Abstract
Bile acids facilitate intestinal nutrient absorption and biliary cholesterol secretion to maintain bile acid homeostasis, which is essential for protecting liver and other tissues and cells from cholesterol and bile acid toxicity. Bile acid metabolism is tightly regulated by bile acid synthesis in the liver and bile acid biotransformation in the intestine. Bile acids are endogenous ligands that activate a complex network of nuclear receptor farnesoid X receptor and membrane G protein-coupled bile acid receptor-1 to regulate hepatic lipid and glucose metabolic homeostasis and energy metabolism. The gut-to-liver axis plays a critical role in the regulation of enterohepatic circulation of bile acids, bile acid pool size, and bile acid composition. Bile acids control gut bacteria overgrowth, and gut bacteria metabolize bile acids to regulate host metabolism. Alteration of bile acid metabolism by high-fat diets, sleep disruption, alcohol, and drugs reshapes gut microbiome and causes dysbiosis, obesity, and metabolic disorders. Gender differences in bile acid metabolism, FXR signaling, and gut microbiota have been linked to higher prevalence of fatty liver disease and hepatocellular carcinoma in males. Alteration of bile acid homeostasis contributes to cholestatic liver diseases, inflammatory diseases in the digestive system, obesity, and diabetes. Bile acid-activated receptors are potential therapeutic targets for developing drugs to treat metabolic disorders.
Key words: Bile acid synthesis, Metabolic regulation, Nuclear receptor, Takeda G protein receptor 5 (TGR5), Farnesoid X receptor (FXR)
INTRODUCTION
Bile acid synthesis is an important pathway for catabolism of cholesterol and is tightly regulated by a complex but integrated network of mechanisms that are not completely understood. Bile acids are detergent molecules that can be highly toxic if accumulated in high concentrations in the liver and other tissues. It is thus necessary to tightly control bile acid synthesis to maintain bile acids at low and constant levels in the circulation pool, including the liver, gallbladder, and intestine. Recent research in mouse models and human patient studies has revealed that bile acids are signaling molecules that activate nuclear receptors and membrane G protein-coupled bile acid receptors to regulate not only the classic bile acid synthesis pathway but also the alternative bile acid synthesis pathways to maintain lipid, glucose, and energy metabolism in the liver, intestine, and adipose tissue. Alteration of bile acid homeostasis affects hepatic metabolic homeostasis, causes inflammation, and contributes to the pathogenesis of metabolic diseases such as nonalcoholic fatty liver disease (NAFLD), diabetes, obesity, and inflammatory bowel diseases (IBDs). Bile acids are required for absorption of fats, steroids, and lipid-soluble vitamins in the intestine for metabolism in the liver1,2 and are signaling molecules that activate nuclear and membrane bile acid receptors to modulate hepatic lipid, glucose, and energy metabolism2–4. It has been known for more than half a century that bile acids are antibacterial agents in the gut that control gut bacteria overgrowth and protect intestinal barrier function. More recent studies of the gut-to-liver axis in bile acid metabolism, composition and pool size, and host metabolism have opened a new era for studying bile acid biology and physiology. This review will focus on recent advances in understanding regulation of bile acid synthesis, metabolism, and homeostasis and the emerging concepts on bile acid signaling crosstalk in metabolic regulation and potential therapeutic drug development for treating NAFLD. Most references cited were published in the last 10 years, and older original references can be found in recent reviews2–7.
BILE ACID BIOLOGY AND PHYSIOLOGY
Enterohepatic Circulation of Bile Acids
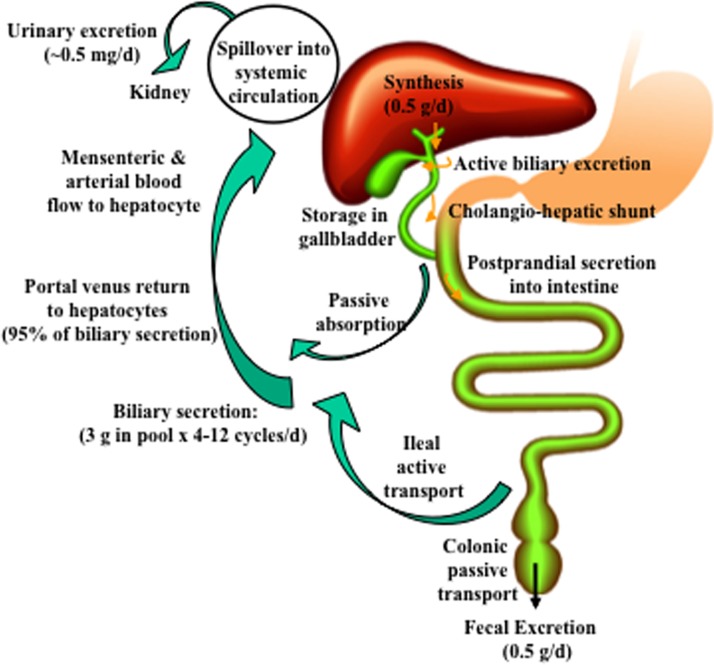
In the liver, conversion of cholesterol to bile acids is the major pathway for catabolism of cholesterol. The liver synthesizes about 0.5 g of bile acids per day (80–90 kg body weight) (Fig. 1). Bile acids are secreted into canaliculi by active transport to form bile. Bile acids are stored in the gallbladder and are released after meal intake into the intestinal tract via the common bile ducts. Small amounts of bile acids are circulated back to hepatocytes via a cholangio-hepatic shunt. In the upper intestine, a small amount of bile acids is passively absorbed, but most bile acids are reabsorbed in the ileum by an active transport system to portal blood circulation and then to the liver. This enterohepatic circulation of bile acids is highly efficient and recovers about 95% of bile acids in the pool. The small amount of bile acid lost in feces (0.5 g/day) is replenished by de novo synthesis in the liver. Enterohepatic circulation of bile acids occurs on average six to eight times a day to maintain a constant bile acid pool size of ∼3 g. Bile acids (∼0.5 mg/day) spilled over into the systemic circulation are excreted into urine.
Figure 1.
Enterohepatic circulation of bile acids. About 0.5 g of bile acids is synthesized per day in an average adult. Bile acids are secreted into bile and stored in the gallbladder. Small amounts of bile acids are circulated from cholangiocytes to the liver via the cholangio-hepatic shunt. After each meal, bile acids are excreted to the intestinal tract. A small amount of bile acids is passively absorbed in the upper intestine into mesenteric and arterial blood flow to hepatocytes. Most bile acids are reabsorbed in the ileum by active transport via apical sodium-dependent bile salt transporter (ASBT) and transported back to the liver via portal blood circulation. A constant bile acid pool (3 g) is circulated 4 to 12 times a day. Approximately 95% of bile acids in bile are recirculated back to the liver, and about 5% (0.5 g) lost in fecal excretion are replenished by de novo synthesis in the liver. A small amount (0.5 mg/day) of bile acid spillover into systemic circulation is cleared in urine.
Bile Acid Synthesis
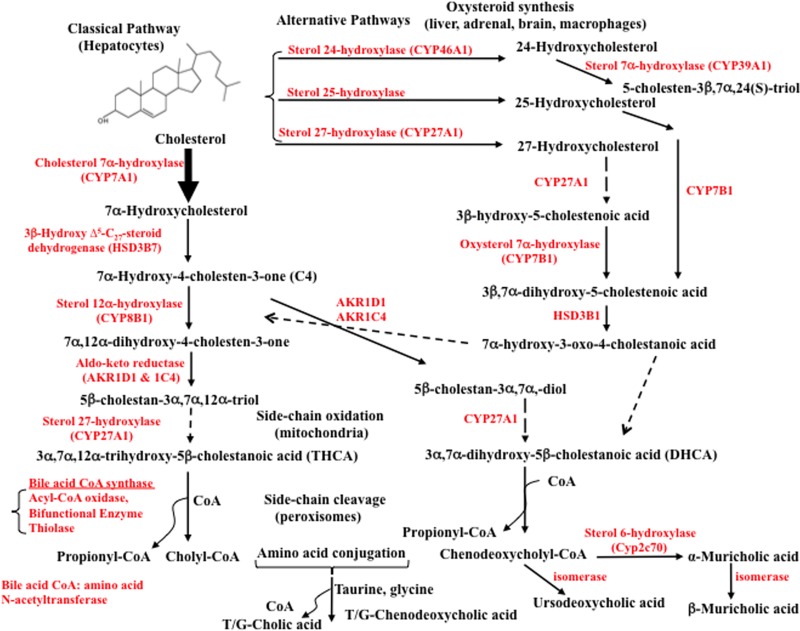
The liver is the only organ that has all the enzymes required for bile acid synthesis (Fig. 2). There are two pathways in the liver: the classic or neutral pathway and the alternative or acidic pathway (all intermediates are cholestenoic acids). The classic pathway starts with modifications of the steroid rings of cholesterol by hydroxylation, isomerization, reduction, and dehydroxylation enzymes located in the cytosol and endoplasmic reticulum, followed by steroid side chain oxidation in the mitochondria and oxidative cleavage of the side chain in peroxisomes. The alternative pathway starts with oxidation of the steroid side chain, then modification of the steroid rings and oxidative cleavage of the steroid side chain.
Figure 2.
Bile acid synthesis pathways. In the liver, cholesterol 7α-hydroxylase (CYP7A1) initiates the classical bile acid synthesis pathway by hydroxylation of the steroid rings at 7α-C for further modifications of the steroid rings, followed by steroid side chain oxidation and cleavage, whereas sterol 27-hydroxylase (CYP27A1) initiates the alternative bile acid synthesis pathway by oxidation of the steroid side chain followed by modifications of the steroid rings and cleavage of the side chain in the classic pathway. CYP27A1 is expressed in most tissues and macrophages. Sterol 25-hydroxylase (a non-CYP450 enzyme) in the liver and steroid 24-hydroxylase (CYP46A1) in the brain also oxidize cholesterol. In the alternative pathways, a nonspecific oxysterol 7α-hydroxylase (CYP7B1) hydroxylates 27-hydroxycholesterol and 25-hydroxycholesterol, whereas a specific sterol 7α-hydroxylase (CYP39A1) hydroxylates 24-hydroxycholesterol. Only the liver has all the enzymes required for the synthesis of cholic acid (CA) and chenodeoxycholic acid (CDCA), the two primary bile acids synthesized in humans (shown on the left, see text for details). The oxidized steroid intermediates (oxysterols) produced in the extrahepatic tissues can be used for bile acid synthesis in the liver. Sterol 12α-hydroxylase (CYP8B1) is required for CA synthesis. Without 12α-hydroxylation, CDCA is synthesized. Following steroid side chain cleavage, cholyl-CoA and chenodeoxycholyl-CoA are conjugated to amino acids, either taurine or glycine. In mice, CDCA is 6α-hydroxylated to form α-muricholic acid (α-MCA) by a sterol-6α-hydroxylase (Cyp2c70) catalyzed reaction. The 7α-OH group in α-MCA is epimerized (isomerized) to a 7β-hydroxyl group to form β-MCA. The 7α-HO group in CDCA can be epimerized to 7β-HO to form ursodeoxycholic acid (UDCA), a highly soluble bile acid in humans and mice.
