 Herein we report the synthesis, base pairing properties and in vivo transliteration of N8-glycosylated 8-aza-deoxyguanosine and 8-aza-9-deaza-deoxyguanosine nucleotides with 8-amino-deoxyinosine, 1-N-methyl-8-amino-deoxyinosine and 7,8-dihydro-8-oxo-deoxy-inosine/adenosine/guanosine as pairing partners.
Herein we report the synthesis, base pairing properties and in vivo transliteration of N8-glycosylated 8-aza-deoxyguanosine and 8-aza-9-deaza-deoxyguanosine nucleotides with 8-amino-deoxyinosine, 1-N-methyl-8-amino-deoxyinosine and 7,8-dihydro-8-oxo-deoxy-inosine/adenosine/guanosine as pairing partners.
Abstract
Herein we report the synthesis of N8-glycosylated 8-aza-deoxyguanosine (N8-8-aza-dG) and 8-aza-9-deaza-deoxyguanosine (N8-8-aza-9-deaza-dG) nucleotides and their base pairing properties with 5-methyl-isocytosine (d-isoCMe), 8-amino-deoxyinosine (8-NH2-dI), 1-N-methyl-8-amino-deoxyinosine (1-Me-8-NH2-dI), 7,8-dihydro-8-oxo-deoxyinosine (8-Oxo-dI), 7,8-dihydro-8-oxo-deoxyadenosine (8-Oxo-dA), and 7,8-dihydro-8-oxo-deoxyguanosine (8-Oxo-dG), in comparison with the d-isoCMe:d-isoG artificial genetic system. As demonstrated by Tm measurements, the N8-8-aza-dG:d-isoCMe base pair formed less stable duplexes as the C:G and d-isoCMe:d-isoG pairs. Incorporation of 8-NH2-dI versus the N8-8-aza-dG nucleoside resulted in a greater reduction in Tm stability, compared to d-isoCMe:d-isoG. Insertion of the methyl group at the N1 position of 8-NH2-dI did not affect duplex stability with N8-8-aza-dG, thus suggesting that the base paring takes place through Hoogsteen base pairing. The cellular interpretation of the nucleosides was studied, whereby a lack of recognition or mispairing of the incorporated nucleotides with the canonical DNA bases indicated the extent of orthogonality in vivo. The most biologically orthogonal nucleosides identified included the 8-amino-deoxyinosines (1-Me-8-NH2-dI and 8-NH2-dI) and N8-8-aza-9-deaza-dG. The 8-oxo modifications mimic oxidative damage ahead of cancer development, and the impact of the MutM mediated recognition of these 8-oxo-deoxynucleosides was studied, finding no significant impact in their in vivo assay.
Introduction
Non-canonical base pairing has been studied by several research groups,1–6 based on their interest to expand the genetic alphabet. A major breakthrough in this field has been reported by Malyshev et al.,7 showing replication of a synthetic hydrophobic base pair (d5SICS–dNAM) in vivo. This base pair system consists of C-nucleosides that are metabolically not easily accessible, where recognition is based on hydrophobic interactions, and no hydrogen-bond interactions (such as in canonical base pairing) are involved. Recently, we have demonstrated that a change in the backbone (HNA replacing DNA) could lead to a reduced recognition of non-canonical bases (i.e. isoCMe and isoG) by natural bases,8 further moving in the direction of a fully orthogonal information system. Here we have investigated new base pair systems from which one of the partners is an N8 substituted purine base and involving hydrogen-bond formation in the recognition process.
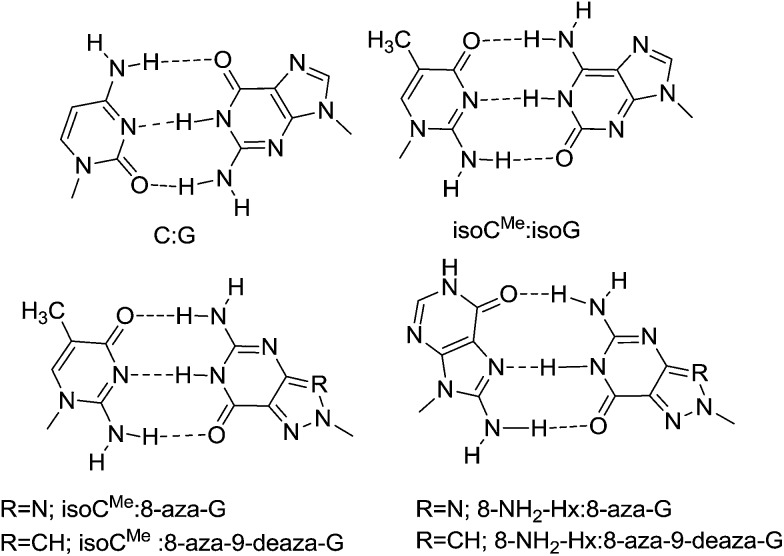
The unnatural isoG:isoCMe base pair9,10 is not adequate to expand the genetic code and has restricted application due to facile tautomerization of isoG which enables it to form base pairs with selected natural nucleobases. Tautomerism leads to promiscuity and heterocycles (via tautomerism) possibly leading to pairing with natural bases, should be avoided when selecting new orthogonal information systems.1 Therefore, we would like to replace the isoG base in the isoG:isoCMe base pair by another heterocycle less prone to tautomerization. In search for new artificial genetic systems efforts have been devoted to the study of C-nucleosides11–13 and nucleosides with an aberrant glycosidic linkage i.e. with an N8 glycosylated nucleobase,14–17 as they demonstrate unexpected pairing behavior and some of them may function as universal bases.18,19 Seela et al., have demonstrated that altering the glycosylation site of a nucleoside from N9 to N8 results in a change of the nucleobase recognition pattern, N8-glycosylated-7-deaza-8-azaguanine behaves like isoguanine20 and forms base pairs with isoCMe and not with cytosine. Therefore, we have investigated N8-glycosylated 8-azaguanine21–23 1a together with its N9-deaza-analogue 1b. Removal of the N9 nitrogen atom (giving 1b) may have an influence on the in vitro and in vivo recognition of the N8-glycosylated purine nucleoside. Potentially, the N8-glycosylated purine nucleoside may be recognized by another purine nucleoside, when Hoogsteen base pairing is involved. To evaluate the potential of involving such Hoogsteen recognition24 in an unnatural base pair, 8-amino-hypoxanthine25,26 2a was selected as the pairing partner for the N8-glycosylated bases (N8-8-aza-dG, N8-8-aza-9-deaza-dG) as shown in Fig. 1. For Hoogsteen pairing in a duplex structure, one of the bases should adopt the ‘syn’ conformation and 8-substituted purines preferentially adopt this conformation.27,28 The 1-Me-8-NH2-dI nucleobase 2b was tested in order to exclude Watson–Crick type recognition in 1a/1b:2a base pairing. Changing the 8-amino group in 2a in an 8-hydroxy group will, first, abolish recognition by 1a and 1b, which would be further demolished by changing the aad system of 2 in the dda system of 5 (the dda system is also present in 1a and 1b) (Fig. 2). Therefore, and to fully understand the base pairing pattern, we have also synthesized and evaluated the 7,8-dihydroxy-8-oxonucleosides with the 6-Oxo293, 6-Oxo-2-amino30–34 4 and 6-amino35–38 5 base moieties. Because of the favored syn conformation of 8-hydroxylated purine, Hoogsteen base pairing may be preferred over Watson–Crick base pairing.39–41 A conclusion is that Watson–Crick type base pairing as well as Hoogsteen type base pairing can be considered when selecting orthogonal base pairs in vitro and in vivo.
Fig. 1. Putative base pair motifs of duplexes with antiparallel strand orientation.
Fig. 2. 2′-Deoxyribo-nucleosides, with modified purine and pyrimidine bases.
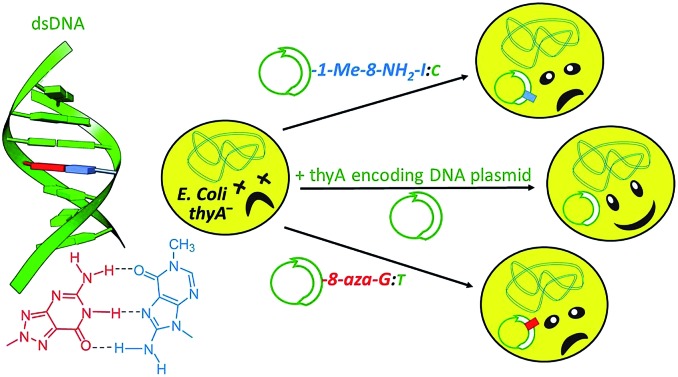
In order to determine the orthogonality of the nucleosides against the biological situation, they were individually analyzed in vivo in an XNA-dependent DNA synthesis assay. For these nucleosides the assay studied the pairing properties of each, with an extension of the study performed for the 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine bases in vivo, the former of which is formed by ROS-mediated damage of DNA.42,43 Both of the 8-oxo-deoxynucleoside bases were analyzed in wild-type and knockout backgrounds of the important 8-oxo repair enzyme MutM.44,45 The assay was to determine whether or not this enzyme in particular was influencing the base pairing results determined by tests, and confirm that the non-natural nucleosides here were not being chemically modified in vivo, thus affecting the results obtained.
Results and discussion
Synthesis of monomers
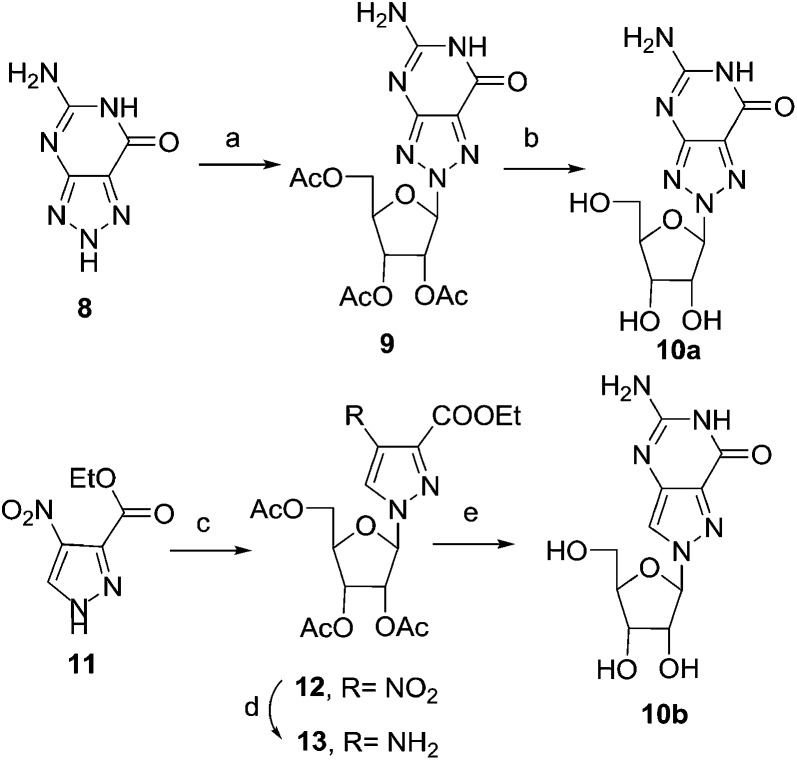
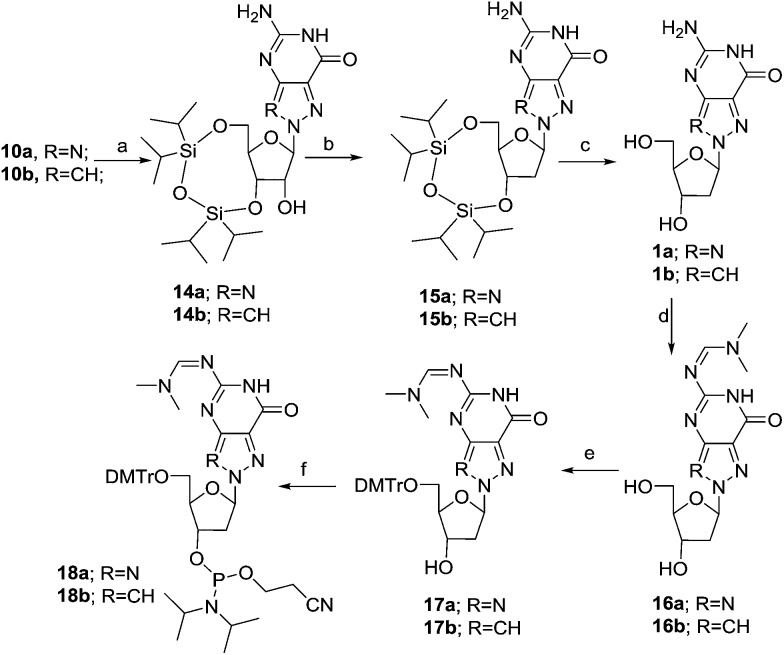
As depicted in Scheme 1, the BF3–Et2O-catalysed glycosylation of commercially available 8-azaguanine (8) with 1,2,3,5-tetra-O-acetyl-β-d-ribofuranose afforded 9 as the major isomer. The assignment of N8 glycosidic bond in 9 was made by comparing spectral data with the known compound 1a in later stage. The cleavage of the acetyl-protecting group was carried out using ammonium hydroxide at room temperature for 8 hours, obtaining compound 10a in 91% yield.46,47 The regioselective protection of the 3′-OH and 5′-OH groups with TIPDSiCl2 gave compound 14a.
Scheme 1. Synthesis of N8-8-aza-G and N8-8-aza-9-deaza-G nucleoside. Reagents and conditions: (a) BF3–Et2O, 1,2,3,5-tetra-O-acetyl-β-d-ribofuranose, CH3CN, 75 °C, 3 h, 54%; (b) NH4OH, rt, 8 h, 91%; (c) BSA, DCE, SnCl4, 1,2,3,5-tetra-O-acetyl-β-d-ribofuranose, rt, 36 h, 85%; (d) H2, 10% Pd/C, MeOH, rt, 8 h, 100%; (e) (i) chloro-formamidine hydrochloride, DMS, 120 °C, 1 h, (ii) NH4OH, rt, 1 h, 42%.
For the deoxygenation of the C2′-hydroxy group, the Barton–McCombie reaction was used. We first converted the nucleoside 14a into the corresponding thiocarbamate, which, after radical reduction in the presence of tributylstannane and AIBN, gave the desired nucleoside 15a (Scheme 2).
Scheme 2. Synthesis of N8-8-aza-dG and N8-8-aza-9-deaza-dG nucleoside. Reagents and conditions: (a) TIPDSiCl2, pyridine, 0 °C to rt, 8 h, 74–77%; (b) (i) TCDI, CH2–Cl2, rt, 8 h; (ii) AIBN, nBu3SnH, toluene, 75 °C, 2 h, 62–68%; (c) TBAF, THF, rt, 4 h, 94–97%; (d) DMF–DMA, MeOH, 50 °C, 1 h, 93–96%; (e) DMTrCl, pyridine, 0 °C to rt, 55 min, 2 h, 76–83%; (f) (i-Pr2N)2POCH2CH2CN, 1H-tetrazole, CH2Cl2, 0 °C to rt, 55 min, 2 h, 69–73%.
