hESCs and demonstrates the differences and/or similarities in the mH2A1.2 nucleosome localization maps. Importantly, comparison of all the genomic sites bound by mH2A1.2 nucleosomes between mouse ESCs (mESCs) and hESCs revealed an overlap more extensive than the overlap between hFBSs and hESCs (33% of ESC sites), suggesting that mH2A1.2 nucleosomes may play similar roles in ESCs in both humans and mice (Fig. 4D). In contrast, the comparison of the respective genomic sites between hFBSs and MEFs revealed only a small overlap (Fig. 4E). Taken together, these data are consistent with the notion that mH2A1.2 plays cell-type-specific roles, which appear to be evolutionarily conserved between humans and mice.
FIG 4.
Distinct mH2A1.2 nucleosome occupancy in human ESCs and human fibroblasts. (A) Numbers of genes occupied by single nucleosomes and polynucleosomes containing mH2A1.2 in regions spanning 5 kb on either side of the TSS in human ESCs and human FBSs. (B) Comparison of numbers of genes occupied by mH2A1.2 single nucleosomes and polynucleosomes in regions spanning 5 kb on either side of the TSS in human ESCs and human FBSs. (C) Snapshots (IGV) of genes bound by mH2A1.2, depicting the cell-type-specific density profiles of human ESCs (blue track) and human FBSs (red track) (compare top and bottom views for each corresponding region). The gene models (black) depict exons as boxes and introns as lines. (D) Comparison of genes occupied by mH2A1.2 single nucleosomes and polynucleosomes in regions spanning 5 kb on either side of the TSS in human and mouse ESCs. (E) Comparison of genes occupied by mH2A1.2 single nucleosomes and polynucleosomes in regions spanning 5 kb on either side of the TSS in hFBSs and MEFs.
The biological processes associated with the mH2A1.2 gene targets in hFBSs are different from those in hESCs (Table 1). More specifically, in hESCs, the mH2A1.2 target genes are mainly implicated in phosphoprotein regulation and various nuclear functions, such as chromatin/chromosome organization and histone core stress response, cell cycle, cell proliferation, and negative regulation of cell development. On the other hand, the genes bound by mH2A1.2 in hFBSs are implicated in glycoprotein regulation, cell differentiation, cell signaling, fibroblast growth factor receptor signaling, and mesenchyme development, including keratins, extracellular matrix, and cell surface molecules. Thus, it seems that in each cell type, mH2A1.2 marks genes that are related to the characteristic phenotype of that cell type. Furthermore, the common mH2A1.2 target genes between these cell types are associated mainly with cell signaling cascades (Table 2). On the other hand, the common mH2A1.2 target genes between hESCs and mESCs are implicated in phosphoprotein regulation, stress response, chromatin organization, cell cycle, etc. (Table 3), while the common mH2A1.2 target genes between hFBSs and MEFs are associated with regulation of gene expression via chromatin alterations and with housekeeping functions (Table 4). Taken together, these data suggest that mH2A1.2 nucleosomes may have cell-type-specific regulatory functions in hFBSs and hESCs but that they share similar functions in hESCs and mESCs. Therefore, mH2A1.2 could block cellular reprogramming by modulating, directly and/or indirectly, the expression of cell-specific genes.
TABLE 1.
GO categories of all mH2A1.2 target genes (within 5 kb on either side of the TSS)a in hESCs and human fibroblasts
| Biological function(s) (GO category) | Log10
P valueb |
|
|---|---|---|
| hESCs | hFBSs | |
| Phosphoprotein | 26.40 | |
| Nucleus | 14.27 | |
| Chromosome, chromosome organization | 13.36 | |
| Chromatin, chromatin assembly or disassembly, chromatin remodeling by hSWI/SNF ATP-dependent complexes | 8.80 | |
| Cell cycle, positive regulation of cell cycle | 8.00 | 1.06 |
| DNA metabolic process | 6.37 | 4.27 |
| Microtubule cytoskeleton, intermediate filament | 5.85 | 1.02 |
| Cell division, regulation of cell division, cell proliferation | 5.85 | |
| Histone core | 5.10 | |
| Protein transport, establishment of protein localization | 5.01 | |
| Protein biosynthesis, negative regulation of macromolecule biosynthetic process, regulation of cellular protein | 4.96 | |
| Nucleosome, histone H2A, histone H2B | 4.77 | |
| Methylation | 4.70 | |
| Translation | 4.44 | |
| Cellular response to stress | 4.43 | |
| Alternative splicing, RNA processing | 4.40 | 3.80 |
| DNA damage, DNA repair | 4.35 | |
| Repressor, negative regulation of transcription | 3.89 | |
| Transcription factor binding, transcription cofactor activity | 3.85 | |
| Ribosome | 3.32 | |
| Transcription, regulation of transcription | 2.29 | 1.10 |
| mRNA transport, posttranscriptional regulation of gene expression | 2.19 | |
| Negative regulation of cell development | 1.62 | |
| Regulation of DNA replication | 1.52 | |
| Ribosome biogenesis, ribosome binding | 1.49 | |
| Positive regulation of specific transcription from RNA polymerase II promoter | 1.44 | |
| Hemopoietic stem cell differentiation | 1.40 | |
| In utero embryonic development, developmental protein, development | 1.33 | 1.82 |
| Positive regulation of cell development, positive regulation of multicellular organism growth, developmental growth | 1.19 | |
| Neuron development, regulation of neurogenesis, regulation of nervous system development, neuron differentiation | 1.05 | 1.68 |
| Glycoprotein | 19.30 | |
| Signal, cell-cell signaling | 12.08 | |
| Secreted | 10.23 | |
| Receptor | 7.24 | |
| Keratin | 4.17 | |
| G-protein-coupled receptor protein signaling pathway | 4.07 | |
| Immune response, defense response | 3.22 | |
| Pattern binding, axonogenesis | 2.89 | |
| Epithelial cell differentiation | 1.66 | |
| Skeletal system development | 1.59 | |
| Extracellular matrix, collagen | 1.54 | |
| Diencephalon development | 1.44 | |
| Positive regulation of biosynthetic process | 1.42 | |
| Cell morphogenesis involved in differentiation | 1.39 | |
| Cell adhesion, cell surface | 1.36 | |
| Fibroblast growth factor receptor binding | 1.29 | |
| Mesenchyme development | 1.23 | |
All genes occupied by mH2A1.2.
The P values indicate the statistical significance of the GO category in the two distinct cell types.
TABLE 2.
GO categories of common mH2A1.2 target genes (within 5 kb on either side of the TSS)a in hESCs and human fibroblasts
| Biological function(s) (GO category) | Log10 P value |
|---|---|
| Intracellular signaling cascade | 2.80 |
| Protein kinase cascade | 2.60 |
| ER to Golgi vesicle-mediated transport | 2.37 |
| Extracellular space | 2.33 |
| Notch signaling pathway | 2.11 |
| Alternative splicing | 1.44 |
| DNA packaging | 1.19 |
| Wnt signaling pathway | 1.17 |
| Positive regulation of cell adhesion | 1.10 |
| Transcription factor binding | 1.07 |
| Negative regulation of cell differentiation | 1.05 |
Common genes in hESCs and hFBSs occupied by mH2A1.2.
TABLE 3.
GO categories of common mH2A1.2 target genes (within 5 kb on either side of the TSS)a in human and mouse ESCs
| Biological function(s) (GO category) | Log10 P value |
|---|---|
| Phosphoprotein | 26.60 |
| Nucleus | 10.02 |
| Repressor | 6.42 |
| Protein biosynthesis | 5.43 |
| RNA processing, RNA splicing | 4.89 |
| Cell cycle | 4.64 |
| Chromosome, chromosome organization | 4.54 |
| Ribosome | 4.36 |
| DNA damage, DNA repair | 4.33 |
| Microtubule cytoskeleton | 4.00 |
| Translation | 3.85 |
| DNA metabolic process | 3.80 |
| Cellular response to stress | 3.60 |
| Protein transport, establishment of protein localization | 3.52 |
| DNA packaging | 3.02 |
| Cell division | 2.92 |
| Transcription factor binding | 2.77 |
| Posttranscriptional regulation of gene expression | 2.74 |
| Chaperone | 2.60 |
| In utero embryonic development | 2.51 |
| Negative regulation of macromolecule metabolic process | 2.47 |
| Chromatin, chromatin organization | 2.46 |
Common genes in hESCs and hFBSs occupied by mH2A1.2.
TABLE 4.