The Classic Bile Acid Synthesis Pathway
The classic pathway of bile acid synthesis is initiated by the rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1) to specifically hydroxylate cholesterol at the 7α position, forming 7α-hydroxycholesterol, which is converted to 7α-hydroxy-4-cholesten-3-one (named C4) by 3β-hydroxy-Δ5-C27-steroid dehydrogenase (HSD3B7)8–10. C4 is the common precursor of cholic acid (CA) and chenodeoxycholic acid (CDCA), two primary bile acids synthesized in human liver. Serum C4 levels reflect the rate of bile acid synthesis in humans11,12. Sterol 12α-hydroxylase (CYP8B1) is a branch point enzyme for CA synthesis. CYP8B1 catalyzes 12α-hydroxylation of C4 to 7α, 12α-dihydroxy-4-cholesten-3-one, which is then converted to 5β-cholestan-3α, 7α, 12α-triol by two aldo-keto-reductases (AKR1D1 and AKR1C4). Without 12α-hydroxylation, C4 is converted to 5β-cholestan-3α, 7α-diol for CDCA synthesis. Mitochondrial sterol 27-hydroxylase (CYP27A1) catalyzes the steroid side chain oxidation of 5β-cholestan-3α, 7α, 12α-triol and 5β-cholestan-3α, 7α-diol to -C27-OH, which is subsequently converted to C27-OOH to form 3α, 7α, 12α-trihydroxy-5β cholestanoic acid (THCA) and 3α, 7α-dihydroxy-5β cholestanoic acid (DHCA), respectively. The very long chain acyl-CoA synthase in persoxisome or sterol side chain cleavage occurs in the peroxisomes. Bile acid-CoA synthase (lyase) activates THCA and DHCA to form acyl-CoAs for peroxisomal β-oxidation by acyl-CoA oxidase and a bifunctional enzyme (enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase), and then a propionyl-CoA is cleaved by thiolase to produce trihydroxycholyl-CoA and dihydroxycholyl-CoA, which are subsequently conjugated to glycine (G) or taurine (T) by bile acid CoA: amino acid N-acyltransferase (BAAT) to form T/G-CA and T/G-CDCA, respectively (Fig. 3A). In mouse liver, most CDCA is converted to α-muricholic acid (α-MCA) by a species-specific sterol 6β-hydroxylase (Cyp2c70), and then the 7α-OH group in α-MCA is epimerized to a 7β-OH group to form β-MCA13. MCAs are the primary bile acids synthesized in mouse liver. In mice and humans, the 7α-OH groups in CDCA can be isomerized to 7β-OH to form ursodeoxycholic acid (UDCA).
Figure 3.
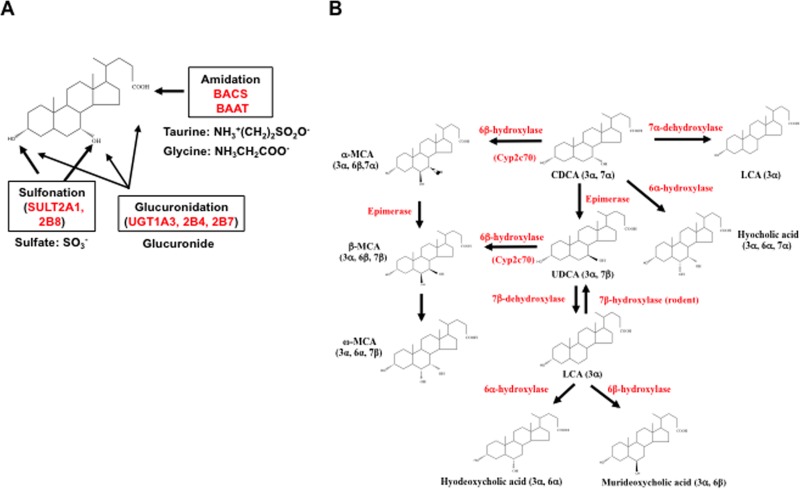
Bile acid conjugation and transformation. (A) Bile acid conjugation. In addition to conjugation to taurine and glycine at the C24-carboxyl group by bile acid Co-A synthase (BACS) and bile acid CoA: amino acid N-acetyltransferase (BAAT), bile acids can be conjugated to sulfate by bile acid sulfotransferase (SULT2A1, SULT2B8) at the C3 and C7 positions, or glucuronidated by UDP-glucuronosyl transferase (UGT1A3, 2B4, and 2B7). (B) Bile acid transformation in enterocytes. Primary bile acids, CDCA, CA, and UDCA, can be converted to other bile acids in the liver and intestine. In the liver, CDCA can be converted to α-MCA, β-MCA, and UDCA as described in Figure 2. In the colon, bacterial bile salt hydratases (dehydroxylases) first deconjugate bile acids to free bile acids, and then gut bacteria converts CA to deoxycholic acid (DCA). CDCA can be converted to lithocholic acid (LCA) by 7α-dehydroxylase and hyocholic acid by 6α-hydroxylase. LCA can be converted to hyodeoxycholic acid by 6α-hydroxylase, or murideoxycholic acid by 6β-hydroxylase. In rodents, LCA can be converted to UDCA by 7β-hydroxylase and UDCA can be converted to CDCA. In the colon, α-MCA and β-MCA can be converted to ω-MCA for fecal excretion.
The Alternative Bile Acid Synthesis Pathways
The alternative bile acid pathway in the liver is initiated by CYP27A1, which converts cholesterol to 27-hydroxycholesterol and then to 3β-hydroxy-5-cholestenoic acid. A nonspecific oxysterol 7α-hydroxylase (CYP7B1) hydroxylates this cholestenoic acid to 3β, 7α-dihydroxy-5-cholestenoic acid, followed by HSD3B1/3B2 to synthesize 7α-hydroxy-3-oxo-4-cholestenoic acid in liver, steroidogenic tissues, and macrophages. In mouse and human liver, sterol 25-hydroylase (CH25H), a non-CYP450 enzyme converts cholesterol to 25-hydroxycholesterol, followed by CYP7B1 to produce 5-cholesten-3β, 7α, 25-triol. In the brain, steroid 24-hydroxylase (CYP46A1) converts cholesterol to 24-hydroxycholesterol, which is then converted to 5-cholesten-3β, 7α, 24(S)-triol by a specific sterol-7α-hydroxylase (CYP39A1) in mouse liver. Oxidized steroid intermediates (oxysterols) formed in the extrahepatic tissues can be transported to the liver for synthesis of bile acids. The alternative pathways produce not only CDCA but also CA.
The Classic Versus Alternative Bile Acid Synthesis Pathways
The classic bile acid synthesis pathway is the major pathway that is regulated by CYP7A1, the only rate-limiting enzyme in bile acid synthesis. The alternative pathways synthesize bile acids in the neonate when CYP7A1 is not expressed. After weaning, CYP7A1 is expressed, and the classic pathway becomes the major pathway for bile acid synthesis in adult liver. It has been suggested that the classic pathway is the predominant pathway for synthesis of ∼80% of bile acids in human livers, whereas the classic and alternative pathways contribute about equally to bile acid synthesis in rodents. Mutations of the CYP7A1 gene in male adult patients only caused mild hypercholesterolemia and premature gallstone diseases, indicating that the alternative bile acid synthesis pathways are activated to produce bile acids when the classic pathway initiated by CYP7A1 is defective14. Mutation of the CYP7B1 gene in a neonate patient caused severe cholestatic liver injury and accumulation of oxysterol metabolites and monohydroxy bile acids, supporting the notion that the alternative bile acid synthesis pathway is important in the neonate15.
Bile Acid Conjugation and Transformation
Bile acids form Na2+ salts (bile salts), which are ionized at physiological pH. Conjugation of bile acids to glycine and taurine at C24 increases bile acid ionization, amphipathic properties, and solubility (Fig. 3A). Bile acids also can be conjugated to glucuronide by the UDP-glucuronosyltransferases UGT 1A1, 2B4, and 2B7 at the C3, C7, or C24 positions. The bile acid sulfotransferases SULT2A1 and SULT2A8 transfer a sulfate group to the C3 or C7 position16. Conjugated bile acids are secreted into bile via the canalicular bile salt export pump (BSEP), forming mixed micelles with cholesterol and phosphatidylcholine in the gallbladder to prevent precipitation of cholesterol and to protect gallbladder epithelium cells from bile acid toxicity. After a meal, cholecystokinin (CCK) released from the pancreas stimulates gallbladder contraction to release bile acids into the gastrointestinal tract. Some bile acids are passively absorbed in the upper intestine, but most are reabsorbed in the ileum and colon where gut microbial bile salt hydrolase (BSH) deconjugates bile acids to free bile acids, then bacterial 7α-dehydroxylase activity converts CA and CDCA to deoxycholic acid (DCA) and lithocholic acid (LCA), respectively17,18 (Fig. 3B). CDCA can be 6α-hydroxylated to hyocholic acid. The 7α-OH groups in CDCA can be isomerized to 7β-OH groups to UDCA, which can be converted to LCA by 7β-dehydroxylase in gut bacteria. Rodents can convert LCA to UDCA by 7α-hydroxylase, to hyodeoxycholic acid by 6α-hydroxylase, or to murideoxycholic acid by 6β-hydroxylase (Fig. 3B). In the colon, α-MCA and β-MCA can be converted to ω-MCA. Secondary bile acids formed in the intestine are excreted into feces, and some are reabsorbed in the colon and are circulated with CA and CDCA in portal blood to the liver, contributing to the circulating bile acid pool. A small amount of LCA (∼1%) reabsorbed in the liver is efficiently sulfur conjugated and excreted into urine.