Subsequently, the removal of the silyl-protecting group, carried out using TBAF in THF at room temperature for 4 hours, afforded compound 1a to 94% yield. The spectral and analytical data of 1a were found to be in accordance with reported data.21 Protection of the amino group of 1a followed by the selective protection of the primary alcohol with 4,4′-dimethoxytrityl chloride (DMTrCl) in pyridine, provided the protected nucleoside 17a,21 which was then converted into phosphoramidite 18a by reaction with bis(diisopropylamino)(2-cyanoethoxy)phosphine in the presence of tetrazole.
The synthesis of the N8-8-aza-9-deaza-dG derivative started from the readily available pyrazole 11. Vorbrüggen reaction of 11 with 1,2,3,5-tetra-O-acetyl-β-d-ribofuranose, in the presence of SnCl4, afforded the desired N1-isomer 12 as the major product (91% yield).48 The reduction of the nitro group of 12, followed by the ring closure with chloroformamidine hydrochloride and deprotection of acetyl groups in one pot, gave the desired compound 10b (Scheme 1). Analogously to what is described for the N8-8-aza-guanine derivatives in Scheme 2, a series of transformations starting from 10b allowed us to obtain the deoxy derivative 1b and finally the corresponding phosphoramidite 18b.
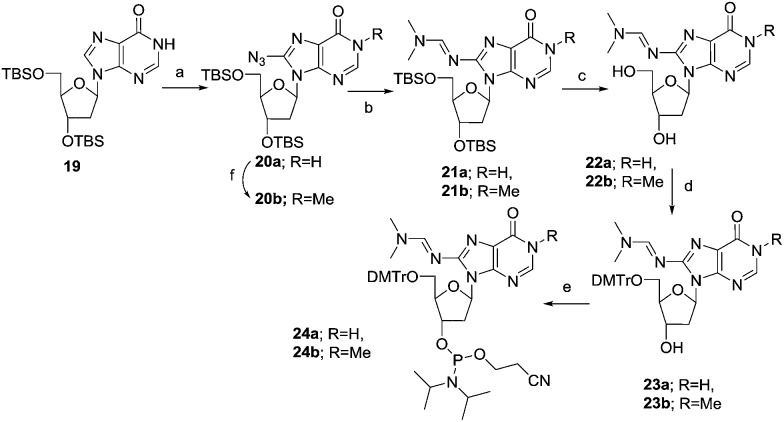
As depicted in Scheme 3, the synthesis of the phosphoramidite of 8-NH2-dI (2a) started from the silylated deoxyinosine 19.49 The compound 19 was treated with tosyl azide and n-butyllithium in anhydrous THF at –78 °C for 2 hours yielding 60% of the protected 8-azido-dI nucleoside 20a. The reduction of the azide group was carried out using sodium borohydride in methanol and further protected with an N,N-dimethylamino methylidene residue using dimethylformamide dimethyl acetal in methanol at 70 °C for 1 hour. The cleavage of the silyl protecting group was carried out using TBAF in THF at room temperature for 12 hours, to obtain compound 22a to 60% yield. Subsequently, compound 22a was tritylated and phosphitylated, furnishing the phosphoramidite 24avia23a. For the synthesis of the 1-Me-8-NH2-dI building block, the N1 methylation of compound 20a was carried out using methyl iodide and sodium hydride to obtain compound 20b in 88% yield. Then similar to the synthesis of 8-NH2-dI phosphoramidite 24a, a series of transformations starting from 20b yielded the corresponding phosphoramidite 24b.
Scheme 3. Synthesis of the phosphoramidite of 8-NH2-dI and 1-Me-8-NH2-dI. Reagents and conditions: (a) TsN3, BuLi, THF, –78 °C, 2 h, 60%; (b) (i) NaBH4, MeOH, 0 °C to rt, 2 h; (ii) DMF–DMA, MeOH, 78 °C, 1 h, 70–73%; (c) TBAF, rt, 3 h, 60–73%; (d) (MeO)2TrCl, pyridine, 0 °C to rt, 12 h, 89–96%; (e) (i-Pr2N)2POC2H4CN, 1H-tetrazole, CH2Cl2, 0 °C to rt, 1 h, 75–77%; (f) Mel, NaH, 0 °C to rt, 12 h, 88%.
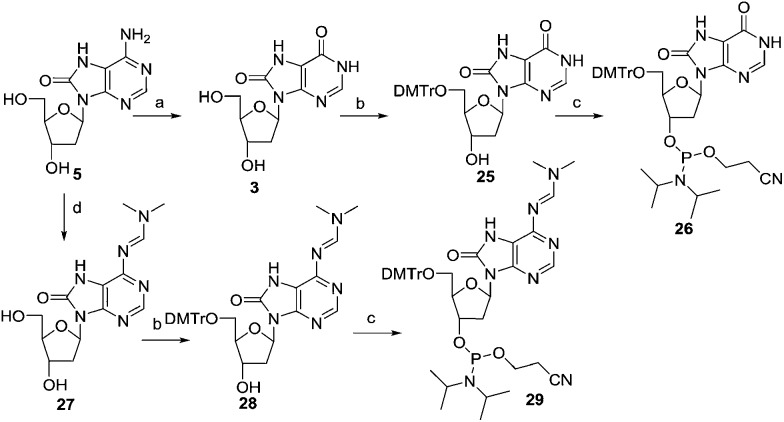
Previously,29 8-Oxo-dI 3 was prepared by enzyme-mediated deamination of 8-Oxo-dA 5. Here, the 8-Oxo-dA505 nucleoside was deaminated by diazotization of the 6-NH2 group using sodium nitrate in acetic acid and water at room temperature for 12 hours, affording 8-Oxo-dI 3 to 77% yield. Subsequently, compound 3 was tritylated followed by phosphitylation, furnishing the phosphoramidite 26via25 (Scheme 4). The spectral and analytical data of 26 were found to be in accordance with that reported.29 The free amino group of 8-Oxo-dA 5 was protected with an N,N-dimethylamino methylidene residue using dimethylformamide dimethyl acetal in methanol. Subsequently, compound 27 was tritylated using DMTrCl in pyridine and DMAP as a catalyst. Furthermore, phosphitylation of the DMTr derivative 28, furnished the phosphoramidite 29.
Scheme 4. Synthesis of the phosphoramidite of 8-Oxo-dI and 8-Oxo-dA. Reagents and conditions: (a) NaNO2, acetic acid: H2O, 12 h, 77%; (b) (MeO)2TrCl, pyridine, 0 °C to rt, 12 h, 69–73%; (c) (i-Pr2N)2POC2H4CN, 1H-tetrazole, CH2Cl2, 0 °C to rt, 1 h, 77–80%; (d) DMF–DMA, MeOH, 3 h, 65 °C, 91%.
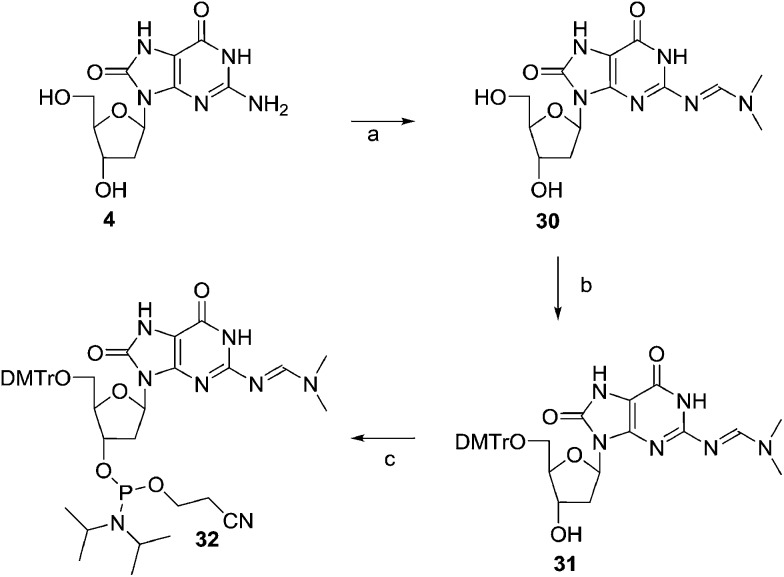
The amino group of 8-Oxo-dG514 was protected with an N,N-dimethylamino methylidene residue using dimethylformamide dimethyl acetal in methanol afforded compound 30. Compound 30 was tritylated and phosphitylated furnishing the required phosphoramidite 32 (Scheme 5).
Scheme 5. Synthesis of the phosphoramidite of 8-Oxo-dG. Reagents and conditions: (a) DMF–DMA, MeOH, 50%; (b) (MeO)2TrCl, pyridine, 0 °C to rt, 12 h, 71%; (c) (i-Pr2N)2POC2H4CN, 1H-tetrazole, CH2Cl2, 0 °C to rt, 1 h, 81%.
Oligonucleotide synthesis and hybridization properties
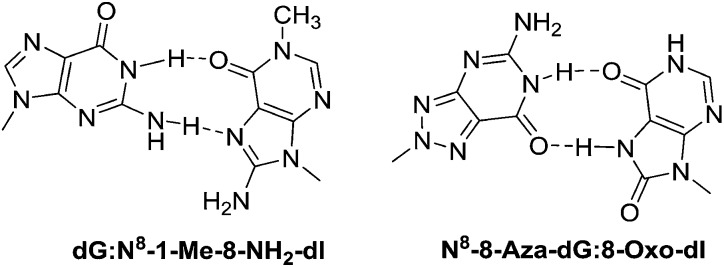
The oligonucleotides were prepared by using the common phosphoramidite method on a solid support employing a DNA synthesizer. The base pairing properties of N8-8-aza-9-deaza-dG and N8-8-aza-dG nucleosides with d-isoCMe,8 8-NH2-dI, 1-Me-8-NH2-dI, 8-Oxo-dI, 8-Oxo-dA, and 8-Oxo-dG were examined by hybridizing oligomers with their complementary strands and determining the Tm of the hybrids by temperature-dependent UV spectroscopy. In order to investigate the base pairing properties, the N8-8-aza-dG 1a, N8-8-aza-9-deaza-dG 1b and d-isoG 7 nucleosides were incorporated in the antiparallel duplex 5′-d(GGT AGC AG*C GGT G)-3′ replacing the G residue. The 8-NH2-dI 2a, 1-Me-8-NH2-dI 2b, 8-Oxo-dI 3, 8-Oxo-dA 5, 8-Oxo-dG 4, and d-isoCMe6 nucleosides were incorporated in the sequence 3′-d(CCA TCG TC*G CCA C)-5′ replacing the C residue. The strength of hybridization was studied by thermal denaturation experiments, which were determined at 260 nm in NaCl (0.1 M) buffer with KH2PO4 (20 mM, pH 7.5) and EDTA (0.1 mM) at a concentration of 4 μm for each strand. The stability of the duplexes was compared to the stability of the natural DNA duplex containing dC:dG (Table 1, entry 1) and unnatural d-isoCMe:d-isoG (Table 1, entry 2). The N8-8-aza-dG:d-isoCMe base pair gave a less stable duplex than the dC:dG (–3.1 °C) pair and d-isoCMe:d-isoG (–5.4 °C). Incorporation of an 8-NH2-dI 2aversus an N8-8-aza-dG nucleoside gave a greater decrease in Tm (–9.5 °C) than incorporation of d-isoCMeversus N8-8-aza-dG nucleoside (–5.4 °C), both decreasing on comparison to the d-isoCMe:d-isoG base pair. Insertion of the methyl group at N1 position of 8-NH2-dI did not affect the duplex stability, which suggests that the base pairing of 8-NH2-dI with N8-8-aza-dG takes place through Hoogsteen base pairing (Fig. 1). Surprisingly, 8-Oxo-dI and 8-Oxo-dG also show similar duplex stability, compared to 8-NH2-dI. As shown in Fig. 3, this could be explained by Wobble type base pairing in which two hydrogen bonds are involved. The sudden decrease in Tm of the duplexes when N8-8-aza-dG is placed opposite to 8-Oxo-dA (–11.2 °C), demonstrated that the C6 acceptor (C O) of 8-Oxo-dI is necessary for base pairing. The experiment proved that the base pair stability for N8-8-aza-dG decreases in the order d-isoCMe > 8-Oxo-dG ≥ 8-NH2-dI ≈ 1-Me-8-NH2-dI ≥ 8-Oxo-dI ≫ 8-Oxo-dA. Approximately similar results were observed for N8-8-aza-9-deaza-dG 1b, which indicates that the N9 nitrogen atom does not play a significant role in duplex stabilization (Table 2).
Table 1. T m values (in °C) of non-self-complementary antiparallel-stranded oligonucleotide duplexes containing dA, dT, dC, dG, 6 (d-isoCMe), and 7 (d-isoG).
| Entry | Duplex | T m (°C) |
| 1 | 5′-d(GGT AGC AGC GGT G)-3′ | 61.3 |
| 3′-d(CCA TCG TCG CCA C)-5′ | ||
| 2 | 5′-d(GGT AGC A7C GGT G)-3′ | 63.6 |
| 3′-d(CCA TCG T6G CCA C)-5′ |
Fig. 3. Putative base pairs between dG:N8-1-Me-8-NH2-dI and N8-8-aza-dG:8-Oxo-dI.
Table 2. T m values (in °C) of non-self-complementary antiparallel-stranded oligonucleotide duplexes containing 1a, 1b {5′-d[GGT AGC A(1a or 1b)C GGT G]-3′} and 6, 2a, 2b, 3, 4, 5 {3′-d[CCA TCG T(6 or 2a or 2b or 3 or 4 or 5)G CCA C]-5′} and ΔTm with respect to entry 1 (Table 1).
| N 8-8-aza-dG (1a) | ΔTm | N 8-8-aza-9-deaza-dG (1b) | ΔTm | |
| d-isoCMe (6) | 58.2 | –3.1 | 58.3 | –3.0 |
| 8-NH2-dI (2a) | 54.1 | –7.2 | 54.4 | –6.9 |
| 1-Me-8-NH2-dI (2b) | 54.0 | –7.3 | 54.0 | –7.3 |
| 8-Oxo-dI (3) | 53.4 | –7.9 | 54.0 | –7.3 |
| 8-Oxo-dG (4) | 54.9 | –6.4 | 55.4 | –5.9 |
| 8-Oxo-dA (5) | 47.0 | –14.3 | 48.6 | –12.7 |
In order to investigate the selectivity of base pairing, the stability of the duplexes with the modified base N8-8-aza-9-deaza-dG, N8-8-aza-dG, d-isoCMe, 8-NH2-dI, 1-Me-8-NH2-dI, 8-Oxo-dI, 8-Oxo-dA, and 8-Oxo-dG and the four canonical bases (A, T, C and G) was investigated by Tm determination (Table 3). This study is done in function of the potential use of the artificial base pairs in vivo. Among the four inosine derivatives (2a, 2b, 3 and 4) the mismatch discrimination (ΔTm) is the lowest for 8-NH2-dI and the highest for 1-Me-8-NH2-dI. For the N8 nucleosides; the N8-8-aza-9-deaza-dG showed better mismatch discrimination than N8-8-aza-dG.
