Summary
Chromosome missegregation during mitosis or meiosis is a hallmark of cancer and the main cause of prenatal death in humans. The gain or loss of specific chromosomes is thought to be random, with cell viability being essentially determined by selection. Several established pathways including centrosome amplification, sister-chromatid cohesion defects, or a compromised spindle assembly checkpoint can lead to chromosome missegregation. However, how specific intrinsic features of the kinetochore—the critical chromosomal interface with spindle microtubules—impact chromosome segregation remains poorly understood. Here we used the unique cytological attributes of female Indian muntjac, the mammal with the lowest known chromosome number (2n = 6), to characterize and track individual chromosomes with distinct kinetochore size throughout mitosis. We show that centromere and kinetochore functional layers scale proportionally with centromere size. Measurement of intra-kinetochore distances, serial-section electron microscopy, and RNAi against key kinetochore proteins confirmed a standard structural and functional organization of the Indian muntjac kinetochores and revealed that microtubule binding capacity scales with kinetochore size. Surprisingly, we found that chromosome segregation in this species is not random. Chromosomes with larger kinetochores bi-oriented more efficiently and showed a 2-fold bias to congress to the equator in a motor-independent manner. Despite robust correction mechanisms during unperturbed mitosis, chromosomes with larger kinetochores were also strongly biased to establish erroneous merotelic attachments and missegregate during anaphase. This bias was impervious to the experimental attenuation of polar ejection forces on chromosome arms by RNAi against the chromokinesin Kif4a. Thus, kinetochore size is an important determinant of chromosome segregation fidelity.
Keywords: kinetochore, mitosis, aneuploidy, centromere, chromosome congression, chromosome segregation, merotelic attachments, Indian muntjac
Graphical Abstract
Highlights
-
•
Centromere/kinetochore functional layers scale proportionally with centromere size
-
•
Kinetochore microtubule binding capacity scales with kinetochore size
-
•
Chromosome congression and bi-orientation are biased by kinetochore size
-
•
Error formation leading to chromosome missegregation is biased by kinetochore size
Drpic et al. use fibroblasts from female Indian muntjac, the mammal with the lowest known chromosome number (2n = 6), to show that chromosome congression and bi-orientation are biased by kinetochore size. Chromosomes with larger kinetochores are also biased to establish erroneous merotelic attachments and missegregate during anaphase. Thus, kinetochore size is an important determinant of chromosome segregation fidelity.
Introduction
Deviation from the normal chromosome number in a given species, also known as aneuploidy, arises through problems in chromosome segregation during mitosis or meiosis. Gain or loss of specific chromosomes can result in stable karyotypes, as in many human trisomies, or represent a permanently unstable condition known as chromosomal instability (CIN), as typically observed in human cancers [1]. The gain/loss of a particular chromosome is believed to be random, with prevalence of particular karyotypes being essentially determined by cell viability and selection. However, we currently do not know whether all chromosomes have the same probability to missegregate.