Bile Acid Pool and Bile Acid Composition
The bile acid pool is defined as the total bile acids circulating in the enterohepatic circulation, including bile acids in the liver (<1%), intestine (∼85%–90%), and gallbladder (∼10%–15%). Bile acids spilled over from the liver to systemic circulation are not counted in the pool. Bile acids in gallbladder bile are from newly synthesized bile acids and bile acids returned to the liver via enterohepatic circulation. Thus, bile acid compositions in gallbladder bile more closely represent bile acid composition in the circulating bile acid pool. In the colon, most bile acids are unconjugated DCA, and most bile acids (∼95%) in the pool are conjugated bile acids. In humans, the bile acid pool consists of CA (∼40%), CDCA (∼40%), and DCA (∼20%), and the ratio of glycine (G)- to taurine (T)-conjugated bile acids is ∼3 to 1, and this bile acid pool is highly hydrophobic. In mice, most bile acids (95%) are taurine conjugated, and the bile acid pool consists of TCA (∼60%) and Tα-MCA and Tβ-MCA (∼40%) and is highly hydrophilic. Increasing bile acid pool size or bile acid hydrophobicity causes cholesterol gallstone disease and cholestatic liver injury. Feeding of mice with CA increases bile acid pool size and hydrophobicity and causes hepatic steatosis and cholestasis. Blockage of biliary bile acid secretion causes accumulation of bile acids in hepatocytes and liver injury. On the other hand, bile acid sequestrants reduce bile acid pool size, stimulate bile acid synthesis, and reduce serum cholesterol in hypercholesterolemic patients and animals.
Interestingly, mice with overexpression of a Cyp7a1 cDNA (Cyp7a1-Tg mice) have an enlarged bile acid pool (2.5-fold increase) consisting mostly of TCDCA (60%) and TMCA (40%) and no TCA due to inhibition of Cyp8b119. In the ileum and colon of Cyp7a1-Tg mice, TMCAs are increased and ceramides are reduced20. These mice are resistant to diet-induced obesity (DIO), likely due to activation of hepatic farnesoid X receptor (FXR) by TCDCA to inhibit lipogenesis, and antagonization of intestinal FXR by TMCAs to reduce ceramide synthesis21. On the other hand, Cyp7a1 −/− mice bred to a pure C57BL/6J genetic background have a reduced bile acid pool size (∼60% of wild type) with a more hydrophilic bile acid pool containing reduced TCA but increased TMCA22. Interestingly, Cyp7a1 −/− mice also have improved glucose tolerance and reduced triglycerides and are resistant to Western high-fat diet (HFD)-induced insulin resistance, likely due to a switch to the alternative bile acid synthesis pathway, less TCA to reduce dietary cholesterol absorption, and more TMCA synthesis to antagonize intestinal FXR activity and reduce ceramide synthesis22. In Cyp8b1 −/− mice, Cyp7a1 expression is increased to increase bile acid pool size with increased TMCAs, and mice are resistant to HFD (60% fat calories)-induced hepatic steatosis23. In Cyp7b1 −/− mice, bile acid pool size and composition are normal, but 25- and 27-hydroxycholesterols are accumulated24. Thus, the existence of two bile acid synthesis pathways is important for switching between two pathways to produce a sufficient bile acid pool size, although bile acid composition may be altered to affect hepatic metabolism.
BILE ACID SIGNALING IN LIVER PATHOPHYSIOLOGY
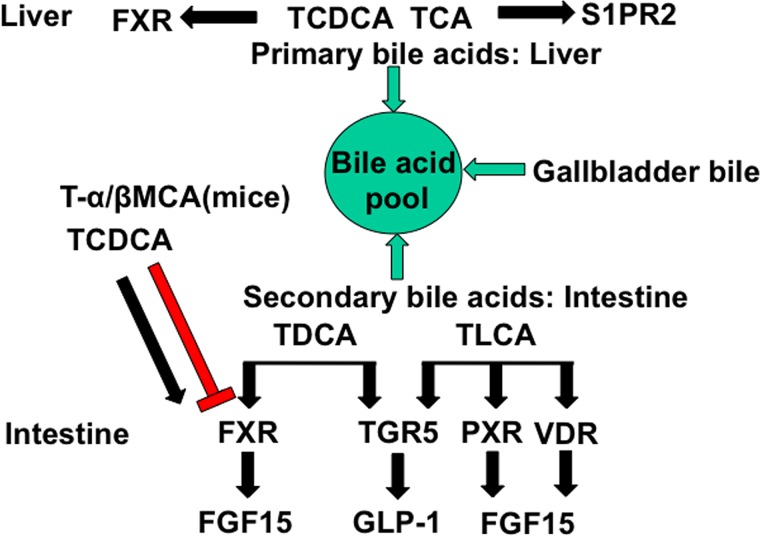
Enterohepatic circulation of bile acids reabsorbs bile acids in the intestine to control bile acid synthesis in the liver by a negative feedback mechanism and maintain a constant bile acid pool. Emerging research in the last two decades has discovered that bile acids are endogenous signaling molecules that activate the nuclear receptors FXR, vitamin D3 receptor (VDR), pregnane X receptor (PXR), and constitutive androstane receptor (CAR), as well as the membrane G protein-coupled receptors Takeda G protein receptor 5 [TGR5; aka G protein bile acid receptor-1 (Gpbar-1)] and sphingosine-1-phosphate receptor 2 (S1PR2) in the gastrointestinal tract (Fig. 4). The circulating bile acid pool consists of primary and secondary bile acids from the liver, gallbladder, and intestine. Primary bile acids are efficacious endogenous ligands of FXR and S1PR2 in the liver. In the intestine, TCDCA activates FXR, whereas TLCA /LCA and DCA activate TGR5, PXR, and VDR. TMCAs are FXR antagonists in the intestine.
Figure 4.
Bile acid-activated receptors in the gastrointestinal system. Bile acid-activated receptors are differentially activated by primary and secondary bile acids in the liver and intestine. The bile acid pool consists both of the primary bile acids TCDCA, TCA, Tα-MCA, and Tβ-MCA, and the secondary bile acids TDCA and TLCA. In the liver, TCDCA activates FXR, whereas TCA activates S1PR2. In the intestine, TCDCA and TDCA activate FXR, whereas Tα-MCA and Tβ-MCA antagonize FXR. TLCA and TDCA activate TGR5, and TLCA activates VDR and PXR.
FXR Signaling
TCDCA is the most potent endogenous FXR agonist with an EC50 of 17 μM. In mice, TCA and TMCAs are the predominant bile acids in the bile acid pool. However, TCA is a very weak FXR agonist (EC50 = ∼590 μM), while T-αMCA (IC50 = 28 μM) and T-βMCA (IC50 = 40 μM) are potent FXR antagonists23 (Fig. 4). Therefore, FXR may not be activated in mouse hepatocytes under physiological conditions. In the intestine, bile acid concentrations are much higher than in hepatocytes, and TCDCA may activate FXR to induce ileum bile acid-binding protein (IBABP), fibroblast growth factor 15 (FGF15), and sinusoidal bile acid efflux transporter organic solute transporter α/β heterodimer (OSTα/β). The secondary bile acids formed in the colon, LCA and DCA, activate TGR5 to stimulate GLP-1 secretion from enteroendocrine L cells, which stimulates insulin secretion from β-cells and improves insulin sensitivity25.
The role of FXR in the regulation of glucose, lipid, and energy metabolism and in improving glucose and insulin sensitivity and obesity has been the subject of many recent reviews2,3,26,27. Contradictory roles of FXR in the regulation of glucose metabolism have been reported recently. Earlier studies have shown that activation of FXR improves glucose homeostasis in diabetic mice28. It has been suggested that activation of FXR induces small heterodimer partner (SHP), which may lower triglycerides by inhibiting liver X receptor (LXR)-induced and steroid response element-binding protein-1c (SREBP-1c)-mediated lipogenesis29. FXR may inhibit fatty acid synthesis by inhibiting glucose-induced nuclear translocation of carbohydrate response element-binding protein (Chrebp), which induces glycolytic genes involved in the conversion of glucose to fatty acids30. In contrast, activation of FXR has been shown to induce fatty acid synthase, implicating bile acid signaling in the stimulation of hepatic lipogenesis31. Furthermore, it was shown that activation of FXR by GW4064 increased body weight and glucose tolerance and reduced energy expenditure in HFD-fed mice (DIO)32, but deficiency of FXR improves glucose homeostasis in DIO mice33. Several recent studies have reported that inhibition of intestinal FXR activity by the antioxidant tempol remodeled the gut microbiome and reduced obesity in mice34. Consistently, deficiency of intestinal FXR protects mice from DIO and diabetes35, and the intestine-selective FXR inhibitor glycine-MCA improved obesity and diabetes in mice36. In contrast, other recent studies report that selective intestinal FXR agonists stimulate FGF15 to promote adipocyte browning, reduce weight, and improve insulin resistance in DIO mice37, and protect against cholestasis38. It is possible that different intestinal FXR agonists alter the gut microbiome differently and result in opposite effects on hepatic metabolism (see Bile Acids and Gut Microbiota, below).
TGR5 Signaling
TGR5 is a Gαs protein-coupled receptor activated by the secondary bile acids TLCA (EC50 = 0.3 μM) and DCA (EC50 = 1 μM). TGR5 is expressed in the epithelial cells of the gastrointestinal system, including intestine, spleen, cholangiocytes, gallbladder, hepatic sinusoidal endothelial cells, and hepatic macrophages (Kupffer cells)39–42. TGR5 may have different functions in different tissues. TGR5 activates cAMP/PKA signaling to stimulate energy metabolism in brown adipose tissue43 and is involved in relaxing and refilling the gallbladder44, secreting glucagon-like peptide 1 (GLP-1) from L cells25, and controlling GI motility45 (Fig. 4). Activation of TGR5 has been shown to protect the liver from bile acid overload during liver regeneration46. Pharmacological activation of TGR5 markedly decreased lipopolysaccharide (LPS)-induced cytokine production in primary macrophages and protected adipose tissue from inflammation and associated insulin resistance through the Akt/mTOR/C/EBP pathway47. Another study reports that activation of TGR5 by a nonsteroidal agonist inhibits LPS-induced TNF-α and IL-12 release in mice and primary human hepatocytes48. In contrast, it has been reported that activation of TGR5 in macrophages promotes bile acid- or LPS-induced inflammation49,50. TGR5 is highly expressed in cholangiocytes, and its expression levels are increased in cholangiocarcinoma cells. In Tgr5 −/− mice, CA feeding- and chronic bile duct ligation-induced cholangiocyte proliferation was significantly reduced, while in wild-type mice, TLCA and TGR5-selective agonists induced cholangiocyte proliferation through increased reactive oxygen species and ERK1/2 phosphorylation51. A recent study shows that TGR5 may be involved in the regulation of CYP7B1, a sexually dimorphic and male-predominant gene in the alternative bile acid synthesis pathway52.