GO categories of common mH2A1.2 targets (within 5 kb on either side of the TSS)a in hFBSs and MEFs
| Biological function(s) (GO category) | Log10 P value |
|---|---|
| l-Alanine transmembrane transporter activity, l-alanine transport | 4.60 |
| Telomere organization | 3.96 |
| Nuclear chromosome | 3.96 |
| Citrullination | 3.80 |
| DNA replication-dependent nucleosome assembly | 3.66 |
| Chromatin silencing at rDNA,b gene silencing by RNA | 3.42 |
| Protein heterotetramerization, protein heterodimerization activity | 3.21 |
| Histone binding, histone fold | 3.07 |
| Nucleosome core, nucleosome, nucleosome assembly | 3.06 |
| l-Proline transmembrane transporter activity | 2.92 |
| Negative regulation of gene expression, epigenetic | 2.92 |
| Positive regulation of gene expression, epigenetic | 2.57 |
| Extracellular exosome | 2.54 |
| Amino acid transport, amino acid transmembrane transporter activity, amino acid transporter, transmembrane | 2.38 |
| Cellular protein metabolic process, cellular amino acid metabolic process | 2.30 |
| Nuclear chromosome, telomeric region | 2.20 |
| Histone H4, conserved site | 2.17 |
| TATA box binding protein associated facor (TAF) | 2.06 |
| Cytokine-cytokine receptor interaction | 1.89 |
| Developmental protein | 1.89 |
| Phosphoprotein | 1.82 |
| Telomere capping | 1.72 |
| Methylation | 1.70 |
| DNA replication-independent nucleosome assembly | 1.62 |
| Alternative splicing | 1.57 |
| Extracellular region | 1.52 |
| Signal-anchor, positive regulation of intrinsic apoptotic signaling pathway | 1.43 |
| Cytosol | 1.41 |
| DNA-templated transcription, initiation | 1.36 |
| Cell membrane | 1.34 |
| Endosome, early endosome | 1.32 |
| Chromosomal rearrangement | 1.21 |
| Beta-catenin–TCF complex assembly | 1.21 |
| CENP-A-containing nucleosome assembly | 1.21 |
| Ubl conjugation | 1.13 |
| Transcriptional repressor activity, RNA polymerase II core promoter-proximal region sequence-specific binding | 1.10 |
| Transcription coactivator binding | 1.06 |
| Regulation of gene silencing | 1.01 |
Common genes in hESCs and hFBSs occupied by mH2A1.2.
rDNA, ribosomal DNA.
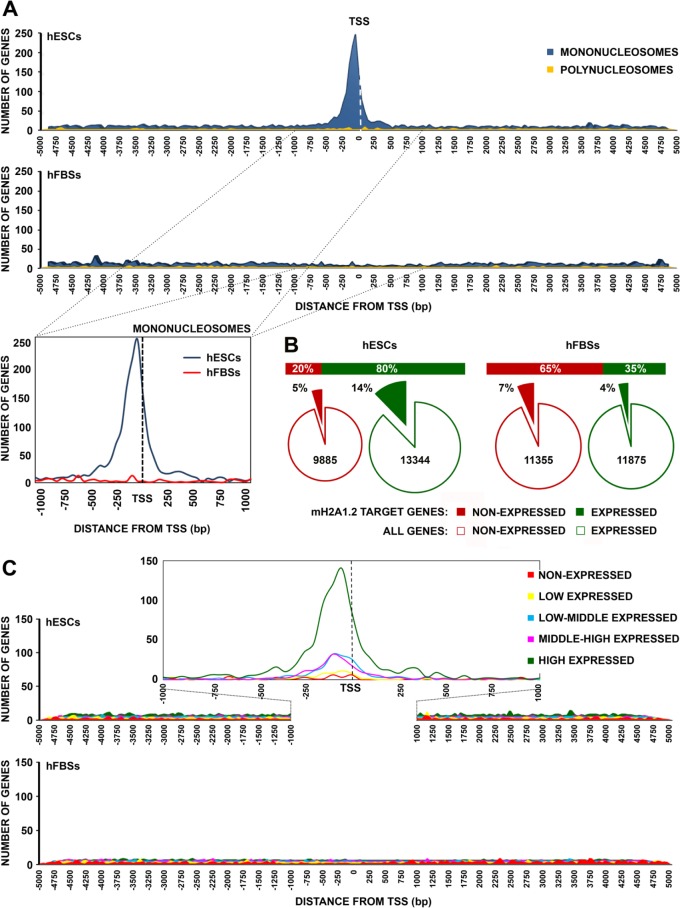
Next, we calculated the average frequency of mH2A1.2 binding to all transcriptional units of the mH2A1.2 targets from 5 kbp upstream to 5 kbp downstream of the TSS in hESCs and hFBSs. Figure 5A shows that single mH2A1.2 nucleosomes preferentially associate with the region spanning 250 bp on either side of the TSS in hESCs. In contrast, singular mH2A1.2 nucleosomes were rare in regions localized farther upstream or downstream of the TSS. On the other hand, in hFBSs, there was no preference for mH2A1.2 nucleosome binding at the TSS of the mH2A1.2 target genes.
FIG 5.
mH2A1.2 nucleosomes are enriched at the promoters of expressed genes in hESCs. (A) (Top) Distribution of mH2A1.2 mononucleosomes and polynucleosomes in the region spanning 5 kb on either side of the TSS of target genes in human ESCs and human FBSs. (Bottom) Distribution of mH2A1.2 mononucleosomes in the region spanning 1 kb on either side of the TSS of target genes in human ESCs and human fibroblasts. (B) (Top) Graph showing the categorization of mH2A1.2 target genes (within 5 kb on either side of TSS) as expressed or nonexpressed genes in human ESCs and human FBSs. (Bottom) Pie charts depicting the percentages of the nonexpressed mH2A1.2 target genes as a fraction of the total number of nonexpressed genes (red line). Similarly, the fractions of expressed mH2A1.2 target genes are depicted compared to the total number of expressed genes (green line). (C) mH2A1.2 is preferentially located at the promoters of active genes in hESCs. mH2A1.2 ChIP aggregated enrichment profiles around TSS in hESCs (top) and hFBSs (bottom). All expressed genes were identified by RNA-seq and divided into five groups according to their expression levels. Each line represents the average number of reads per transcript plotted relative to the TSS for each expression group. At the top is an enlargement of the region within 1 kb on either side of the TSS of all mH2A1.2 targets in hESCs. mH2A1.2 is depleted from the promoters of all expression categories in hFBSs (bottom).
Subsequently, all mH2A1.2 target genes in the two cell types were divided into quartiles according to their expression levels, as determined by RNA sequencing, while genes with zero reads were classified into a fifth group (33, 34). The genes in the last group, together with the genes of the first quartile (with very low expression values), were considered nonexpressed genes. As depicted in Fig. 5B, we discovered that most (65%) of the mH2A1.2 target genes in hFBSs were not expressed, whereas most (80%) of the mH2A1.2 target genes in hESCs were expressed. Importantly, our analysis also revealed that an expressed gene in hESCs had a higher probability (P < 0.001) of being a target of mH2A1.2 than a nonexpressed gene in hFBSs (14% and 7%, respectively), as determined by performing a chi-square test. Figure 5C shows that the mH2A1.2 nucleosome binding frequencies were significantly higher for the promoters of mid-level or highly expressed genes than for the promoters of low-level or nonexpressed genes in hESCs. In addition, as the expression levels of the genes increased, so did the number of genes in which mH2A1.2 nucleosomes were located in the TSS area. Importantly, the relative positions of the mH2A1.2 nucleosomes appeared to be similar in all expressed genes regardless of their actual expression levels, whereas mH2A1.2 nucleosomes were rare at TSS of nonexpressed genes.
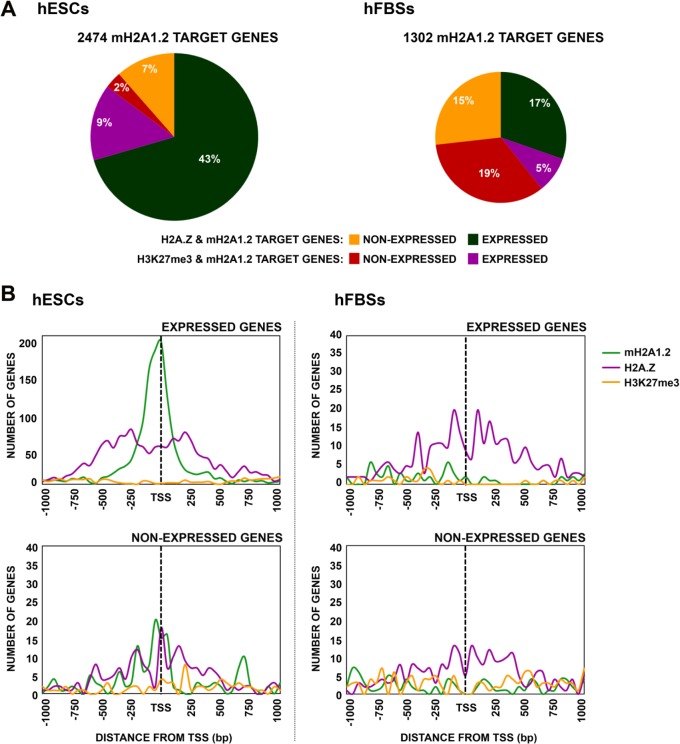
To investigate the relationship of the histone variants mH2A1.2 and H2A.Z with the repressive histone marker H3K27me3, we compared our mH2A1.2 ChIP-seq data with those for H2A.Z and H3K27me3 from hESCs and hFBSs, as determined by the ENCODE project (http://genome.ucsc.edu/). Remarkably, our comparative analysis revealed that 50% of all of our mH2A1.2 target genes in hESCs are also targets for H2A.Z nucleosomes. Of these, 86% correspond to expressed genes (Fig. 6A, left), while in hFBSs, the H2A.Z nucleosomes are found in ∼30% of the mH2A1.2 target genes without a preference for expressed or nonexpressed genes (Fig. 6A, right). On the other hand, the repressive histone modification H3K27me3 marker is found in only 11% and 24% of all mH2A1.2 target genes in hESCs and hFBSs, respectively (Fig. 6A). Taken together, these data support the notion that mH2A1.2 and H2A.Z nucleosomes share target genes expressed in hESCs, whereas H3K27me3 marks nonexpressed mH2A1.2 target genes in hFBSs.