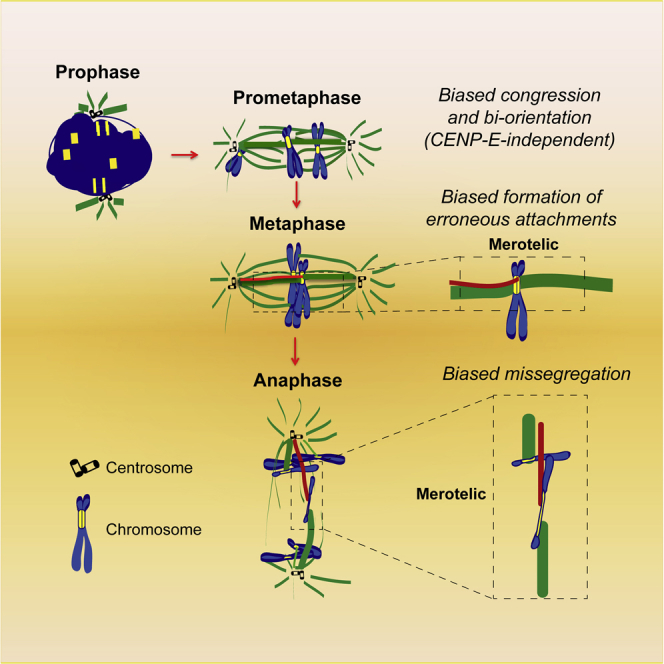
The kinetochore is a vital component required for chromosome segregation in eukaryotes because it establishes the interface with mitotic spindle microtubules. In mammals, the initial contacts between mitotic spindle microtubules and kinetochores take place during prometaphase, after nuclear envelope breakdown (NEB). Scattered chromosomes then align at the spindle equator by a process known as chromosome congression [2]. When chromosomes are favorably positioned between the spindle poles they establish end-on kinetochore-microtubule attachments and congress after bi-orientation. More peripheral chromosomes are laterally transported along spindle microtubules toward the equator by the plus-end-directed kinetochore motor CENP-E (kinesin-7) [3, 4]. Intriguingly, some metazoans, such as C. elegans, congress all their chromosomes in the absence of a CENP-E ortholog, but their kinetochores extend along the entire chromosome length [5]. How kinetochore size impacts the mechanisms of chromosome congression and segregation remains unknown.
Kinetochore size varies among different animal and plant species [5, 6, 7, 8, 9, 10, 11], between different chromosomes within a given species (including humans) [6, 12, 13, 14, 15, 16, 17], and in response to microtubule attachments throughout mitosis [18, 19]. Kinetochore size is primarily determined by the length of α-satellite DNA, the presence of CENP-B boxes, and the extent of incorporation of CENP-A at centromeres [20, 21, 22, 23]. Additionally, vertebrate kinetochores have an expandable module formed by proteins involved in spindle assembly checkpoint (SAC) signaling, motor proteins (e.g., CENP-E and cytoplasmic dynein), and microtubule regulating proteins (e.g., CLASPs) located in the fibrous corona [16, 18, 24, 25]. How the different centromere and kinetochore functional layers scale with centromere size has not been elucidated.
More recently, computational modeling predicted that adaptive changes in kinetochore size and shape play a critical role in chromosome orientation and error prevention during spindle assembly in human cells [19]. However, the physiological relevance of kinetochore size in chromosome segregation has not been experimentally evaluated due to technical limitations, as even the largest human kinetochores are not resolvable by conventional light microscopy in living cells.
To overcome these limitations, here we took advantage of the unique cytological features of the Indian muntjac, a small deer whose females have the lowest known chromosome number (2n = 6) in mammals [26]. Due to centromere-telomere and centromere-centromere tandem fusions during evolution [27], Indian muntjac chromosomes are large and morphologically distinct, with one pair of acrocentric chromosomes (chromosomes 3+X) containing an unusually large compound kinetochore [8, 26, 28]. We show that Indian muntjac cells are amenable for both pharmacological inhibition and genetic manipulation by RNAi. These capacities, combined with high-resolution live-cell and fixed-cell microscopy, allowed us to demonstrate that chromosome congression and segregation in this placental mammal are not random and are strongly biased by kinetochore size. The implications for chromosome segregation fidelity in metazoans are discussed.
