Abstract
Accumulation of mitochondrial DNA (mtDNA) mutations has been proposed to contribute to the initiation and progression of tumors. By using high-throughput sequencing strategies, we measured 33 specimens including 11 hepatocellular carcinoma (HCC) tissues, 11 corresponding adjacent tissues, and 11 normal liver tissues. We identified 194 single nucleotide variants (SNVs; including insert and deletion) in 33 liver tissues, and 13 somatic novel mutations were detected, including 7 mutations in the coding region. One of the seven somatic mutations (T7609C, 91.09%) is synonymous, which does not change amino acid coding; the other four somatic mutations (T6115C, 65.74%; G8387A, 12.23%; G13121A, 93.08%; and T14180C, 28.22%) could result in amino acid substitutions, potentially leading to mitochondrial dysfunction. Furthermore, two mutations in tRNA might influence amino acid transportation. Consistent with a previous study, we also found that mtDNA copy number was significantly reduced in HCC tissues. Therefore, we established a mitochondrial genome depletion cell line ρ0 and revealed that mtDNA loss reduced proliferation and migration in HCC cells but promoted their resistance to 5-fluorouracil. Our results suggested that somatic mtDNA mutations may cause mitochondrial dysfunction and affect chemoresistance of HCC cells. These new identified somatic mutations may serve as a reference for future studies of cancer mitochondrial genomes.
Key words: Mitochondrial DNA (mtDNA) mutations, Hepatocellular carcinoma (HCC), High-throughput sequencing
INTRODUCTION
Mitochondria, which are bacterium-sized organelles, are observed in all nucleated cells and are the principal generators of cellular adenosine triphosphate (ATP)1,2. Mitochondrial DNA (mtDNA) is a multicopy, 16,569-bp circular double strand DNA (dsDNA) molecule, which contains 37 genes: 13 of these genes encode 13 polypeptides of respiratory enzyme complexes, and the remaining 24 genes encode 22 transfer RNAs and 2 ribosomal RNAs (12S and 16S) used for protein synthesis in mtDNA3,4. Because of the absence of histone proteins, defective DNA repair mechanisms, and proximity to large amounts of ROS, mtDNA is especially vulnerable to mutation compared with nuclear DNA5,6.
Apart from inherited mtDNA disorders associated with defects in oxidative energy metabolism7–10, it is emerging that acquired somatic mutations in the mtDNA are contributing to neurodegenerative diseases, aging, and cancer11–14. In 1998, Polyak et al. made a landmark report that somatic mtDNA point mutations existed in human colorectal tumors but not in the normal, healthy tissues from the same individuals15. Since then, increasing studies have described the high frequency of mtDNA mutations in several other types of human cancers, including both solid tumors and leukemia16–20. Different from other types of cancer, hepatocellular carcinoma (HCC) is at least partially accompanied by chronic inflammation caused by viral infection. Nishikawa and his colleagues demonstrated that the frequency of mtDNA mutations was markedly increased in both noncancerous and cancerous liver specimens in HCC patients compared with control liver tissues21. It is suggested that the mutation of mtDNA in HCC is associated with viral infection. Furthermore, HCC had a high frequency of somatic mutations in the displace (D-loop) and coding region of mtDNA compared with the corresponding noncancerous and control liver tissues. In 2004, Lee and colleagues found that about 40% of the HCCs carried somatic mutations in the D-loop, and in 2010 they reported that about 25% of the HCCs carried somatic mutations in the coding region of mtDNA22,23. Although considerable efforts have been made to investigate the mtDNA alteration in HCC, standard DNA sequencing techniques restrict profound detection of mtDNA alteration in large-scale samples from different cohorts.
Many pathogenic mutations occurred in a proportion of mtDNA copies within a single cell, resulting in a heteroplasmy situation with a mixture of mutated and wild-type genomes24. In the presence of heteroplasmy, there is a threshold level of mutation, under which mutated genomes are functionally recessive for both the disease and biochemical phenotypes25. Alternatively, the concept of homoplasmy describes an ideal state in which all mtDNAs from an individual, tissue, or even a cell are identical. However, present evidence indicates that mtDNA is constantly suffering mutation26, so a few mutations will be present at a low level and thereby might not be easily detected in tissue homogenate or peripheral blood samples. Therefore, accurate sequencing would help us understand the relationship between mtDNA mutations and disease progression.
In this study, we searched somatic mutations in the mtDNA of HCC patients by high-throughput sequencing strategies and identified seven novel mutations in the coding region of mtDNA in HCC tissues. In addition, an in vitro ρ0 cell model of mitochondrial dysfunction was established to elucidate the possible role of mtDNA in the progression of HCC.