FIG 6.
The histone variants mH2A1.2 and H2A.Z cooccupy TSS of highly expressed genes in ESCs. (A) Percentages of all mH2A1.2 target genes cobound by H2A.Z or marked by the H3K27me3 repressive mark in hESCs and hFBSs. (B) (Top) Distribution of H2A.Z nucleosomes and H3K27me3 modification in the region spanning 1 kb on either side of the TSS of expressed mH2A1.2 target genes in hESCs and hFBSs. (Bottom) Distribution of H2A.Z nucleosomes and H3K27me3 modification in the region spanning 1 kb on either side of the TSS of nonexpressed mH2A1.2 target genes in hESCs and hFBSs.
Next, we determined the exact positions of H2A.Z nucleosomes relative to the mH2A1.2 nucleosomes for the expressed and nonexpressed genes in hESCs and hFBSs. As seen in Fig. 6B (top left), we found that two H2A.Z nucleosomes flank the single mH2A1.2 nucleosome located at the TSS of expressed genes in hESCs, whereas the presence of mH2A1.2 and H2A.Z nucleosomes was rare in these regions in low-level or nonexpressed hESC genes (Fig. 6B, bottom left). Interestingly, despite the lower number of target genes, the relative distribution of H2A.Z in hFBSs is similar to that in hESCs in either expressed or nonexpressed genes (Fig. 6B, right), thus differing from that of mH2A1.2 (compare Fig. 5C with 6B).
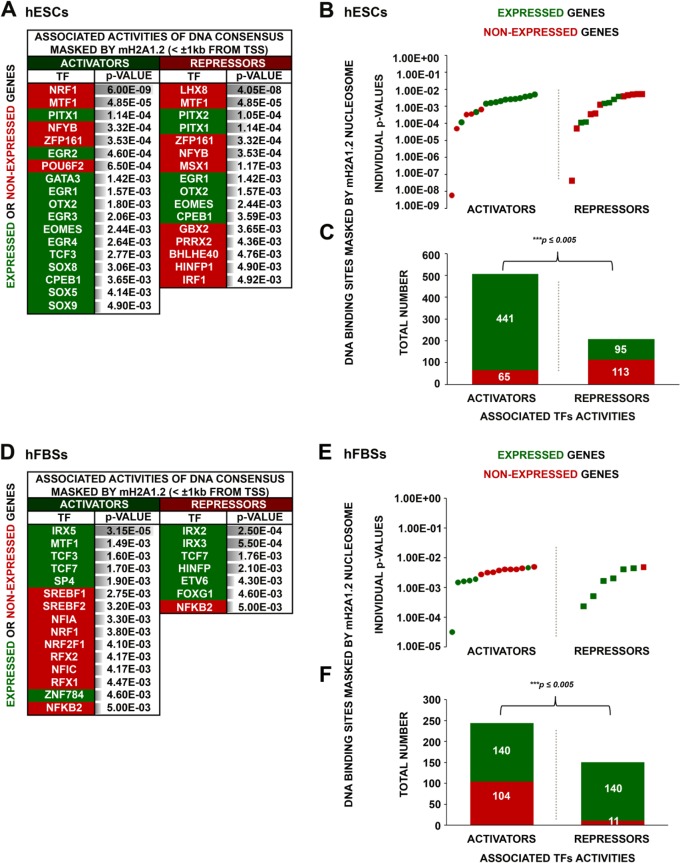
Next, we compared the DNA sequences underlying mH2A1.2 nucleosomes in expressed and nonexpressed genes in hESCs and discovered that in both cases, mH2A1.2 nucleosomes mask activator and repressor binding sites (Fig. 7A and B). Remarkably, mH2A1.2 nucleosomes mask the transcriptional activator's NRF1 binding sites in repressed genes with a probability much higher (Fig. 7A, P values) than the probability of masking other activator binding sites in either expressed or nonexpressed genes in hESCs (Fig. 7A and B). Similarly, mH2A1.2 nucleosomes mask the transcriptional repressor's LHX8 binding sites in repressed genes. Paradoxically, the total number of activator binding sites masked by mH2A1.2 nucleosomes in expressed genes is significantly higher than the respective number of repressor binding sites (Fig. 7C). On the other hand, in hFBSs, repressor binding sites are masked in expressed genes, whereas there is no preference for the masking of activator binding sites in expressed or nonexpressed genes (Fig. 7D and E). In addition, the total number of activator binding sites masked by mH2A1.2 in nonexpressed genes is higher than the respective number of repressor binding sites (Fig. 7F). Taken together, these data imply that mH2A1.2 nucleosomes play a direct bifunctional role in positive and negative control of transcription.
FIG 7.
Distinct transcriptional-regulatory elements are masked by mH2A1.2 nucleosomes in hESCs and hFBSs. (A) Main TF DNA binding consensus motifs corresponding to either activators or repressors identified within the footprint of the mH2A1.2 mononucleosomes for expressed or nonexpressed genes in hESCs. The relative enrichment (P value) for each predicted motif is shown. (B) Scatterplot depicting the probabilities calculated for each predicted motif (individual P values) corresponding to activators or repressors found in either expressed or nonexpressed genes in hESCs. (C) Sums of all activator and repressor putative DNA binding sites masked by mH2A1.2 mononucleosomes in expressed and nonexpressed genes in hESCs illustrating a higher proportion of activator masking sites in expressed genes. (D) Same as panel A, but for hFBSs. (E) Same as panel B, but for hFBSs. (F) Same as panel C, but for hFBSs, except that in hFBSs a higher proportion of repressor binding sites are masked in expressed genes.
Identification of an mH2A1.2-regulated gene-regulatory pathway active in MET during cellular reprogramming.
Next, we examined the 197 genes that are commonly bound by mH2A1.2 in hESCs and hFBSs (Fig. 4B). We found that 96 genes (49%) were not expressed in both cell types, whereas 77 genes (39%) were expressed in both hFBSs and hESCs. Intriguingly, the nonexpressed genes belong to a class of effectors associated with the negative regulation of gene expression via chromatin alterations (Table 5), whereas the expressed genes appear to control housekeeping functions, such as cell signaling and splicing (Table 6).
TABLE 5.
GO categories of common mH2A1.2 target (within 5 kb on either side of the TSS) nonexpressed genesa in human ESCs and human fibroblasts
| Biological function(s) (GO category) | Log10 P value |
|---|---|
| Chromatin silencing at rDNA | 54.23 |
| Telomere organization, nuclear chromosome, telomeric region | 50.66 |
| Negative regulation of gene expression, epigenetic | 49.41 |
| DNA replication-dependent nucleosome assembly | 47.74 |
| Positive regulation of gene expression, epigenetic | 46.35 |
| Nucleosome core, nucleosome, nucleosome assembly | 44.64 |
| Protein heterotetramerization, protein heterodimerization activity | 43.85 |
| Citrullination | 43.51 |
| Histone fold, histone binding | 40.51 |
| Gene silencing by RNA | 38.89 |
| Histone H4, conserved site | 31.06 |
| TATA box binding protein associated factor (TAF) | 29.46 |
| Telomere capping | 26.27 |
| DNA replication-independent nucleosome assembly | 25.31 |
| DNA-templated transcription, initiation | 22.96 |
| Histone H3 | 22.30 |
| Beta-catenin-TCF complex assembly | 21.80 |
| CENP-A-containing nucleosome assembly | 21.80 |
| Double-strand break repair via nonhomologous end joining | 19.36 |
| Extracellular region | 18.96 |
| Regulation of gene silencing | 17.51 |
| Methylation | 16.52 |
| Chromosomal rearrangement | 11.43 |
Common nonexpressed genes in hESCs and hFBSs occupied by mH2A1.2.
TABLE 6.