Mechanisms of Bile Acid-Activated Receptors in the Regulation of Bile Acid Synthesis
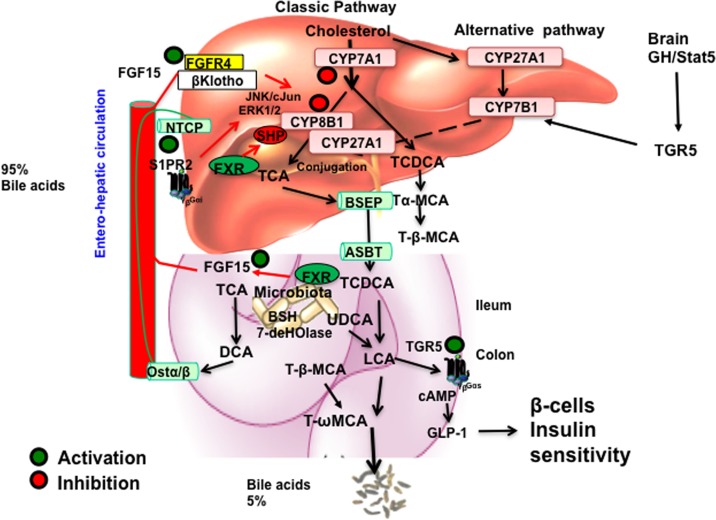
A complex network of signaling pathways regulates bile acid synthesis (Fig. 5). Bile acid inhibition of CYP7A1 is a classic feedback mechanism whereby the end products of a metabolic pathway inhibit the first enzyme in the pathway. The mechanism of bile acid feedback regulation has been studied over 50 years, but the molecular mechanism is still not clear. Two mechanisms have been proposed (Fig. 5). In the liver, activation of FXR by agonists induces SHP to inhibit HNF4 and LRH-1 trans-activation of CYP7A1 and CYP8B1 gene expression. In the intestine, activation of FXR by agonists induces FGF15 (or human FGF19), which is transported via portal blood circulation to hepatocytes to activate FGF receptor 4(FGFR4)/β-Klotho complex. The complex inhibits CYP7A1 and CYP8B1 gene transcription via an unknown mechanism that may involve the cJun and ERK1/2 pathways of the mitogen-activated protein kinase (MAPK). This intestine-to-liver signaling pathway may be a more physiologically relevant mechanism for bile acid feedback regulation of bile acid synthesis. TCA activation of S1PR2 may stimulate ERK1/2 signaling to inhibit CYP7A1 and CYP8B1 gene transcription, but the role of S1PR2 in bile acid feedback regulation has not been studied. Activation of TGR5 may regulate expression of CYP7B1, and TGR5 signaling in the brain may stimulate growth hormone secretion to activate STAT5 signaling in hepatocytes, which induces gene transcription of the male-predominant CYP7B1 to regulate bile acid composition52.
Figure 5.
Mechanisms of bile acid regulation of bile acid synthesis. In the liver, activation of FXR by agonists induces SHP to inhibit CYP7A1 and CYP8B1 gene transcription. TCA activates S1PR2 and may inhibit CYP7A1 via the ERK1/2 pathway. In the intestine, TCDCA activates FXR to induce FGF15, which is transported to hepatocytes to activate FGFR4/β-Klotho receptor, which stimulates the JNK/cJun and ERK1/2 pathways to inhibit CYP7A1 and CYP8B1. In the brain, growth hormone (GH) activates STAT5 signaling to activate TGR5 signaling to induce CYP7B1 in male mice and regulate the alternative bile acid synthesis pathway. In the intestine, TLCA activates TGR5 in L cells to stimulate GLP-1 secretion, which increases insulin sensitivity by stimulating insulin secretion from pancreatic β-cells.
CIRCADIAN RHYTHM IN BILE ACID METABOLISM
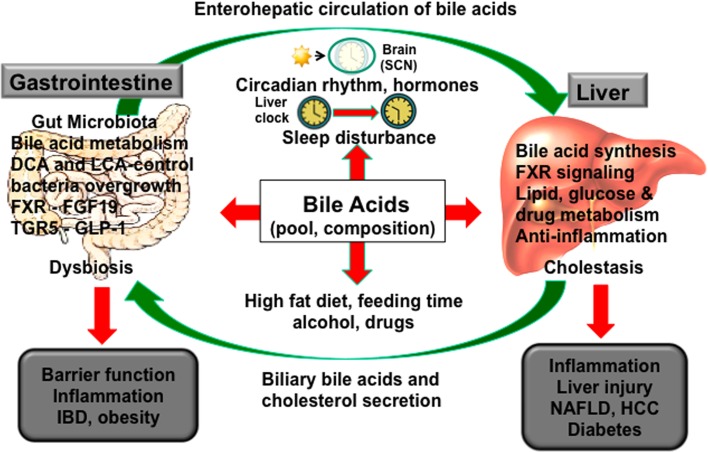
Figure 6 illustrates the close linkage between the liver and gut in bile acid synthesis and metabolism, which is modulated by circadian rhythms27. The central circadian clock is located in the hypothalamic suprachiasmatic nucleus (SCN), which generates biological rhythms via a transcription–translation feedback loop of core clock genes encoding transcriptional repressors Period 1/2/3 (Per1/2/3) and cryptochrome 1/2 (Cry1/2), and transcriptional activators brain and muscle ARNT-like protein 1 (Bmal1) and circadian locomotor output cycles kaput (Clock). The central clock evolved due to the rotation of the Earth around the sun every 24 h. The peripheral clocks are synchronized to the central clock via the core clock genes Per/Cry and Bmal1/Clock, which regulate transcription of thousands of other clock-controlled genes. A secondary regulatory loop of the core clock is composed of reverse-erythroblastosis α (Rev-erbα) and retinoid acid-related receptor α (RORα). The peripheral clocks are influenced and entrained by neuronal input to tissues, hormones, physiological activity, and environmental factors such as diets, time of feeding, drugs, and alcohol53–59. Rev-erbα and RORα regulate CYP7A1 and CYP8B1 in bile acid synthesis to exhibit a distinct circadian rhythm of bile acid synthesis in mice60. Fasting and refeeding cycles and nutrients regulate bile acid synthesis. Feeding rapidly induces Cyp7a1 but represses Cyp8b1, whereas fasting represses Cyp7a1 but induces Cyp8b1. In rodents, Cyp7a1 expression peaks 2 h into the dark phase and rapidly decreases in the light phase27. In humans, bile acid synthesis shows a distinct diurnal rhythm, which peaks at 1:00 pm and 9:00 pm, and declines at midnight61. Bile acid synthesis varies in individuals, is lower in females compared to males, and is correlated to serum triglycerides and circulating FGF19 levels62. Circulating FGF19 has a pronounced diurnal rhythm, peaking 90–120 min after the postprandial rise of serum bile acids63. Activation of intestinal FXR induces FGF15 during the late postprandial state64. Circadian disruptions, such as shift work or chronic sleep disorders, contribute to cardiovascular diseases65, digestive diseases66, and metabolic syndrome67–69. A recent study in mice shows that even short-term sleep disruption of 6 h during the day for 5 days significantly suppressed core clock gene (Clock and Per1/2, Rev-erbα), the genes involved in hepatic lipid metabolism (Srebp1, hepatocyte nuclear factor 4α), Cyp7a1 regulators (hepatocyte nuclear factor 4α and D-site-binding protein), and Cyp7a1 expression69. Disruption of circadian rhythms alters bile acid homeostasis, causes liver and intestine inflammation, and alters the gut microbiome and contributes to cholestatic liver injury, NAFLD, diabetes, and IBDs and dysbiosis27,70,71.
Figure 6.
The gut-to-liver axis and circadian rhythms in bile acid metabolism. Bile acid synthesis in the liver is controlled in part by circadian expression of CYP7A1 expression. Bile acids control the gut microbiota population, and gut bacteria regulate bile acid metabolism, bile acid composition, and enterohepatic circulation of bile acids. Sleep disruption, high-fat diet (HFD), alcohol, and drugs alter the central clock in the hypothalamic suprachiasmatic nucleus (SCN) of the brain to desynchronize the peripheral clocks in the liver and intestine. Disruption of circadian rhythms alters bile acid homeostasis, causes liver and intestine inflammation and dysbiosis, and contributes to cholestatic liver injury, nonalcoholic fatty liver disease (NAFLD), diabetes, and inflammatory bowel diseases.