Results
Indian Muntjac Centromere and Kinetochore Functional Layers Scale Proportionally with Centromere Size
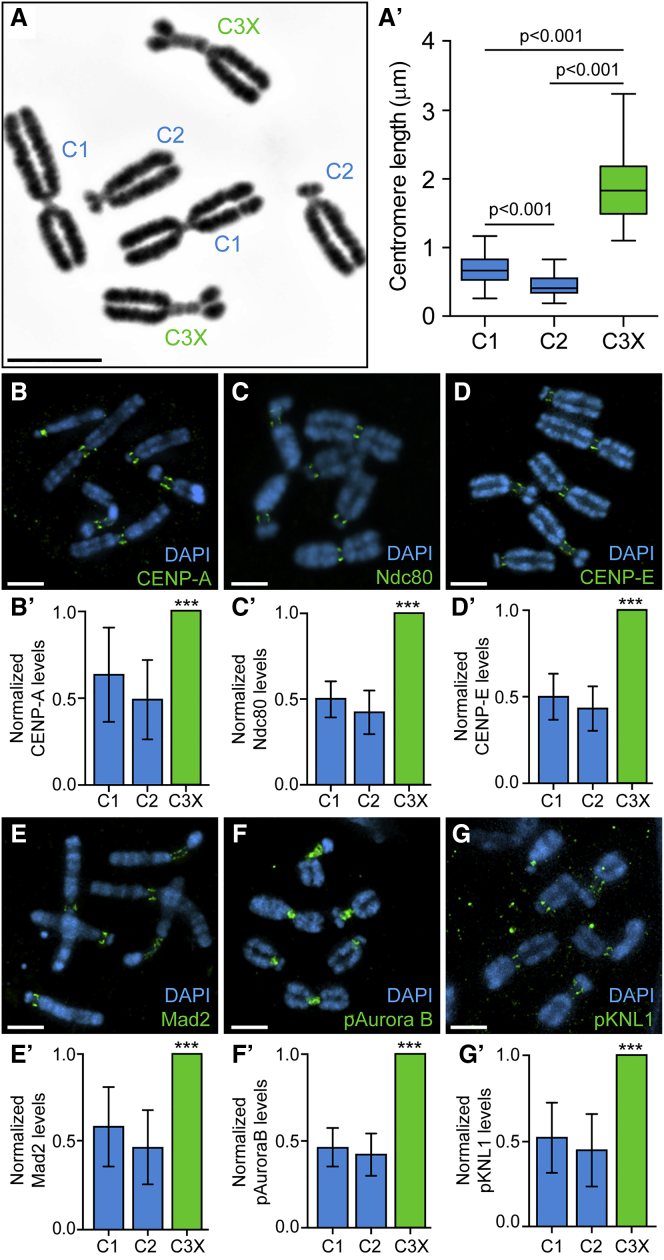
To characterize Indian muntjac kinetochores, we started by measuring their centromere length using chromosome spreads from female hTERT-immortalized primary fibroblasts [29]. In agreement with previous reports [8, 28], chromosome 3+X contained a distinctively large centromere, measuring 1.87 ± 0.47 μm (mean ± SD; n = 85 kinetochores, 40 cells) (Figures 1A and 1A′). The centromeres of chromosomes 1 and 2 were significantly smaller, measuring 0.68 ± 0.20 μm (n = 90 kinetochores) and 0.45 ± 0.15 μm (n = 77 kinetochores), respectively (Figures 1A and 1A′). To determine how different structural and functional centromere and kinetochore proteins scale with centromere length, we used fluorescence microscopy in chromosome spreads. Namely, we quantified the total levels of inner and outer kinetochore proteins involved in kinetochore assembly and end-on kinetochore-microtubule attachments (CENP-A and Ndc80/Hec1, respectively), SAC response (Mad2), as well as fibrous corona motor proteins (CENP-E). Additionally, we have also quantified the levels of active Aurora B on centromeres [30] and of one of its phosphorylated kinetochore substrates (pKNL1 [31]). We found that all these proteins scaled proportionally with centromere size in the absence of microtubules (Figures 1B–1G and 1B′–1G′).
Figure 1.
Centromere and Kinetochore Functional Layers Scale Proportionally with Centromere Size
(A) Normal karyotype of female Indian muntjac fibroblasts. Scale bar, 5 μm.
(A′) Centromere length of Indian muntjac chromosomes (box-whisker plots, n = 40 cells, ∼80 kinetochores per chromosome type, Mann-Whitney rank-sum test, p < 0.001 for all comparisons).
