Abstract
Soft tissue sarcoma of the tongue represents a very rare head and neck cancer with connective tissue features, and the genetics underlying this rare cancer are largely unknown. There are less than 20 cases reported in the literature thus far. Here, we reported the first whole-exome characterization (>×200 depth) of an undifferentiated sarcoma of the tongue in a 31-year-old male. Even with a very good sequencing depth, only 19 nonsynonymous mutations were found, indicating a relatively low mutation rate of this rare cancer (lower than that of human papillomavirus (HPV)-positive head and neck cancer). Yet, among the few genes that are somatically mutated in this HPV-negative undifferentiated tongue sarcoma, a noticeable deleterious frameshift mutation (with a very high allele frequency of >93%) of a gene for DNA replication and repair, namely POLDIP2 (DNA polymerase delta interacting protein 2), and two recurrent mutations of the adipogenesis and adipocyte differentiation gene RETSAT (retinol saturase), were identified. Thus, somatic events likely affecting adipogenesis and differentiation, as well as potential stem mutations to POLDIP2, may be implicated in the formation of this rare cancer. This identified somatic whole-exome sequencing profile appears to be distinct from that of other reported adult sarcomas from The Cancer Genome Atlas, suggesting a potential unique genetic profile for this rare sarcoma of the tongue. Interestingly, this low somatic mutation rate is unexpectedly found to be accompanied by multiple tumor protein p53 and NOTCH1 germline mutations of the patient’s blood DNA. This may explain the very early age of onset of head and neck cancer, with likely hereditary predisposition. Our findings are, to our knowledge, the first to reveal a unique genetic profile of this very rare undifferentiated sarcoma of the tongue.
Keywords: Head and neck, Sarcoma of the tongue, low mutational burden, POLDIP2 and RETSAT mutations
Introduction
Soft tissue sarcomas of the head and neck are an uncommon heterogeneous group of malignancies 1, with the most common subtype being malignant fibrous histiocytoma 2, 3. Yet, undifferentiated sarcoma of the tongue represents an even rarer tumor within this heterogeneous group of sarcomas with limited treatment options. Thus far in the literature, only about 20 cases have been reported 4. At the time of writing, the underlying genetic aberrations of this rare undifferentiated soft tissue cancer of the tongue remains unknown. Here, we describe a case report of an undifferentiated sarcoma of the tongue with associated pathological analysis and whole-exome sequencing (WES) of the tumor to identify the underlying genetic changes and attempt to determine if there exists any potential druggable mutations for treatment.
Case report
A 31-year-old Chinese male who had a 1-pack-year smoking history, was a non-drinker, had no significant past medical history, no exposure to radiation, no family history of carcinomas, particularly no family history of members with young onset malignancies, presented in December of 2016 with an anterior tongue mass. Patient biopsy revealed it to be an undifferentiated sarcoma. Further magnetic resonance imaging (MRI) demonstrated a lesion localized to the tongue that involved the extrinsic tongue musculature. Subsequently the patient underwent a total glossectomy with bilateral selective neck dissection levels I–III with an anterolateral thigh-free-flap reconstruction in January 2017. The pathological findings from the surgical specimen are further described below. WES analyses were performed on the fresh surgically resected tumor (tumor content, >50%) and a paired blood sample. Adjuvant chemotherapy and radiation were recommended for this patient, but both options were declined because of concerns regarding long term toxicity and the effects on speech and swallowing. As of October 2017, the patient was disease-free with no documented recurrences on repeated MRI of the oral cavity and neck.
Pathological findings
The tumor mass measured grossly 7 × 7 × 5.5 cm on examination following resection. On microscopic examination, the tumor was composed of mostly spindle cells arranged in short fascicles or a vague storiform pattern. Frequent mitotic figures and prominent tumoral necrosis were seen. No osseous, chondroid or rhadomyomatous differentiation was seen on haematoxylin and eosin staining ( Figure 1– Figure 3). The closest margins were 2 mm and located at the postero-inferior, right and left margins.