Table 3. T m values (in °C) of antiparallel-stranded oligonucleotide duplexes containing 1a, 1b {5′-d[GGT AGC A(1a or 1b)C GGT G]-3′} and 2a, 2b, 3, 4, 5 {3′-d[CCA TCG T(2a, or 2b or 3 or 4, or 5)G CCA C]-5′} hybridized against complementary oligonucleotides with natural bases.
| A | T | C | G | ΔTm a | |
| N 8-8-aza-dG (1a) | 47.1 | 53.8 | 46.9 | 54.6 | 7.7 |
| N 8-8-aza-9-deaza-dG (1b) | 46.3 | 53.0 | 47.0 | 55.8 | 9.5 |
| 8-NH2-dI (2a) | 50.7 | 51.6 | 53.1 | 52.9 | 2.4 |
| 1-Me-8-NH2-dI (2b) | 46.2 | 43.7 | 43.1 | 52.1 | 9.0 |
| 8-Oxo-dI (3) | 55.8 | 50.3 | 53.5 | 47.9 | 7.9 |
| 8-Oxo-dG (4) | 54.7 | 50.0 | 57.0 | 54.9 | 7.0 |
| 8-Oxo-dA (5) | 45.8 | 54.7 | 44.5 | 51.2 | 10.2 |
aIn this case ΔTm corresponds to the difference between lowest and highest Tm (mismatch discrimination).
The selectivity of the 1a:2a base pair is low, as the Tm of the duplexes containing 1a:2a are only 1.0–3.4 °C higher than the Tm of the duplexes of 2a and the natural bases (A, T, C and G). The selectivity of the 1a:2b base pair is much better, due to a large drop in the Tm of duplexes containing the 2b:A, 2b:T and 2b:C base pairs. However, discrimination with G (2b:G base pair) stays low. In case of 8-Oxo-dI 3, the stability of the 3:A is higher than of 3:1a (which is similar than 3:C). Hence the mismatch discrimination is highest with the G base (3:G). 8-Oxo-dG (4) shows better base pairing with C (4:C) and with G (4:C), than 8-Oxo-dI (3:C and 3:G). Base pairing of 8-Oxo-dG with canonical bases has been described previously.32 Base pair stability of 4:A and 4:G is the same as of 4:1a. The base pair stability of 8-Oxo-dA versus canonical bases decreases in the order of T > G ≫ A > C. The results with 1b are similar to the results with 1a, except that 1b:5 is somewhat more stable than 1a:5 and 1b:G is somewhat more stable than 1a:G. The result, however, could be sequence dependent. Based on the Tm data, the 1-Me-8-NH2-dI:N8-8-aza-dG (or N8-8-aza-9-deaza-dG) base pair is the most discriminative versus A, T, and C, except versus G. However this does not mean that it would be the best orthogonal base pair in vivo.
Recognition of modified bases in vivo
A range of modified oligonucleosides were chemically synthesized with flanking DNA backbones and subsequently analysed for their biological characteristics in vivo. These oligonucleotides included separately each of 1-Me-8-NH2-dI, 8-NH2-dI, N8-8-aza-dG, N8-8-aza-9-deaza-dG, 8-Oxo-dI, 8-Oxo-dA and 8-Oxo-dG. The DNA backbone represented the 18-base long coding strand encoding the active site of the essential ThyA enzyme, whereby the nucleosides indicated replaced DNA within various codons (Fig. 4).
Fig. 4. The general structure of the mosaic test oligos. The 18 bases derive from the wild-type active site of E. coli thyA. Their positions and encoded amino acids are shown, including the catalytically essential Cys residue at 146. The various positions within this sequence where the modified nucleosides were tested within are bolded and underlined.
XNA-dependent DNA synthesis in vivo
We investigated the relationship between the nature of the chemical modifications of the 8-derivatives and the response of the E. coli genetic system to base pair with sufficient recognition and selectivity to enable survival in vivo. The aim in this study was to identify which of these modified nucleosides lacked such recognition, and possessed poor base-pairing ability with the canonical bases so as to approach orthogonality away from the biological system.
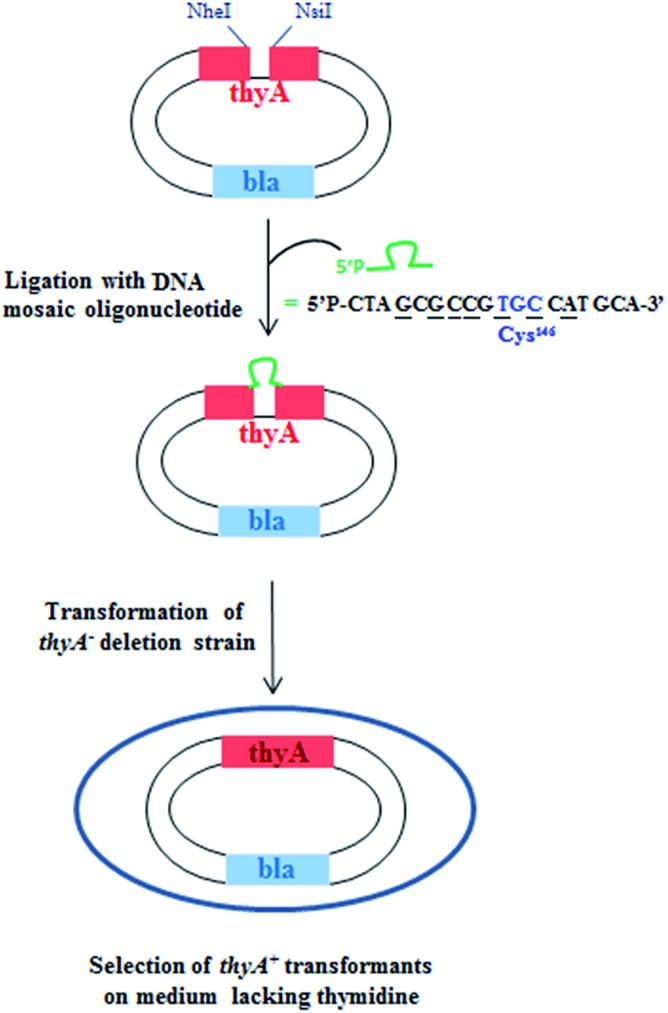
The xenobiotic nucleosides were tested using the published and established gapped-vector assay.52,53 In summary, DNA base/modified nucleoside mosaic oligomers were ligated enzymatically into a gapped form of the inactive thyA gene, which encodes an essential section of the ThyA active site. The inactive thyA gene was located on a plasmid vector (the pAK1/pAK2 heteroduplex) together with the bla ampicillin resistance gene permitting selection on two criteria. Once ligated and circularized, the plasmid was transformed into a strain of E. coli lacking thyA, a lethal growth phenotype in thymidine (dT) deficient media (Fig. 5).
Fig. 5. Selection screen to test DNA oligonucleotides bearing modified base(s) in templates for DNA synthesis in vivo.
The completion of the thyA gene finalizes the plasmid, which permits bacteria survival in the dT deficient media. The ratio between bacterial colony numbers in ±dT media indicates the success of the mosaic xenobiotic oligonucleotide to serve as a template for DNA polymerization. The mosaic oligonucleotides all took the form of the six middle codons that encode the active site of ThyA around the catalytically essential cysteine (codon TGC or TGT). The forms of each of the oligomers tested follow, including the nucleosides and their positions within the oligomer (Table 4).
Table 4. Sequence of the oligomers used in the XNA-dependent DNA synthesis experiments.
| Sequence (5′–3′) | X |
| d(P-CTA XCG CCG TGC CAT GCA) | 1-Me-8-NH2-dI, 8-NH2-dI, 8-Oxo-dI, N8-8-aza-dG, or N8-8-aza-9-deaza-dG |
| d(P-CTA GCG CCG XGC CAT GCA) | 1-Me-8-NH2-dI, 8-NH2-dI, 8-Oxo-dI, N8-8-aza-dG, or N8-8-aza-9-deaza-dG |
| d(P-CTA GCG CCG TGX CAT GCA) | 1-Me-8-NH2-dI, 8-NH2-dI, 8-Oxo-dI, N8-8-aza-dG, or N8-8-aza-9-deaza-dG |
| d(P-CTA GCG CCG TGC CXT GCA) | 1-Me-8-NH2-dI, 8-NH2-dI, 8-Oxo-dI, N8-8-aza-dG, or N8-8-aza-9-deaza-dG |
| d(P-CTA GCX CCT TGT CAT GCA) | 8-Oxo-dI, N8-8-aza-dG, N8-8-aza-9-deaza-dG, 8-Oxo-dA, or 8-Oxo-dG |
| d(P-CTA GCX CXT TGT CAT GCA) | 8-Oxo-dI, N8-8-aza-dG, N8-8-aza-9-deaza-dG8-Oxo-dA, or 8-Oxo-dG |
| d(P-CTA GCX XCT TGT CAT GCA) | 8-Oxo-dI, N8-8-aza-dG, N8-8-aza-9-deaza-dG, 8-Oxo-dA, or 8-Oxo-dG |
| d(P-CTA GCX XXT TGT CAT GCA) | 8-Oxo-dI, N8-8-aza-dG, 8-Oxo-dA, or 8-Oxo-dG |
The thyA plasmid genes from sixteen clones selected on dT deficient media were sequenced for each mosaic oligonucleotide tested (Fig. 6).
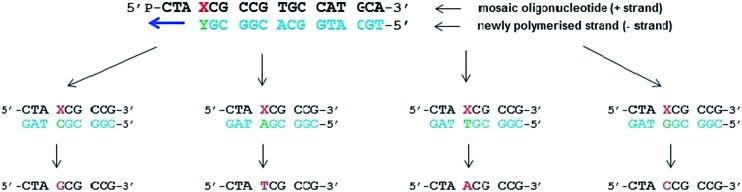
Fig. 6. Fate of mosaic oligomers in vivo. The first step in vivo is the polymerisation of the complementary DNA strand followed by canonical replication. Different possible incorporations of canonical nucleotides facing the base modified DNA nucleoside during the synthesis of the first DNA strand and the resulting coding strand are presented.
1-Me-8-NH2-dI and 8-NH2-dI
The assay and interpretation of the 8-NH2-dI and 1-Me-8-NH2-dI and nucleotides in vivo follows (Fig. 7).
Fig. 7. The in vivo interpretation of the 8-NH2-dI nucleotide (top) and 1-Me-8-NH2-dI nucleotide (bottom) within the essential thyA gene. The normalized ratio is the experimentally derived average number of thymidine-prototrophic colonies (bla+ thyA+) from the average total number of colonies (bla+ thyA– and bla+ thyA+). The modified section of the oligomer sequence indicates the position/s of the 8-NH2-dI or 1-Me-8-NH2-dI nucleotide as appropriate.
Analysis of the 1-Me-8-NH2-dI nucleotide in vivo reveals faint recognition as G and even less as T, with nothing apparent as C or A. Comparison to the related inosine-derivative 8-NH2-dI, which lacks the N1 methyl group, shows that recognition is entirely as G (and none with the other canonical bases), demonstrating that the presence of this 1-Me group is sufficient to reduce recognition as G but increase slightly recognition as T.
The 8-NH2-dI nucleotide base pairs the canonical bases in vivo preferentially of the order C ⋙ T = G = A, whereas the methylated form 1-Me-8-NH2-dI nucleotide appears to base pair preferentially in the order C ≫ A > G = T. These findings agree with the in vitro Tm data interpretations (Table 3), indicating perseverance of the recognition patterns between terminal stabilization in the artificial base pairs and enzymatic polymerization in vivo. Sequencing analysis was performed on purified plasmids from positive transformants and in both experiments revealed only wild-type sequences, indicating no forced changes of the genetic code by the bases.
N 8-8-aza-dG and N8-8-aza-9-deaza-dG
The assay and interpretation of the two N8 nucleosides, N8-8-aza-dG and N8-8-aza-9-deaza-dG nucleotides in vivo follow (Fig. 8 and 9).
Fig. 8. The in vivo interpretation of the N8-8-aza-dG nucleotide within the essential thyA gene. The normalized ratio is the experimentally derived average number of thymidine-prototrophic colonies (bla+ thyA+) from the average total number of colonies (bla+ thyA– and bla+ thyA+). The modified section of the oligomer sequence indicates the position of the N8-8-aza-dG nucleotide/s.
Fig. 9. The in vivo interpretation of the N8-8-aza-9-deaza-dG nucleotide within the essential thyA gene. The normalized ratio is the experimentally derived average number of thymidine-prototrophic colonies (bla+ thyA+) from the average total number of colonies (bla+ thyA– and bla+ thyA+). The modified section of the oligomer sequence indicates the position of the N8-8-aza-9-deaza-dG nucleotide/s.
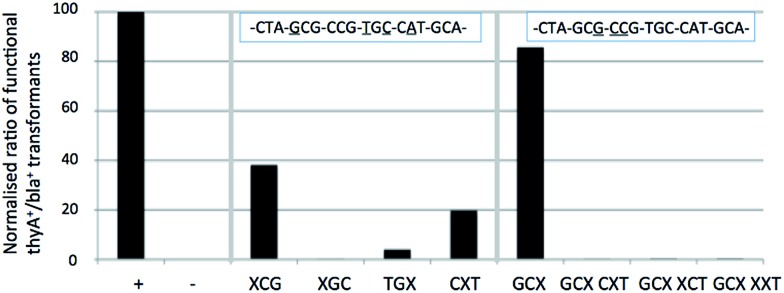
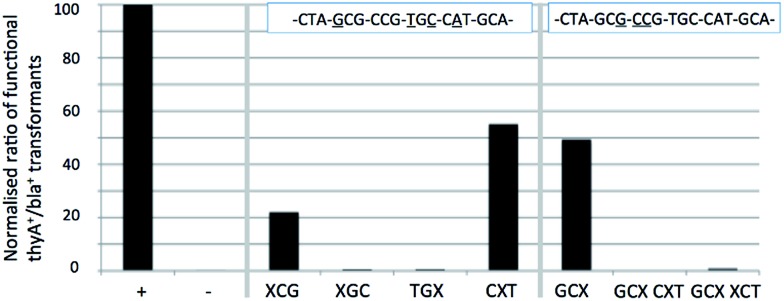
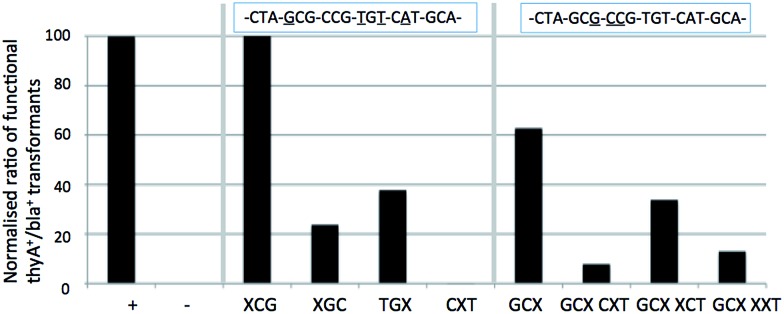
Analysis shows that N8-8-aza-dG appears to behave as an A or C nucleotide, and lacks any recognition as a T or G nucleotide. Sequencing of positive colonies revealed the presence of incorporated A's into the XCG codon (changing the amino acid to Thr from Ala, tolerated to approximately 30% of comparable wild-type survival through DNA control tests). In the GCX codon changes were also observed, with mispairing resulting in the inclusion of silent C and A residues (tolerated to 100% as wild-type) for an Ala residue in the enzyme primary structure. The N8-8-aza-dG nucleoside base pairs in the order of T ≫ G > C = A. The N8-8-aza-9-deaza-dG nucleotide appears to behave similarly to the derivative N8-8-aza-dG, with an interpreted base pairing preferentiality of T ≫ G = C = A, and similarly a proportion of changes to A nucleotides as observed through sequencing, also in the XCG codon (tolerated to Thr).