BILE ACIDS AND THE GUT MICROBIOTA
The gut-to-liver axis plays a critical role in the regulation of metabolic homeostasis and in preventing metabolic diseases72. Bile acids regulate the gut microbiome structure by controlling gut bacteria overgrowth and protecting the epithelial barrier via FXR signaling73. FXR signaling shapes the gut microbiota and controls hepatic lipid metabolism74,75. Gut bacteria metabolize bile acids to regulate bile acid composition and bile acid hydrophobicity. Intestinal bile acids are antimicrobial agents that control gut microbial overgrowth by altering membrane lipid composition, solubilizing membranes, and dissociating integral membrane proteins and damaging membrane integrity. Gut microbes deconjugate bile acids and convert primary bile acids to secondary bile acids in the colon. Gram-negative bacteria have a higher bile acid tolerance than Gram-positive bacteria. BSH activity is high in Gram-positive bacteria, such as Lactobacillus, Enterococcus, Bacteroides, and Clostridium. BSH deconjugates glycine- and taurine-conjugated bile acids to release glycine and taurine, which are metabolized to ammonia and CO2, and also sulfite from taurine. Bile acids are carbon sources for energy metabolism in some Gram-negative anaerobic bacteria. Feeding a CA-containing diet increases the gut Firmicutes-to-Bacteriodetes ratio76,77 and causes dysbiosis and contributes to liver inflammation78. Dietary saturated fats increase TCA and promote bile-tolerant and sulfur-producing Bilophila wadsworthia to increase proinflammatory cytokines and colitis in Il10 −/− mice79. Bacteria capable of converting primary bile acids to secondary bile acids have been isolated from humans and rodents. The bile acid inducible (Bai) operon in bile acid dehydroxylating gut bacteria have been mapped in Clostridium scindens, Clostridium hylemonase, Clostridium hiranonis, and Clostridium sordellii 18. The baiE gene encodes the bile acid 7α-dehydroxylase activities, whereas the baiI gene may encode the bile acid 7β-dehydroxylase. Bacterial hydroxy-steroid dehydrogenases (HSD) epimerize the hydroxyl groups at 3-, 7-, and 12-C of bile acids. The α- and β-HSD activities in Clostridium absonum epimerize the 7α-HO group of CDCA to the 7β-HO group of UDCA, increasing the solubility and decreasing the toxicity of bile acids. However, the 7β-dehydroxylase activities in C. scindens and Clostridium hiranonase and Bacteroides can convert UDCA back to LCA. Antibiotics alter the gut microbiota and impair the production of secondary bile acids, which are protective of gut barrier function. Bacteria with 7α-dehydroxylase activity (i.e., C. scindens) may be used to increase secondary bile acids and alter gut bile acid composition to protect against Clostridium difficile infection in patients with antibiotic-associated diarrhea and colitis80.
Recent studies have implicated Cyp8b1 in dyslipidemia, insulin resistance81, and atherosclerosis82. In diabetic patients, bile acid pool size and the serum ratio of 12α-hydroxylated bile acids to non-12α-hydroxylated bile acids are increased. Cyp8b1 −/− mice, germ-free mice and antibiotic-treated mice share similar metabolic phenotypes: they all have increased Cyp7a1 expression and bile acid synthesis, enlarged bile acid pool, and are resistant to HFD-induced hepatic steatosis due to antagonism of intestinal FXR activity by TMCAs and increased Cyp7a1 expression23,83. Antibiotics modulate the gut microbiota and decrease Cyp8b1 expression to alter bile acid profiles in mice and protect against diet-induced metabolic disorders84. Germ-free mice have increased Cyp7b1 expression and increased TMCA23. Bile acids induce UCP-1 in brown adipose tissue to stimulate energy metabolism in mice housed at thermoneutrality85. Reducing ambient temperature attenuated DIO in mice by increasing brown adipose tissue thermogenesis, reshaping gut microbiome, and changing bile acid profile similar to germ-free mice86. Another study reports that Cyp7b1 may play a role in cold-induced thermogenesis87. Thus, cold-induced conversion of cholesterol to bile acids, by increasing the alternative pathway and altering bile acid composition, may shape the gut microbiome and promote adaptive thermogenesis in brown adipose tissues in mice.
Male Fxr −/− mice are more susceptible to Western diet (WD)-induced hepatic steatosis, insulin resistance, and obesity compared to female mice. Bile acids and the microbiota may play a role in mediating sex differences in hepatic inflammation, dysbiosis, and NAFLD in mice88–90. The sex differences in WD-induced hepatic steatosis, insulin resistance, bile acids, and microbiota profiles are FXR dependent89. Male Fxr −/− mice have more severe hepatic inflammation and steatosis compared to female mice, and WD feeding aggravates hepatic steatosis more in these male mice88. Normally, WD increases biliary secretion of bile acids and reshapes the gut microbiota in obesity by increasing Firmicutes and decreasing Bacteriodetes 91. WD decreases Firmicutes and increases Proteobacteria in Fxr −/− mice92. The incidence of streptozotocin (STZ)–HFD-induced HCC is significantly higher in male mice than in female mice. Metagenomic analysis showed differences in gut bacteria that are involved in bile acid metabolism between normal male and female mice. STZ-HFD treatment amplified the observed sex difference in gut microbiota. Female mice have a higher ratio of Firmicutes to Bacteriodetes compared to male mice. STZ-HFD treatment reduced Firmicutes and Bacteriodetes and markedly increased Proteobacteria in female mice, but much less so in male mice. It was found that STZ-HFD treatment caused more intrahepatic retention of hydrophobic bile acids (TCA, TCDCA, and TLCA) in male mice compared to female mice. STZ-HFD treatment strongly reduced Cyp7b1 mRNA expression, consistent with a much higher increase in TCA in male than female mice, and an altered gut microbiota between male and female mice.
Either activation or antagonizing intestinal FXR signaling resulted in reducing weight and increasing insulin sensitivity in DIO mice34–37. The underlying mechanisms could be different. The intestine FXR antagonist Gly-MCA and the intestine-selective FXR agonist fexaramine may shape the gut microbiome differently, resulting in opposite effects on insulin resistance and obesity in mice. It seems clear that both intestinal FXR/FGF15 signaling and TGR5/GLP-1 signaling modulate hepatic metabolism. The gut-to-liver axis is critically involved in whole-body lipid, glucose, and energy metabolism.
BILE ACID SIGNALING IN LIVER PATHOBIOLOGY
FXR and TGR5 in Anti-Inflammation
Bile acids have different effects on cell proliferation and inflammation in different cell types and tissues51. CA feeding causes hepatocyte proliferation. More hydrophilic bile acids TCA and UDCA protect against apoptosis, while the hydrophobic bile acids TLCA and GCDCA cause hepatic apoptosis and liver injury. Activation of FXR and TGR5 signaling in mice by their respective agonists has been shown to protect against inflammation in the liver and intestine by inhibiting NF-κB-induced inflammatory cytokine production and liver inflammation93–100. FXR agonists have been shown to protect against inflammation in cholestasis, cirrhosis, atherosclerosis, and IBD and are in clinical trials to treat NAFLD and primary biliary cirrhosis96,101,102. FXR signaling also inhibits dextran sodium sulfate-induced IBDs103,104. TGR5 agonist INT777 has been used to reduce body weight and stimulate GLP-1 secretion to improve insulin sensitivity25, and attenuates atherosclerosis by reducing macrophage inflammation99 and liver ischemia and reperfusion injury105. The dual FXR and TGR5 agonist INT-767 has been shown to switch M1 proinflammatory responses to M2 anti-inflammatory responses and alleviate NAFLD in db/db mice106 and atherosclerosis in Apoe and Ldl receptor double knockout mice107.
Cholestatic Liver Diseases
Cholestasis is a pathological condition where normal bile flow out of the liver is reduced or disrupted by tumors or inflammation, leading to intrahepatic accumulation of bile acids108. Accumulation of cytotoxic bile acids activates NF-kB-mediated proinflammatory cytokine production. High levels of toxic bile acids damage the bile duct epithelium and elevate biliary pressure to rupture the bile duct and expose hepatocytes to high concentrations of bile acids, inflammatory infiltration, and leads to hepatocyte cell death. Cholestasis could result from genetic defects in canalicular transporters (i.e., BSEP), mechanical obstruction of bile duct by gallstones or tumors, factors associated with pregnancy [intrahepatic cholestasis of pregnancy (ICP)], autoimmune destruction of the bile ducts in and outside of the liver, or drug-induced liver toxicity109,110. Mutations in the FXR gene have been shown to cause progressive familial intrahepatic cholestasis in human patients111,112. Primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) are two common types of progressive chronic cholestasis diagnosed in humans. The etiology of PBC and PSC involves genetic, immune, and environmental factors. PBC is an autoimmune disease with high prevalence in females and is caused by inflammation in the small intrahepatic bile duct and impaired hepatic bile acid secretion and accumulation. PSC is an idiopathic liver disease characterized by chronic liver inflammation and progressive destruction of intrahepatic and extrahepatic bile ducts in men. Selective activation of intestinal FXR has been shown to increase FGF15, repress Cyp7a1 and bile acid pool, and protect against intrahepatic cholestasis in bile duct-ligated mice and Mdr2 −/− mice38.
BILE ACID THERAPY
UDCA (Ursodiol™) has been used as a bile acid replacement therapy to reduce bile acid toxicity for patients with bile acid synthesis deficiency, gallstone dissolution, and digestive diseases for several decades113. UDCA activates PKC and MAPK signaling to alleviate liver injury in cholestasis. UDCA increases hepatobiliary secretion and promotes biliary HCO3 − secretion to protect hepatocytes and cholangiocytes from hydrophobic bile acid-induced liver injury114–116. However, UDCA is not effective in treating patients with PSC. Nor-ursodeoxycholic acid (norUDCA), a derivative of UDCA with shorter side-chain C23, has been shown to increase HCO3 − secretion and improve sclerosing cholangitis in the Mdr2 −/− model of cholangiopathy116,117.
FXR Agonists
A synthetic CDCA derivative, 6-ethyl-CDCA (OCA, INT-747, OCALIVA®) is a selective FXR agonist (EC50 = 0.099 μM) with therapeutic potential for treating cholestasis118,119. OCA treatment showed effective protection in experimental cholestasis models120. Recent clinical trials reported that OCA significantly improved liver tests in patients with PBC121,122 and reduced serum alkaline phosphatase, GGT, and alanine aminotransferase activities121. OCA improved nonalcoholic steatohepatitis (NASH) scores in clinical trials and is a promising therapy for NASH123,124. The intestine-restricted FXR agonist fexaramine (EC50 = 25 μM) has been shown to promote adipose tissue browning to reduce weight and diabetes in DIO mice37.
TGR5 Agonists
Activation of TGR5 stimulates GLP-1 secretion from enteroendocrine L cells, which leads to stimulation of insulin synthesis and secretion from pancreatic β-cells125–127. It also protects cholangiocytes from bile acid toxicity in cholestasis and is a potential therapy for cholestasis125. However, TGR5 agonists may also promote proliferation, apoptosis, and progression of cholangiocarcinoma51. Several potent TGR5-selective agonists are in development for treating metabolic diseases128,129. A synthetic bile acid derivative, INT-777 [6α-ethyl-23(S)-methyl-CA, EC50 = 0.82 μM] is a TGR5-selective agonist and has been shown to protect intestinal barrier function and immune response to experimental colitis95,130, reduce macrophage inflammation by activating cAMP signaling to inhibit NF-kB activity and inflammatory cytokine production, and reduce plaque formation and atherosclerosis in Ldlr −/− mice99. TGR5 agonists induced NO production and reduced monocyte adhesion in vascular endothelial cells131. A non-bile acid and potent TGR5 agonist has been shown to stimulate GLP-1 secretion and lower glucose132,133. Other non-bile acid TGR5 agonists have been designed to treat type 2 diabetes133,134.