MATERIALS AND METHODS
Human HCC Tissues and DNA Extraction
HCC samples, including adjacent and normal liver tissues, were obtained and histologically confirmed from 11 patients with their informed consent at the Shanghai Eastern Hepatobiliary Surgery Hospital. According to a protocol approved by the medical ethics committee, all the tissues were kept in liquid nitrogen immediately after surgical resection. Total DNA of the tissues was extracted by the Allprep DNA/RNA/Protein Mini kit (QIAGEN) according to the manufacturer’s instructions. The final DNA was dissolved in ddH2O and frozen at −30°C.
mtDNA Copy Number Variation
mtDNA copy number variation (CNV) was conducted by a relative quantification PCR analysis. Chr16-CNV and Chr2-CNV are internal control genes on autosome, whose copy number is considered to be two in normal diploid cells. mtDNA-CNV-1 and mtDNA-CNV-2 are specific amplification fragments on mtDNA. Relative copy number is calculated by 2−ΔCt, ΔCt = Ct(target) − Ct (internal control). Relative copy number to the median was used to revise PCR efficiency.
High-Throughput Sequencing
Two independent sets of a long PCR amplification system were applied to enrich the target region and prepare the sequencing library. The libraries were then assessed with Illumina Hiseq system. All the sequencing work was conducted by Geneskies Biotechnologies Inc. Single nucleotide variants were detected by GATK HaplotypeCaller (https://software.broadinstitute.org/gatk/best-practices/) and VarScan (http://varscan.sourceforge.net/). A multiple protein sequence alignment was performed on COBALT database (https://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?LINK_LOC=BlastHomeLink).
Cell Culture
HCC SK-HEP-1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin and incubated in a 5% CO2 chamber at 37°C. ρ0 cells were derived from SK-HEP-1 cells by passaging in the presence of 100 ng/ml EtBr, 100 mg/L pyruvate, and 50 mg/L uridine for more than 20 generations27. PCR analysis was performed to characterize the depletion of mtDNA using primers specific for human mtDNA: COX1, F-5′-CCTAGGGATAACAGCGCAAT-3′ (forward) and 5′-TAGAAGAGCGATGGTGAGAG-3′ (reverse), and internal control primers: GAPDH, 5′-ACCACAGTCCATGCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′ (reverse).
Cell Proliferation Assay
Cell counting kit-8 (CCK-8) assay and EdU assay were performed to determine cell proliferation. Three replicated wells were measured for each time point. After each well was incubated with 10 μl of CCK-8 reagent (Dojindo) for 3 h at 37°C, the optical density (OD) 450 nm of each well was measured using a microplate reader. The experiment was repeated independently three times. EdU was assessed by the EdU DNA proliferation in vitro detection (RiboBio) according to the manufacturer’s instructions.
To determine in vitro cytotoxicity, SK-HEP-1 cells and ρ0 cells were initially seeded in 96-well plates at a concentration of 1 × 104 cells (three wells per group) and then incubated for 24 h. Cells were treated with 16 μM 5-fluorouracil for another 24, 48, and 72 h. Cells were also treated with 5-fluorouracil at a gradient concentration of 0, 2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 μM, and then 10 μl of CCK-8 solution was added to each well for further incubation for 3 h in 5% CO2 at 37°C. The OD value was measured by a microplate reader at 450-nm wavelengths.
Cell Migration Assay
Transwell system and wound healing migration assays (6 wells, 24 wells, 8-μm pore size with polycarbonate membrane; Corning) were performed according to the manufacturer’s protocol. In the Transwell system, SK-HEP-1 and ρ0 cells (1 × 105) were seeded in the inserts of each well. The inserts were filled with 300 μl of DMEM supplemented with 1% FBS. The bottom of the well was filled with 700 μl of DMEM containing 10% FBS, and the cells were allowed to migrate for 6 h at 37°C in a humidified atmosphere containing 5% CO2. The membrane was fixed in methanol and then stained with crystal violet solution. After cells on the upper side of the membrane were removed by cotton swabs, cells on the bottom side of the membrane were taken and counted under a light microscope. For the wound healing assay, SK-HEP-1 and ρ0 cells (1 × 105) were seeded in six wells. Scratches were performed with a sterile 10-μl pipette tip, and at 0 and 24 h photographs were taken.
Data Analysis
Association between somatic mutations and the clinicopathologic characteristics of HCC were analyzed using Fisher’s exact test using the Statistical Program for Social Sciences (SPSS) program package. A value of p < 0.05 was considered statistically significant.