We hypothesized that the N9 nitrogen atom may assist Hoogsteen base pairing, although the similar findings in vivo between the two N8 nucleosides suggests the N9 nitrogen atom does not offer a significant role in recognition, agreeing with the previously indicated findings in vitro for this pair with regards to duplex stabilization. Modifications in recognition primarily occur on the XCG, CXT and GCX codons, but the influence remains marginal, as other codons are not affected, indicating little difference in vivo of the recognition of these two N8-glycosylated nucleosides. These findings do not discredit the in vitro Tm data, where N8-8-aza-dG showed worse mismatch discrimination than its N9-deaza-analogue. The general appearance of the in vivo results indicate an agreement to the in vitro findings, where general mismatch is comparably lower in N8-8-aza-9-deaza-dG except towards T, which appears easier to pair. Between the two nucleosides, the N8-8-aza-9-deaza-dG is the most orthogonal of this pair, and one of the most orthogonal of all of those tested in this study.
8-Oxo-dI, 8-Oxo-dG and 8-Oxo-dA
The assay and interpretation of the 8-Oxo-dI, 8-Oxo-dG and 8-Oxo-dA nucleotides in vivo follow (Fig. 10 and 11).
Fig. 10. The in vivo interpretation of the 8-Oxo-dG nucleotide (left) and 8-Oxo-dA nucleotide (right) within the essential thyA gene. The normalized ratio is the experimentally derived average number of thymidine-prototrophic colonies (bla+ thyA+) from the average total number of colonies (bla+ thyA– and bla+ thyA+). The modified section of the oligomer sequence indicates the position of the 8-Oxo-dG and 8-Oxo-dA nucleotide/s, for single (left) and double (right) codons for each.
Fig. 11. The in vivo interpretation of the 8-Oxo-dI nucleotide within the essential thyA gene. The normalized ratio is the experimentally derived average number of thymidine-prototrophic colonies (bla+ thyA+) from the average total number of colonies (bla+ thyA– and bla+ thyA+). The modified section of the oligomer sequence indicates the position of the 8-Oxo-dI nucleotide/s, for single (top) and double (bottom) codons.
The response of the 8-Oxo-dI nucleotides reveals the worst in vivo mismatch discrimination of all of the nucleotides tested here. Wide base-pairing capabilities with the canonical bases is shown by the apparent survival rates recovered through the tests. As with the previous tests, sequencing was performed of the now thymidine-prototrophic colonies (bla+thyA+), showing widespread changes to the gene. In the XCG codon, over 80% of the 8-Oxo-dI nucleotides had become T, changing the amino acid to Ser (tolerated lightly compared to wild-type Ala). Similarly, the codons GCX and (GCX) XCT returned around 15% each changes to T (silent change). This nucleotide has a base-pairing preferential order of A = C > G ≫ T, which support Hoogsteen recognition in vivo.
The in vitro findings and hypothesis of 8-Oxo-dI base pairing stabilities of the order A > C > G agree with the in vivo data. As with all of the nucleotides tested, the contribution of the oligo sequence is important as it imposes restrictions on the result obtained that is not observable in the in vitro work, underlining the importance of these vital tests in the study of modified nucleotides.
The 8-Oxo-dG and 8-Oxo-dA nucleotides show different patterns of base-pairing discrimination. For 8-Oxo-dG, recognition as an G appears strongest, with weak evidence it behaves as a T, C or A. Base-pairing preferentiality in vivo appears in the order of C > T > A ≫ G, with an aversion to base-pairing G also shown in the sequencing data across the double (GCX) XCT codons, which showed changes towards G, T and A in descending order. The finding of mismatches are largely further agreed upon by the in vitro dataset, although differences imposed by the primary sequence structure are observed in these tests.
The 8-Oxo-dA nucleotide in contrast shows a much cleaner and less promiscuous base-pairing profile than 8-Oxo-dG. The in vivo data indicates that this nucleotide is strongly recognized as an A, affording a base-pairing order of T ≫ A ≫ G = C. Sequencing of the positive clones showed widespread changes to A, especially within the GCX codon (tolerating the silent change) and in (GCX) XCT which promoted a change to Thr (with poor in vivo survival compared to wild-type). As with the other nucleotides, the base pairing stability determined in vitro of T > G ≫ A > C agrees well with the findings in vivo, possibly with sequence dependent effects.
It was suspected that the DNA repair pathways of the host test strain of E. coli might have been interacting with and perhaps modifying the 8-oxo modifications of these compounds. It is widely reported that the gene MutM (encoding the MutM DNA glycosylase) can recognize 8-Oxo-dG and remove it from DNA towards protection against mutagenic lesions.44,45
Accordingly, both 8-Oxo-dG and 8-Oxo-dA nucleotides were tested in the same assay in the equivalent strain of E. coli that had been modified in the MutM locus, deleting the gene. On following the same experimental procedure it was revealed that there was in fact no significant difference between the assay response in the strain with and the strain without MutM, indicating that the impact of MutM in this assay format was negligible. This demonstrated that the known mutagenic base (8-Oxo-dG) was not being modified in vivo during our assay by MutM, and therefore not modifying our findings.
Conclusions
We have developed an efficient route for the synthesis of oligonucleotides containing N8-8-aza-dG, N8-8-aza-9-deaza-dG, 8-NH2-dI, 1-Me-8-NH2-dI, 8-Oxo-dI, 8-Oxo-dA, and 8-Oxo-dG nucleotides on solid phase support by using phosphoramidite building blocks 18a, 18b, 24a, 24b, 26, 29, and 32 respectively. The base pairing and mismatch discrimination of N8-8-aza-9-deaza-dG, N8-8-aza-dG with d-isoCMe, 8-NH2-dI, 1-Me-8-NH2-dI, 8-Oxo-dI, 8-Oxo-dA, and 8-Oxo-dG in duplex DNA with antiparallel chain orientation were determined by Tm measurements. The new base pairs form less stable duplexes compared to the dC:dG and d-isoCMe:d-isoG pairs. Incorporation of 8-NH2-dI versus N8-8-aza-dG nucleoside gives a greater decrease in Tm than of d-isoCMeversus N8-8-aza-dG. Insertion of the methyl group at N1 position of 8-NH2-dI does not affect duplex stability, which suggests that H-bonding with N8-8-aza-dG takes place via Hoogsteen base pairing. As could be expected there is no straight correlation between duplex stability (Tm measurement) and base pair recognition in vivo. For example, N8-8-aza-dG is best recognized by G in vitro (Tm) and by T in vivo (E. coli). This, of course, reflects the contribution of the polymerase in base-pairing recognition.
From all of the tests in vivo, the compounds belonging to the family of 8-amino-deoxyinosines (1-Me-8-NH2-dI and 8-NH2-dI) and N8-8-aza-9-deaza-dG have shown to be the most orthogonal from the E. coli genetic system and the base-pairing rules of the canonical bases (A, T, G and C). The 8-aza-deoxyguanosines (N8-8-aza-dG and N8-8-aza-9-deaza-dG) were recognized largely as A (base pairing T), although the 9-deaza modifications affected general pairing with canonical bases and afforded more orthogonality against the unsubstituted N8-8-aza-dG. The 9-deaza modifications did not change base pairing preferentiality, maintaining the findings observed in vitro.
In testing the 8-Oxo-dG and 8-Oxo-dA nucleosides, a side test was performed after deleting the repair enzyme MutM. This revealed that at least this enzyme was not modifying the 8-oxo-deoxynucleosides and therefore not giving an incorrect view into the orthogonality of the nucleoside (as if it had been corrected to a G through the repair mechanism).
Between the in vitro and in vivo tests it appears that a base pairing of either of the N8-glycosylated nucleosides against 8-NH2-dI or 1-Me-8-NH2-dI may identify a near orthogonal pair. Poor responses in vivo for these two pairs of nucleosides suggest that they are not strongly pairing with the canonical bases. The biggest problem foreseeable in this pair is the tendency for these bases to lack strong discrimination against the canonical base G, although these pairs would be discriminative against A, T and C.
Experimental section
1H, 13C and 31P NMR spectra were recorded on 300 MHz (1H NMR, 300 MHz; 13C NMR, 75 MHz; 31P NMR, 121 MHz) or 500 MHz (1H NMR, 500 MHz; 13C NMR, 125 MHz) or 600 MHz (1H NMR, 600 MHz; 13C NMR, 150 MHz) spectrometers. 2D NMRs (H-COSY, HSQC and HMBC) were used for the assignment of all the intermediates and final compounds. Mass spectra were acquired on a quadrupole orthogonal acceleration time-of-flight mass spectrometer. Column chromatographic separations were carried out by gradient elution with suitable combination of two/three solvents and silica gel (100–200 mesh or 230–400 mesh). Solvents for reactions were distilled prior to use: THF and toluene from Na/benzophenone; CH2Cl2 and CH3CN from CaH2; Et3N and pyridine from KOH.
5-Amino-2-(2,3,5-O-triacetyl-β-d-ribofuranosyl)-2H-[1,2,3]triazolo[4,5-d]pyrimidin-7-one (9)
β-d-Ribofuranose-1,2,3,5-tetraacetate (8.06 g, 23.01 mmol) was added to a solution of compound 8 (3.50 g, 25.31 mmol) in acetonitrile (115 mL). The reaction mixture was heated to 75 °C and boron trifluoride diethyl etherate (3.03 mL, 24.16 mmol) was added. After 3 h the volatiles were removed in vacuo and the residue was purified by flash chromatography (hexane/EtOAc 7 : 3) to give the product 9 (5.10 g, 54%). 1H NMR (300 MHz, DMSO-d6): δ 11.01 (br s, 1H, NH), 6.67 (br s, 2H, NH2), 6.32 (d, J = 2.8 Hz, 1H, H-1′), 5.84–5.75 (m, 1H, H-2′), 5.68–5.49 (m, 1H, H-3′), 4.54–4.33 (m, 2H, H-4′, H-5a′), 4.07 (dd, J = 12.1, 4.8 Hz, 1H, H-5b′), 2.09 (s, 6H, CH3), 1.98 (s, 3H, CH3); 13C NMR (75 MHz, DMSO-d6): δ 169.9 (C O), 169.5 (C O), 169.2 (C O), 159.6 (C7), 156.5 (C3a), 154.6 (C5), 128.0 (C7a), 93.6 (C1′), 80.0 (C4′), 73.2 (C3′), 70.3 (C2′), 62.3 (C5′), 20.4, 20.3 (CH3); HRMS (ESI+) calcd for C15H18N6NaO8 [M + Na]+ 433.1078, found 433.1076.
5-Amino-2-(β-d-ribofuranosyl)-2H-[1,2,3]triazolo[4,5-d]pyrimidin-7-one (10a)46
Concentrated ammonium hydroxide (48 mL) solution was added to compound 9 (5 g, 12.19 mmol) and the mixture was stirred for 8 h at room temperature. The aqueous solution was dried by lyophilization. The residue was purified by flash column chromatography (CH2Cl2/MeOH 8.5 : 1.5) to yield the title compound 10a as a white solid (3.17 g, 91%). 1H NMR (300 MHz, DMSO-d6): δ 10.96 (br s, 1H, NH), 6.57 (br s, 2H, NH2), 5.87 (d, J = 3.8 Hz, 1H, H-1′), 5.60 (br s, 1H, OH), 5.24 (br s, 1H, OH), 4.79–4.77 (m, 1H, OH), 4.49–4.48 (m, 1H, H-2′), 4.25–4.21 (m, 1H, H-3′), 4.00–3.97 (m, 1H, H-4′), 3.60–3.56 (m, 1H, H-5a′), 3.50–3.44 (m, 1H, H-5b′); 13C (75 MHz, DMSO-d6): δ 159.6 (C7), 156.8 (C3a), 154.5 (C5), 127.5 (C7a), 96.8 (C1′), 86.2 (C4′), 74.6 (C3′), 70.8 (C2′), 62.2 (C5′); HRMS (ESI+) calcd for C9H12N6NaO5 [M + Na]+ 307.0761, found 307.0772.
5-Amino-2-[3,5-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-β-d-ribofuranosyl]-2H-[1,2,3]triazolo[4,5-d] pyrimidin-7-one (14a)
To a suspension of the intermediate 10a (3 g, 10.55 mmol), in anhydrous pyridine (52 mL) under argon atmosphere, 1,3-dichloro-1,1,3,3-tetraisopropyldisiloxane (3.63 mL, 11.61 mmol) at 0 °C was added and then the mixture was stirred at room temperature for 8 h. The reaction was quenched by the addition of MeOH. The solvents were evaporated and the product 14a (4.3 g, 77%) was isolated by flash column chromatography (hexane/EtOAc 1 : 1). 1H NMR (600 MHz, DMSO-d6): δ 11.09 (br s, 1H, NH), 6.62 (br s, 2H, NH2), 5.89 (s, 1H, H-1′), 5.75 (d, J = 4.6 Hz, 1H, OH), 4.89 (dd, J = 8.5, 4.6 Hz, 1H, H-3′), 4.44 (t, J = 3.7 Hz, 1H, H-2′), 4.05–3.99 (m, 1H, H-5a′), 3.95–3.79 (m, 2H, H-4′, H-5b′), 1.12–0.87 [m, 28H, 4 × CH(CH3)2]; 13C NMR (151 MHz, DMSO-d6): δ 159.7 (C7), 156.8 (C3a), 154.7 (C5), 127.5 (C7a), 96.2 (C1′), 81.4 (C4′), 74.4 (C3′), 71.5 (C2′), 61.6 (C5′), 17.4, 17.3, 17.2, 17.1, 17.0, 16.96, 16.91, 12.8, 12.5, 12.3, 12.2, 12.1 [CH(CH3)2]; HRMS (ESI+) calcd for C21H38N6NaO6Si2 [M + Na]+ 527.2463, found 527.2469.
5-Amino-2-[3,5-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-2-deoxy-β-d-ribofuranosyl]-2H-[1,2,3]triazolo[4,5-d]pyrimidin-7-one (15a)
Compound 14a (2 g; 3.80 mmol) was treated with 1,1′-thiocarbonyldiimidazole (0.676 g; 3.80 mmol) in dry CH2Cl2 (38 mL) for 8 h at room temperature under argon atmosphere. The solvent was evaporated and the residue was dissolved in CH2Cl2 (20 mL) and washed with H2O (10 mL). The aqueous layer was extracted twice with CH2Cl2 (10 mL); the combined organic layers were dried over Na2SO4 and evaporated. The crude product obtained as slightly yellow foam, was used without further purification in the next step. The crude product was dissolved in dry toluene (76 mL), AIBN (0.125 g; 0.76 mmol) and Bu3SnH (2.56 mL; 9.50 mmol) were added under argon atmosphere and the mixture was degassed with argon for 15 min. Afterwards, the reaction was heated at 75 °C for 2 h. The solution was evaporated in vacuo, and the product was purified by flash chromatography (hexane/EtOAc 1 : 1). The compound 15a was obtained as white solid (1.20 g, 62% over two steps). 1H NMR (600 MHz, DMSO-d6): δ 11.46 (s, 1H, NH), 6.87 (s, 2H, NH2), 6.30 (d, J = 7.5 Hz, 1H, H-1′), 5.12 (m, 1H, H-3′), 3.91–3.79 (m, 2H, H-4′, H-5a′), 3.72 (dd, J = 12.3, 8.8 Hz, 1H, H-5b′), 2.79 (m, 1H, H-2a′), 2.64–2.50 (m, 1H, H-2b′), [m, 28H, 4 × CH(CH3)2]. 13C NMR (151 MHz, DMSO-d6): δ 159.6 (C7), 157.0 (C3a), 155.1 (C5), 127.3 (C7a), 91.0 (C1′), 84.8 (C4′), 72.6 (C3′), 63.6 (C5′), 40.1 (C2′), 17.3, 17.3, 17.2, 17.1, 16.9, 16.83, 16.81, 12.7, 12.5, 12.2, 12.1, 12.0 [CH(CH3)2]. HRMS (ESI+) calcd for C21H39N6O5Si2 [M + H]+ 511.2514, found 511.2517.