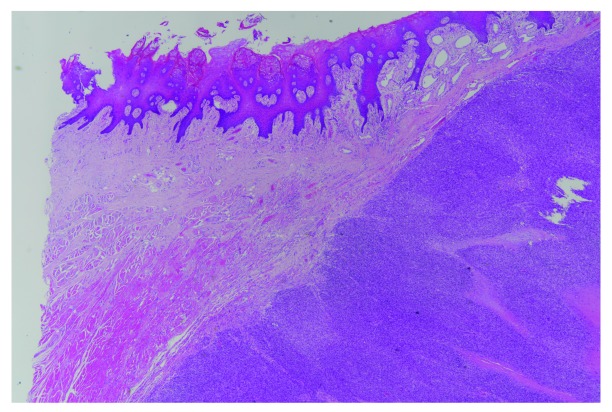
Figure 1. Sarcoma forming cellular fascicles is seen in the stroma of the tongue, focally invading the intrinsic muscle (left upper corner).
The surface squamous epithelium shows no dysplastic field change or connection with sarcoma. (H&E stain; magnification, ×20).
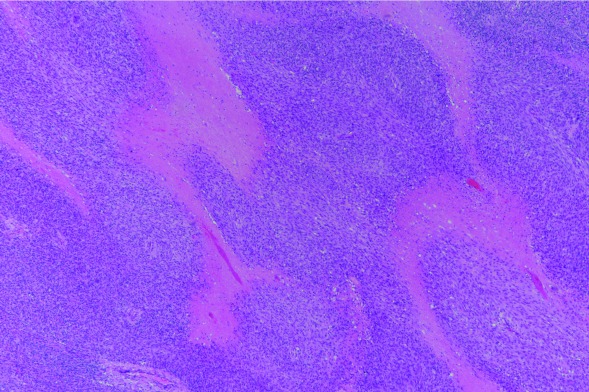
Figure 2. Tumor necrosis (pink foci) was prominent in the undifferentiated sarcoma.
(H&E stain; magnification, ×40).
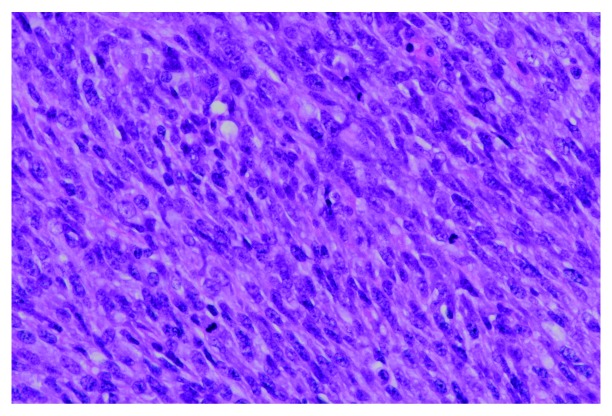
Figure 3. The sarcoma tumor cells possessed enlarged, elongated, pleomorphic and hyperchromatic nuclei, distinct nucleoli and small amount of cytoplasm.
Mitotic figures were frequently noted. No osseous, chondroid or rhadomyomatous differentiation was seen. (H&E stain; magnification, ×400).
Further immunohistochemical studies showed that the tumor cells were positive for vimentin and p16; had focal weak positivity for SMA; had moderate positivity for TLE-1; and were negative for p63, EMA, AE1/AE3 antibodies, c-Kit, S100, CD31, CD34, desmin, myogenin, melan A, HMB45 antibody, ALK-1, human herpesvirus 8 (HHV8), CD99 (MIC-2) and SRY-box 10 (SOX10). Calponin was equivocal. No loss of integrase interactor 1 (INI1) staining was noted. PCR for high-risk human papillomavirus (HPV) DNA was negative. The Molecular Break-Apart FISH test for SS18 translocation was negative. Overall the pathological features demonstrated a high-grade sarcoma with no definite line of differentiation, consistent with an undifferentiated sarcoma.
Mutational profile of the patient’s tumor
WES at >200x depth (Illumina HiSeq 4000) was performed to determine the genetic aberrations underlying this rare form of head and neck cancer. Normal DNA from the patient’s blood was also subjected to WES at the same depth. Surprisingly, WES revealed only 19 non-synonymous mutations in the tumor. The very low rate of non-synonymous mutations is uncommon in HPV-negative and HPV-positive head and neck squamous cell carcinoma (HNSCC) 5, and is slightly lower than that of reported sarcomas.