RESULTS
High-Frequency and Novel mtDNA Mutations Were Found in the Exonic Region
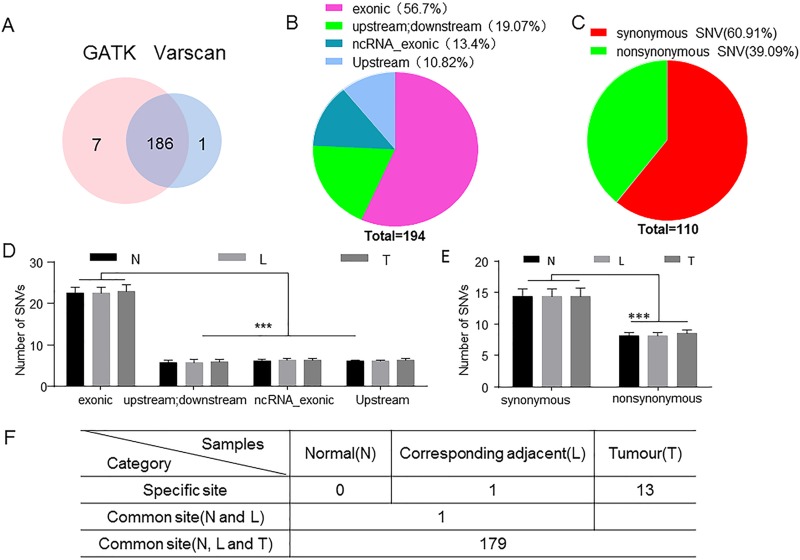
The acquired mtDNA sequence of each sample was processed by GATK and VarScan, respectively. In total, 186 single nucleotide variants (SNVs; including insert and deletion) in both GATK and VarScan, 7 only in GATK and 1 in VarScan, were detected (Fig. 1A). Generally, GATK is sensitive to insert/deletion (InDel) mutations, so GATK could compensate VarScan detection. Therefore, we combined the results from GATK and VarScan.
Figure 1.
Analysis and distribution of detected mitochondrial DNA (mtDNA) mutations. (A) A total of 194 single nucleotide variants (SNVs; including insert and deletion) were detected by GATK and VarScan; 186 SNVs were found in both GATK and VarScan, 7 SNV were discovered only in GATK, and 1 was discovered in VarScan. (B) mtDNA mutations were distributed in different regions. There are 56.7% SNV mutations that were located in the exonic region and 43.3% mutations that were located in other regions. (C) Synonymous mutations and nonsynonymous mutations accounted for 60.91% and 39.09% in the exonic region, respectively. (D, E) Mutations in the exonic region were significantly higher than those in the other regions, and synonymous mutations were significantly more than nonsynonymous mutations. (F) To further analyze these mutations, 179 mutations were observed in three groups, and 13 mutations only in the hepatocellular carcinoma (HCC) tissues. Two-way ANOVA was used for statistical analyses of the distribution of mtDNA mutations. N, normal; L, corresponding adjacent; T, tumor. ***p ≤ 0.0001 compared with the indicated groups.
To further analyze these mutations, we found that most of these SNV mutations are located in the exonic region (110/194, 56.7%) (Fig. 1B). Among the 110 mutations, 60.91% were synonymous and 39.09% were nonsynonymous. Nonsynonymous mutation might potentially result in substitution of amino acid residue and consequently alter the structure and function of protein (Fig. 1C). Additionally, 13.4% (26/194) of mutations were found in the exonic region of noncoding RNA (nc-RNA exonic). Also, 10.82% (21/194) of mutations occurred upstream and 19.07% (37/194) of mutations were detected upstream of one gene but downstream of another gene (Fig. 1B). There were no significant differences between different regions of SNV mutation counts in these three groups (HCC, adjacent, and normal) (Fig. 1D). Mutations in the exonic region were significantly higher than those in the other regions, and synonymous SNVs were significantly more than nonsynonymous SNVs (Fig. 1D and E). Although no significant difference was found in the mutation counts of different tissue specimens from each individual with HCC, mtDNA mutations were more frequently found in the exonic region (Fig. 1F).