FXR and TGR5 Dual Agonist
The dual FXR and TGR5 agonist INT-767 (EC50 = 0.03 μM for FXR and EC50 = 0.63 μM for TGR5) is a 6-ethyl-CDCA derivative with a C23-sulfate group. INT-767 has been shown to improve diabetes and hepatic steatosis and decrease inflammation in db/db and DIO mice106,135,136. INT-767 reduces liver injury in Mdr2 −/− mice137. A recent study showed that INT-767 stimulates intracellular [Ca2+] and cAMP activity to stimulate GLP-1 secretion and improve glucose and lipid metabolism138. Interestingly, INT-767 reduced expression of the genes in the classic bile acid synthesis pathway while inducing those in the alternate pathway to decrease TCA and increase TMCAs. TCA is highly efficacious in intestinal absorption of dietary cholesterol, and TMCA antagonizes intestinal FXR signaling. Surprisingly, OCA and INT-767 induce expression of intestinal Tgr5 and pro-hormone convertase 1/3, which converts proglucagon to GLP-1 in the ileum. Consistently, Fxr −/− and Tgr5 −/− mice have reduced GLP-1 secretion. This study uncovered a novel mechanism in which activation of FXR induces Tgr5 gene expression and increases [Ca2+] and cAMP activity to stimulate GLP-1 secretion and improve hepatic glucose and lipid metabolism. Another FXR and TGR5 dual agonist has been designed to reduce weight and treat diabetes139. Activation of both FXR and TGR5 may represent an effective therapy for treating hepatic steatosis, obesity, and diabetes138.
CONCLUSIONS
Recent advances in the discovery of the role of bile acids in cell signaling and metabolism have contributed enormously to the current understanding of the mechanism of regulation of bile acid homeostasis and hepatic metabolism. Bile acid signaling via FXR regulates hepatic metabolism, and intestinal FXR and TGR5 signaling crosstalk modulates the gut microbiome, bile acid homeostasis, and host metabolism. Gender differences in bile acids and gut microbiome in diet-induced steatosis, NASH, and hepatocellular carcinoma have been unveiled and need to be further studied to identify differentially regulated hormones, bacteria, bile acids, and metabolic pathways responsible for increased prevalence of these diseases in males89,90. The emerging research of bile acid metabolism and homeostasis has been translated to drug therapy targeted to treating metabolic disorders. Bile acid receptors are signaling integrators, and bile acid derivatives are in clinical trials for treating dyslipidemia, NAFLD, diabetes, and cardiovascular diseases6,26. Bariatric surgeries are effective in reducing weight and improving insulin resistance in obese patients, and FXR and TGR5 signaling have been implicated in increasing serum bile acids, GLP-1, and FGF19 after bariatric surgery140–142. However, the underlying molecular mechanism of bile acid signaling in improving diabetes after bariatric surgery is not clear and needs to be elucidated to develop therapeutic strategies to cure diabetes and NAFLD.
ACKNOWLEDGMENTS
This research was supported by NIH grants DK44442 and DK58379.
Footnotes
The authors declare no conflicts of interests.
REFERENCES
- 1. Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res. 2009;50(Suppl):S120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chiang JY. Bile acids: Regulation of synthesis. J Lipid Res. 2009;50:1955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89(1):147–91. [DOI] [PubMed] [Google Scholar]
- 4. Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thomas C, Auwerx J, Schoonjans K. Bile acids and the membrane bile acid receptor TGR5—Connecting nutrition and metabolism. Thyroid 2008;18(2):167–74. [DOI] [PubMed] [Google Scholar]
- 6. Arab JP, Karpen SJ, Dawson PA, Arrese M, Trauner M. Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology 2017;65(1):350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11(1):55–67. [DOI] [PubMed] [Google Scholar]
- 8. Chiang JY. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr Rev. 2002;23(4):443–63. [DOI] [PubMed] [Google Scholar]
- 9. Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:1370174. [DOI] [PubMed] [Google Scholar]
- 10. Chiang JY. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40(3):539–51. [DOI] [PubMed] [Google Scholar]
- 11. Axelson M, Aly A, Sjovall J. Levels of 7 alpha-hydroxy-4-cholesten-3-one in plasma reflect rates of bile acid synthesis in man. FEBS Lett. 1988;239(2):324–8. [DOI] [PubMed] [Google Scholar]
- 12. Honda A, Yamashita K, Numazawa M, Ikegami T, Doy M, Matsuzaki Y, Miyazaki H. Highly sensitive quantification of 7alpha-hydroxy-4-cholesten-3-one in human serum by LC-ESI-MS/MS. J Lipid Res. 2007;48(2):458–64. [DOI] [PubMed] [Google Scholar]
- 13. Takahashi S, Fukami T, Masuo Y, Brocker CN, Xie C, Krausz KW, Wolf CR, Henderson CJ, Gonzalez FJ. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J Lipid Res. 2016;57(12):2130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pullinger CR, Eng C, Salen G, Shefer S, Batta AK, Erickson SK, Verhagen A, Rivera CR, Mulvihill SJ, Malloy MJ, Kane JP. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 2002;110(1):109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Setchell KDR, Schwarz M, O’Connell NC, Lund EG, Davis DL, Lathe R, Thompson HR, Weslie Tyson R, Sokol RJ, Russell DW. Identification of a new inborn error in bile acid synthesis: Mutation of the oxysterol 7a-hydroxylase gene causes severe neonatal liver disease. J Clin Invest. 1998;102(9):1690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng L, Yuen YL, Xu J, Liu X, Chan MY, Wang K, Fong WP, Cheung WT, Lee SS. Identification and characterization of a novel PPARalpha-regulated and 7alpha-hydroxyl bile acid-preferring cytosolic sulfotransferase mL-STL (Sult2a8). J Lipid Res. 2017;58(6):1114–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–59. [DOI] [PubMed] [Google Scholar]
- 18. Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016;7(1):22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li T, Owsley E, Matozel M, Hsu P, Novak CM, Chiang JY. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology 2010;52(2):678–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi Y, Jiang C, Cheng J, Krausz KW, Li T, Ferrell JM, Gonzalez FJ, Chiang JY. Bile acid signaling in lipid metabolism: Metabolomic and lipidomic analysis of lipid and bile acid markers linked to anti-obesity and anti-diabetes in mice. Biochim Biophys Acta 2015;1851:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie C, Jiang C, Shi J, Gao X, Sun D, Sun L, Wang T, Takahashi S, Anitha M, Krausz KW, and others. An intestinal farnesoid X receptor-ceramide signaling axis modulates hepatic gluconeogenesis in mice. Diabetes 2017;66(3):613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferrell JM, Boehme S, Li F, Chiang JY. Cholesterol 7{alpha}-hydroxylase-deficient mice are protected from high fat/high cholesterol diet-induced metabolic disorders. J Lipid Res. 2016;57:1144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–35. [DOI] [PubMed] [Google Scholar]
- 24. Li-Hawkins J, Lund EG, Turley SD, Russell DW. Disruption of the oxysterol 7alpha-hydroxylase gene in mice. J Biol Chem. 2000;275(22):16536–42. [DOI] [PubMed] [Google Scholar]
- 25. Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, and others. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Porez G, Prawitt J, Gross B, Staels B. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease: Thematic review series: New lipid and lipoprotein targets for the treatment of cardiometabolic diseases. J Lipid Res. 2012;53(9):1723–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66(4):948–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci USA 2006;103(4):1006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113(10):1408–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caron S, Huaman Samanez C, Dehondt H, Ploton M, Briand O, Lien F, Dorchies E, Dumont J, Postic C, Cariou B, and others. Farnesoid X receptor inhibits the transcriptional activity of carbohydrate response element binding protein in human hepatocytes. Mol Cell Biol. 2013;33(11):2202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matsukuma KE, Bennett MK, Huang J, Wang L, Gil G, Osborne TF. Coordinated control of bile acids and lipogenesis through FXR-dependent regulation of fatty acid synthase. J Lipid Res. 2006;47(12):2754–61. [DOI] [PubMed] [Google Scholar]
- 32. Watanabe M, Horai Y, Houten SM, Morimoto K, Sugizaki T, Arita E, Mataki C, Sato H, Tanigawara Y, Schoonjans K, and others. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem. 2011;286(30):26913–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prawitt J, Abdelkarim M, Stroeve JH, Popescu I, Duez H, Velagapudi VR, Dumont J, Bouchaert E, van Dijk TH, Lucas A, and others. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes 2011;60(7):1861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, Cai J, Qi Y, Fang ZZ, Takahashi S, and others. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125(1):386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang C, Xie C, Lv Y, Li J, Krausz KW, Shi J, Brocker CN, Desai D, Amin SG, Bisson WH, and others. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015;6:10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, Yoshihara E, Perino A, Jacinto S, Lukasheva Y, and others. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med. 2015;21(2):159–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Modica S, Petruzzelli M, Bellafante E, Murzilli S, Salvatore L, Celli N, Di Tullio G, Palasciano G, Moustafa T, Halilbasic E, and others. Selective activation of nuclear bile acid receptor FXR in the intestine protects mice against cholestasis. Gastroenterology 2012;142(2):355–65 e1–4. [DOI] [PubMed] [Google Scholar]
- 39. Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, and others. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. [DOI] [PubMed] [Google Scholar]
- 40. Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun. 2002;298(5):714–9. [DOI] [PubMed] [Google Scholar]
- 41. Keitel V, Cupisti K, Ullmer C, Knoefel WT, Kubitz R, Haussinger D. The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology 2009;50:861–70. [DOI] [PubMed] [Google Scholar]
- 42. Keitel V, Ullmer C, Haussinger D. The membrane-bound bile acid receptor TGR5 (Gpbar-1) is localized in the primary cilium of cholangiocytes. Biol Chem. 2010;391(7):785–9. [DOI] [PubMed] [Google Scholar]
- 43. Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, and others. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006;439:484–9. [DOI] [PubMed] [Google Scholar]