GO categories of common mH2A1.2 target (within 5 kb on either side of the TSS) expressed genesa in human ESCs and human fibroblasts
| Biological function(s) (GO category) | Log10 P value |
|---|---|
| Cytosol | 4.10 |
| Phosphoprotein | 4.02 |
| Neuroblastoma breakpoint family (NBPF) domain | 2.74 |
| Membrane | 2.62 |
| Alternative splicing | 2.25 |
| Nucleus | 1.96 |
| Regulation of macroautophagy | 1.89 |
| DNA replication, recombination, and repair | 1.44 |
| Kinase | 1.41 |
| Proteasome-mediated ubiquitin-dependent protein catabolic process | 1.36 |
| Beta-catenin destruction complex | 1.29 |
| Late endosome | 1.12 |
| Nucleus-transcribed mRNA catabolic process, nonsense-mediated decay | 1.11 |
| Oxidation-reduction process | 1.10 |
| Centromere | 1.10 |
| UBL conjugation pathway | 1.09 |
| Regulation of signal transduction by p53 class mediator | 1.08 |
| Cell surface receptor signaling pathway | 1.05 |
| Ligase activity | 1.04 |
| Translation initiation | 1.00 |
Common expressed genes in hESCs and hFBSs occupied by mH2A1.2.
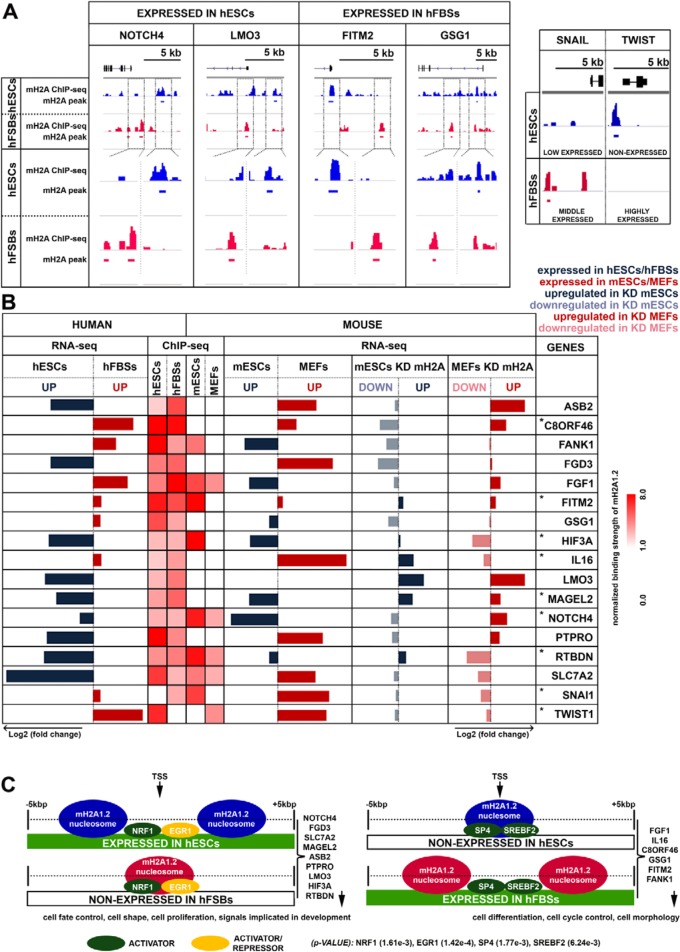
Remarkably, we discovered that the remaining 24 commonly mH2A1.2-bound genes displayed opposite expression patterns in hESCs and hFBSs. Among these, 15 genes displayed the most significant opposite expression patterns, and 6 of these genes were expressed in hFBSs and not in hESCs (IL-16, FGF1, GSG1, FANK1, FITM2, and C8ORF46 genes), whereas 9 genes were expressed in hESCs but not in hFBSs (NOTCH4, FGD3, SLC7A2, MAGEL2, ASB2, PTPRO, LMO3, HIF3A, and RTBDN genes) (Fig. 8). In addition, 7 out of the 15 genes displayed similar expression patterns in both mESCs and hESCs, implying that they may have an evolutionarily conserved function (Fig. 8B). Furthermore, mH2A1.2-containing nucleosomes bound at distinct sites at the promoters of these genes (Fig. 8A), masking mainly activator binding sites (Fig. 8C). For example, in the case of FITM2, the mH2A1.2 nucleosome masked the activator sites of SP4 and SREBF2 in the nonexpressing hESCs, whereas the same sites were nucleosome free in the expressing hFBSs (Fig. 8A and C). On the other hand, in the case of NOTCH4, the mH2A1.2 nucleosome masked the NRF1/EGR1 activator sites in the nonexpressing hFBSs, whereas the same sites were nucleosome free in the expressing hESCs (Fig. 8A and C). In general, the region spanning 2 kbp on either side of the TSS of the expressed genes in hFBSs was free of mH2A1.2 nucleosomes, whereas the opposite was true for the same region in hESCs. Reciprocally, the region spanning 2 kbp on either side of the TSS of genes expressed in hESCs was free of nucleosomes, whereas the same regions containing activator binding sites were masked by mH2A1.2 nucleosomes in the nonexpressed genes in hFBSs (Fig. 8C).
FIG 8.
mH2A1.2 nucleosomes occupy the promoters of a specific set of genes and direct opposite expression programs in hESCs and hFBSs. (A) Snapshots of ChIP-seq profiles (IGV) of genes bound by mH2A1.2 depicting the distinct nucleosome positioning in hESCs (blue track) and hFBSs (red track) (compare top and bottom views for each corresponding region). The ChIP-seq profiles of the key MET regulators, SNAI1 and TWIST1, are shown on the right. The gene models (black) depict exons as boxes and introns as lines. (B) Comparison of the expression levels of the commonly bound mH2A1.2 target genes and the key regulators of MET (SNAI1 and TWIST1 [bottom]) as determined by RNA-seq in hESCs and hFBSs. The heat map (right) displays the ChIP-seq data for mH2A1.2 nucleosomes (within 5 kb on either side of the TSS) of the common target genes with opposite expression patterns in hESCs and hFBSs and the corresponding analysis of the same genes in mESCs and MEFs. The right part of the table displays our RNA-seq analysis in WT mESCs and MEFs, as well as in mH2A1.2-KD mESCs and MEFs. The asterisks denote genes with similar expression patterns in both hESCs/mESCs and in hFBSs/MEFs. (C) Relative positioning of mH2A1.2 nucleosomes in the region within 5 kb on either side of the TSS of the common target genes in hESCs and hFBSs that are differentially expressed in these cell types.
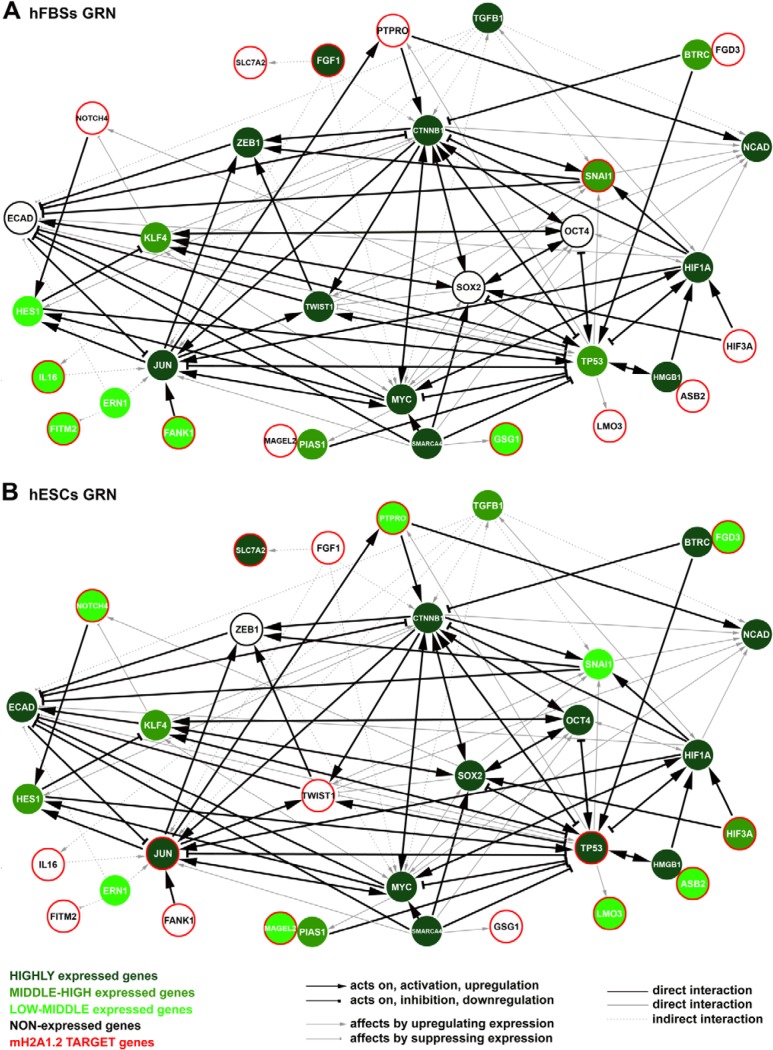
Although our studies on mH2A1.2 nucleosome occupancy provide various mechanistic insights, they do not provide a global view of how mH2A1.2 blocks cellular reprogramming. To do so, we integrated gene interaction data from publicly available databases with gene expression data from all available databases, along with our ChIP-seq and RNA-seq experiments, and we reconstructed a GRN centered around genes that are key factors of cellular reprogramming and direct downstream targets of mH2A1.2. The integration process resulted in the establishment of distinctive GRNs for hESCs and hFBSs composed of 33 nodes and 86 edges (Fig. 9). We focused on key connector nodes with known roles in MET, like SNAI1, TWIST1, TGFB1, HIF3A, etc., and discovered high interconnectivity of mH2A1.2 with transcription factors previously known to block reprogramming, like p53 (35); to stabilize cell fate, like JUN (36); to regulate the cadherin switch, like SNAI1 and TWIST1 (20, 37); and others. These data are consistent with the existence of highly regulated nodes defined by the actions of a diverse set of genes with modulatory and effector activities.