(B–G) Immunofluorescence of Indian muntjac chromosome spreads (blue) and the centromere and kinetochore proteins (green) CENP-A (B), Ndc80 (C), CENP-E (D), Mad2 (E), pAurora B (F), and pKNL1 (G). Scale bars, 5 μm.
(B′–G′) Respective quantification of protein levels at Indian muntjac kinetochores, relative to chromosome 3+X (C3X) for CENP-A (B′), Ndc80 (C′), CENP-E (D′), Mad2 (E′), pAurora B (F′), and pKNL1 (G′) (mean ± SD, n ≥ 37 cells per condition, ∼100 kinetochores per chromosome type, ∗∗∗p < 0.001 relative to controls, Mann-Whitney rank-sum test).
Indian Muntjac Kinetochores Show Standard Structural Organization and Their Microtubule Binding Capacity Scales with Kinetochore Size
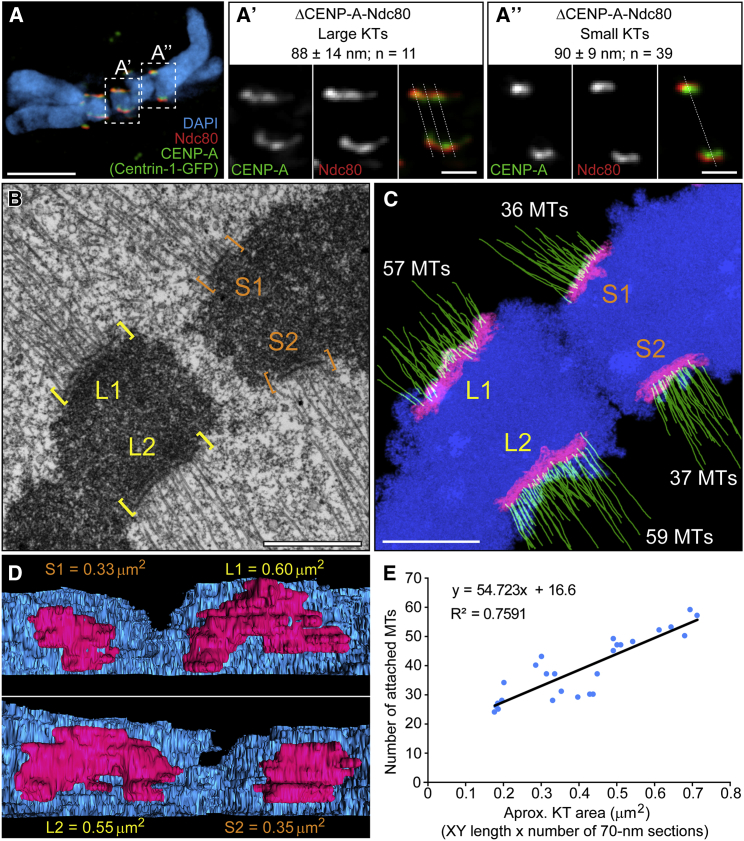
To get insight into the structural organization of Indian muntjac kinetochores, we measured the corresponding distances between CENP-A and Ndc80/Hec1 (also known as Delta [32]) in small and large kinetochores from aligned bi-oriented chromosomes (Figure 2A). We found that intra-kinetochore distances in small and large kinetochores were identical, suggesting similar molecular organization (Figures 2A′ and 2A″). It is noteworthy that scanning each large kinetochore in three different places resulted in a Delta range that was similar to the one observed in small kinetochores, consistent with a repetitive sub-unit organization [33].
Figure 2.
Indian Muntjac Kinetochores Show Typical Structural Organization and Their Microtubule Binding Capacity Scales with Kinetochore Size
(A) Selected optical planes from an Indian muntjac fibroblast stably expressing Centrin-1-GFP to label the centrioles (green), showing kinetochore pairs for C3X (A′) and neighbor chromosome with smaller (A″) centromere. The inner and outer parts of the kinetochores were delineated by CENP-A (green) and Ndc80/Hec1 (red). DNA was counterstained with DAPI (blue).