Somatic Mutations in HCC on Coding Region Potentially Affect Mitochondrial Oxidative Phosphorylation (OXPHOS)
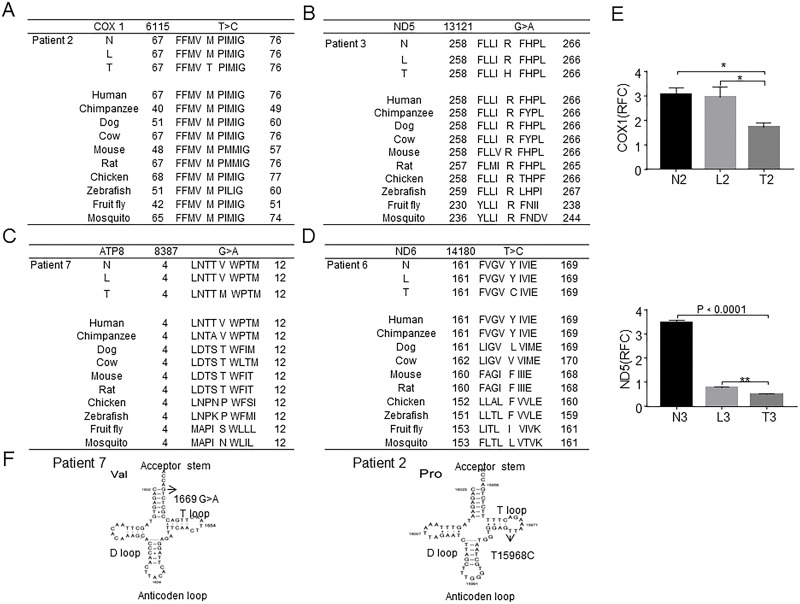
Among the 13 somatic mutations, 7 of mtDNA in 5 HCC sample (5/11, 45.5%) mutations existed in the coding region. One of them (T7609C, 91.09%) was synonymous polymorphisms, and the other four somatic mutations (T6115C, 65.74%; G8387A, 12.23%; G13121A, 93.08%; and T14180C, 28.22%) could result in amino acid substitutions, which might potentially lead to mitochondrial dysfunction (Table 1). The T6115C mutation in cytochrome c oxidase subunit 1 (COX1) gene causes a substitution of amino acid residue from methionine to threonine, and the G13121A mutation in NADH dehydrogenase subunit 5 (ND5) gene results in an amino acid substitution from arginine to histidine. Both the missense mutations occurred at the highly conserved region of mtDNA, which potentially affect the function of protein and mitochondrial OXPHOS (Fig. 2A and B). Although the G8387A and T14180C mutations also could cause amino acid substitution, they did not occur at the highly conserved region of mtDNA (Fig. 2C and D). The expression of COX1 and ND5 in tumor tissues was lower than in adjacent and normal liver tissues (Fig. 2E). Additionally, one mutation in tRNAval (G1669A, 42.93%) from the acceptor stem and another one mutation in tRNApro (T15968C, 28.29%) from the T-loop were detected, which might influence the amino acid transport (Fig. 2F). These mutations, which we present here, have never been reported, especially in coding region to potentially affect mitochondrial oxidative phosphorylation (OXPHOS).
Table 1.
Summary of the Mitochondrial DNA (mtDNA) Mutations in the 33 HCC Clinical Samples
| SNV No. | Gene | Position | Ref. Allele | Alt. Allele | Gene Region | Function | Predicted Protein Variants | Alt. Freq. (0.1–0.9) | Alt. Freq. ≥0.9 | Novelty* |
|---|---|---|---|---|---|---|---|---|---|---|
| SNV00164 | RNR1 | 72 | T | C | Upstream | T10 (90.99%) | − | |||
| SNV00118 | RNR1 | 316 | G | C | Upstream | T3 (15.15%) | + | |||
| SNV00119 | RNR1 | 319 | T | C | Upstream | T3 (34.61%) | + | |||
| SNV00102 | tRNA-Val | 1,669 | G | A | ncRNA_exonic | T7 (42.93%) | + | |||
| SNV00149 | COX1 | 5,894–5,894 | – | C | Upstream; downstream | T4 (10.27%) | + | |||
| SNV00150 | COX1 | 6,115 | T | C | Exonic | Nonsynonymous SNV | M71T | T2 (65.74%) | + | |
| SNV00167 | COX2 | 7,609 | T | C | Exonic | Synonymous SNV | G8G | T4 (91.09%) | + | |
| SNV00174 | ATP8 | 8,387 | G | A | Exonic | Nonsynonymous SNV | V8M | T7 (12.23%) | + | |
| SNV00026 | ND5 | 13,121 | G | A | Exonic | Nonsynonymous SNV | R262H | T3 (93.08%) | + | |
| SNV00035 | ND6 | 14,180 | T | C | Exonic | Nonsynonymous SNV | Y165C | T6 (28.22%) | + | |
| SNV00066 | tRNA-Pro | 15,968 | T | C | ncRNA_exonic | T2 (28.29%) | + | |||
| SNV00078 | tRNA-Pro | 16,183–16,183 | – | C | Upstream; downstream | T6 (11.23%) | + | |||
| SNV00101 | tRNA-Pro | 16,537 | C | T | Upstream; downstream | T2 (64.40%) | – |
Ref Allele: Revised Cambridge reference sequence (rCRS) of the human mitochondrial DNA: NC_012920 gi:251831106.
Novelty refers to no reports from the mitomap database.
Figure 2.