The somatically mutated genes were, in the order of allele frequencies of these 19 mutational events, POLDIP2 (DNA polymerase delta interacting protein 2), tubulin gamma complex associated protein 3 TUBGCP3,, mutated in colorectal cancers ( MCC), TUBA3D (tubulin alpha 3d), DDX11 (DEAD/H-box helicase 11), CWF19L2 (CWF19 like 2, cell cycle control), ZNF91 (zinc finger protein 91), RETSAT (retinol saturase) (2 mutations), PRR21 (proline rich 21), TAS2R46 (taste 2 receptor member 46), FAM186A (family with sequence similarity 186 member A), HEATR5A (HEAT repeat containing 5A), VPS4B (vacuolar protein sorting 4 homolog B), PRAMEF12 (PRAME family member 12), FAM170B (family with sequence similarity 170 member B), BBS4 (Bardet-Biedl syndrome 4), ARHGAP5 (Rho GTPase activating protein 5) and ATAD3B (ATPase family, AAA domain containing 3B) ( Table 1). Strikingly, based on the functional annotation of these mutated genes, five of the top seven mutated genes with high allele frequencies (>10%) were known to be involved in DNA replication, and mitosis ( Table 1). This suggests that major somatic mutations of this rare tumor appear to affect DNA replication and mitosis, consistent with an aggressive phenotype. Of note, the patient’s tumor carried a 93% allele frequency of the POLDIP2 S28fs mutation, which is a hotspot mutation in multiple cancers. In addition, the MCC gene was also mutated in this rare tumor ( MCC p.G20S) at a high allele frequency of 28%, indicating its likely role as a driver event for tumorigenesis. Interestingly, the PRR21 mutation is also found to be mutated at high frequencies in TCGA HNSCC cohort with a hotspot mutation S86Gfs*291 and M48Tfs*329 mutation, while in this tumor, PRR21 mutation occurred at G114C, a position near to these two hotspot mutations in HNSCC.
Table 1. List of the 19 non-synonymous mutations found in the undifferentiated sarcoma with their mutation type, allele frequency and potential function.
| Gene | Chromosome
position |
Mutation type | Nucleotide change | Amino acid
change |
Allele
Frequency |
Full gene name | Potential gene function |
|---|---|---|---|---|---|---|---|
| POLDIP2 | chr17:26,684,390 | Frameshift
insertion |
c.83dupG | Ser28fs | 93% | DNA polymerase delta
interacting protein 2 |
DNA replication and DNA
repair |
| TUBGCP3 | chr13:113,201,852 | Frameshift
deletion |
c.1235_
1249delCGCGCGACTTTCCCA |
Thr412_
Pro416del |
50% | Tubulin gamma complex
associated protein 3 |
largely unknown, tubulin
gamma may be involved in mitosis |
| MCC | chr5:112824054 | Missense | c.58G>A | Gly20Ser | 28% | Mutated in colorectal
cancers |
negative regulator of cell
cycle |
| TUBA3D | chr2:132237643 | Missense
variant & splice region variant |
c.377C>T | Ala126Val | 15% | Tubulin alpha 3d | tubulin family, transport,
mitosis |
| DDX11 | chr12:31237978 | Missense | c.556C>T | Arg186Trp | 14% | DEAD/H-box helicase 11 | putative RNA helicases |
| CWF19L2 | chr11:107325241 | Missense | c.274G>A | Glu92Lys | 13% | CWF19 like 2, cell cycle
control |
likely in cell cycle control |
| ZNF91 | chr19:23544852 | Missense | c.929C>A | Ala310Asp | 11% | Zinc finger protein 91 | unknown |
| RETSAT | chr2:85570857 | Missense | c.1598C>T | Ala533Val | 9% | Retinol saturase | promote adipogenesis and
favors normal adipocyte differentiation |
| PRR21 | chr2:240982060 | Missense | c.340G>T | Gly114Cys | 9% | Proline rich 21 | unknown |
| FAM186A | chr12:50746486 | Missense | c.4129A>C | Thr1377Pro | 7% | Family with sequence
similarity 186 member A |
unknown |
| HEATR5A | chr14:31792790 | Missense | c.3768G>A | Met1256Ile | 7% | HEAT repeat containing 5A | unknown |
| RETSAT | chr2:85570849 | Missense | c.1606G>A | Gly536Arg | 6% | Retinol saturase | promote adipogenesis and
favors normal adipocyte differentiation |
| VPS4B | chr18:61060779 | Missense | c.1096C>T | Arg366Cys | 6% | Vacuolar protein sorting 4
homolog B |
intracellular protein
trafficking |
| PRAMEF12 | chr1:12835741 | Missense | c.343A>G | Ile115Val | 5% | PRAME family member 12 | unknown |
| FAM170B | chr10:50340041 | Missense | c.469G>T | Ala157Ser | 5% | Family with sequence
similarity 170 member B |