(A′ and A″) Higher-magnification views of C3X (A′) and smaller kinetochores (A″). Dashed lines denote where intra-kinetochore distances were measured. Scale bars, 5 μm (A) and 1 μm (A′ and A″). Differences between large and small kinetochores were not statistically significant (t test). KT, kinetochore.
(B) Single electron microscopy section from consecutive series highlighting the standard organization of the Indian muntjac centromere and kinetochore plates. L1 and L2 correspond to the plates on chromosome C3X; S1 and S2 correspond to the plates on a neighboring chromosome with smaller kinetochores. Scale bar, 2 μm.
(C) Z projection of the entire volume of the corresponding series shown in Figure S1. K fibers on the C3X chromosome comprise a larger number of microtubules (green). Kinetochore plates (magenta) and chromosomes (blue) are indicated. Scale bar, 1 μm. MT, microtubule.
(D) Surface-rendered model of the volume shown in Figure S1. C3X kinetochores are approximately twice as large as in chromosomes with smaller kinetochores.
(E) Quantification of the number of attached microtubules as a function of the approximate kinetochore area. Plot shows serial-section electron microscopy data from 26 kinetochores from 13 chromosomes and 3 cells.
See also Figure S1.
Next, we performed serial-section electron microscopy of metaphase Indian muntjac chromosomes. Both small and large kinetochores displayed expected trilaminar plates adjacent to centromeric heterochromatin and end-on attached microtubules (Figures 2B and S1). Manual tracing and projection of all microtubules whose ends terminate at the kinetochores demonstrated that the large kinetochores from chromosome 3+X bind more microtubules than smaller kinetochores in Indian muntjac (Figures 2C and S1). 3D surface rendering of entire kinetochore volumes indicated that the size differences among metaphase Indian muntjac chromosomes are maintained upon microtubule attachments and that large attached kinetochores were often slightly bent in response to spindle forces (Figure 2D). Quantification of the total number of attached microtubules per kinetochore from 26 serial-sectioned kinetochores (13 chromosomes, 3 cells) revealed a strong positive correlation between the number of kinetochore microtubules and the respective kinetochore area (Figure 2E).