5-Amino-2-(2-deoxy-β-d-ribofuranosyl)-2H-[1,2,3]triazolo[4,5-d]pyrimidin-7-one (1a)21
Compound 15a (1.1 g; 2.15 mmol) was dissolved in dry THF (14 mL) and tetrabutylammonium fluoride solution 1.0 M in THF (6.46 mL, 6.46 mmol) was added. The reaction was stirred for 3 h at room temperature. The solvent was evaporated in vacuo, and the crude residue was purified by flash chromatography (CH2Cl2/MeOH 9 : 1) to yield the product 1a as white solid (0.560 g, 97%). Spectral and analytical data were in agreement with previous report.21
2-(2-Deoxy-β-d-ribofuranosyl)-5-(dimethylaminomethylidene)-2H-[1,2,3]triazolo[4,5-d]pyrimidin-7-one (16a)21
To a solution of compound 1a (0.50 g, 1.86 mmol) in MeOH (12 mL) was added dimethylformamide dimethyl acetal (499 μL, 3.73 mmol), and the mixture was heated at 50 °C for 1 h. The reaction was cooled to room temperature and concentrated under reduced pressure. The crude residue was purified by flash column chromatography (CH2Cl2/MeOH 95 : 5) to yield the compound 16a as a white solid (0.56 g, 93%). Spectral and analytical data were in agreement with previous report.21
2-(5-O-Dimethoxytrityl-2-deoxy-β-d-ribofuranosyl)-5-(dimethylaminomethylidene)-2H-[1,2,3]triazolo[4,5-d]pyrimidin-7-one (17a)21
The compound 16a (0.50 g, 1.55 mmol) was co-evaporated with dry pyridine twice under argon atmosphere and then dissolved in dry pyridine (15 mL). 4,4′-Dimethoxytrityl chloride (0.524 g, 1.55 mmol) in CH2Cl2 (1.5 mL) was slowly added drop wise under argon atmosphere at 0 °C, then the mixture was stirred at room temperature for 2 h. The reaction was quenched by the addition of MeOH and the solvents were evaporated. The residue was dissolved in CH2Cl2 and washed with H2O, the organic layers were dried on Na2SO4 and evaporated under argon atmosphere. The compound 17a was isolated by column chromatography (CH2Cl2/MeOH/TEA 97 : 2 : 1) as a white solid (0.81 g, 83%). Spectral and analytical data were in agreement with previous report.21
2-(5-O-Dimethoxytrityl-2-deoxy-β-d-ribofuranosyl)-5-(dimethylaminomethylidene)-2H-[1,2,3]triazolo[4,5-d]pyrimidin-7-one-4-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (18a)
To a stirred solution of 17a (0.320 g, 0.511 mmol) and 1 M solution of bis(diisopropylamino)(2-cyanoethoxy)phosphine (767 μL, 0.767 mmol) in anhydrous CH2Cl2 (4 mL), under argon atmosphere and at 0 °C, was added 0.45 M solution of 1H-terazole (1.13 mL, 0.562 mmol) dropwise. After 10 min, the ice bath was removed and reaction mixture was allowed to stir at room temperature for 45 min. The reaction mixture was diluted with CH2Cl2 and was washed with 1 M TEAB solution. The extracts were dried over Na2SO4 and concentrated in vacuo. The crude mixture was purified by flash column chromatography (hexane/acetone/TEA, 65 : 34 : 1) to afford amidite 18a (0.310 g, 73% yield). 31P NMR (121 MHz, CDCl3) δ 149.3, 149.2. HRMS (ESI+) calcd for C42H53N9O7P [M + H]+ 826.3799, found 826.3818.3
Ethyl 1-(2,3,5-O-triacetyl-β-d-ribofuranosyl)-4-nitro-1H-pyrazole-3-carboxylate (12)
To the compound 11 (1.85 g, 10 mmol) in 66 mL of 1,2-dichloroethane, N,O-bis(trimethylsilyl) acetamide (4.9 mL, 20 mmol) was added and the reaction was stirred for 30 minutes at room temperature. Afterwards, β-d-ribofuranose-1,2,3,5-tetraacetate (3.18 g, 10 mmol) was added into the reaction mixture, followed by 1 M solution of stannic chloride in CH2Cl2 (8 mL, 8 mmol) and the resulting mixture was stirred at room temperature for 36 h. The reaction solution was quenched with cold saturated aq. NaHCO3 (20 mL) and extracted with CH2Cl2. The organics were washed with sat. aq. NaHCO3 (2 × 15 mL) and brine (2 × 15 mL), dried over MgSO4, filtered, concentrated, and purified by flash column chromatography (hexane/EtOAc 7 : 3) to give compound 12 (3.8 g, 85%) as light yellow oil. 1H NMR (300 MHz, CDCl3): δ 8.47 (s, 1H, H-5), 5.91 (d, J = 2.9, 1H, H-1′), 5.70 (dd, J = 5.2, 2.9 Hz, 1H, H-2′), 5.45 (dd, J = 6.2, 5.2 Hz, 1H, H-3′), 4.47–4.22 (m, 4H, H-4′, H-5a′, OCH2), 4.25 (dd, J = 12.5, 3.6 Hz, 1H, H-5b′), 2.15 (s, 6H, CH3), 2.07 (s, 3H, CH3), 1.38 (t, J = 7.1 Hz, 3H, CH3); 13C (75 MHz, CDCl3): δ 170.2, 169.2, 169.0 (C O), 159.8 (C O), 140.1 (C4), 134.2 (C3), 129.3 (C5), 92.5 (C1′), 80.6 (C4′), 74.2 (C3′), 69.5 (C2′), 62.5 (C5′), 62.0 (CH2O), 20.5, 20.2, 13.8 (CH3); HRMS (ESI+) calcd for C17H21N3O11 [M + Na]+ 466.1068, found 466.1063.
Ethyl 4-amino-1-(2,3,5-O-triacetyl-β-d-ribofuranosyl)-1H-pyrazole-3 (13)
To a solution of compound 12 (3.7 g, 8.35 mmol) in MeOH (41 mL) was added 10% Pd/C (88 mg), and the mixture was stirred at room temperature for 8 h under a hydrogen atmosphere (1 atm). The catalyst was removed by filtration over celite. The filtrate was concentrated and compound 13 (3.45 g, 100%) was used as such in the next step. 1H NMR (300 MHz, DMSO-d6): δ 7.38 (s, 1H, H-5), 5.99 (d, J = 4.2 Hz, 1H, H-1′), 5.70 (dd, J = 5.2, 2.9 Hz, 1H, H-2′), 5.45 (dd, J = 6.2, 5.2 Hz, 1H, H-3′), 4.47–4.22 (m, 4H, H-4′, H-5a′, OCH2), 4.25 (dd, J = 3.6, 12.5 Hz, 1H, H-5b′), 2.15 (s, 6H, CH3), 2.07 (s, 3H, CH3), 1.28 (t, J = 7.1 Hz, 3H, CH3); 13C (75 MHz, DMSO-d6): δ 170.1, 169.5, 169.3, 162.8 (C O), 135.7 (C4), 130.6 (C3), 116.2 (C5), 90.7 (C1′), 79.6 (C4′), 72.9 (C3′), 70.4 (C2′), 62.8 (C5′), 59.8 (CH2O), 20.5, 20.3, 14.3 (CH3); HRMS (ESI+) calcd for C17H23N3O9 [M + Na]+ 436.1326, found 436.1331.
5-Amino-2-(β-d-ribofuranosyl)-2H-pyrazolo[4,3-d]pyrimidin-7-one (10b)
A mixture of 13 (1.2 g, 2.9 mmol), chloroformamidine hydrochloride (0.84 g, 7.26 mmol) and dimethylsulfone (1.37 g, 14.5 mmol), was heated for 1 hour at 120 °C in an open flask with magnetic stirring. After the mixture was cooled to room temperature, H2O (5 mL) was added. The solution was then neutralized with NH4OH, stirred for 1 h and then the aqueous solution was dried by lyophilization. The crude residue was purified by flash column chromatography (CH2Cl2/MeOH 8 : 2) to yield the compound 10b as a white solid (0.35 g, 42%). 1H NMR (300 MHz, DMSO-d6): δ 10.95 (br s, 1H, NH), 8.00 (s, 1H, H-9), 6.07 (br s, 2H, NH2), 5.72 (d, J = 4.1 Hz, 1H, H-1′), 5.54 (d, J = 5.0 Hz, 1H, OH), 5.19 (m, 1H, OH), 4.98 (t, J = 5.8 Hz, 1H, OH), 4.31–4.29 (m, 1H, H-2′), 4.14–4.09 (m, 1H, H-3′), 3.95–3.91 (m, 1H, H-4′), 3.65–3.60 (m, 1H, H-5a′), 3.54–3.33 (m, 1H, H-5b′); 13C (75 MHz, DMSO-d6): δ 157.3 (C7), 151.2 (C5), 139.3 (C3a), 133.4 (C7a), 119.5 (C3), 94.9 (C1′), 85.4 (C4′), 75.1 (C3′), 70.3 (C2′), 61.4 (C5′); HRMS (ESI+) calcd for C10H13N5O5 [M + Na]+ 306.0809, found 306.0810.
5-Amino-2-[3,5-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-β-d-ribofuranosyl]-2H-pyrazolo[4,3-d]pyrimidin-7-one (14b)
Reaction of compound 10b (0.8 g, 2.82 mmol), in anhydrous pyridine (14 mL) under argon atmosphere with 1,3-dichloro-1,1,3,3-tetraisopropyldisiloxane (970 μL, 3.11 mmol), was carried out as described for compound 14a and purified by flash column chromatography (CH2Cl2/MeOH 95 : 5) affording compound 14b as white solid (1.1 g, 74%) by 1H NMR (300 MHz, DMSO-d6): δ 10.60 (br s, 1H, NH), 7.83 (s, 1H, H-9), 5.96 (br s, 2H, NH2), 5.79 (s, 1H, H-1′), 5.69 (d, J = 4.4 Hz, 1H, OH), 4.59 (dd, J = 8.2, 4.6 Hz, 1H, H-3′), 4.29 (t, J = 4.4 Hz, 1H, H-2′), 4.03–3.98 (m, 2H, H-4′, H-5a′), 3.90–3.87 (m, 1H, H-5b′), 1.05–0.94 [m, 28H, 4 × CH(CH3)2]; 13C (75 MHz, DMSO-d6): δ 156.9 (C7), 150.9 (C5), 139.1 (C3a), 133.9 (C7a), 119.8 (C3), 94.0 (C1′), 80.9 (C4′), 75.8 (C2′), 70.1 (C3′), 60.3 (C5′), 17.3, 17.2, 17.1, 17.1, 16.9, 16.8, 13.0, 12.8, 12.7, 12.4, 12.3, 12.2 [CH(CH3)2]; HRMS (ESI+) calcd for C22H40N5O6Si2 [M + H]+ 526.2511, found 526.2507.
5-Amino-2-[3,5-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-2-deoxy-β-d-ribofuranosyl]-2H-pyrazolo[4,3-d]pyrimidin-7-one (15b)
Compound 14b (1.2 g; 2.28 mmol) was treated with 1,1′-thiocarbonyldiimidazole (0.406 g; 2.28 mmol) in dry CH2Cl2 (22 mL) for 8 h at 35 °C then with AIBN (75 mg; 0.456 mmol) and Bu3SnH (1.54 mL; 5.70 mmol) in dry toluene (45 mL), was carried out as described for compound 15a and purified by flash column chromatography (1% to 3% MeOH in CH2Cl2). The compound 15b was obtained as white powder (0.790 g, 68% over two steps). 1H NMR (500 MHz, DMSO-d6): δ 10.83 (br s, 1H, NH), 7.82 (s, 1H, H-9), 6.16 (d, J = 7.3 Hz, 1H, H-1′), 6.13 (s, 2H, NH2), 4.97 (m, 1H, H-3′), 3.85–3.79 (m, 2H, H-4′, H-5a′), 3.74 (dd, J = 12.4, 8.3 Hz, 1H, H-5b′), 2.74–2.70 (m, 1H, H-2a′), 2.50–2.45 (m, 1H, H-2b′), 1.05–0.94 [m, 28H, 4 × CH(CH3)2]; 13C NMR (125 MHz, DMSO-d6): δ 157.0 (C7), 151.0 (C5), 139.2 (C3a), 133.6 (C7a), 119.7 (C3), 88.6 (C1′), 84.8 (C4′), 72.1 (C3′), 63.3 (C5′), 40.1 (C2′), 17.4, 17.3, 17.2, 17.1, 17.0, 16.9, 16.8, 13.1, 12.8, 12.5, 12.3, 12.0 [CH(CH3)2]; HRMS (ESI+) calcd for C22H40N5O5Si2 [M + H]+ 510.2562, found 510.2561.
5-Amino-2-(2-deoxy-β-d-ribofuranosyl)-2H-pyrazolo[4,3-d]pyrimidin-7-one (1b)
Reaction of compound 15b (0.8 g; 1.57 mmol) with tetrabutylammonium fluoride in THF (4.71 mL, 4.71 mmol) in dry THF (9.4 mL) and was carried out as described for compound 1a and purified by flash column chromatography (CH2Cl2/MeOH 8 : 2) to yield the product 1b as white solid (0.395 g, 94%). 1H NMR (300 MHz, DMSO-d6): δ 10.38 (br s, 1H, NH), 7.95 (s, 1H, H-9), 6.14 (t, J = 4.1 Hz, 1H, H-1′), 5.94 (br s, 2H, NH2), 5.28 (d, J = 4.3 Hz, 1H, OH), 4.87 (t, 1H, J = 5.5 Hz, OH), 4.37–4.34 (m, 1H, H-3′), 3.86–3.82 (m, 1H, H-4′), 3.57–3.50 (m, 1H, H-5a′), 3.46–3.40 (m, 1H, H-5b′), 2.62–2.54 (m, 1H, H-2a′), 2.32–2.28 (m, 1H, H-2b′); 13C (75 MHz, DMSO-d6): δ 157.3 (C7), 150.9 (C5), 139.2 (C3a), 133.1 (C7a), 119.3 (C3), 90.6 (C1′), 88.1 (C4′), 70.5 (C3′), 61.8 (C5′), 40.0 (C2′); HRMS (ESI+) calcd for C10H14N5O4 [M + H]+ 268.1040, found 268.1055.