Six somatic mutations in HCC in the coding region potentially affect mitochondrial oxidative phosphorylation (OXPHOS). (A) T6115C mutation in the coding region was detected in HCC patient 2. The mutation caused a methionine in cytochrome c oxidase subunit 1 (COX1) to be replaced with threonine. (B) The G13121A mutation was detected in HCC patient 3. The mutation caused arginine in NADH dehydrogenase subunit 5 (ND5) to be replaced with histidine. These somatic mutations occurred at the evolutionarily highly conserved region. (C, D) G8387A and T14180C were detected in HCC patients 7 and 6, respectively, and these mutations do not occur at the highly conserved region. (E) The expressions of COX1 and ND5 in the tumor were lower than that in the adjacent and normal liver tissues. (F) Somatic mtDNA mutations G1669A and T15968C in HCC patients 7 and 2. The G1669A mutation occurred at the acceptor stem of the tRNAval gene, and T15968C mutation occurred in the T-loop of the tRNApro gene. Student’s t-test was used for statistical analyses of the expression of COX1 and ND5. *p ≤ 0.001 compared with the indicated groups.
Association Between Somatic mtDNA Mutations and Clinicopathologic Features in HCC Patients
Compared with mtDNA sequence deposited in GenBank (NC_012920.1), 194 mtDNA mutations were found in all these 33 examined tissue specimens from individuals with HCC. Most of the mtDNA alterations obtained from the tumor tissue specimens were also apparent in the paired adjacent and normal tissue specimens, which is consistent with a previous study21. However, there were no significant associations between somatic mtDNA mutations and the HCC clinicopathologic features (Table 2). Notably, we observed that larger tumor size of HCC developed with more somatic mtDNA mutations (p = 0.137), which may show significant association in a larger number of HCC patients.
Table 2.
Clinicopathologic Features in HCC Patients With and Without Mutation in the Entire mtDNA
| Characteristics | No. of Patients | Somatic Mutation | p Value | |
|---|---|---|---|---|
| Negative (n = 5) | Positive (n = 6) | |||
| Age | 0.303 | |||
| <60 | 7 | 4 | 3 | |
| ≥60 | 4 | 1 | 3 | |
| Gender | 0.819 | |||
| Male | 7 | 3 | 4 | |
| Female | 4 | 2 | 2 | |
| Tumor size | 0.137 | |||
| ≤50 cm2 | 7 | 2 | 5 | |
| ≥50 cm2 | 4 | 3 | 1 | |
| Differentiation | 0.535 | |||
| Poor differentiated | 3 | 2 | 1 | |
| Moderate differentiated | 4 | 1 | 3 | |
| Well differentiated | 4 | 2 | 2 | |
| HBV | 0.887 | |||
| Negative | 2 | 1 | 1 | |
| Positive | 9 | 4 | 5 | |
| TNM stage | 0.621 | |||
| II | 8 | 4 | 4 | |
| III | 3 | 1 | 2 | |
| AFP | 0.740 | |||
| <1,210 μg/L | 6 | 3 | 3 | |
| >1,210 μg/L | 5 | 2 | 3 | |
mtDNA Copy Number Was Significantly Lower in HCC Patients
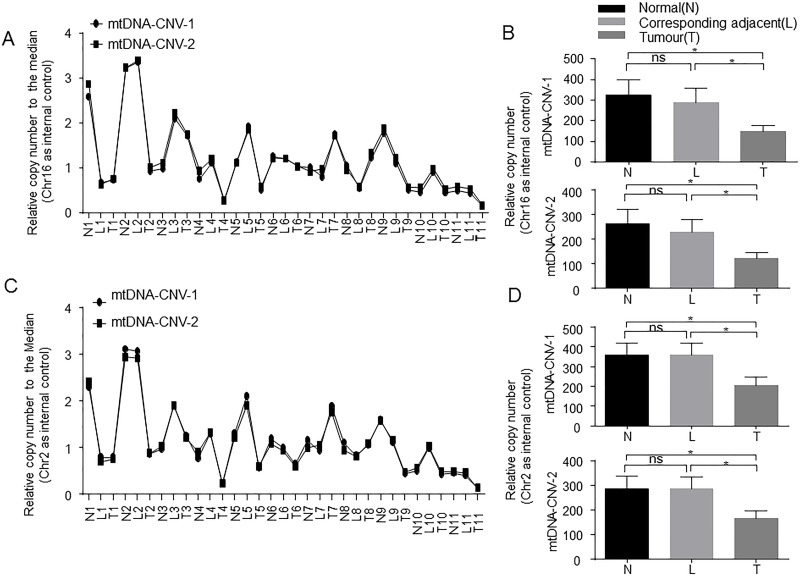
mtDNA CNV has been described in many different types of cancers28. To evaluate whether the abundance of mtDNA was altered within the tumor tissues of HCC patients, we analyzed the mtDNA copy number of the HCC tissues and adjacent and normal liver tissues by a relative quantification PCR analysis. We chose chromosome 16 or chromosome 2 as internal control and used two primers to assess mtDNA copy number. Relative copy number to the median was analyzed to revise PCR efficiency. We found that the results of the two pieces of target fragment on mtDNA showed a similar trend, which indicated that our results are convincing (Fig. 3A and C). Importantly, we found that the mean copy number of mtDNA in HCC patients was significantly lower than that of the adjacent and normal liver tissues, which is consistent with previous results observed in other types of cancers. There was no significant difference between the adjacent and normal liver tissues (Fig. 3B and D).