- 44. Li T, Holmstrom SR, Kir S, Umetani M, Schmidt DR, Kliewer SA, Mangelsdorf DJ. The G protein-coupled bile acid receptor, TGR5, stimulates gallbladder filling. Mol Endocrinol. 2011;25(6):1066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alemi F, Poole DP, Chiu J, Schoonjans K, Cattaruzza F, Grider JR, Bunnett NW, Corvera CU. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 2013;144(1):145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pean N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, Branchereau S, Spraul A, Guettier C, Humbert L, and others. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology 2013;58:1451–60. [DOI] [PubMed] [Google Scholar]
- 47. Perino A, Pols TW, Nomura M, Stein S, Pellicciari R, Schoonjans K. TGR5 reduces macrophage migration through mTOR-induced C/EBPbeta differential translation. J Clin Invest. 2014;124(12):5424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hogenauer K, Arista L, Schmiedeberg N, Werner G, Jaksche H, Bouhelal R, Nguyen DG, Bhat BG, Raad L, Rauld C, and others. G-protein-coupled bile acid receptor 1 (GPBAR1, TGR5) agonists reduce the production of proinflammatory cytokines and stabilize the alternative macrophage phenotype. J Med Chem. 2014;57(24):10343–54. [DOI] [PubMed] [Google Scholar]
- 49. Lou G, Ma X, Fu X, Meng Z, Zhang W, Wang YD, Van Ness C, Yu D, Xu R, Huang W. GPBAR1/TGR5 mediates bile acid-induced cytokine expression in murine Kupffer cells. PLoS One 2014;9(4):e93567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mobraten K, Haugbro T, Karlstrom E, Kleiveland CR, Lea T. Activation of the bile acid receptor TGR5 enhances LPS-induced inflammatory responses in a human monocytic cell line. J Recept Signal Transduct Res. 2015;35(5):402–9. [DOI] [PubMed] [Google Scholar]
- 51. Reich M, Deutschmann K, Sommerfeld A, Klindt C, Kluge S, Kubitz R, Ullmer C, Knoefel WT, Herebian D, Mayatepek E, and others. TGR5 is essential for bile acid-dependent cholangiocyte proliferation in vivo and in vitro. Gut 2016;65(3):487–501. [DOI] [PubMed] [Google Scholar]
- 52. Donepudi AC, Boehme S, Li F, Chiang JY. G-protein-coupled bile acid receptor plays a key role in bile acid metabolism and fasting-induced hepatic steatosis in mice. Hepatology 2017;65(3):813–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002;109(3):307–20. [DOI] [PubMed] [Google Scholar]
- 54. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–21. [DOI] [PubMed] [Google Scholar]
- 55. Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA 2009;106(50):21453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17(11):2100–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science 2010;330(6009):1349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, and others. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20(6):1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pathak P, Li T, Chiang JY. Retinoic acid-related orphan receptor alpha regulates diurnal rhythm and fasting induction of sterol 12alpha-hydroxylase in bile acid synthesis. J Biol Chem. 2013;288(52):37154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Galman C, Angelin B, Rudling M. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology 2005;129(5):1445–53. [DOI] [PubMed] [Google Scholar]
- 62. Galman C, Angelin B, Rudling M. Pronounced variation in bile acid synthesis in humans is related to gender, hypertriglyceridaemia and circulating levels of fibroblast growth factor 19. J Intern Med. 2011;270(6):580–8. [DOI] [PubMed] [Google Scholar]
- 63. Lundasen T, Galman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260(6):530–6. [DOI] [PubMed] [Google Scholar]
- 64. Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, Xu HE, Shulman GI, Kliewer SA, Mangelsdorf DJ. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 2011;331(6024):1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Morris CJ, Purvis TE, Hu K, Scheer FA. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc Natl Acad Sci USA 2016;113(10):E1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Konturek PC, Brzozowski T, Konturek SJ. Gut clock: Implication of circadian rhythms in the gastrointestinal tract. J Physiol Pharmacol. 2011;62(2):139–50. [PubMed] [Google Scholar]
- 67. Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, and others. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005;308(5724):1043–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Maury E, Hong HK, Bass J. Circadian disruption in the pathogenesis of metabolic syndrome. Diabetes Metab. 2014;40(5):338–46. [DOI] [PubMed] [Google Scholar]
- 69. Ferrell JM, Chiang JY. Short-term circadian disruption impairs bile acid and lipid homeostasis in mice. Cell Mol Gastroenterol Hepatol. 2015;1(6):664–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. Circadian disorganization alters intestinal microbiota. PLoS One 2014;9(5):e97500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ferrell JM, Chiang JY. Circadian rhythms in liver metabolism and disease. Acta Pharm Sin B 2015;5(2):113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24(1):41–50. [DOI] [PubMed] [Google Scholar]
- 73. Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, and others. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA 2006;103(10):3920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang L, Xie C, Nichols RG, Chan SH, Jiang C, Hao R, Smith PB, Cai J, Simons MN, Hatzakis E, and others. Farnesoid X receptor signaling shapes the gut microbiota and controls hepatic lipid metabolism. mSystems 2016;1(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gonzalez FJ, Jiang C, Patterson AD. An intestinal microbiota-farnesoid X receptor axis modulates metabolic disease. Gastroenterology 2016;151(5):845–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011;141(5):1773–81. [DOI] [PubMed] [Google Scholar]
- 77. Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014;146(6):1513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. De Minicis S, Rychlicki C, Agostinelli L, Saccomanno S, Candelaresi C, Trozzi L, Mingarelli E, Facinelli B, Magi G, Palmieri C, and others. Dysbiosis contributes to fibrogenesis in the course of chronic liver injury in mice. Hepatology 2014;59(5):1738–49. [DOI] [PubMed] [Google Scholar]
- 79. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012;487(7405):104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, Schurch CM, McCoy KD, Kuehne SA, Minton NP, and others. Functional intestinal bile acid 7alpha-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front Cell Infect Microbiol. 2016;6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li-Hawkins J, Gafvels M, Olin M, Lund EG, Andersson U, Schuster G, Bjorkhem I, Russell DW, Eggertsen G. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest. 2002;110(8):1191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Slatis K, Gafvels M, Kannisto K, Ovchinnikova O, Paulsson-Berne G, Parini P, Jiang ZY, Eggertsen G. Abolished synthesis of cholic acid reduces atherosclerotic development in apolipoprotein E knockout mice. J Lipid Res. 2010;51(11):3289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hu X, Bonde Y, Eggertsen G, Rudling M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J Intern Med. 2014;275(1):27–38. [DOI] [PubMed] [Google Scholar]
- 84. Zhang Y, Limaye PB, Renaud HJ, Klaassen CD. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol. 2014;277(2):138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zietak M, Kozak LP. Bile acids induce uncoupling protein 1-dependent thermogenesis and stimulate energy expenditure at thermoneutrality in mice. Am J Physiol Endocrinol Metab. 2016;310(5):E346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zietak M, Kovatcheva-Datchary P, Markiewicz LH, Stahlman M, Kozak LP, Backhed F. Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab. 2016;23(6):1216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Worthmann A, John C, Ruhlemann MC, Baguhl M, Heinsen FA, Schaltenberg N, Heine M, Schlein C, Evangelakos I, Mineo C, and others. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat Med. 2017;23(7):839–49. [DOI] [PubMed] [Google Scholar]
- 88. Jena P, Sheng L, Liu H-X, Kalanetra KM, Mirsoian A, Murphy WJ, French SW, Krishnan VK, Mils DA, Wan Y-JY. Western diet-induced dysbiosis in FXR knockout mice causes persistent hepatic inflammation post antibiotic treatment. Am J Pathol. 2017;187(8):1800–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sheng L, Jena PK, Liu H-X, Kalanetra KM, Gonzalez FJ, French SW, Krishnan VK, Miller DA, and Wan Y-JV. Gender differences in bile acids and mcirobiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci Rep. 2017;7(1):1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Xie G, Wang X, Zhao A, Yan J, Chen W, Jiang R, Ji J, Huang F, Zhang Y, Lei S, and others. Sex-dependent effects on gut microbiota regulate hepatic carcinogenic outcomes. Sci Rep. 2017;7:45232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O’Toole PW, Cotter PD. The gut microbiota and its relationship to diet and obesity: New insights. Gut Microbes 2012;3(3):186–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008;3(4):213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fiorucci S, Cipriani S, Mencarelli A, Renga B, Distrutti E, Baldelli F. Counter-regulatory role of bile acid activated receptors in immunity and inflammation. Curr Mol Med. 2010;10(6):579–95. [DOI] [PubMed] [Google Scholar]
- 94. Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology 2011;54(4):1421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One 2011;6(10):e25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183(10):6251–61. [DOI] [PubMed] [Google Scholar]
- 97. Renga B, Migliorati M, Mencarelli A, Fiorucci S. Reciprocal regulation of the bile acid-activated receptor FXR and the interferon-gamma-STAT-1 pathway in macrophages. Biochim Biophys Acta 2009;1792(6):564–73. [DOI] [PubMed] [Google Scholar]
- 98. Guo C, Qi H, Yu Y, Zhang Q, Su J, Yu D, Huang W, Chen WD, Wang YD. The G-protein-coupled bile acid receptor Gpbar1 TGR5) inhibits gastric inflammation through antagonizing NF-kappaB signaling pathway. Front Pharmacol. 2015;6:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, Rizzo G, Gioiello A, Adorini L, Pellicciari R, and others. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14(6):747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Perino A, Schoonjans K. TGR5 and Immunometabolism: Insights from physiology and pharmacology. Trends Pharmacol Sci. 2015;36(12):847–57. [DOI] [PubMed] [Google Scholar]