2-(2-Deoxy-β-d-ribofuranosyl)-5-(dimethylaminomethylidene)-2H-pyrazolo[4,3-d]pyrimidin-7-one (16b)
Reaction of compound 1b (0.430 g, 1.61 mmol) with dimethylformamide dimethyl acetal (430 μL, 3.22 mmol) in MeOH (10 mL) was carried out as described for compound 16a and purified by flash column chromatography (CH2Cl2/MeOH 95 : 5) to yield the compound 16b as a white solid (0.50 g, 96%). 1H NMR (500 MHz, DMSO-d6): δ 11.11 (br s, 1H, NH), 8.54 (s, 1H, N CH), 8.16 (s, 1H, H-9), 6.19 (t, J = 6.1 Hz, 1H, H-1′), 5.32 (d, J = 4.3 Hz, OH), 4.92 (t, J = 5.4 Hz, 1H, OH), 4.38 (m, 1H, H-3′), 3.87–3.85 (m, 1H, H-4′), 3.62–3.51 (m, 1H, H-5a′), 3.49–3.40 (m, 1H, H-5b′), 3.12 (s, 3H, NCH3), 3.00 (s, 3H, NCH3), 2.62–2.57 (m, 1H, H-2a′), 2.34–2.29 (m, 1H, H-2b′); 13C NMR (125 MHz, DMSO-d6): δ 158.0 (N CH), 157.4 (C7), 155.1 (C5), 138.3 (C3a), 134.6 (C7a), 121.0 (C3), 90.7 (C1′), 88.2 (C4′), 70.4 (C3′), 61.7 (C5′), 40.4 (NCH3), 39.8 (C2′), 34.5 (NCH3); HRMS (ESI+) calcd for C13H19N6O4 [M + H]+ 323.1462, found 323.1455.
2-(5-O-Dimethoxytrityl-2-deoxy-β-d-ribofuranosyl)-5-(dimethylaminomethylidene)-2H-pyrazolo[4,3-d]pyrimidin-7-one (17b)
The reaction of compound 16b (0.450 g, 1.4 mmol) in dry pyridine (14 mL) and 4,4′-dimethoxytrityl chloride (0.473 g, 1.4 mmol) was carried out as described for compound 17a and purified by flash column chromatography (CH2Cl2/MeOH/TEA 97 : 2 : 1), affording compound 17b as a white solid (0.665 g, 76%). 1H NMR (500 MHz, DMSO-d6): δ 11.20 (br s, 1H, NH), 8.53 (s, 1H, N CH), 8.12 (s, 1H, H-9), 7.35–7.08 (m, 9H, ArH), 6.80–6.73 (m, 4H, ArH), 6.26 (dd, J = 6.7, 3.8 Hz, 1H, H-1′), 5.35 (s, 1H, OH), 4.46–4.43 (m, 1H, H-3′), 4.00–3.84 (m, 1H, H-4′), 3.71 (s, 3H, OCH3), 3.69 (s, 3H, OCH3), 3.13 (s, 3H, NCH3), 3.10–3.04 (m, 2H, H-5a′,b′), 3.01 (s, 3H, NCH3), 2.85–2.67 (m, 1H, H-2a′), 2.34–2.30 (m, 1H, H-2b′); 13C NMR (125 MHz, DMSO-d6): δ 158.0 (N CH), 157.9, 157.5, 157.4 (Ar, C7), 155.2, 155.1 (Ar, C5), 144.9, 135.7, 135.6, 135.5, 134.7, 134.6 (C7a, C3a, Ar), 129.8, 129.5, 127.7, 126.5, 121.4, 113.1, 91.1 (Ar), 89.9 (C1′), 85.9 (C4′), 85.3 (CPh3), 70.3 (C3′), 63.9 (C5′), 55.0 (2 × OCH3), 40.5 (NCH3), 40.0 (C2′), 39.8 (NCH3); HRMS (ESI+) calcd for C34H37N6O6 [M + H]+ 625.2768, found 625.2748.
2-(5-O-Dimethoxytrityl-2-deoxy-β-d-ribofuranosyl)-5-(dimethylaminomethylidene)-2H-pyrazolo[4,3-d]pyrimidin-7-one-3-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (18b)
The reaction of compound 17b (0.250 g, 0.40 mmol) and 1 M bis(diisopropylamino)(2-cyanoethoxy)phosphine (600 μL, 0.60 mmol) and 0.45 M 1H-terazole (880 μL, 0.440 mmol) in anhydrous CH2Cl2 (4 mL), was carried out as described for compound 18a and purified by flash column chromatography (hexane/acetone/TEA, 55 : 44 : 1) afforded amidite 18b (0.230 g, 69% yield). 31P NMR (121 MHz, CDCl3) δ 149.1, 148.8. HRMS (ESI+) calcd for C43H53N8O7P [M + H]+ 825.3847, found 825.3849.
8-Azido-3′-5′-O-di(tertbutyldimethylsilyl)-2′-deoxyinosine (20a)
To a solution of compound 19 (1.0 g, 2.08 mmol) in dry THF (40 mL) under argon atmosphere at –78 °C was added n-BuLi (2.5 M solution in hexane, 4.2 mL, 10.4 mmol) and the solution was stirred 1 h at –78 °C. p-Toluenesulfonyl azide (0.1 M in toluene, 13.7 mL, 6.24 mmol) was added drop wise and the reaction mixture was stirred for 2 h at –78 °C and quenched with NH4Cl. After removing volatiles, the residue was extracted with CH2Cl2. The combined organic layers were washed with water, brine and dried over Na2SO4 and the solvent was removed under reduced pressure and the residue was purified by flash column chromatography (CH2Cl2/MeOH 50/1) to yield compound 20a as light yellow solid (0.65 g, 60%). 1H NMR (300 MHz, CDCl3): δ 13.5 (s, 1H, NH), 8.02 (s, 1H, H-2), 6.20 (t, J = 6.9 Hz, 1H, H-1′), 4.69–4.74 (m, 1H, H-3′), 3.89–3.92 (m, 1H, H-4′), 3.82 (dd, J = 10.8, 6.5 Hz, 1H, H-5a′), 3.69 (dd, J = 10.8, 4.7 Hz, 1H, H-5b′), 3.25 (quint, J = 6.8 Hz, 1H, H-2a′), 2.14–2.21 (m, 1H, H-2b′), 0.92 (s, 9H, CCH3), 0.86 (s, 9H, CCH3), 0.12 (s, 6H, SiCH3), 0.019 (s, 3H, SiCH3), –0.006 (s, 3H, SiCH3); 13C-NMR (75 MHz, CDCl3): δ 158.4 (C6), 149.3, 145.7, 143.7 (C4, C8, C2), 123.1 (C5), 87.9, 83.7 (C4′, C1′), 72.3 (C3′), 62.8 (C5′), 37.4 (C2′), 26.0, 25.9 (CH3), 18.5, 18.1 (CCH3), –4.5, –4.6, –5.2, –5.3 (SiCH3). HRMS: calcd for C22H40N7O4Si2 [M + H]+ 522.2675, found 522.2682.
8-Amino-N8-(dimethylaminomethylidene)-3′-5′-O-di(tertbutyldimethylsilyl)-2′-deoxyinosine (21a)
To a pre-cooled solution of compound 20a (600 mg, 1.15 mmol) in MeOH (6 mL) was added NaBH4 (0.218 g, 5.75 mmol) portion wise at 0 °C. The reaction mixture was allowed to warm to room temperature and stirred for 2 h. After the completion of the reaction, excess NaBH4 was carefully quenched with saturated NH4Cl (2 mL) and the solvents were removed under reduced pressure. The crude product was used without further purification for the next step. The solution of crude product in N,N-dimethylforamide-dimethylacetal (5 mL) was heated at 70 °C for 1 h. After removing solvent, the crude residue was purified by flash column chromatography (CH2Cl2/MeOH 30/1) to yield compound 21a (448 mg, 70%) as light yellow solid. 1H NMR (300 MHz, CDCl3): δ 13.4 (s, 1H, NH), 8.71 (s, 1H, N CH), 7.75 (s, 1H, H-2), 6.64 (t, J = 7.3 Hz, 1H, H-1′), 4.71–4.72 (m, 1H, H-3′), 3.83–3.92 (m, 2H, H-4′, H-5a′), 3.68 (dd, J = 4.2, 9.7 Hz, 1H, H-5b′), 3.39 (quint, J = 5.9 Hz, 1H, H-2a′), 2.08–2.15 (m, 1H, H-2b′), 0.91 (s, 9H, CCH3), 0.86 (s, 9H, CCH3), 0.10 (s, 6H, SiCH3), –0.01 (s, 3H, SiCH3), –0.02 (s, 3H, SiCH3); 13C-NMR (75 MHz, CDCl3): δ 158.7, 157.6, 155.8, 148.7, 141.6 (C6, C N, C2, C8, C4), 123.1 (C5), 87.4, 83.1 (C1′, C4′), 73.2 (C3′), 63.4 (C5′), 40.9 (NCH3), 36.9 (C2′), 34.8 (NCH3), 26.0, 25.9 (CH3), 18.5, 18.1 (CCH3), –4.5, –5.1, –5.2 (SiCH3). HRMS: calcd for C25H47N6O4Si2 [M + H]+ 551.3191, found 551.3198.
8-Amino-N8-(dimethylaminomethylidene)-2′-deoxyinosine (22a)
To a solution of compound 21a (0.55 g, 1.0 mmol) in THF (10 mL) was added TBAF (1 M in THF, 2.2 mL, 2.2 mmol) and the solution was stirred overnight at room temperature. After removing volatiles, the crude residue was purified by flash column chromatography (CH2Cl2/MeOH 20/1) to yield compound 22a (0.19 g, 60%) as a white solid. 1H NMR (300 MHz, DMSO-d6): δ 8.56 (s, 1H, N CH), 7.82 (s, 1H, H-2), 6.50 (t, J = 6.4 Hz, 1H, H-1′), 4.41–4.42 (m, 1H, H-3′), 3.82 (br s, 1H, H-4′), 3.64 (dd, J = 11.8, 4.2 Hz, 1H, H-5a′), 3.48 (dd, J = 11.7, 4.3 Hz, 1H, H-5b′), 3.14 (s, 3H, NCH3), 3.02 (s, 4H, NCH3, H-2a′), 1.98–2.05 (m, 1H, H-2b′); 13C-NMR (75 MHz, DMSO-d6): δ 159.0, 156.8, 153.0, 147.3, 145.1 (C6, C N, C2, C8, C4), 122.3 (C5), 87.7, 82.8 (C1′, C4′), 71.6 (C3′), 62.5 (C5′), 40.2 (NCH3), 37.4 (C2′), 34.2 (NCH3). HRMS: calcd for C13H19N6O4 [M + H]+ 323.1462, found 323.1474.
8-Amino-N8-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-2′-deoxyinosine (23a)
Reaction of compound 22a (0.16 g, 0.50 mmol) with 4,4′-dimethoxytrityl chloride (0.168 g, 0.50 mmol) in dry pyridine/DMF (16 mL, 7/1 v/v) was carried out as described for compound 17a and purified by flash column chromatography (CH2Cl2/MeOH/TEA 80/2/1) to yield compound 23a (0.3 g, 96%) as white solid. 1H NMR (500 MHz, DMSO-d6): δ 8.53 (s, 1H, N CH), 7.93 (s, 1H, H-2), 7.27 (d, J = 7.3 Hz, 2H, ArH), 7.30–7.27 (m, 8H, ArH), 6.85–6.74 (m, 4H, ArH), 6.52 (t, J = 7.2 Hz, 1H, H-1′), 5.25 (d, J = 5.25 Hz, OH), 4.47–4.43 (m, 1H, H-3′), 3.97–3.70 (m, 1H, H-4′), 3.72 (s, 3H, OCH3), 3.71 (s, 3H, OCH3), 3.451–3.26 (m, 2H, H-H-5′), 3.14–3.09 (m, 4H, CH3, H-2a′), 2.94 (s, 3H, NCH3), 2.14–2.06 (m, 1H, H-2b′). 13C NMR (125 MHz, DMSO-d6) δ 158.3, 157.4, 156.0, 154.9 (Ar, C6, C N, C2, C8, C4), 147.0, 145.5, 136.2, 136.1, 130.1, 130.0, 128.0, 127.9, 126.8 (Ar), 122.0 (C5), 113.3, 113.3 (Ar), 86.0, 85.6 [C1′, C(CH3)3], 82.5 (C4′), 71.8 (C3′), 64.9 (C5′), 55.4 (OCH3), 36.9, 34.7, 33.7 (2NCH3, C2′). HRMS: calcd for C34H37N6O6 [M + H]+ 625.2769, found 625.2770.
8-Amino-N8-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-2′-deoxyinosine-3′-(2-cyanoethyl-N,N-diisopropyl)-phosphoramidite (24a)
Reaction of compound 23a (0.2 g, 0.32 mmol) with 1 M bis(diisopropylamino)(2-cyanoethoxy)phosphine (1 M in CH3CN, 0.96 mL, 0.96 mmol) and 1H-tetrazole (0.45 M in CH3CN, 1.56 mL, 0.70 mmol) in anhydrous CH2Cl2 (10 mL), was carried out as described for compound 18a and purified by flash column chromatography (CH2Cl2/MeOH/TEA 80/2/1) to yield compound 24a (0.20 g, 75%) as white solid. 31P NMR (121 MHz, CDCl3) δ 148.5, 148.0. HRMS: calcd for C13H19N6O4 [M + H]+ 323.1462, found 323.1474.
8-Azido-3′-5′-O-di(tertbutyldimethylsilyl)-N1-methyl-2′-deoxyinosine (20b)
To a solution of sodium hydride (344 mg, 8.62 mmol) in dry THF (30 mL), compound 20a (3.0 g, 5.75 mmol) in THF (50 mL) was added drop wise at 0 °C and stirred for 15 min. Methyl iodide (0.72 mL, 11.50 mmol) was added slowly to the reaction mixture at 0 °C. The reaction mixture was stirred at room temperature for 12 h and then NH4Cl (5 mL) was added to the reaction mixture to quench the excess sodium hydride and the reaction mixture was evaporated to dryness. The residue obtained was extracted with CH2Cl2 (2 × 100 mL), washed with water (2 × 15 mL), brine (10 mL) and dried over anhydrous Na2SO4 and the solvent was removed under reduced pressure and the residue was purified by flash column chromatography (n-hexane/ethyl acetate 7/3) to yield compound 20b as a yellow solid (2.71 g, 88%). 1H NMR (500 MHz, CDCl3) δ 7.87 (s, 1H, H-2), 6.16 (t, J = 6.9 Hz, 1H, H-1′), 4.74–4.69 (m, 1H, H-3′), 3.90–3.86 (m, 1H, H-4′), 3.79 (dd, J = 10.9, 6.4 Hz, 1H, H-5b′), 3.68 (dd, J = 10.9, 4.6 Hz, 1H, H-5a′), 3.63 (s, 3H, NCH3), 3.18 (pent, J = 6.9 Hz, 1H, H-2b′), 2.15 (ddd, J = 10.8, 6.9, 3.9 Hz, 1H, H-2a′), 0.92 (s, 9H, CCH3), 0.85 (s, 9H, CCH3), 0.11 (s, 6H, SiCH3), 0.02 (s, 3H, SiCH3), –0.01 (s, 3H, SiCH3); 13C NMR (125 MHz, CDCl3) δ 156.1 (C6), 147.7 (C4), 146.0 (C2), 145.1 (C8), 122.9 (C5), 87.6 (C4′), 83.4 (C1′), 72.1 (C3′), 62.7 (C5′), 37.3 (C2′), 34.4 (NCH3), 26.0, 25.9 (CCH3), 18.4, 18.1 (CH3C), –4.5, –4.6, –5.2, –5.3 (CH3Si); HRMS (ESI+): calcd for C23H41N7O4Si2 [M + H]+ 536.2831; found 536.2832.