unknown |
| BBS4 | chr15:73023650 | Missense | c.716T>C | Ile239Thr | 5% | Bardet-Biedl syndrome 4 | Bardet-Biedl syndrome
gene family, associated with severe pigmentary retinopathy, obesity, polydactyly, renal malformation and cognitive disability. |
| TAS2R46 | chr12:11214601 | Missense | c.293T>C | Leu98Pro | 4% | Taste 2 receptor member 46 | Taste receptor |
| ARHGAP5 | chr14:32561340 | Missense | c.1465G>A | Glu489Lys | 4% | Rho GTPase activating
protein 5 |
Negatively regulates RHO
GTPases in cytoskeleton changes |
| ATAD3B | chr1:1431060 | Missense | c.1810T>C | Tyr604His | 3% | ATPase family, AAA domain
containing 3B |
Interact with matrix nucleoid
complexes |
Most strikingly, among all the somatic mutations identified in this patient, we found that recurrent somatic events of RETSAT ( RETSAT p.A533V and p.G536R mutations), a gene known to be important for the promotion of adipogenesis and normal adipocyte differentiation. It is plausible that RETSAT mutations may affect adipocyte adipogenesis and differentiation, related to the sarcoma features of this rare tumor.
Gene copy number analysis was also performed and a total of 221 somatic copy number alterations (CNA) events were identified ( Figure 4 and Supplementary Table 1). The somatic CNA detected in this patient did not harbor any of these common HNSCC CNA events including losses of chr. 3p and 8p, as well as focal amplification or gains of chr. 3q26/28, 5p15, and 8q24 5. The common CNA events in sarcomas including aberrations of the MDM2-p53 and p16-cyclin dependent kinase 4 (CDK4)-retinoblastoma-associated protein pathways, deletion of TP53 (tumor protein p53), CDKN2A (cyclin dependent kinase inhibitor 2A), as well as chr. 12q13-q15 and CDK4 amplification were also absent in this patient 6. These results suggest that this unique tumor is likely distinct from HNSCC and adult soft tissue sarcomas.
Figure 4. A Circos plot showing somatic genetic variations.
An ideogram of a normal karyotype is shown in the outermost ring. Chromosomes are segmented into contiguous bins of 1 Mb in size. The second ring from outside shows CNVs at corresponding chromosomal positions. Distinct heights of red dots indicate different inferred copy numbers. Regions that are not covered by any dots have a copy number of two. The third ring shows SNV density. Purple dots with different heights indicate SNV densities of the corresponding regions. The innermost ring shows positions of genes with non-synonymous mutations found in the undifferentiated sarcoma. CNV, copy number variation; SNV, single nucleotide variant.
Multiple germline mutations in key tumor suppressor genes
Given the young age of this patient, we also carefully examined potential aberrations in his germline mutational profile by WES of his blood DNA. Importantly, we found that this patient carried multiple germline mutational events of TP53, including a missense mutation of TP53 p.P72R (100% allele frequency), 2 mutations at the immediate 5’-UTR, likely in the promoter region of TP53 (c.-112G>A; and c.-123C>G; both with 100% allele frequencies), as well as two other intron variants with unknown effects ( Table 2). The TP53 germline events likely explain the early age of onset for cancer for this patient. In addition, a 5’-UTR mutation, likely at the promoter region of CDKN2A, was identified (c.*193G>C), with an allele frequency of 100%. A CASP8 (caspase 8) missense mutation, CASP8 p.L14R, was also found in this patient’s blood with a 46.7% allele frequency. However, the potential function of this mutation is unclear. Lastly, this patient carries as many as 23 germline variants of NOTCH1; however, none of these variants are non-synonymous mutations, and the effects of these variants are largely unknown. No telomerase reverse transcriptase promoter or exon mutation was found. Lastly, no germline gene copy number alterations were observed for any critical HNSCC-associated oncogenes or suppressor genes ( Supplementary Table 2)
Table 2. List of germline mutations identified by gene, chromosome position, mutation type, nucleotide change, amino acid change and allele frequency.