Figure 3.
mtDNA copy number was significantly lower in HCC patients. (A) mtDNA copy number was detected by a relative quantification PCR analysis. mtDNA-CNV-1 and mtDNA-CNV-2 were used as two primers, and Chr16 was used as internal control. (B) Quantification of mtDNA copy number in three groups of liver tissues. (C) Chr2 was used as internal control to analyze mtDNA copy number. (D) Quantification of mtDNA copy number in three groups of liver tissues. N, normal; L, corresponding adjacent; T, tumor. *p ≤ 0.05 compared with indicated groups.
Mitochondrial Depletion Decreased Proliferation and Migration But Is Resistant to 5-Fluorouracil Cytotoxicity in the Hepatocellular Carcinoma Cell Line SK-HEP-1
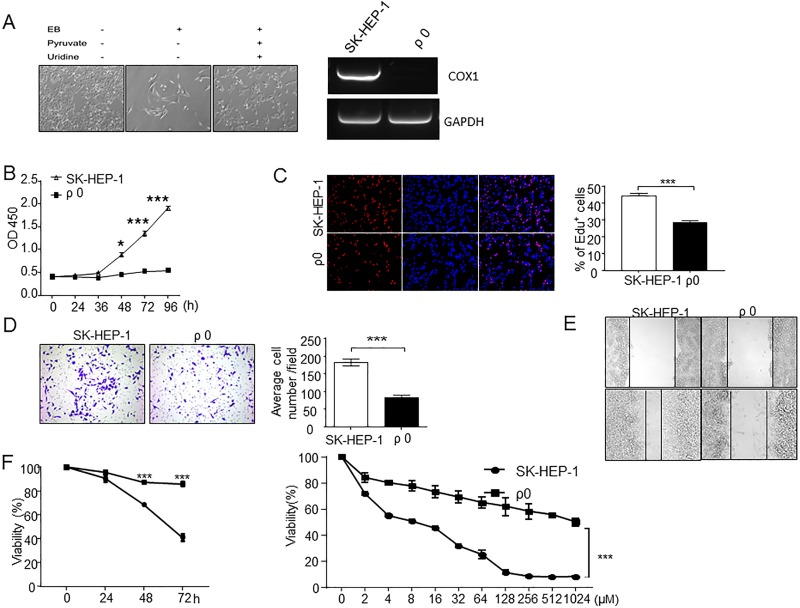
To elucidate the potential role of mitochondrial dysfunction in HCC progression, we used an in vitro model of mtDNA-depleted ρ0 cells. ρ0 cells, with long spindle-shaped fibrocyte-like adherent growth, are derived from SK-HEP-1 and require pyruvate and uridine as supplements (Fig. 4A). COX1 is one component of proto-transporting complexes encoded by mtDNA. Relative gene expression of COX1 was characterized to confirm mtDNA depletion (Fig. 4A). Compared to their parental SK-HEP-1 cells, ρ0 cells have reduced growth rates (Fig. 4B and C) and decreased migration by in vitro Transwell migration assay (Fig. 4D and E). We next examined the sensitivity to 5-fluorouracil and found that the ρ0 cells were more resistant to 5-fluorouracil (Fig. 4F), which suggests that the depletion or dysfunction of mitochondrial may also affect the chemoresistance of HCC cells.
Figure 4.
EtBr-induced mtDNA-depleted ρ0 cells decreased cell proliferation and migration but increased 5-fluorouracil resistance. (A) Representative pictures of ρ0 cells, which survive only in the presence of 100 mg/L pyruvate and 50 mg/L uridine. Semiquantitative PCR of COX1 was analyzed to characterize the depletion of mtDNA in ρ0 cells. (B, C) Cell proliferation was detected by cell counting kit-8 (CCK-8) analysis and EdU. Compared to SK-HEP-1, SK-HEP-1-derived ρ0 cells had significantly lower growth rate. (D, E) Transwell migration assays and wound healing assay in SK-HEP-1 and mitochondrial depletion ρ0 cells. The migration in ρ0 was less than in SK-HEP-1 cells. (D) Left: 200× magnification; right: quantification analysis. (F) Left: SK-HEP-1 and ρ0 cells treated with 16 μM 5-fluorouracil for 24, 48, and 72 h; right: SK-HEP-1 and ρ0 cells treated with 5-fluorouracil at gradient concentrations of 0, 2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 μM for 72 h. The viable cells were counted using CCK-8 assay. ***p ≤ 0.001 compared with the indicated groups.