FIG 9.
mH2A1.2-centered gene regulatory networks in hESCs and hFBSs define cell fates. GRNs for hFBSs (A) and hESCs (B) were constructed by integrating the experimentally verified interactions of key molecules of cellular reprogramming with direct downstream targets of mH2A1.2 based on our ChIP-seq and RNA-seq analyses.
Our GRN representations connect the specific set of 15 mH2A1.2 target genes identified by ChIP-seq that display opposite expression patterns in hFBS and hESCs with known regulators of cell fate determinants. Nearly all previously known regulators of hFBS and hESC identity were predicted to be under the combined control of two or more regulators (Fig. 9), some of which are directly regulated by mH2A1.2 (e.g., JUN and TP53 in hESCs and SNAI1 in hFBSs) and/or indirectly regulated, as H2A1.2 regulates their regulators. The visualization of our hFBS and hESC GRNs revealed an architecture in which most of the key cell fate regulators are localized at the center of each network, whereas the mH2A1.2 target genes are placed at the peripheries of the networks. Thus, mH2A1.2 appears to function as a gatekeeper of cell fate, a notion consistent with its previously defined role in generating robustness in gene expression (3).
DISCUSSION
We report here that mH2A1.2 is a negative regulator of human cell reprogramming by blocking MET. We provided a comprehensive analysis of the genomic sites occupied by mH2A1.2 nucleosomes in hFBSs and in hESCs and how they affect the reprogramming of hFBSs to pluripotency. We have reconstructed mH2A1.2-centered GRNs that support the hFBS and hESC fates. Our data show high complexity of mH2A1.2-centered GRNs in hFBSs and hESCs, with distinct factors playing critical roles in specifying each cell type by promoting and/or limiting progression to alternative cell fates. We propose that the exact positions of mH2A1.2 nucleosomes in specific genes with key regulatory roles guarantee the robustness of both networks.
Previous studies have demonstrated that mH2A1.2 acts as a barrier to cellular reprogramming of both mouse embryonic fibroblasts and human keratinocytes (23–25). However, human keratinocytes represent an epithelial cell type, which, in contrast to hFBSs, may not need to go through MET to be reprogrammed (38). Here, we demonstrated that mH2A1.2 blocks reprogramming of hFBSs by affecting both the efficiency and the kinetics of the process. We demonstrated that in mH2A1-KD cells the N-CAD-to-E-CAD switch occurs earlier than in WT cells and that mH2A1.2 overexpression blocks the activation of E-CAD expression without affecting N-CAD expression. Furthermore, we showed that E-CAD is not a direct target of mH2A1.2 but that its expression is affected by a gene-regulatory network influenced by mH2A1.2 nucleosomes controlling the expression of key regulatory genes, like the SNAI1 gene.
The presence of mH2A1.2 nucleosomes correlates with nonexpressed genes in hFBSs, while in hESCs, it is related to expressed genes. As the expression levels of the hESCs genes increase, so does the number of the genes bearing mH2A1.2 nucleosomes around their TSS. Therefore, other factors, such as chromatin modifications or other histone variants and/or transcription factors, may play an essential role in the deposition of mH2A1.2 on the genomes of different cell types. Our analyses revealed that H2A.Z-containing nucleosomes flank the singular mH2A1.2 nucleosomes at the TSS of expressed genes in hESCs, whereas in hFBSs, the two histone variants appear to be segregated. In general, in hFBSs, mH2A1.2 nucleosomes mask activator binding sites in nonexpressed genes, whereas in expressed genes they mask repressor binding sites. Consistent with this is the observation that mH2A1.2 nucleosomes colocalize with the repressive histone marker H3K27me3 at pluripotency-related genes in MEFs (25). Similarly, we found that mH2A1.2 and H3K27me3 coexist in nonexpressed genes in hFBSs, in agreement with previous results in human keratinocytes (24).
We inferred that mH2A1.2-centered cell-specific GRNs are key components in understanding biological processes. The hFBS and the hESC cellular phenotypes are established and maintained by the temporal regulation and dynamics of networks of coregulated genes. Key regulators in the two networks are mH2A1.2 target genes involved in maintaining the hFBS and hESC identities. Our data are consistent with the notion that the differential expression of these key genes results in the establishment of distinct GRNs that uniquely determine the hFBS and hESC phenotypes. Specifically, the SNAI1 gene is an mH2A1 target gene in hFBSs, and its expression is maintained at steady-state levels by mH2A1.2, since mH2A1.2-KD decreases its expression (Fig. 8B). Furthermore, in hFBSs, the GRN implies that the mH2A1.2-dependent robust expression of FGF1 and SNAI1 reinforces the mesenchymal phenotype, along with the sustained expression of TWIST1 and ZEB1, all of which are required for E-CAD repression (20, 29, 30, 37, 39–43). At the same time, the network ensures that HIF3A and NOTCH4 (44, 45) are repressed in an mH2A1.2-dependent manner, and thus, the pluripotency factors OCT4 and SOX2 remain repressed. In summary, our hFBS network suggests that the high expression levels of JUN, which inhibits reprogramming by stabilizing the hFBS cell state (35), are maintained by the direct and/or the indirect effects of the mH2A HIF3A, SLC7A2, FANK1, IL-16, FGF1 and SNAI1 target genes (46–51).
The hESC state is characterized by the absence of TWIST1 expression due to the presence of mH2A1.2 nucleosomes and, consequently, by the lack of ZEB1 expression, all of which lead to derepression of E-CAD expression (52). Notably, the knockdown of mH2A1.2 does not significantly affect the expression of TWIST1 in mESCs (Fig. 8B). Thus, the low levels of SNAI1 expression in the inhibition of E-CAD transcription are counterbalanced by the higher expression levels of KLF4, which induce the transcription of E-CAD (53). In hESCs, the mH2A1.2-dependent expression of HIF3A and NOTCH4 contributes to the expression of KLF4, which regulates the expression of E-CAD and induces the expression of the pluripotent OCT4 and SOX2 genes. In addition, the mH2A1.2-suppressed FGF1 expression leads to attenuation of the TGFB1 pathway required for the expression of ZEB1 and TWIST1. Notably, TP53 and JUN genes constitute mH2A1.2 target genes in hESCs displaying high levels of expression. p53 blocks MET (35), although its transient suppression increases reprogramming efficiency (54). In agreement with this, p53 suppresses the expression of OCT4 and SOX2 (55). Therefore, our network suggests that MYC may act as an intermediate between the mH2A1.2 targets JUN and TP53, as JUN increases the expression of MYC, whereas p53 suppresses MYC expression (Fig. 9A and B).
Our GRNs provide essential insights into the factors and the role of mH2A1.2 nucleosomes in establishing specific cell fates. However, the actual molecular mechanism by which this could occur remains elusive because of the limited specificity displayed by metazoan transcription factors and by the extensive cross talk of signaling pathways (1). Thus, the robustness of regulatory networks must depend on increased specificity of molecular recognition events leading to reduction of noise and erroneous alterations in gene expression. Furthermore, as regulatory systems grow in size and complexity with evolution, the number of potentially noisy interactions grows as well. As a result, biological systems have developed constraints to minimize noisy interactions and increase robustness. In this regard, our GRNs provide clues concerning the outer limits of the network and how they are defined to ensure reliable gene expression. We note that cross-interactions between components of our GRNs and other regulators could disturb the robustness of the networks and lead to erroneous gene expression with significant consequences for cell fate determination. We found that the presence of mH2A1.2 nucleosomes in the regulatory regions of GRN genes that either “feed” the network or play important roles in on/off switches guarantees less expression noise and, thus, reliable gene expression. Furthermore, our finding that the ESC-specific deposition of mH2A1.2 nucleosomes at the promoters of JUN and TP53 genes, which are known to inhibit reprogramming (35, 36) and which have multiple specific regulatory partners (hubs) in our GRN, ensures that mH2A1.2 can buffer their expression variability to avoid nonspecific interactions and the subsequent loss of the stemness phenotype, since JUN and p53 could function as cross talk interaction points with other, irrelevant networks. We propose that the presence of mH2A1.2 nucleosomes reduces cross talk by restricting the access of transcription factors exclusively to cognate sites.