| Gene | Chromosome
position |
Mutation type | Nucleotide change | Amino acid
change |
Allele
frequency |
|---|---|---|---|---|---|
| CASP8 | Chr. 2: 202,122,995 | Missense | c.41A>G | Lys14Arg | 46.7 |
| CASP8 | Chr. 2: 202,141,512 | Intron | c.838-38C>T | . | 62.0 |
| CASP8 | Chr. 2: 202,151,163 | Intron | c.1482-19A>G | . | 50.4 |
| CASP8 | Chr. 2: 202,152,162 | 3_prime_UTR | c.*845T>A | . | 76.9 |
| CDKN2A | Chr. 9: 21,968,199 | 3_prime_UTR | c.*193G>C | . | 100.0 |
| HRAS | Chr. 11: 534,242 | Synonymous | c.81T>C | His27His | 44.7 |
| NOTCH1 | Chr. 9: 139,391,636 | Synonymous | c.6555C>T | Asp2185Asp | 99.4 |
| NOTCH1 | Chr. 9: 139,396,408 | Intron | c.5473-43T>C | . | 100.0 |
| NOTCH1 | Chr. 9: 139,396,690 | Intron | c.5384+34G>A | . | 100.0 |
| NOTCH1 | Chr. 9: 139,397,707 | Synonymous | c.5094C>T | Asp1698Asp | 99.4 |
| NOTCH1 | Chr. 9: 139,400,406 | Intron | c.4015-73G>A | . | 100.0 |
| NOTCH1 | Chr. 9: 139,400,904 | Intron | c.4014+75G>A | . | 100.0 |
| NOTCH1 | Chr. 9: 139,401,504 | Intron | c.3644-79C>T | . | 85.7 |
| NOTCH1 | Chr. 9: 139,402,663 | Intron | c.3325+21A>G | . | 100.0 |
| NOTCH1 | Chr. 9: 139,402,908 | Intron | c.3172-71A>G | . | 100.0 |
| NOTCH1 | Chr. 9: 139,403,240 | Intron | c.3171+81_3171+82insC | . | 100.0 |
| NOTCH1 | Chr. 9: 139,403,268 | Intron | c.3171+54A>G | . | 100.0 |
| NOTCH1 | Chr. 9: 139,403,554 | Intron | c.2970-31A>G | . | 100.0 |
| NOTCH1 | Chr. 9: 139,405,261 | Splice_region_& intron | c.2588-4G>A | . | 100.0 |
| NOTCH1 | Chr. 9: 139,407,452 | Intron | c.2467+21G>A | . | 100.0 |
| NOTCH1 | Chr. 9: 139,407,932 | Synonymous | c.2265T>C | Asn755Asn | 100.0 |
| NOTCH1 | Chr. 9: 139,410,177 | Intron | c.1670-9A>G | . | 100.0 |
| NOTCH1 | Chr. 9: 139,410,424 | Intron | c.1669+9T>C | . | 100.0 |
| NOTCH1 | Chr. 9: 139,410,589 | Intron | c.1556-43T>C | . | 100.0 |
| NOTCH1 | Chr. 9: 139,410,679 | Intron | c.1556-133A>G | . | 100.0 |
| NOTCH1 | Chr. 9: 139,411,714 | Intron | c.1555+10A>G | . | 100.0 |
| NOTCH1 | Chr. 9: 139,411,880 | Intron | c.1442-43C>T | . | 100.0 |
| NOTCH1 | Chr. 9: 139,412,197 | Splice_region_& intron | c.1441+7C>T | . | 100.0 |
| NOTCH1 | Chr. 9: 139,418,260 | Synonymous | c.312T>C | Asn104Asn | 46.2 |
| TP53 | Chr. 17: 7,578,115 | Intron | c.672+62A>G | . | 100.0 |
| TP53 | Chr. 17: 7,578,645 | 5_prime_UTR | c.-112G>A | . | 100.0 |
| TP53 | Chr. 17: 7,579,472 | Missense | c.215C>G | Pro72Arg | 100.0 |
| TP53 | Chr. 17: 7,579,643 | Intron | c.96+41_97-54delACCTGGAGGGCTGGGG | . | 84.4 |
| TP53 | Chr. 17: 7,579,801 | 5_prime_UTR | c.-123C>G | . | 100.0 |
Discussion
The development of soft tissue sarcomas in the oral cavity and head and neck region in general are rare, with the development of an undifferentiated sarcoma in the oral cavity of an even lower likelihood 7, 8. A US population database analysis of head and neck sarcoma patients identified that the median age of adults affected was 55–59 years old, most commonly affecting the skin and soft tissues 2. This contrasts with the young age of our patient at presentation (31 years). The presence of a rare soft tissue sarcoma in a young patient provided the evidence to evaluate for germline mutations in addition to the somatic mutations, leading to the discovery of multiple germline TP53 mutations, as well as multiple CDKN2A, and potentially multiple NOTCH1 mutations of unclear functions. Notably, a CASP8 mutation was also found in his germline. It is important to note that all these genes are known to be somatically mutated in HNSCC 5, 9.