DISCUSSION
In this study, we identified somatic mutations in HCC by high-throughput sequencing strategies. We identified 194 SNVs throughout mtDNA in 33 specimens; 180 mutations existed in tumors and paired corresponding adjacent and normal liver tissues. Such integrated mutations might be a result of single nucleotide polymorphism from germline or chronic insult of hepatitis. Although there was no significant association of clinicopathologic features with the mtDNA mutations, two of seven (28.6%) somatic mutations occurred in the evolutionarily conserved coding regions, which suggests that they may result in mitochondrial dysfunction in HCC cells. Consistent with previous studies, we observed decreased mtDNA copy number and detected several novel mutations in HCC. In addition, evidence from our in vitro experiments revealed that mitochondrial depletion results in reduced proliferation and migration but 5-fluorouracil resistance, which further confirmed that the depletion or dysfunction of mitochondria may also affect the proliferation and chemoresistance of HCC cells.
Heteroplasmies throughout mtDNA are common in mammalians. Generally, there are approximately 100 mitochondria in each cell, and each mitochondrion contains 2–10 copies of mtDNA29. Thus, mtDNA mutations often show heteroplasmic with a mixture of normal and mutant mtDNA copies. Several studies have detected mtDNA mutation in HCC patients by direct Sanger sequencing and PCR-RFLP analysis21–23,30,31. However, these methods are restrained by the limited number of targets. High-throughput sequencing technology allows for screening the mitochondrial genome and simultaneously detecting the level of mtDNA heteroplasmy at all sites across the mtDNA genome in a reliable and cost-effective manner over large numbers of cohorts32. We identified 13 novel somatic mutations in HCC tissues, and 11 of them have never been reported in other diseases (Table 2). These mutations specific to HCC suggest their potential role in HCC progression.
Alterations of mtDNA have been demonstrated in both solid tumors and leukemia. How these mtDNA mutations contribute to the development of the tumor remains unknown. Exchange of normal and altered mtDNA with pathogenic mutations could result in transformation of cancer cell phenotypes33–35. Owing to the heteroplasmics of mtDNA, no techniques or methods are available to manipulate mtDNA (such as knocking out the gene or gene transfer) directly to obtain the mutated mtDNA except by isolating primary cells from tumor tissues. Cybrids were generated by repopulating ρ0 cells devoid of mtDNA with altered mtDNA derived from enucleated cells. Therefore, the mtDNA transfer technique to construct cybrids is very valuable to investigate interaction of mtDNA mutations with various phenotypes.
The requirement of functional mitochondria for cancer cells has been confirmed by the establishment of ρ0 cells through growth in ethidium bromide36. The resulting ρ0 cancer cells have reduced growth rates. In our experiments, the proliferation of ρ0 cells derived from SK-HEP-1 was significantly reduced. Furthermore, the improved resistance of ρ0 cells against 5-fluorouracil is consistent with a previous report in gastric cells37 and suggests that the mtDNA mutation observed in cancers is one of the important causes of the resistance against chemotherapeutics frequently observed in differentiated cancers.
In conclusion, using high-throughput sequencing strategies and setting up a ρ0 cells line, we have detected novel mtDNA mutations in patients with HCC and preliminarily demonstrated that mitochondrial dysfunction may affect proliferation and chemoresistance in HCC cells. In this study, we established ρ0 cells and identified four somatic nonsynonymous mutations (T6115C, 65.74%; G8387A, 12.23%; G13121A, 93.08%; and T14180C, 28.22%) could result in amino acid substitutions, and tRNA mutations could cause amino acid substitution. Future studies, by adopting the ρ0 cells to construct these mutations cybrids, could help understand the functions of these mutations in the progression of HCC.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81670562 to X. Kong, 81670598 to Q. Xia, and 81372233 to H. Wu ) and by a grant from the Committee of Science and Technology of Shanghai Municipal Government (16401970600-03) to X. Kong. We thank Dr. Li-Wei Dong (an associate investigator in the Second Military Medical University, Shanghai, P.R. China) for providing HCC liver tissues.