We have previously demonstrated that composite mH2A1.2 nucleosomes containing the transcription factor NRF-1 can buffer transcriptional noise by blocking low-affinity and unwanted access of transcription factors to “undesired” regulatory elements, which could interfere with gene expression programs (3). Here, we also showed that mH2A1.2 nucleosomes are bound with extreme cell type specificity to regulatory regions of genes that play key roles in the establishment of cell fate. Transcription factors like NRF-1 and LHX8 facilitate the assembly of such high-affinity bound mH2A1.2 nucleosomes. We postulate that the reconstructed GRNs acquire functional robustness, at least via the ability of strategically positioned mH2A1.2 nucleosomes on key regulatory genes to minimize variability in gene expression despite the existence of extensive cross talk between the various factors perturbing the normal system.
MATERIALS AND METHODS
Cell culture.
Primary human fibroblasts (XY) were isolated from adult forearm skin by biopsy, cultured, and expanded in fibroblast medium consisting of Dulbecco's modified Eagle's medium (Sigma) supplemented with 15% fetal bovine serum (Gibco), 2 mM l-GlutaMAX (Gibco), 0.1 mM minimal essential medium (MEM) nonessential amino acids (Gibco), 1% penicillin-streptomycin (Gibco), and 0.1 mM beta-mercaptoethanol (AppliChem).
The human ESC line HuES3 (XY) (56) was maintained on a feeder layer of mitomycin C (Sigma)-inactivated mouse embryonic fibroblasts. Human ESCs and human iPSCs derived from human fibroblasts were grown in ESC medium consisting of Dulbecco's modified Eagle's medium, nutrient mixture F-12 (Gibco) supplemented with 20% knockout serum replacement (Gibco), 2 mM l-GlutaMAX, 0.1 mM MEM nonessential amino acids, 1% penicillin-streptomycin, 0.1 mM beta-mercaptoethanol, and 10 ng/ml recombinant human fibroblast growth factor-basic (bFGF) (Pepro Tech).
Viral constructs and lentivirus production.
The cassette containing the 4 human Yamanaka factors (OSKM, linked by 2A peptides, T2A, P2A, and E2A, respectively) (57) was cloned into a Dox-inducible lentiviral vector. Briefly, the cassette was amplified by PCR, using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific), from the pLM-CMV-fsv2a vector (57) with the following primers: F, 5′-GAGAGAATTCGTTATGGCGGGACACCTGG-3′, and R, 5′-GATTGAATTCTCACATGTGTGAGAGGGGC-3′. The PCR product was digested with AgeI/SalI enzymes and subsequently ligated into the FUW-TetO vector (Addgene; 20321) digested by the same enzymes. The reverse tetracycline transactivator M2rtTA was expressed under a constitutively active human ubiquitin C promoter from the FUW-M2rtTA vector (Addgene; 20342).
The shRNAs targeting exon 9 of the mH2A1 gene affect the expression of both mH2A1 isoforms, mH2A1.1 and mH2A1.2 (shmH2A1), and the nonsense targeted sequences (scramble) were cloned into the TRC2-pLKO.1 vector (Sigma) in the AgeI/EcoRI multiple-cloning site. The same shRNAs were cloned into another TRC2-pLKO.1 vector in which GFP was cloned before the puromycin sequence, as previously described (58). The sense/antisense primers that were used for shmH2A1 were pLKO.1 mH2A1 sense, 5′-CCGGCCAGTTACTTCGTGTCTACAACTCGAGTTGTAGACACGAAGTAACTGGTTTTTG-3′, and pLKO.1 mH2A1 antisense, 5′-AATTCAAAAACCAGTTACTTCGTGTCTACAACTCGAGTTGTAGACACGAAGTAACTGG-3′, and for scramble they were pLKO.1 scramble sense, 5′-CCGGAACAGTCGCGTTTGCGACTGGCTCGAGCCAGTCGCAAACGCGACTGTTTTTTTG-3′, and pLKO.1 scramble antisense, 5′-AATTCAAAAAAACAGTCCGTTTGCGACTGGCTCGAGCCAGTCGCAAACGCGACTGTT-3′.
The construct that bears the cassette containing mH2A1.2-myc-His-internal ribosome entry site (IRES)-GFP (mH2A1.2/myc-tag) or no insert (empty) was previously described (3). To generate the H2A-myc-his-IRES-GFP lentivirus, we subcloned the H2A gene obtained by PCR using Q5 High-Fidelity DNA polymerase (New England BioLabs) from human genomic DNA with primers F, 5′-GAGAAGCGCTAGTTATGTCTGGGCGTGGC-3′, and R, 5′-ACCGGTCAGATCTTCTTCAGAAATAAGTTTTTGTTCCTTGCCCTTCGCCTTGTGGTGGCTC-3′.
For the production of viral particles, 293T cells were cotransfected by the classical calcium-phosphate method at 20 to 30% confluence with a mixture of viral plasmids bearing the gene of interest and packaging constructs, such psPAX2 (Addgene; 12260) and pMD2G (Addgene; 12259). The medium was replaced 24 h after transfection, and the virus-containing supernatant was collected at 72 h, filtered through a 0.45-μm filter, and used for lentiviral infection.
iPSC derivation.
Human fibroblasts were seeded 24 h before transduction in a 10-cm dish. Twenty-four hours later, the cells were infected with a mixture of virus supernatant containing the OSKM factors, M2rtTa, and either the shmH2A1 or scramble or either the mH2A1.2/myc-tag or empty vector in a ratio of 1:1:1 in the presence of 6 μg/ml Polybrene. The culture medium was changed after 24 h, and 2 days later, the human fibroblasts were replated on Matrigel (Corning)-coated dishes. Day 0 was determined when Dox (Sigma) was added to the medium. It was added until day 30. Until day 7, the cells were grown in fibroblast medium. From day 8 to day 19, the cells were cultured in ESC medium supplemented with 15% HyClone (Gibco), 5% knockout serum replacement, while until day 30 they were cultured in ESC medium. In each case, the medium was refreshed every 48 h.
RNA extraction and cDNA synthesis.
Total RNA was purified with TRIzol reagent (Invitrogen) and treated with DNase (Roche) to remove genomic DNA contamination. The RNA concentration was determined spectrophotometrically at 260 nm, while the quality of purification was determined by the 260/280-nm ratio (1.7 to 2.0), which indicates high RNA quality. One microgram of total RNA was used for reverse transcription reactions with Superscript II (Invitrogen) and oligo(dT) primers, according to the manufacturer's instructions.
Quantitative real-time PCR assay.
Duplicates or triplicates of each sample were assayed by quantitative real-time PCR using KAPA SYBR green qPCR master mix (KAPA Biosystems) and analyzed with a CFX96 real-time PCR system (Bio-Rad). The primers used for each target were as follows: for N-CAD, F, 5′-TCCCATCATAATCACAGATTCGG-3′, and R, 5′-CAGCACAAGGATAAGCAGGA-3′; for E-CAD, F, 5′-CCAGAATAAAGACCAAGTGACCAC-3′, and R, 5′-CCTCCGAAGAAACAGCAAGAG-3′; for SNAI1, F, 5′-ATGCCGCGCTCTTTCCTCG-3′, and R, 5′-GCCAGGACAGAGTCCCAGA-3′; for NANOG, F, 5′-TACCTCAGCCTCCAGCAGAT-3′, and R, 5′-CCTTCTGCGTCACACCATT-3′; and for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), F, 5′-GAAGGTGAAGGTCGGAGTCA-3′, and R, 5′-TTGAGGTCAATGAAGGGGTC-3′. The GAPDH gene was used as a reference gene for normalization.
Alkaline phosphatase staining and immunocytochemistry.
In order to detect AP activity, iPS colonies were fixed with 4% formaldehyde for 10 min at 4°C and, after washing with phosphate-buffered saline (PBS), were incubated for 15 min at room temperature (RT) in AP buffer (100 mM Tris-HCl, 100 mM NaCl, 5 mM MgCl2, 0.05% Tween 20, pH 9.5). iPSC colonies were stained with nitroblue tetrazolium (NBT)/BCIP (5-bromo-4-chloro-3-indolylphosphate) (Roche) diluted in AP buffer for 30 min in the dark. AP+ cells were detected and counted to calculate the efficiency of reprogramming.
For immunocytochemistry, cells were fixed with 4% paraformaldehyde for 15 min at RT, washed with PBS, and treated with PBS containing 10% fetal bovine serum, 0.1% Triton X-100 for 1 h at RT. Primary antibody to E-CAD (sc-21791; Santa Cruz; 1:50 dilution) was incubated overnight at 4°C. Secondary antibody, Alexa Fluor 568-conjugated goat anti-mouse IgG (Abcam; 1:500 dilution), was incubated for 1 h at RT. Nuclei were stained with 1 mg/ml DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen).
Western blotting.