Interestingly, the somatic mutational profile of this tumor was rather distinct from both HNSCC and sarcomas, with a relatively low number of non-synonymous mutations. Furthermore, unlike the genetic profile of soft tissue sarcomas, our sarcoma patient lacked the common somatic mutations in TP53, PTEN (phosphatase and tensin homolog) and CDKN2A 6, 10. Moreover, his tumor also lacked the most common somatic mutations of HNSCC, such as TP53, NOTCH1, CKDN2A, PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha) and FAT1 (FAT atypical cadherin 1) 5, 11, 12. This was partly contributed to by his heavy germline mutations of some of these key tumor suppressor genes, including TP53 and CDKN2A, which are commonly mutated in both sarcoma and HNSCC, usually somatically. Furthermore, his germline events also carried CASP8 and NOTCH1 mutations, which are commonly somatically mutated in HNSCC rather than in the germline. This difference in mutation profile in conjunction with the young age of the patient led us to consider heavy germline mutations as a possible underlying cause of this rare undifferentiated sarcoma of the tongue.
Germline mutations in TP53 are associated with familial clustering of early onset carcinomas, including pre-menopausal breast cancer, soft tissue sarcomas, adrenal cortical carcinomas or choroid plexus carcinomas, which together are classified as Li–Fraumeni syndrome based on Chompret criteria 13– 16. Li–Fraumeni-like syndrome represents a subset of patients with familial clustering that do not meet the criteria for Li–Fraumeni syndrome 15. However, de novo germline mutations in TP53 have been described, proven and are thought to be relatively common 17, 18. Our case highlights the importance of considering TP53 mutations in early onset soft tissue sarcomas, even in those without familial histories, as this offers the potential to appropriately manage the patient and possibly the family.
The development of soft tissue sarcomas in an uncommon location in a young patient indicated the possibility of germline events, despite the lack of a family history of carcinomas at a young age. The early detection of these mutations can be useful in treating these patients, given the consideration that radiation wherever possible should be avoided to prevent the development of radiation-induced second malignancies 19– 22. Regarding the surveillance for second primary cancers, full-body MRI examinations and positron emission tomography have been recommended, as opposed to exposure to radiation with computed tomography 19. This case highlights the need for vigilance and consideration of possible hereditary predisposition syndromes or de novo germline mutations in patients with rare tumours at a young age and with atypical genetic profiles.
Methods
Tissue collection
Tumor tissue and blood sample were collected from the patient under written informed consent according to The Joint Chinese University of Hong Kong – New Territories East Cluster Clinical Research Ethics Committee, Hong Kong SAR (protocol number CRE-2015.396). The tumor mass was freshly frozen for DNA extraction using the QIAGEN DNeasy Blood and Tissue kit. Blood DNA was extracted from patient’s buffy coat in a similar manner. DNA samples were then quantified and quality-assessed using a bioanalyzer.
Exome sequencing and variant calling
Genomic DNA of the patient’s tumor and buffy coat were used for WES using the Agilent SureSelect Human All Exon V5 Kit, with sequencing performed using the Illumina HiSeq 4000 platform (Macrogen, Korea) with a goal coverage of ×200 for tumor and ×100 for the blood (buffy coat) sample of the same patient.