8-Amino-N8-(dimethylaminomethylidene)-3′-5′-O-di(tertbutyldimethylsilyl)-N1-methyl-2′-deoxy-inosine (21b)
The reaction of compound 20b (2.5 g, 4.67 mmol) with NaBH4 (882 mg, 23.33 mmol) in MeOH (26 mL) and then with N,N-dimethylformamide-dimethylacetal (15 mL) was carried out as described for compound 21a, and purified by flash column chromatography (CH2Cl2/MeOH 15/1) to yield yellow solid (1.92 g, 73%). 1H NMR (500 MHz, DMSO-d6) δ 8.56 (s, 1H, CH N), 8.18 (s, 1H, H-2), 6.47 (t, J = 7.0 Hz, 1H, H-1′), 4.71 (dt, J = 6.7, 3.6 Hz, 1H, H-3′), 3.80 (dd, J = 10.4, 6.7 Hz, 1H, H-5b′), 3.77–3.72 (m, 1H, H-4′), 3.59 (dd, J = 10.4, 4.9, 1H, H-5a′), 3.47 (s, 3H, NCH3), 3.28–3.21 (m, 1H, H-2b′), 3.15 (s, 3H, NCH3), 3.02 (s, 3H, NCH3), 2.09 (ddd, J = 10.9, 7.0, 3.6 Hz, 1H, H-2a′), 0.90 (s, 9H, CCH3) 0.83 (s, 9H, CCH3), 0.11 (s, 6H, SiCH3), –0.01 (s, 3H, SiCH3), –0.03 (s, 3H, SiCH3). 13C NMR (125 MHz, DMSO-d6) δ 157.1 (C N), 155.7 (C6), 154.6 (C4), 146.8 (C8), 145.5 (C2), 121.7 (C5), 86.3 (C4′), 81.9 (C1′), 72.5 (C3′), 62.8 (C5′), 40.2 (NCH3), 36.2 (C2′), 34.3 (NCH3), 33.4 (NCH3), 25.7 (CH3), 18.0 (CCH3), 17.7 (CCH3), –4.6 (SiCH3), –4.7 (SiCH3). HRMS (ESI+): calcd for C23H43N5O4Si2 [M + H]+ 565.3348; found 565.3358.
8-Amino-N8-(dimethylaminomethylidene)-N1-methyl-2′-deoxyinosine (22b)
The reaction of compound 21b (565 mg, 1.0 mmol) with TBAF (1 M in THF, 2.2 mL, 2.2 mmol) in THF (10 mL) was carried out as described for compound 22a and purified by flash column chromatography (CH2CI2/MeOH, 9 : 1) to give 22b (246 mg, 73%) as a pale yellow solid. 1H NMR (500 MHz, DMSO-d6) δ 8.56 (s, 1H, CH N), 8.21 (s, 1H, H-2), 6.49 (t, J = 6.4 Hz, 1H, H-1′), 5.25 (s, 1H, OH), 4.96 (s, 1H, OH), 4.43 (br s, 1H, H-3′), 3.81 (br s, 1H, H-4′), 3.64 (dd, J = 11.4, 4.6 Hz, 1H, H-5b′), 3.47 (br s, 4H, NCH3, H-5a′), 3.14 (s, 3H, NCH3), 3.09–2.90 (m, 4H, NCH3, H-2b′), 2.05 (ddd, J = 8.8, 6.4, 2.0 Hz, 1H, H-2a′); 13C NMR (125 MHz, DMSO-d6) δ 157.1 (C N), 155.5 (C6), 154.4 (C8), 146.5 (C4), 145.6 (C2), 121.7 (C5), 87.5 (C4′), 82.5 (C1′), 71.3 (C3′), 62.3 (C5′), 40.3 (NCH3), 37.1 (NCH3), 34.4 (C2′), 33.4 (NCH3); HRMS (ESI+): calcd for C14H20N6O4 [M + Na]+ 359.1438; found 359.1444.
8-Amino-N8-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-N1-methyl-2′-deoxyinosine (23b)
Reaction of 22b (420 mg, 1.25 mmol) with 4,4′-dimethoxytrityl chloride (465 mg, 1.37 mmol) in dry pyridine (45 mL), in dry CH2Cl2 (5 mL) was carried out as described for compound 17a and purified by flash column chromatography (CH2Cl2/MeOH/TEA 80/2/1) to yield compound 23b (710 mg, 89%) as white solid. 1H NMR (500 MHz, CDCl3) δ 8.68 (s, 1H, N CH), 7.45 (s, 1H, H-2), 7.41 (d, J = 7.2 Hz, 2H, Ar), 7.31–7.15 (m, 7H, Ar), 6.78–6.73 (m, 4H, Ar), 6.66 (t, J = 6.9 Hz, 1H, H-1′), 4.85–4.80 (m, 1H, H-3′), 4.11–4.06 (m, 1H, H-4′), 3.77 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 3.54 (s, 3H, NCH3), 3.52–3.45 (m, 1H, H-5b′), 3.32–3.25 (m, 2H, H-2b′, H-5b′), 3.07 (m, 3H, NCH3), 3.02 (m, 3H, NCH3), 2.28 (ddd, J = 12.3, 6.9, 4.5 Hz, 1H, H-2a′); 13C NMR (125 MHz, CDCl3) δ 158.5 (Ar), 157.5 (N C), 156.7 (C6), 155.4 (C8), 146.9 (C4), 145.0 (Ar), 143.7 (C2), 136.3, 136.1, 130.2, 128.2, 127.8, 126.7 (Ar), 122.8 (C5), 113.1 (Ar), 86.3 (CPh3), 85.5 (C4′), 82.6 (C1′), 73.6 (C3′), 64.6 (C5′), 55.3 (OCH3), 40.9 (NCH3), 37.2 (C2′), 34.8, 34.1 (NCH3); HRMS (ESI+): calcd for C35H38N6O6 [M + H]+ 639.2925; found 639.2926.
8-Amino-N8-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-N1-methyl-2′-deoxyinosine-3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (24b)
The reaction of compound 23b with 1 M bis(diisopropylamino)(2-cyanoethoxy)phosphine (1.18 mL, 1.40 mmol) and 0.45 M 1H-tetrazole (1.45 mL, 0.77 mmol) in CH2Cl2 (25 mL) was carried out as described for compound 18a and purified by flash column chromatography (CH2Cl2/MeOH/TEA 40/1/1) to yield compound 24b (0.380 g, 77%) as white solid. 31P NMR (202 MHz, CDCl3) δ 148.6, 148.2. HRMS (ESI+): calcd for C44H55N8O7P [M + H]+ 839.4003; found 839.4014.
7,8-Dihydro-8-oxo-2′-deoxyinosine (3)29
A solution of NaNO2 (516 mg, 7.48 mmol) in (2 mL) of water was added to a stirred solution of 1 g (3.74 mmol) of compound 5 in 50 mL of 95% aqueous acetic acid. The reaction mixture was stirred overnight. The solvent was removed under reduced pressure and the oily residue was purified by flash column chromatography (CH2CI2/MeOH, 4 : 1) to give 3 (780 mg, 77%) as a white solid. Spectral and analytical data were in agreement with previous report.29
7,8-Dihydro-5′-O-dimethoxytrityl-8-oxo-2′-deoxyinosine (25)29
Reaction of compound 3 (350 mg, 1.35 mmol) with 4,4′-dimethoxytrityl chloride (486 mg, 1.44 mmol) in dry pyridine (70 mL) was carried out as described for compound 17a and purified by flash column chromatography (CH2CI2/MeOH/TEA, 95 : 4 : 1) to give 25 (510 mg, 69%) as a white foam. Spectral and analytical data were in agreement with previous report.29
7,8-Dihydro-5′-O-dimethoxytrityl-8-oxo-2′-deoxyinosine-3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (26)29
The reaction of compound 25 (400 mg, 0.70 mmol) with 1 M bis(diisopropylamino)(2-cyanoethoxy)phosphine (1.40 mL, 1.40 mmol) and 0.45 M 1H-tetrazole (1.7 mL, 0.77 mmol) in CH2Cl2 (25 mL) was carried out as described for compound 18a and purified by flash column chromatography (n-hexane/acetone/TEA, 39 : 60 : 1) to give 26 (430 mg, 80%) as a white foam. Spectral and analytical data were in agreement with previous report.29
7,8-Dihydro-N6-(dimethylaminomethylidene)-8-oxo-2′-deoxyadenosine (27)
To a suspension of compound 5 (1 g, 3.74 mmol) in MeOH (35 mL), N,N-dimethylformamide dimethyl acetal (2.5 mL, 18.7 mmol) was added. The reaction mixture was stirred at 65 °C for 3 h. The reaction mixture was evaporated to dryness and purified by flash column chromatography (CH2Cl2/MeOH : TEA, 90 : 9 : 1) to give 27 (1.1 g, 91%) as a white solid. 1H NMR (500 MHz, DMSO-d6) δ 8.72 (s, 1H, HC N), 8.19 (s, 1H, H-2), 6.17 (dd, J = 8.2, 6.5 Hz, 1H, H-1′), 5.20 (s, 1H, OH), 5.08 (s, 1H, OH), 4.42–4.38 (m, 1H, H-3′), 3.83–3.79 (m, 1H, H-4′), 3.62 (dd, J = 11.7, 4.5 Hz, 1H, H-5b′), 3.47 (br s, 1H, H-5a′), 3.14 (s, 1H, NCH3), 3.10 (s, 1H, NCH3), 3.10–2.95 (m, 1H, H-2b′), 2.01 (ddd, J = 9.2, 6.5, 2.6 Hz, H-2a′); 13C NMR (125 MHz, DMSO-d6) δ 156.0 (C N), 152.2 (C8), 149.6 (C2), 148.9 (C6), 148.3 (C4), 111.7 (C5), 87.5 (C4′), 81.4 (C1′), 71.4 (C3′), 62.4 (C5′), 40.4 (NCH3), 36.0 (C2′), 34.3 (NCH3); HRMS (ESI+): calcd for C13H18N6O4 [M + H]+ 323.1462; found 323.1455.
7,8-Dihydro-N6-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-8-oxo-2′-deoxy adenosine (28)
The reaction of compound 27 (600 mg, 1.86 mmol) with 4,4′-dimethoxytrityl chloride (693 mg, 2.05 mmol) in dry pyridine (60 mL), was carried out as described for compound 17a and purified by flash column chromatography (CH2CI2/MeOH/TEA, 95 : 4 : 1) to give 28 (850 mg, 73%) as a white foam. 1H NMR (600 MHz, DMSO-d6) δ 8.70 (s, 1H, HC N), 8.04 (s, 1H, H-2), 7.36–7.34 (m, 2H, Ar), 7.24–7.14 (m, 7H, Ar), 6.71–6.81 (m, 4H, Ar), 6.17 (dd, J = 7.2, 6.2 Hz, 1H, H-1′), 4.52 (br s, 1H, H-3′), 3.94–3.88 (m, 1H, H-4′), 3.71 (s, 3H, OCH3), 3.69 (s, 3H, OCH3), 3.20–3.05 (m, 9H, 2 × NCH3, H-5a′,b′, H-2b′), 2.09 (ddd, J = 12.7, 7.2, 4.8 Hz, 1H, H-2a′). 13C NMR (150 MHz, DMSO-d6) δ 157.9, 157.8, 155.8, 152.3, 149.7, 148.5, 145.1 (C2, C4, C8, C6, C N, Ar), 135.8, 135.6, 129.7, 129.5, 128.9, 127.6, 127.6, 127.41, 126.4, 113.0, 112.9, 112.7 (Ar), 111.7 (C5), 85.2 (CPh3), 85.1 (C4′), 80.7 (C1′), 71.0 (C3′), 64.2 (C5′), 55.0, 54.9 (OCH3), 40.4 (NCH3), 35.6 (C2′), 34.3 (NCH3); HRMS (ESI+): calcd for C34H36N6O6 [M + H]+ 625.2768; found 625.2776.
7,8-Dihydro-N6-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-8-oxo-2′-deoxyadenosine-3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (29)
The reaction of compound 28 (900 mg, 1.44 mmol), 1 M bis(diisopropylamino)(2-cyanoethoxy)phosphine (2.88 mL, 2.88 mmol) and 0.45 M solution of 1H-tetrazole (3.5 mL, 1.58 mmol) in CH2Cl2 (30 mL) was carried out as described for compound 18a and purified by flash column chromatography (n-hexane/acetone/TEA, 49 : 50 : 1) to give 29 (920 mg, 77%) as a white foam. 31P NMR (202 MHz, DMSO-d6) δ 147.4, 146.8. HRMS (ESI+): calcd for C43H53N8O7P [M + H]+ 825.3847; found 825.3842.
7,8-Dihydro-N2-(dimethylaminomethylidene)-8-oxo-2′-deoxyguanosine (30)
To a suspension of compound 4 (1 g, 3.53 mmol) in MeOH (25 mL), N,N-dimethylformamide dimethyl acetal (4.7 mL, 35.3 mmol) was added. The reaction mixture was stirred at 25 °C for 12 h. The resulting precipitate was isolated, and was washed with CH2Cl2 to give 30 (600 mg, 50%) as a white solid. 1H NMR (600 MHz, DMSO-d6) δ 8.46 (s, 1H, N CH), 6.10 (t, J = 7.3 Hz, 1H, H-1′), 5.16 (s, 1H, OH), 4.75 (s, 1H, OH), 4.39–4.33 (m, 1H, H-3′), 3.76–3.71 (m, 1H, H-4′), 3.56 (dd, J = 11.5, 4.6 Hz, 1H, H-5b′), 3.44 (dd, J = 11.5, 4.2 Hz, 1H, H-5a′), 3.12 (s, 3H, NCH3), 3.00 (s, 3H, NCH3), 2.99–2.91 (m, 1H, H-2b′), 1.94 (ddd, J = 9.6, 6.7, 2.7 Hz, 1H, H-2a′); 13C NMR (150 MHz, DMSO-d6) δ 157.8 (C N), 156.4 (C6), 152.2 (C8), 151.7 (C2), 145.6 (C4), 102.2 (C5), 87.3 (C4′), 81.1 (C1′), 71.2 (C2′), 62.3 (C5′), 40.7 (NCH3), 36.0 (C2′), 34.6 (NCH3); HRMS (ESI+): calcd for C13H18N6O5 [M + Na]+ 361.1231; found 361.1227.