The cells were counted and lysed with sodium dodecyl sulfate (SDS) loading buffer (50 mM Tris-HCl, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol, pH 6.8). After heating at 100°C for 5 min, protein extracts were separated by SDS-15% polyacrylamide gel electrophoresis and transferred to a nitrocellulose blotting membrane (GE Healthcare). The blots were blocked with 5% skim milk in PBS-0.1% Tween 20 (PBST) for 1 h at RT. The primary antibodies anti-mH2A1.2 (1:5,000) (59) and anti-GAPDH (1:10,000) (Ambion) were diluted in PBST containing 2% skim milk and incubated at 4°C overnight. Secondary antibodies conjugated to horseradish peroxidase (HRP), anti-rabbit IgG–HRP (Cell Signaling) and anti-mouse IgG–HRP (Cell Signaling), and diluted in Tris-buffered saline with Tween 20 (TBST) (1:5,000) were incubated for 1 h at RT. Signals were detected with Immobilon Western chemiluminescent HRP substrate (Millipore). Following the scanning of the images, Gel Analyzer software version I was used to quantify the intensity of the bands. GAPDH was used as a reference protein for normalization.
Isolation of mononucleosomes (S1) and polynucleosomes (S2).
Human fibroblasts and human ESCs were grown to 90% confluence in 5- by 15-cm dishes and 10- by 10-cm dishes, respectively. The cells were resuspended in 2 ml of N-ChIP buffer I (0.3 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM Tris-HCl, 0.5 mM dithiothreitol [DTT], pH 7.5). Next, the cells were lysed by adding 2 ml of N-ChIP buffer II (0.3 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM Tris-HCl, 0.4% NP-40, 0.5 mM DTT, pH 7.5), and the nuclei were purified with a sucrose cushion by layering lysed cells on 8 ml of N-ChIP buffer III (1.2 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM Tris-HCl, 0.5 mM DTT, pH 7.5), followed by centrifugation at 3,000 rpm for 30 min. The supernatant was discarded, and the nuclear pellet was resuspended in 1 ml of MNase digestion buffer (0.32 M sucrose, 50 mM Tris-HCl, 4 mM MgCl2, 1 mM CaCl2, pH 7.5). The chromatin was digested by adding 1.5 μl micrococcal nuclease (NEB; 2,000 gel units/μl) for 5 min at 37°C. The reaction was stopped by adding EDTA (pH 8.0) and EGTA to a final concentration of 10 mM, and the suspension was centrifuged at 10,000 rpm for 10 min. The supernatant S1 is the soluble fraction containing mainly mononucleosomes and a small amount of di- and trinucleosomes. Optionally, the pellet was suspended in 1 ml dialysis buffer (1 mM Tris-HCl, pH 7.5, 0.2 mM EDTA) and dialyzed against 1 liter of dialysis buffer overnight at 4°C. The suspension was centrifuged at 10,000 rpm for 10 min, and the supernatant, S2, containing higher-molecular-weight chromatin (polynucleosomes, i.e., dinucleosomes and higher) was retained. Fifty microliters of S1 chromatin was transferred to a new 1.5-ml tube, and after adding 2.5 μl 10% SDS, it was stored at 4°C for the input control.
N-ChIP.
Mononucleosome (S1) extracts were diluted 1:4 in N-ChIP incubation buffer (50 mM NaCl, 50 mM Tris-HCl, 5 mM EDTA, pH 7.5) with either the appropriate antibody (2 μg affinity-purified anti-mH2A1.2 raised against mH2A1.2-specific residues in rabbit [59]) or IgG control and incubated overnight at 4°C. Next, the appropriate amount of equilibrated protein G Dynabeads (Life Technologies) was added and incubated for 2 h at 4°C. The beads were collected on a magnetic rack and washed two times in washing buffer A (50 mM Tris-HCl, 10 mM EDTA, 75 mM NaCl, pH 7.5), washing buffer B (50 mM Tris-HCl, 10 mM EDTA, 125 mM NaCl, pH 7.5), and washing buffer C (50 mM Tris-HCl, 10 mM EDTA, 175 mM NaCl, pH 7.5). Bound chromatin was eluted with proteinase K in proteinase K digestion buffer (20 mM HEPES, 1 mM EDTA, 0.5% SDS, pH 7.9) at 50°C for 30 min. Finally, the DNA was purified with Ampure XP beads in 30% polyethylene glycol (PEG) 8000), 1.25 M NaCl and then subjected to library preparation for Illumina sequencing.
Illumina library preparation.
ChIP-DNA (from N-ChIP) and input DNA (3) were used to generate Illumina sequencing libraries (60) with minor modifications. Briefly, Illumina adapters were ligated to polished ends of DNA fragments and the libraries were enriched by limited amplification (as determined by quantitative PCR [qPCR]) (59). The libraries were sequenced with Illumina NextSeq500 at the Greek Genome Center.
High-throughput sequencing data analysis.
The quality of the reads was estimated with FastQC. The first and last 5 nucleotides of each read were trimmed with the BOWTIE 0.12.7 trim option. The reads were mapped to hg19 using BOWTIE 0.12.7 (parameters: m = 4; v = 2) and then shuffled to be normalized to their input sequences and to equilibrate the sequencing depth of the experiments. A total of 7 × 106 and 10 × 106 reads were used for peak calling for hESCs and hFBSs, respectively. The aligned sequences were attributed to significantly enriched genomic regions using MACS1.4.2 (no model; sf = 75 bp; keep dup = 1) (31) and SICER 1.1 (window size = 150; fragment size = 75; effective genome fraction = 0.75; gap size = 150; false-discovery rate [FDR] = 100) algorithms (32). The total numbers of identified peaks were 19,464 peaks for hESCs and 27,532 peaks for hFBSs. Intersections of data sets via BEDTOOLS were performed in order to compare the mH2A1.2 targets among the cell types. In addition, intersecting the mH2A1.2 peaks obtained by MACS with the wide genomic interval bound by mH2A1.2, obtained by SICER, distinguished the mH2A1.2 mononucleosomes from mH2A1.2 polynucleosomes (<800). The relative genomic enrichment of ChIP peaks was detected using the CEAS package 1.0.2 (default parameters) (61). The ontologies of the respective genes were identified though the DAVID bioinformatics database (62).
Data corresponding to H3K27me3 and H2A.Z occupancy in the H1 ES cell line (male) and fibroblast BJ cell line (male) or NHDF cells (female) were downloaded from ENCODE (http://genome.ucsc.edu/).
Gene expression analysis.
The published RNA-seq data for the human ESC line HuES3 (33) and for fibroblasts isolated from human forearm skin (34) (SRR2453370 and SRR1014764, respectively) were downloaded from GEODATASETS (http://www.ncbi.nlm.nih.gov/gds) and reanalyzed. Alignment of the reads was performed with TopHat v1.3.1 (default parameters) (63). The quantification of the overlap of the aligned reads with all known genes was performed with HtSeq-count version 0.5.3p3 (default parameters) (64). The genes were divided into quartiles according to their expression values, while genes with zero expression were classified into a separate, fifth group.
Motifs and gene ontology (GO) analyses.
The annotation and categorization of DNA motifs enriched in expressed and nonexpressed genes in both human ESCs and human fibroblasts were performed as follows. First, all regions of 150 bp within 1 kb on either side of the TSS for either expressed or nonexpressed genes in each cell type bound by mH2A1.2, as defined by MACS-SICER analysis, were converted to Fasta files. Subsequently, motifs enriched for each category were identified with MEME Suite 4.11.2 (http://meme-suite.org/) using the default parameters (63). The associated activities of TFs (activators, repressors, or both) were determined from the Genecards database (http://www.genecards.org/).
Network analysis.
Ingenuity pathway analyses (IPA) (Ingenuity Systems) were used to identify experimentally verified interactions among the master regulators of epithelial-mesenchymal transition (EMT)/MET (SNAI1, TWIST1, and ZEB1), the characteristic transmembrane proteins for epithelial and mesenchymal cell types (E-CAD and N-CAD, respectively), the four Yamanaka factors (OCT4, SOX2, KLF4, and MYC), and the 15 common target genes of mH2A1.2 with differential expression between the two cell types (hFBSs and hESCs) (expressed in hFBSs: FGF1, GSG1, FITM2, FANK1, and IL-16) (expressed in hESCs: NOTCH4, SLC7A2, FGD3, MAGEL2, ASB2, PTPRO, LMO3, HIF3A, RTBDN, and C80RF46).
Accession number(s).
Raw sequence data were submitted under the study accession number SRP105049 with sample accession numbers SRR5469273, SRR5469274, SRR5469275, and SRR5469276 to the Sequence Read Archive (SRA) database (https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?cmd=show&f=main&m=main&s=main).
ACKNOWLEDGMENTS
We thank Kostas Vekrellis for providing biopsy specimens of human skin fibroblasts, Rebecca Matsas for providing the human ES cell line HUES3, Stefanos Tsiftsoglou for help with the reprogramming technique, and Georgia Dermentzaki for the TRC2-pLKO.1-GFP plasmid vector. We also thank members of the Thanos laboratory for critical reading of the manuscript and useful advice during the work.
This work was supported by grants to D.T. from the Greek Secretariat for Research and Technology (Cooperative Grants SYNERGASIA I number 969 and Excellence Award ARISTEIA I number 1567) and from the European Committee and the KMW offsets program.