Upon sequencing, all reads (FASTQ files) were mapped to the hg19 human reference genome assembly with Burrows-Wheeler Alignment Tool (version 0.7.12). Variant calling of single nucleotide variants (SNVs) and indels was performed using the Genome Analysis Toolkit (version v3.4.0) HaplotypeCaller pipeline. Called variants were annotated with SnpEff (version 4.3), the exome sequencing data have been deposited into European Nucleotide Archive (ENA) with accession number PRJEB25783. Somatic mutations were defined as mutations found in the tumor tissue of the patient but not in the patient’s blood. All identified recurrent mutations were confirmed in Integrative Genomics Viewer (IGV) (version 2.4) as a final check of the calling (100% accurate).
Copy number alterations (CNAs)
CNAs were analyzed from segmented WES data using the Control-FREEC copy number and genotype caller (version 11.0) in both somatic and germline mode. In both modes, captured genomic regions were characterized using the SureSelect Human All Exon V5 bed file. Somatic CNA analyses were run by referencing tumor WES data to the paired blood WES data, whereas germline CNA analysis were run by referencing blood WES data to the hg19 human genome assembly. Identified regions were annotated with ANNOVAR (version 2017-07-17) and SnpEff. Segments with CNA were manually confirmed with an IGV check. Genetic variations were visualized using Circos (version 0.69) with “chromosomes_unites = 1000000”.
Immunohistochemical staining
The patient’s tumor tissue was FFPE-preserved and sectioned (5–8 µm) for immunohistochemical and H&E staining. Immunohistochemical staining was performed with the following antibodies: Melan A (M7196, Dako, Denmark), HMB34 (M0634, Dako, Denmark), ALK-1 (M7195, Dako, Denmark), HHV8 (NCL-HHV8-LNA, Novo, UK), MIC-2 (M3601, Dako, Denmark), SOX-10 (ACI3099, Biocare, USA), Calp (M3556, Dako, Denmark), INI1 (612110, DB transduction, USA), Vimentin (M0725, Dako, Denmark), p16 (805-4713, Ventana, USA), SMA (M0851, Dako, Denmark), p53 (M7001, Dako, Denmark), TLE1 (401M-16, Cell Marque, USA), p63 (M7317, Dako, Denmark), EMA (MS348, Thermo, UK), AE1/3 (M3515, Dako, Denmark), c-kit (A4502, Dako, Denmark), S100 (NCL-S100p, Leica, UK), CD31 (M0823, Dako, Denmark), CD34 (NCL-L-END, Leica, UK), Desmin (M0760, Dako, Denmark) and Myogenin (M3559, Dako, Denmark). Results were examined by an experienced pathologist for the presence or absence of the target proteins. Pictures were taken under a light microscope at ×20, ×40 and ×400 magnification.
HPV analysis
The tumor cells from the formalin-fixed, paraffin-embedded sections were isolated for DNA extraction. The DNA was subjected to PCR analysis using the consensus primers GP5+/6+, as previously documented 23.
Consent
Written informed consent for publication of their clinical details and accompanying images was obtained from the patient.
Data availability
The exome sequencing data have been deposited into European Nucleotide Archive (ENA) with accession number PRJEB25783.