- 101. Hageman J, Herrema H, Groen AK, Kuipers F. A role of the bile salt receptor FXR in atherosclerosis. Arterioscler Thromb Vasc Biol. 2010;30(8):1519–28. [DOI] [PubMed] [Google Scholar]
- 102. Mencarelli A, Fiorucci S. FXR an emerging therapeutic target for the treatment of atherosclerosis. J Cell Mol Med. 2010;14(1–2):79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, and others. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011;60(4):463–72. [DOI] [PubMed] [Google Scholar]
- 104. Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 2008;48(5):1632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yang H, Zhou H, Zhuang L, Auwerx J, Schoonjans K, Wang X, Feng C, Lu L. Plasma membrane-bound G protein-coupled bile acid receptor attenuates liver ischemia/reperfusion injury via the inhibition of toll-like receptor 4 signaling in mice. Liver Transpl. 2017;23(1):63–74. [DOI] [PubMed] [Google Scholar]
- 106. McMahan RH, Wang XX, Cheng LL, Krisko T, Smith M, El Kasmi K, Pruzanski M, Adorini L, Golden-Mason L, Levi M, and others. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J Biol Chem. 2013;288(17):11761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Miyazaki-Anzai S, Masuda M, Levi M, Keenan AL, Miyazaki M. Dual activation of the bile acid nuclear receptor FXR and G-protein-coupled receptor TGR5 protects mice against atherosclerosis. PLoS One 2014;9(9):e108270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ghonem NS, Assis DN, Boyer JL. Fibrates and cholestasis. Hepatology 2015;62(2):635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jacquemin E. Progressive familial intrahepatic cholestasis. Clin Res Hepatol Gastroenterol. 2012;36(Suppl 1):S26–35. [DOI] [PubMed] [Google Scholar]
- 110. Zollner G, Trauner M. Mechanisms of cholestasis. Clin Liver Dis. 2008;12(1):1–26, vii. [DOI] [PubMed] [Google Scholar]
- 111. Chen F, Ananthanarayanan M, Emre S, Neimark E, Bull LN, Knisely AS, Strautnieks SS, Thompson RJ, Magid MS, Gordon R, and others. Progressive familial intrahepatic cholestasis, type 1, is associated with decreased farnesoid X receptor activity. Gastroenterology 2004;126(3):756–64. [DOI] [PubMed] [Google Scholar]
- 112. Gomez-Ospina N, Potter CJ, Xiao R, Manickam K, Kim MS, Kim KH, Shneider BL, Picarsic JL, Jacobson TA, Zhang J, and others. Mutations in the nuclear bile acid receptor FXR cause progressive familial intrahepatic cholestasis. Nat Commun. 2016;7:10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lioudaki E, Ganotakis ES, Mikhailidis DP. Lipid lowering drugs and gallstones: A therapeutic option? Curr Pharm Des. 2011;17(33):3622–31. [DOI] [PubMed] [Google Scholar]
- 114. Hohenester S, Wenniger LM, Paulusma CC, van Vliet SJ, Jefferson DM, Elferink RP, Beuers U. A biliary HCO3- umbrella constitutes a protective mechanism against bile acid-induced injury in human cholangiocytes. Hepatology 2012;55(1):173–83. [DOI] [PubMed] [Google Scholar]
- 115. Prieto J, Garcia N, Marti-Climent JM, Penuelas I, Richter JA, Medina JF. Assessment of biliary bicarbonate secretion in humans by positron emission tomography. Gastroenterology 1999;117(1):167–72. [DOI] [PubMed] [Google Scholar]
- 116. Beuers U. Drug insight: Mechanisms and sites of action of ursodeoxycholic acid in cholestasis. Nat Clin Pract Gastroenterol Hepatol. 2006;3(6):318–28. [DOI] [PubMed] [Google Scholar]
- 117. Halilbasic E, Fiorotto R, Fickert P, Marschall HU, Moustafa T, Spirli C, Fuchsbichler A, Gumhold J, Silbert D, Zatloukal K, and others. Side chain structure determines unique physiologic and therapeutic properties of norursodeoxycholic acid in Mdr2-/- mice. Hepatology 2009;49(6):1972–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, Willson TM. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem. 2002;45(17):3569–72. [DOI] [PubMed] [Google Scholar]
- 119. Pellicciari R, Costantino G, Camaioni E, Sadeghpour BM, Entrena A, Willson TM, Fiorucci S, Clerici C, Gioiello A. Bile acid derivatives as ligands of the farnesoid X receptor. Synthesis, evaluation, and structure-activity relationship of a series of body and side chain modified analogues of chenodeoxycholic acid. J Med Chem. 2004;47(18):4559–69. [DOI] [PubMed] [Google Scholar]
- 120. Fiorucci S, Clerici C, Antonelli E, Orlandi S, Goodwin B, Sadeghpour B, Sabatino G, Russo G, Castellani D, Willson TM, and others. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor (FXR) ligand, in estrogen induced cholestasis. J Pharmacol Exp Ther. 2005;313:604–12. [DOI] [PubMed] [Google Scholar]
- 121. Hirschfield GM, Mason A, Luketic V, Lindor K, Gordon SC, Mayo M, Kowdley KV, Vincent C, Bodhenheimer HC Jr, Pares A, and others. Efficacy of obeticholic acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic acid. Gastroenterology 2015;148(4):751–61 e8. [DOI] [PubMed] [Google Scholar]
- 122. Nevens F, Andreone P, Mazzella G, Strasser SI, Bowlus C, Invernizzi P, Drenth JP, Pockros PJ, Regula J, Beuers U, and others. A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N Engl J Med. 2016;375(7):631–43. [DOI] [PubMed] [Google Scholar]
- 123. Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, Adorini L, Sciacca CI, Clopton P, Castelloe E, and others. Efficacy and safety of the farnesoid x receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology 2013;145(3):574–82 e1. [DOI] [PubMed] [Google Scholar]
- 124. Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, and others. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 2015;385(9972):956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5: A valuable metabolic target. Dig Dis. 2011;29(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. 2005;329(1):386–90. [DOI] [PubMed] [Google Scholar]
- 127. Kumar DP, Asgharpour A, Mirshahi F, Park SH, Liu S, Imai Y, Nadler JL, Grider JR, Murthy KS, Sanyal AJ. Activation of transmembrane bile acid receptor TGR5 modulates pancreatic islet alpha cells to promote glucose homeostasis. J Biol Chem. 2016;291(13):6626–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sato H, Macchiarulo A, Thomas C, Gioiello A, Une M, Hofmann AF, Saladin R, Schoonjans K, Pellicciari R, Auwerx J. Novel potent and selective bile acid derivatives as TGR5 agonists: Biological screening, structure-activity relationships, and molecular modeling studies. J Med Chem. 2008;51(6):1831–41. [DOI] [PubMed] [Google Scholar]
- 129. Tiwari A, Maiti P. TGR5: An emerging bile acid G-protein-coupled receptor target for the potential treatment of metabolic disorders. Drug Discov Today 2009;14(9–10):523–30. [DOI] [PubMed] [Google Scholar]
- 130. Pellicciari R, Sato H, Gioiello A, Costantino G, Macchiarulo A, Sadeghpour BM, Giorgi G, Schoonjans K, Auwerx J. Nongenomic actions of bile acids. Synthesis and preliminary characterization of 23- and 6,23-alkyl-substituted bile acid derivatives as selective modulators for the G-protein coupled receptor TGR5. J Med Chem. 2007;50(18):4265–8. [DOI] [PubMed] [Google Scholar]
- 131. Kida T, Tsubosaka Y, Hori M, Ozaki H, Murata T. Bile acid receptor TGR5 agonism induces NO production and reduces monocyte adhesion in vascular endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33(7):1663–9. [DOI] [PubMed] [Google Scholar]
- 132. Duan H, Ning M, Chen X, Zou Q, Zhang L, Feng Y, Zhang L, Leng Y, Shen J. Design, synthesis, and antidiabetic activity of 4-phenoxynicotinamide and 4-phenoxypyrimidine-5-carboxamide derivatives as potent and orally efficacious TGR5 agonists. J Med Chem. 2012;55(23):10475–89. [DOI] [PubMed] [Google Scholar]
- 133. Duan H, Ning M, Zou Q, Ye Y, Feng Y, Zhang L, Leng Y, Shen J. Discovery of intestinal targeted TGR5 agonists for the treatment of type 2 diabetes. J Med Chem. 2015;58(8):3315–28. [DOI] [PubMed] [Google Scholar]
- 134. Zambad SP, Tuli D, Mathur A, Ghalsasi SA, Chaudhary AR, Deshpande S, Gupta RC, Chauthaiwale V, Dutt C. TRC210258, a novel TGR5 agonist, reduces glycemic and dyslipidemic cardiovascular risk in animal models of diabesity. Diabetes Metab Syndr Obes. 2013;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Rizzo G, Passeri D, De Franco F, Ciaccioli G, Donadio L, Orlandi S, Sadeghpour B, Wang XX, Jiang T, Levi M, and others. Functional characterization of the semisynthetic bile acid derivative INT-767, a dual farnesoid X receptor and TGR5 agonist. Mol Pharmacol. 2010;78(4):617–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. D’Amore C, Di Leva FS, Sepe V, Renga B, Del Gaudio C, D’Auria MV, Zampella A, Fiorucci S, Limongelli V. Design, synthesis, and biological evaluation of potent dual agonists of nuclear and membrane bile acid receptors. J Med Chem. 2014;57(3):937–54. [DOI] [PubMed] [Google Scholar]
- 137. Baghdasaryan A, Claudel T, Gumhold J, Silbert D, Adorini L, Roda A, Vecchiotti S, Gonzalez FJ, Schoonjans K, Strazzabosco M, and others. Dual farnesoid X receptor/TGR5 agonist INT-767 reduces liver injury in the Mdr2-/- (Abcb4-/-) mouse cholangiopathy model by promoting biliary HCO output. Hepatology 2011;54(4):1303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Pathak P, Liu H, Boehme S, Xie C, Krausz KW, Gonzalez F, Chiang JYL. Farnesoid X receptor induces Takeda G-protein receptor 5 crosstalk to regulate bile acid synthesis and hepatic metabolism. J Biol Chem. 2017;292(26):11055–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Carino A, Cipriani S, Marchiano S, Biagioli M, Santorelli C, Donini A, Zampella A, Monti MC, Fiorucci S. BAR502, a dual FXR and GPBAR1 agonist, promotes browning of white adipose tissue and reverses liver steatosis and fibrosis. Sci Rep. 2017;7:42801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Steinert RE, Peterli R, Keller S, Meyer-Gerspach AC, Drewe J, Peters T, Beglinger C. Bile acids and gut peptide secretion after bariatric surgery: A 1-year prospective randomized pilot trial. Obesity (Silver Spring) 2013;21(12):E660–8. [DOI] [PubMed] [Google Scholar]
- 141. Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, Wilson-Perez HE, Sandoval DA, Kohli R, Backhed F, and others. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 2014;509:183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. McGavigan AK, Garibay D, Henseler ZM, Chen J, Bettaieb A, Haj FG, Ley RE, Chouinard ML, Cummings BP. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut 2015;66(2):226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]