REFERENCES
- 1.Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 2.Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavigne MD, Vatsellas G, Polyzos A, Mantouvalou E, Sianidis G, Maraziotis I, Agelopoulos M, Thanos D. 2015. Composite macroH2A/NRF-1 nucleosomes suppress noise and generate robustness in gene expression. Cell Rep 11:1090–1101. doi: 10.1016/j.celrep.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Agelopoulos M, Thanos D. 2006. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J 25:4843–4853. doi: 10.1038/sj.emboj.7601364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. 2012. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skene PJ, Henikoff S. 2013. Histone variants in pluripotency and disease. Development 140:2513–2524. doi: 10.1242/dev.091439. [DOI] [PubMed] [Google Scholar]
- 8.Campos EI, Reinberg D. 2009. Histones: annotating chromatin. Annu Rev Genet 43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 9.Talbert PB, Henikoff S. 2010. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Adachi K, Scholer HR. 2012. Directing reprogramming to pluripotency by transcription factors. Curr Opin Genet Dev 22:416–422. doi: 10.1016/j.gde.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Niwa H. 2014. The pluripotency transcription factor network at work in reprogramming. Curr Opin Genet Dev 28:25–31. doi: 10.1016/j.gde.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Smith ZD, Nachman I, Regev A, Meissner A. 2010. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol 28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. 2012. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, Bar-Nur O, Cheloufi S, Stadtfeld M, Figueroa ME, Robinton D, Natesan S, Melnick A, Zhu J, Ramaswamy S, Hochedlinger K. 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadtfeld M, Hochedlinger K. 2010. Induced pluripotency: history, mechanisms, and applications. Genes Dev 24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. 2010. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Shu X, Pei D. 2014. The function and regulation of mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr Opin Genet Dev 28:32–37. doi: 10.1016/j.gde.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pehrson JR, Fried VA. 1992. MacroH2A, a core histone containing a large nonhistone region. Science 257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 22.Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. 2005. Structural characterization of the histone variant macroH2A. Mol Cell Biol 25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasque V, Gillich A, Garrett N, Gurdon JB. 2011. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J 30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrero MJ, Sese B, Kuebler B, Bilic J, Boue S, Marti M, Izpisua Belmonte JC. 2013. Macrohistone variants preserve cell identity by preventing the gain of H3K4me2 during reprogramming to pluripotency. Cell Rep 3:1005–1011. doi: 10.1016/j.celrep.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 25.Gaspar-Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Leu NA, Leroy G, Liu S, Costanzi C, Valle-Garcia D, Schaniel C, Lemischka I, Garcia B, Pehrson JR, Bernstein E. 2013. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat Commun 4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. 2010. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dell'Orso S, Wang AH, Shih HY, Saso K, Berghella L, Gutierrez-Cruz G, Ladurner AG, O'Shea JJ, Sartorelli V, Zare H. 2016. The histone variant MacroH2A1.2 is necessary for the activation of muscle enhancers and recruitment of the transcription factor Pbx1. Cell Rep 14:1156–1168. doi: 10.1016/j.celrep.2015.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefkova K., Prochazkova J., Pachernik J. 2015. Alkaline phosphatase in stem cells. Stem Cells Int 2015:628368. doi: 10.1155/2015/628368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. 2000. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 30.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, Lee G, Rinn JL, Meissner A, Park PJ, Hochedlinger K. 2015. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung M, Jin SG, Zhang X, Xiong W, Gogoshin G, Rodin AS, Pfeifer GP. 2015. Longitudinal epigenetic and gene expression profiles analyzed by three-component analysis reveal down-regulation of genes involved in protein translation in human aging. Nucleic Acids Res 43:e100. doi: 10.1093/nar/gkv473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Han Q, Peng T, Peng M, Wei B, Li D, Wang X, Yu S, Yang J, Cao S, Huang K, Hutchins AP, Liu H, Kuang J, Zhou Z, Chen J, Wu H, Guo L, Chen Y, Chen Y, Li X, Wu H, Liao B, He W, Song H, Yao H, Pan G, Chen J, Pei D. 2015. The oncogene c-Jun impedes somatic cell reprogramming. Nat Cell Biol 17:856–867. doi: 10.1038/ncb3193. [DOI] [PubMed] [Google Scholar]
- 37.Peinado H, Olmeda D, Cano A. 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Weinberg RA. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, de Herreros AG. 2011. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem 286:12024–12032. doi: 10.1074/jbc.M110.168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raju R, Palapetta SM, Sandhya VK, Sahu A, Alipoor A, Balakrishnan L, Advani J, George B, Kini KR, Geetha NP, Prakash HS, Prasad TS, Chang YJ, Chen L, Pandey A, Gowda H. 2014. A network map of FGF-1/FGFR signaling system. J Signal Transduct 2014:962962. doi: 10.1155/2014/962962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mori S, Kodaira M, Ito A, Okazaki M, Kawaguchi N, Hamada Y, Takada Y, Matsuura N. 2015. Enhanced expression of integrin alphavbeta3 induced by TGF-beta is required for the enhancing effect of fibroblast growth factor 1 (FGF1) in TGF-beta-Induced epithelial-mesenchymal transition (EMT) in mammary epithelial cells. PLoS One 10:e0137486. doi: 10.1371/journal.pone.0137486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. 2009. A small-molecule inhibitor of TGF-beta signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell 5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maherali N, Hochedlinger K. 2009. TGFbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol 19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiba S. 2006. Notch signaling in stem cell systems. Stem Cells 24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 45.Noggle SA, Weiler D, Condie BG. 2006. Notch signaling is inactive but inducible in human embryonic stem cells. Stem Cells 24:1646–1653. doi: 10.1634/stemcells.2005-0314. [DOI] [PubMed] [Google Scholar]
- 46.Ambrosetti D, Holmes G, Mansukhani A, Basilico C. 2008. Fibroblast growth factor signaling uses multiple mechanisms to inhibit Wnt-induced transcription in osteoblasts. Mol Cell Biol 28:4759–4771. doi: 10.1128/MCB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang SL, Wu C, Xiong ZF, Fang X. 2015. Progress on hypoxia-inducible factor-3: its structure, gene regulation and biological function. Mol Med Rep 12:2411–2416. doi: 10.3892/mmr.2015.3689. [DOI] [PubMed] [Google Scholar]
- 48.Imai T, Horiuchi A, Wang C, Oka K, Ohira S, Nikaido T, Konishi I. 2003. Hypoxia attenuates the expression of E-cadherin via up-regulation of SNAIL in ovarian carcinoma cells. Am J Pathol 163:1437–1447. doi: 10.1016/S0002-9440(10)63501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo D, Wang J, Li J, Post M. 2011. Mouse snail is a target gene for HIF. Mol Cancer Res 9:234–245. doi: 10.1158/1541-7786.MCR-10-0214. [DOI] [PubMed] [Google Scholar]
- 50.McMahon S, Charbonneau M, Grandmont S, Richard DE, Dubois CM. 2006. Transforming growth factor beta1 induces hypoxia-inducible factor-1 stabilization through selective inhibition of PHD2 expression. J Biol Chem 281:24171–24181. doi: 10.1074/jbc.M604507200. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Song W, Hu T, Zhang N, Miao S, Zong S, Wang L. 2011. Fank1 interacts with Jab1 and regulates cell apoptosis via the AP-1 pathway. Cell Mol Life Sci 68:2129–2139. doi: 10.1007/s00018-010-0559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Yori JL, Johnson E, Zhou G, Jain MK, Keri RA. 2010. Kruppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J Biol Chem 285:16854–16863. doi: 10.1074/jbc.M110.114546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasmussen MA, Holst B, Tumer Z, Johnsen MG, Zhou S, Stummann TC, Hyttel P, Clausen C. 2014. Transient p53 suppression increases reprogramming of human fibroblasts without affecting apoptosis and DNA damage. Stem Cell Reports 3:404–413. doi: 10.1016/j.stemcr.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M, He Y, Dubois W, Wu X, Shi J, Huang J. 2012. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell 46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, Melton DA. 2004. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med 350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 57.Papapetrou EP, Sadelain M. 2011. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat Protoc 6:1251–1273. doi: 10.1038/nprot.2011.374. [DOI] [PubMed] [Google Scholar]
- 58.Dermentzaki G, Paschalidis N, Politis PK, Stefanis L. 2016. Complex effects of the ZSCAN21 transcription factor on transcriptional regulation of alpha-synuclein in primary neuronal cultures and in vivo. J Biol Chem 291:8756–8772. doi: 10.1074/jbc.M115.704973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ford E, Nikopoulou C, Kokkalis A, Thanos D. 2014. A method for generating highly multiplexed ChIP-seq libraries. BMC Res Notes 7:312. doi: 10.1186/1756-0500-7-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 61.Shin H, Liu T, Manrai AK, Liu XS. 2009. CEAS: cis-regulatory element annotation system. Bioinformatics 25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 62.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 63.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]