REFERENCES
- 1. Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: Roles of inherited and somatic mutations. Nat Rev Genet. 2012;13(12):878–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abu-Amero KK, Alzahrani AS, Zou M, Shi Y. High frequency of somatic mitochondrial DNA mutations in human thyroid carcinomas and complex I respiratory defect in thyroid cancer cell lines. Oncogene 2005;24(8):1455–60. [DOI] [PubMed] [Google Scholar]
- 4. Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature 1981;290(5806):457–65. [DOI] [PubMed] [Google Scholar]
- 5. Croteau DL, Bohr VA. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J Biol Chem. 1997;272(41):25409–12. [DOI] [PubMed] [Google Scholar]
- 6. Wallace DC. Mitochondrial diseases in man and mouse. Science 1999;283(5407):1482–8. [DOI] [PubMed] [Google Scholar]
- 7. Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46(3):428–33. [PMC free article] [PubMed] [Google Scholar]
- 8. Howell N, Bindoff LA, McCullough DA, Kubacka I, Poulton J, Mackey D, Taylor L, Turnbull DM. Leber hereditary optic neuropathy: Identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet. 1991;49(5):939–50. [PMC free article] [PubMed] [Google Scholar]
- 9. Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 1990;61(6):931–7. [DOI] [PubMed] [Google Scholar]
- 10. van den Ouweland JM, Lemkes HH, Ruitenbeek W, Sandkuijl LA, de Vijlder MF, Struyvenberg PA, van de Kamp JJ, Maassen JA. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet. 1992;1(5):368–71. [DOI] [PubMed] [Google Scholar]
- 11. Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18(23):6927–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wallace DC. Mitochondrial genetics: A paradigm for aging and degenerative diseases? Science 1992;256(5057):628–32. [DOI] [PubMed] [Google Scholar]
- 13. Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012;12(10):685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20(3):291–3. [DOI] [PubMed] [Google Scholar]
- 16. Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 2000;287(5460):2017–9. [DOI] [PubMed] [Google Scholar]
- 17. He L, Luo L, Proctor SJ, Middleton PG, Blakely EL, Taylor RW, Turnbull DM. Somatic mitochondrial DNA mutations in adult-onset leukaemia. Leukemia 2003;17(12):2487–91. [DOI] [PubMed] [Google Scholar]
- 18. Jerónimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene 2001;20(37):5195–8. [DOI] [PubMed] [Google Scholar]
- 19. Kirches E, Krause G, Warich-Kirches M, Weis S, Schneider T, Meyer-Puttlitz B, Mawrin C, Dietzmann K. High frequency of mitochondrial DNA mutations in glioblastoma multiforme identified by direct sequence comparison to blood samples. Int J Cancer 2001;93(4):534–8. [DOI] [PubMed] [Google Scholar]
- 20. Lee HC, Huang KH, Yeh TS, Chi CW. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J Gastroenterol. 2014;20(14):3950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishikawa M, Nishiguchi S, Shiomi S, Tamori A, Koh N, Takeda T, Kubo S, Hirohashi K, Kinoshita H, Sato E, Inoue M. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res. 2001;61(5):1843–5. [PubMed] [Google Scholar]
- 22. Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res. 2004;547(1–2):71–8. [DOI] [PubMed] [Google Scholar]
- 23. Yin PH, Wu CC, Lin JC, Chi CW, Wei YH, Lee HC. Somatic mutations of mitochondrial genome in hepatocellular carcinoma. Mitochondrion 2010;10(2):174–82. [DOI] [PubMed] [Google Scholar]
- 24. Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: Heredity, heteroplasmy and disease. Trends Genet. 1997;13(11):450–5. [DOI] [PubMed] [Google Scholar]
- 25. Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum Mol Genet. 1994;3(1):13–9. [DOI] [PubMed] [Google Scholar]
- 26. Taylor RW, Barron MJ, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, Taylor GA, Plusa SM, Needham SJ, Greaves LC, Kirkwood TB, Turnbull DM. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112(9):1351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA-depleted cells against cell death: Role of mitochondrial superoxide dismutase. J Biol Chem. 2004;279(9):7512–20. [DOI] [PubMed] [Google Scholar]
- 28. Lin CS, Lee HT, Lee MH, Pan SC, Ke CY, Chiu AW, Wei YH. Role of mitochondrial DNA copy number alteration in human renal cell carcinoma. Int J Mol Sci. 2016;17(6):814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136(3):507–13. [DOI] [PubMed] [Google Scholar]
- 30. Okochi O, Hibi K, Uemura T, Inoue S, Takeda S, Kaneko T, Nakao A. Detection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patients. Clin Cancer Res. 2002;8(9):2875–8. [PubMed] [Google Scholar]
- 31. Tamori A, Nishiguchi S, Nishikawa M, Kubo S, Koh N, Hirohashi K, Shiomi S, Inoue M. Correlation between clinical characteristics and mitochondrial D-loop DNA mutations in hepatocellular carcinoma. J Gastroenterol. 2004;39(11):1063–8. [DOI] [PubMed] [Google Scholar]
- 32. Ye F, Samuels DC, Clark T, Guo Y. High-throughput sequencing in mitochondrial DNA research. Mitochondrion 2014;17:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008;320(5876):661–4. [DOI] [PubMed] [Google Scholar]
- 34. Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders D, Hosseini SY, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA 2005;102(3):719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005;65(5):1655–63. [DOI] [PubMed] [Google Scholar]
- 36. Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. 1991;88(23):10614–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hung WY, Wu CW, Yin PH, Chang CJ, Li AF, Chi CW, Wei YH, Lee HC. Somatic mutations in mitochondrial genome and their potential roles in the progression of human gastric cancer. Biochim Biophys Acta 2010;1800(3):264–70. [DOI] [PubMed] [Google Scholar]