The Molecular Landscape Required to Establish Functional Kinetochore-Microtubule Attachments Is Conserved in Indian Muntjac
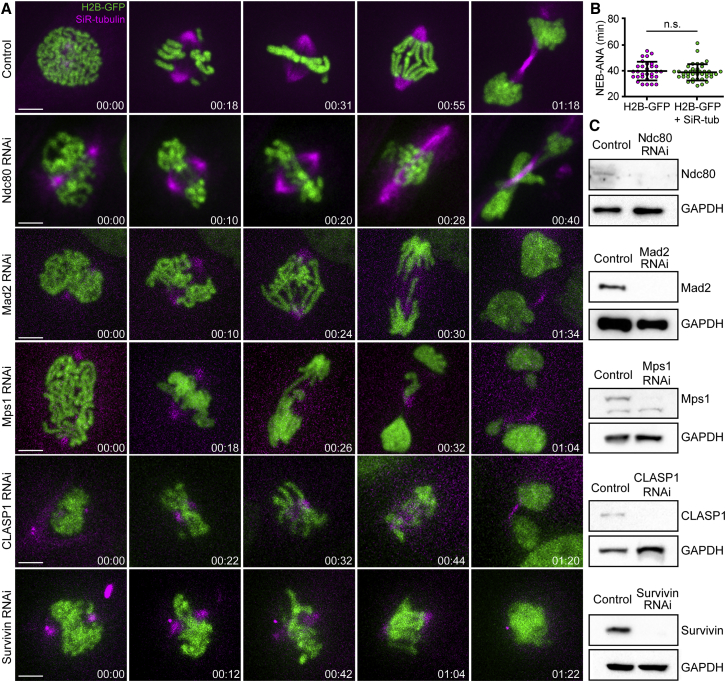
To investigate whether Indian muntjac kinetochores are functionally equivalent to other placental mammals, such as humans, we used RNAi to deplete representative centromere and kinetochore components involved in the establishment of end-on kinetochore-microtubule attachments (Ndc80 complex), SAC response (Mad2 and Mps1), the regulation of kinetochore-microtubule dynamics (CLASP1), and error correction (chromosomal passenger complex). To visualize chromosomes and microtubules in living cells, we stably expressed histone H2B-GFP and used 20–50 nM SiR-tubulin [34], which did not interfere with normal mitotic progression and chromosome segregation (Figures 3A and 3B; Video S1). Depletion of Ndc80 resulted in massive chromosome missegregation (Figures 3A and 3C; Video S1). As expected, Mad2 or Mps1 depletion accelerated the onset of anaphase and led to the formation of lagging chromosomes, whereas CLASP1 RNAi resulted in short spindles (Figures 3A and 3C; Video S1). Finally, depletion of the chromosomal passenger complex protein Survivin caused an overall defect in spindle assembly and incapacity to segregate chromosomes during anaphase, followed by cytokinesis failure and polyploidy (Figures 3A and 3C; Video S1). Overall, these results suggest that the molecular landscape required to establish functional kinetochore-microtubule attachments is conserved between Indian muntjac and humans.
Figure 3.
The Molecular Landscape Required to Establish Functional Kinetochore-Microtubule Attachments Is Conserved in Indian Muntjac
(A) Live-cell imaging of Indian muntjac fibroblasts stably expressing H2B-GFP to visualize the chromosomes (green) and treated with 50 nM SiR-tubulin to label spindle microtubules (magenta). Ndc80, Mad2, Mps1, Clasp1, and Survivin were knocked down by RNAi. Scale bars, 5 μm. Time, hr:min.