7,8-Dihydro-N2-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-8-oxo-2′-deoxy guanosine (31)
The reaction of compound 30 (480 mg, 1.42 mmol) with 4,4′-dimethoxytrityl chloride (528 mg, 1.56 mmol) in dry pyridine (60 mL) was carried out as described for compound 17a and purified by flash column chromatography (CH2CI2/MeOH/TEA, 97 : 2 : 1) to give 31 (650 mg, 71%) as a white foam. 1H NMR (600 MHz, DMSO-d6) δ 11.53 (s, 1H, NH), 10.82 (s, 1H, NH), 8.33 (s, 1H, C N), 7.36–7.31 (m, 2H, Ar), 7.25–7.15 (m, 7H, Ar), 6.72–6.82 (m, 4H, Ar), 6.13 (dd, J = 7.9, 5.0 Hz, 1H, H-1′), 5.24 (d, J = 4.9 Hz, 1H, OH), 4.46 (quin, J = 4.9 Hz, 1H, H-3′), 3.81–3.85 (m, 1H, H-4′), 3.72, 3.71 (OCH3), 3.20 (dd, J = 9.9, 7.5 Hz, 1H, H-5b′), 3.07 (dd, J = 9.9, 3.3 Hz, 1H, H-5a′), 3.02, 2.99 (NCH3), 2.90–2.96 (m, 1H, H-2b′), 2.09 (ddd, J = 13.5, 7.9, 5.0 Hz, 1H, H-2a′); 13C NMR (150 MHz, DMSO-d6) δ 157.9, 157.8, 157.4, 156.0, 152.1, 151.6, 145.5, 145.1 (C6, C2, C N, C8, C4, Ar), 135.8, 135.7, 129.6, 129.5, 127.7, 127.6, 126.4, 113.0, 112.9 (Ar), 112.7 (C5), 102.2 (CPh3), 85.1 (C4′), 80.3 (C1′), 70.9 (C3′), 64.5 (C5′), 54.9, 54.9 (OCH3), 40.6 (NCH3), 36.4 (C2′), 34.6 (NCH3); HRMS (ESI+): calcd for C34H36N6O7 [M + Na]+ 663.2537; found 663.2537.
7,8-Dihydro-N2-(dimethylaminomethylidene)-5′-O-dimethoxytrityl-8-oxo-2′-deoxyguanosine-3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (32)
The reaction of compound 31 (440 mg, 0.68 mmol), 1 M bis(diisopropylamino)(2-cyanoethoxy)phosphine (1.37 mL, 1.37 mmol) and 0.45 M solution of 1H-tetrazole (1.7 mL, 0.75 mmol) in CH2Cl2 (20 mL) was carried out as described for compound 18a and purified by flash column chromatography (n-hexane/acetone/TEA, 49 : 50 : 1) to give 32 (470 mg, 81%) as a white foam. 31P NMR (202 MHz, DMSO-d6) δ 147.8, 147.6. HRMS (ESI+): calcd for C43H53N8O8P [M + Na]+ 863.3616; found 863.3620.
Oligonucleotide synthesis
Oligonucleotide assembly was performed with an Expedite DNA synthesizer (Applied Biosystems) by using the phosphoramidite approach. The oligomers were deprotected and cleaved from the solid support by treatment with aqueous ammonia (30%) for 2 h. After gel filtration on a NAP-25 column (Sephadex G25-DNA grade; Pharmacia) with water as eluent, the crude mixture was analysed by using a Mono-Q HR 5/5 anion exchange column, after which purification was achieved by using a Mono-Q HR 10/100 GL column (Pharmacia) with the following gradient system: A = 10 mM NaClO4 in 15% CH3CN, pH 7.4, B = 600 mM NaClO4 in 15% CH3CN, pH 7.4. The low-pressure liquid chromatography system consisted of a Merck-Hitachi L-6200A intelligent pump, a Mono-Q HR 10/100 GL column (Pharmacia), an Uvicord SII 2138 UV detector (Pharmacia-LKB) and a recorder. The product-containing fraction was desalted on a NAP-25 column and lyophilised. Analysis by mass spectrometry followed.
UV melting experiments
Oligomers were dissolved in a buffer solution containing NaCl (0.1 M), potassium phosphate (0.02 M, pH 7.5) and EDTA (0.1 mM). The concentration was determined by measuring the absorbance in Milli-Q water at 260 nm at 80 °C. The following extinction coefficients were used: dA, ε = 15 000; dT, ε = 8500; C, ε = 7100; dG, ε = 12 100; N8-8-aza-dG, ε = 2200; N8-8-aza-9-deaza-dG, ε = 2500; 8-NH2-dI, ε = 6800; 1-Me-8-NH2-dI, ε = 7600; d-isoCMe, ε = 6300; d-isoG, ε = 4600; 8-Oxo-dI, ε = 7500; 8-Oxo-dA, ε = 11 100; 8-Oxo-dG, ε = 5900. The concentration for each strand was 4 μM in all experiments. Melting curves were determined with a Varian Cary 100 BIO spectrophotometer. Cuvettes were maintained at constant temperature by water circulation through the cuvette holder. The temperature of the solution was measured with a thermistor that was directly immersed in the cuvette. Temperature control and data acquisition were carried out automatically with an IBM-compatible computer by using Cary WinUV thermal application software. A quick heating and cooling cycle was carried out to allow proper annealing of both strands. The samples were then heated from 10 to 80 °C at a rate of 0.2 °C min–1, and were cooled again at the same speed. Melting temperatures were determined by plotting the first derivative of the absorbance as a function of temperature; data plotted were the average of two runs. Up and down curves showed identical Tm values.
In vivo assay of XNA-oligonucleotides
The oligomers were dissolved in MilliQ water to 100 mM. Before assay, they were diluted ten-fold in the same medium. Testing was performed with a gapped heteroduplex vector produced through the enzymatic digestion and PCR assisted denaturation and hybridization of the pAK1 and pAK2 plasmids.45 A mix of equimolar (25 ng each) purified pAK1 (NheI and NsiI cut) and purified pAK2 (two-fold EcoRI cut and dephosphorylated) was diluted into 10 mM Tris–HCl (pH 7.5) with 100 mM NaCl. The mixture was denatured at 95 °C for 5 min before cooling to ambient temperature over 2 h, before water dialysis on 0.05 μM nitrocellulose filters (Millipore) for 30 min. The oligomers (0.02 pmol), as well as a positive d(CTAGCGCCGTGCCATGCA) and negative d(CTAGCGCCG···CATGCA) oligomer controls, were added separately to the dialyzed heteroduplex mixture diluted into 1× DNA ligase T4 reaction buffer (NEB) for 20 μL per sample. The mixture was denatured at 85 °C as before. Ligation was performed by adding 5 U T4 DNA ligase (NEB) supplemented with 1 mM ATP to the samples before overnight incubation (16 °C). Ligated mixtures were dialyzed as before, and transformed by electroporation into electro-competent E. coli K12 (ΔthyA:aadA). Incubation of the electroporated bacteria was made at 37 °C for 1 hour, before plating 100 μL of a serial dilution of the suspension onto Muller-Hinton (MH) media containing 100 μg mL–1 ampicillin (spreading the 100 and 10–1 dilutions) and onto the same media supplemented additionally with 0.3 mM thymidine (10–3 and 10–4 dilutions).
In vivo assay of control DNA-oligonucleotides
Control DNA oligomers were designed by modifying the 18-long standard oligomer to incorporate single changes at the codons tested. All DNA oligomers were ordered at Eurofins MWG and dissolved in MilliQ water to a concentration of 100 mM before dilution ten-fold for testing. Assay of these oligos followed that of the XNA-oligonucleotides, following the same protocol as above.
Data analysis and sequencing
As before,8 the total numbers of thymidine-prototrophic colonies and ampicillin resistant colonies were taken from the plate counts, and the ratio of the former over the latter was calculated. Positive thymidine-prototrophic colonies were restreaked twice to MH ampicillin, before growth in 2 mL liquid cultures of the same medium. Plasmids were recovered by Miniprep (Qiagen) and the plasmids sequenced using an oligo for pAK1 d(AACAGTGGCGCGCCTGG).
Generation of the ΔMutM strain
The MutM gene was deleted from the E. coli K12 strain (ΔthyA:aadA) through pKD46 meditation recombination, using a deletion construct containing flanking regions of the gene around an apramycin resistance cassette (aac(3)IV). Recombination was made by the published procedure.54 Deletion was confirmed by external primers through colony-PCR.
Supplementary Material
Acknowledgments
We thank Luc Baudemprez for technical assistance and Chantal Biernaux for editorial help. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement no. ERC-2012-ADG_20120216/320683, from FWO Vlaanderen, Belgium (G078014N) and from the KU Leuven Research Council (OT/1414/128). Mass spectrometry was made possible by the support of the Hercules Foundation of the Flemish Government (grant 20100225-7).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sc03474d
References
- Benner S. A. Acc. Chem. Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- Liu H., Gao J., Lynch S. R., Saito Y. D., Maynard L., Kool E. T. Science. 2003;302:868–871. doi: 10.1126/science.1088334. [DOI] [PubMed] [Google Scholar]
- Hirao I., Kimoto M., Yamashige R. Acc. Chem. Res. 2012;45:2055–2065. doi: 10.1021/ar200257x. [DOI] [PubMed] [Google Scholar]
- Malyshev D. A., Dhami K., Quach H. T., Lavergne T., Ordoukhanian P., Torkamani A., Romesberg F. E. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12005–12010. doi: 10.1073/pnas.1205176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashige R., Kimoto M., Takezawa Y., Sato A., Mitsui T., Yokoyama S., Hirao I. Nucleic Acids Res. 2012;40:2793–2806. doi: 10.1093/nar/gkr1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput J. C., Yu H., Zhang S. Chem. Biol. 2012;19:1360–1371. doi: 10.1016/j.chembiol.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Malyshev D. A., Dhami K., Lavergne T., Chen T., Dai N., Foster J. M., Corrêa I. R., Romesberg F. E. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bande O., Abu El Asrar R., Braddick D., Dumbre S., Pezo V., Schepers G., Pinheiro V. B., Lescrinier E., Holliger P., Marlière P., Herdewijn P. Chem.–Eur. J. 2015;21:5009–5022. doi: 10.1002/chem.201406392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. C., Sherrill C. B., Marshall D. J., Moser M. J., Prudent J. R. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer C. Y., Moroney S. E., Benner S. A. Biochemistry. 1993;32:10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- Ogawa A. K., Wu Y., Berger M., Schultz P. G., Romesberg F. E. J. Am. Chem. Soc. 2000;122:8803–8804. [Google Scholar]
- Benner S. A., Sismour A. M. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool E. T. Acc. Chem. Res. 2002;35:936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- Koh J. S., Dervan P. B. J. Am. Chem. Soc. 1992;114:1470–1478. [Google Scholar]
- Kazimierczuk Z., Binding U., Seela F. Helv. Chim. Acta. 1989;72:1527–1536. [Google Scholar]
- He J., Seela F. Org. Biomol. Chem. 2003;1:1873–1883. doi: 10.1039/b301608k. [DOI] [PubMed] [Google Scholar]
- Seela F., Zulauf M., Debelak H. Helv. Chim. Acta. 2000;83:1437–1453. [Google Scholar]
- He J., Seela F. Helv. Chim. Acta. 2002;85:1340–1354. [Google Scholar]
- Seela F., Debelak H. Nucleic Acids Res. 2000;28:3224–3232. doi: 10.1093/nar/28.17.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seela F., Kröschel R. Org. Biomol. Chem. 2003;1:3900–3908. doi: 10.1039/b309485p. [DOI] [PubMed] [Google Scholar]
- Seela F., Lampe S. Helv. Chim. Acta. 1994;77:1003–1017. [Google Scholar]
- Seela F., Lampe S. Helv. Chim. Acta. 1993;76:2388–2397. [Google Scholar]
- Meyer R. B., Revankar G., Cook P. D., Ehler K., Schweizer M., Robins R. J. Heterocycl. Chem. 1980;17:159–169. [Google Scholar]
- Cubero E., Avinó A., de la Torre B. G., Frieden M., Eritja R., Luque F. J., González C., Orozco M. J. Am. Chem. Soc. 2002;124:3133–3142. doi: 10.1021/ja011928+. [DOI] [PubMed] [Google Scholar]
- Cubero E., Güimil-García R., Luque F. J., Eritja R., Orozco M. Nucleic Acids Res. 2001;29:2522–2534. doi: 10.1093/nar/29.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eritja R. and Garcia R., US Pat., US 2003/0135040 A l, 2003.
- Uesugi S., Ikehara M. J. Am. Chem. Soc. 1977;99:3250–3253. doi: 10.1021/ja00452a008. [DOI] [PubMed] [Google Scholar]
- Nikolova E. N., Zhou H., Gottardo F. L., Alvey H. S., Kimsey I. J., Al-Hashimi H. M. Biopolymers. 2013;99:955–968. doi: 10.1002/bip.22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka N., Greenberg M. M. Nucleic Acids Res. 2005;33:1637–1643. doi: 10.1093/nar/gki305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodepudi V., Iden C. R., Johnson F. Nucleosides Nucleotides. 1991;10:755–761. [Google Scholar]
- Bodepudi V., Shibutani S., Johnson F. Chem. Res. Toxicol. 1992;5:608–617. doi: 10.1021/tx00029a004. [DOI] [PubMed] [Google Scholar]
- Koizume S., Kamiya H., Inoue H., Ohtsuka E. Nucleosides, Nucleotides Nucleic Acids. 1994;13:1517–1534. [Google Scholar]
- Wang Y., Wang Y. Chem. Res. Toxicol. 2006;19:837–843. doi: 10.1021/tx060032l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S., Takeshita M., Grollman A. P. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Guy A., Duplaa A. M., Harel P., Téoule R. Helv. Chim. Acta. 1988;71:1566–1572. [Google Scholar]
- Komatsu H., Ichikawa T., Nakai M., Takaku H. Nucleosides Nucleotides. 1992;11:85–95. [Google Scholar]
- Petraccone L., Duro I., Erra E., Randazzo A., Virno A., Giancola C. Nucleosides, Nucleotides Nucleic Acids. 2007;26:675–679. doi: 10.1080/15257770701490597. [DOI] [PubMed] [Google Scholar]
- Esposito V., Randazzo A., Virgilio A., Cozzuto L., Mayol L. Nucleosides, Nucleotides Nucleic Acids. 2005;24:783–788. doi: 10.1081/ncn-200060156. [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M., Bodepudi V., Shibutani S., Eisenberg M., Johnson F., Grollman A. P., Patel D. J. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- McAuley-Hecht K. E., Leonard G. A., Gibson N. J., Thomson J. B., Watson W. P., Hunter W. N., Brown T. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- Beard W. A., Batra V. K., Wilson S. H. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2010;703:18–23. [Google Scholar]
- Wood M. L., Esteve A., Morningstar M. L., Kuziemko G. M., Essigmann J. M. Nucleic Acids Res. 1992;20:6023–6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen A. P., Murphy J. A. Free Radical Biol. Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Pham L., Cruz C., Miller J. H. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou J., Kasai H., Shibutani S., Chung M. H., Laval J., Grollman A. P., Nishimura S. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R. D., Montgomery J. A. J. Med. Chem. 1976;19:1186–1191. doi: 10.1021/jm00232a004. [DOI] [PubMed] [Google Scholar]
- Davoll J. J. Chem. Soc. 1958:1593–1599. [Google Scholar]
- Townsend L. B., Chemistry of nucleosides and nucleotides, Plenum Press, New York, 1994. [Google Scholar]
- Buff M. C., Schäfer F., Wulffen B., Müller J., Pötzsch B., Heckel A., Mayer G. Nucleic Acids Res. 2010:1148. doi: 10.1093/nar/gkp1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatgilialoglu C., Navacchia M. L., Postigo A. Tetrahedron Lett. 2006;47:711–714. [Google Scholar]
- Kannan A., Burrows C. J. J. Org. Chem. 2010;76:720–723. doi: 10.1021/jo102187y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochet S., Kaminski P. A., van Aerschot A., Herdewijn P., Marlière P. C. R. Biol. 2003;326:1175–1184. doi: 10.1016/j.crvi.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Pezo V., Liu F. W., Abramov M., Froeyen M., Herdewijn P., Marlière P. Angew. Chem., Int. Ed. Engl. 2013;52:8139–8143. doi: 10.1002/anie.201303288. [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.