Funding Statement
Dr Stanley Ho Medical Foundation (to JYKC); General Research Fund from the Research Grant Council, Hong Kong (#1711484 and #17121616 to VWYL); Theme-based Research from the Research Grant Council, Hong Kong (T12-401/13-R to VWYL); Hong Kong Cancer Fund (to VWYL); and the Direct Grant for Research (#2016.095), the Chinese University of Hong Kong (to VWYL and JYKC).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
Supplementary material
Supplementary Table 1. Somatic copy number variations obtained from DNA taken from the patient’s tumor.References
- 1. Cormier JN, Pollock RE: Soft tissue sarcomas. CA Cancer J Clin. 2004;54(2):94–109. 10.3322/canjclin.54.2.94 [DOI] [PubMed] [Google Scholar]
- 2. Peng KA, Grogan T, Wang MB: Head and neck sarcomas: analysis of the SEER database. Otolaryngol Head Neck Surg. 2014;151(4):627–633. 10.1177/0194599814545747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shuman AG, Brennan MF, Palmer FL, et al. : Soft tissue sarcoma of the head & neck: nomogram validation and analysis of staging systems. J Surg Oncol. 2015;111(6):690–695. 10.1002/jso.23868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nguyen A, Vaudreuil A, Haun P, et al. : Clinical Features and Treatment of Fibrous Histiocytomas of the Tongue: A Systematic Review. Int Arch Otorhinolaryngol. 2018;22(1):94–102. 10.1055/s-0037-1602819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Network: Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Research Network. Electronic address: elizabeth.demicco@sinaihealthsystem.ca, Cancer Genome Atlas Research Network: Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell. 2017;171(4):950–965.e28. 10.1016/j.cell.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ariyoshi Y, Shimahara M, Omura K, et al. : Epidemiological study of malignant tumors in the oral and maxillofacial region: survey of member institutions of the Japanese Society of Oral and Maxillofacial Surgeons, 2002. Int J Clin Oncol. 2008;13(3):220–228. 10.1007/s10147-007-0756-9 [DOI] [PubMed] [Google Scholar]
- 8. Chang AE, Chai X, Pollack SM, et al. : Analysis of clinical prognostic factors for adult patients with head and neck sarcomas. Otolaryngol Head Neck Surg. 2014;151(6):976–983. 10.1177/0194599814551539 [DOI] [PubMed] [Google Scholar]
- 9. Stransky N, Egloff AM, Tward AD, et al. : The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jour G, Scarborough JD, Jones RL, et al. : Molecular profiling of soft tissue sarcomas using next-generation sequencing: A pilot study toward precision therapeutics. Hum Pathol. 2014;45(8):1563–1571. 10.1016/j.humpath.2014.04.012 [DOI] [PubMed] [Google Scholar]
- 11. Agrawal N, Frederick MJ, Pickering CR, et al. : Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stransky N, Egloff AM, Tward AD, et al. : The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li FP, Fraumeni JF, Jr, Mulvihill JJ, et al. : A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48(18):5358–5362. [PubMed] [Google Scholar]
- 14. Malkin D, Li FP, Strong LC, et al. : Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–1238. 10.1126/science.1978757 [DOI] [PubMed] [Google Scholar]
- 15. Olivier M, Hollstein M, Hainaut P: TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. 10.1101/cshperspect.a001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez KD, Noltner KA, Buzin CH, et al. : Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27(8):1250–1256. 10.1200/JCO.2008.16.6959 [DOI] [PubMed] [Google Scholar]
- 17. Chompret A, Brugières L, Ronsin M, et al. : P53 germline mutations in childhood cancers and cancer risk for carrier individuals. Br J Cancer. 2000;82(12):1932–1937. 10.1054/bjoc.2000.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lalloo F, Varley J, Moran A, et al. : BRCA1, BRCA2 and TP53 mutations in very early-onset breast cancer with associated risks to relatives. Eur J Cancer. 2006;42(8):1143–1150. 10.1016/j.ejca.2005.11.032 [DOI] [PubMed] [Google Scholar]
- 19. Agir H, MacKinnon C, Tan ST: Li-Fraumeni syndrome: a case with 4 separate primary sarcomas and 5 sequential free flaps in the maxillofacial region. J Oral Maxillofac Surg. 2008;66(8):1714–1719. 10.1016/j.joms.2007.09.015 [DOI] [PubMed] [Google Scholar]
- 20. Varley J: TP53, hChk2, and the Li-Fraumeni syndrome. Methods Mol Biol. 2003;222:117–129. 10.1385/1-59259-328-3:117 [DOI] [PubMed] [Google Scholar]
- 21. Hisada M, Garber JE, Fung CY, et al. : Multiple primary cancers in families with Li-Fraumeni syndrome. J Natl Cancer Inst. 1998;90(8):606–611. 10.1093/jnci/90.8.606 [DOI] [PubMed] [Google Scholar]
- 22. Limacher JM, Frebourg T, Natarajan-Ame S, et al. : Two metachronous tumors in the radiotherapy fields of a patient with Li-Fraumeni syndrome. Int J Cancer. 2001;96(4):238–242. 10.1002/ijc.1021 [DOI] [PubMed] [Google Scholar]
- 23. Jacobs MV, Snijders PJ, van den Brule AJ, et al. : A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol. 1997;35(3):791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]