(B) Mitotic timing of Indian muntjac fibroblasts stably expressing H2B-GFP with or without addition of 50 nM SiR-tubulin. There is no statistically significant difference in mitotic timing from NEB to anaphase onset (ANA) in the presence or absence of SiR-tubulin (Mann-Whitney rank-sum test, p = 0.591). n.s., not significant.
(C) Protein lysates obtained after RNAi were immunoblotted with an antibody specific to each protein of interest. GAPDH was used as loading control.
See also Video S1.
Any Chromosome May Use Either the CENP-E-Dependent or -Independent Pathway to Congress, Regardless of Kinetochore Size
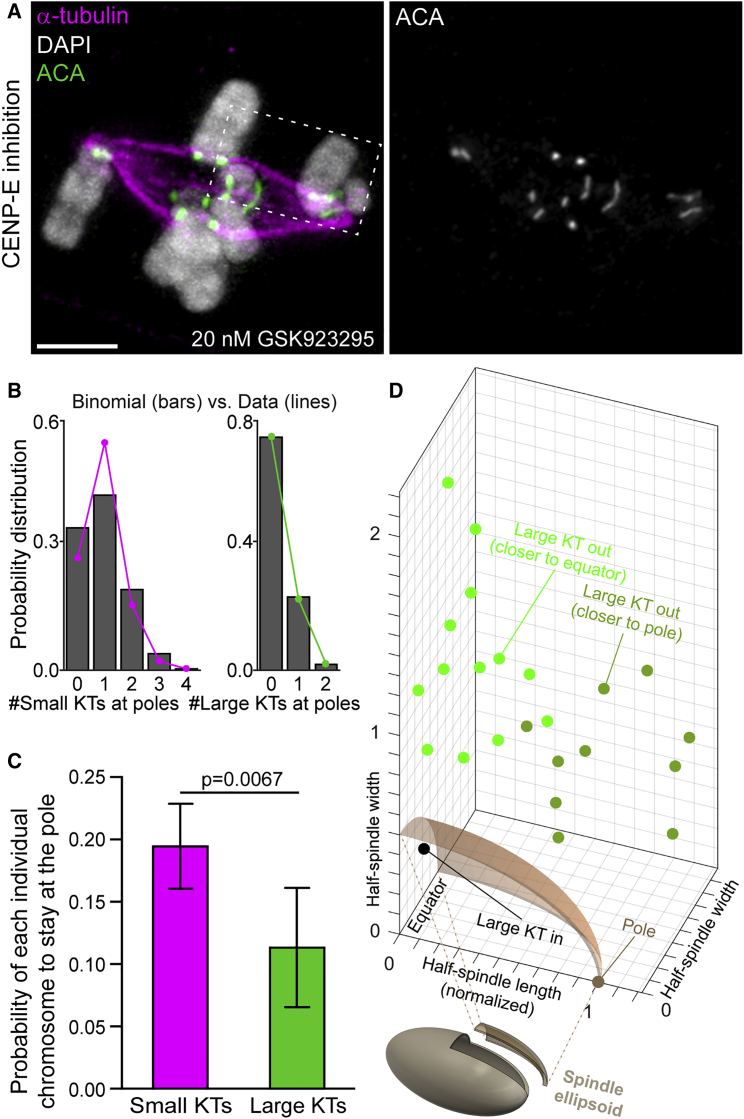
Next, we investigated whether chromosome congression in Indian muntjac fibroblasts also relied on motor-dependent and -independent pathways. To do so, we inhibited the kinetochore motor CENP-E for 1 hr with 20 nM GSK923295 [35], which more than doubled the normal frequency of mitotic cells with chromosomes at the poles, consistent with inhibition of CENP-E (Figure S2) [4]. Importantly, increasing the amount of CENP-E inhibitor by an order of magnitude did not result in further increase of mitotic cells with chromosomes at the poles, suggesting full inhibition of CENP-E motor activity at 20 nM, without displacing endogenous CENP-E from kinetochores (Figure S2).
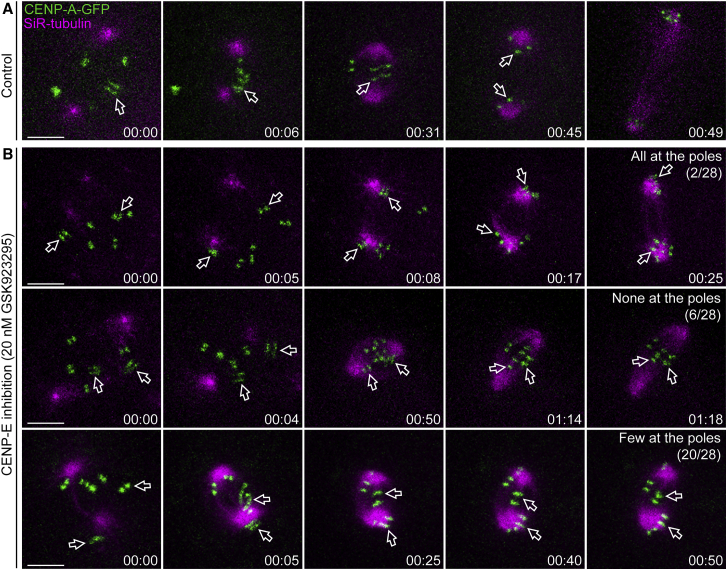
To directly test the implications of kinetochore size for chromosome congression, we followed mitosis in Indian muntjac fibroblasts stably expressing CENP-A-GFP after CENP-E inhibition (Figure 4; Video S2). We found three different scenarios: (1) very few cells showed all chromosomes at the poles (2/28 cells, six independent experiments); (2) some cells aligned all their chromosomes at the metaphase plate soon after NEB (6/28 cells, six independent experiments); and (3) most cells showed at least one chromosome, either with a small or large kinetochore, that remained at the poles (20/28 cells, six independent experiments). These data demonstrate that any chromosome may use either the CENP-E-dependent or -independent pathway to congress, regardless of kinetochore size.
Figure 4.
Any Chromosome May Use Either the CENP-E-Dependent or CENP-E-Independent Pathway to Congress, Regardless of Kinetochore Size
(A) Control Indian muntjac fibroblasts stably expressing CENP-A-GFP (green) and treated with 20 nM SiR-tubulin (magenta). Scale bar, 5 μm. Time, hr:min.
(B) Indian muntjac fibroblasts stably expressing CENP-A-GFP (green) and treated with 20 nM SiR-tubulin (magenta) after CENP-E inhibition with 20 nM GSK923295. Scale bars, 5 μm. Time, hr:min. n = 28 cells, pool of six independent experiments.
The arrows indicate the position of large kinetochores from C3X chromosomes. See also Figure S2 and Video S2.
Chromosome Congression and Bi-orientation in Indian Muntjac Are Biased by Kinetochore Size
To determine the number of chromosomes with small or large kinetochores at the pole after CENP-E inhibition, we performed immunofluorescence in fixed cells (Figure 5A). We found that the number of chromosomes with small or large kinetochores at the pole followed an almost perfect binomial distribution (Figure 5B). This indicated that the fate of each individual chromosome was largely independent of the other chromosomes of the same class and that the state of the chromosomes with small kinetochores did not influence the state of the chromosomes with large kinetochores, and vice versa. Most strikingly, the probability of each individual chromosome with small kinetochores to stay at the pole was approximately twice the probability of a chromosome with a large kinetochore: 0.19 ± 0.034 versus 0.11 ± 0.048, respectively (mean ± SD) (Figure 5C).
Figure 5.
Chromosome Congression and Bi-orientation in Indian Muntjac Are Biased by Kinetochore Size
(A) Immunofluorescence of an Indian muntjac fibroblast after CENP-E inhibition showing chromosomes (DAPI; white in merged image), kinetochores (ACA, white; green in merged image), and microtubules (α-tubulin; magenta in merged image). Scale bar, 5 μm.
(B) Quantification of the number of chromosomes with small or large kinetochores at the pole after CENP-E inhibition by immunofluorescence in fixed cells (magenta and green lines) and respective theoretical prediction based on a binomial distribution (gray bars).
(C) Probability of each individual chromosome with small or large kinetochores to stay at the pole upon CENP-E inhibition (arbitrary units) (mean ± SD, n = 621 cells, six independent experiments, p = 0.0067, t test).
(D) 4D (x, y, z, t) tracking of chromosomes with large kinetochores after CENP-E inhibition to determine their position relative to the poles at NEB and the forming mitotic spindle (see dashed box in A for reference). Note that chromosomes with large kinetochores are randomly distributed relative to the equator and the spindle poles.
In human cells, >96% of the chromosomes relying on CENP-E for congression are normally excluded from the spindle region and locate closer to one of the spindle poles at NEB [4]. To exclude that the observed bias for Indian muntjac chromosomes with large kinetochores to align independent of CENP-E was due to a tendency to localize in the spindle region and/or equidistantly to the spindle poles at NEB, we performed four-dimensional (4D, x, y, z, t) tracking of chromosomes with large kinetochores after CENP-E inhibition in living cells (n = 23 large kinetochore pairs, 13 cells). We found that 22/23 Indian muntjac chromosomes with large kinetochores were excluded from the spindle ellipsoid region and were nearly randomly positioned along the spindle axis at NEB (45% of the kinetochores were closer to the poles versus 55% of the kinetochores that were closer to the spindle equator; Figure 5D; Video S3). Overall, these data indicate that chromosomes with a larger kinetochore rely less on CENP-E motor activity and are biased to congress after bi-orientation, independent of chromosome positioning relative to the spindle region and poles at NEB.
Chromosomes with Larger Kinetochores Are More Prone to Establish Erroneous Merotelic Attachments that Result in Non-random Missegregation
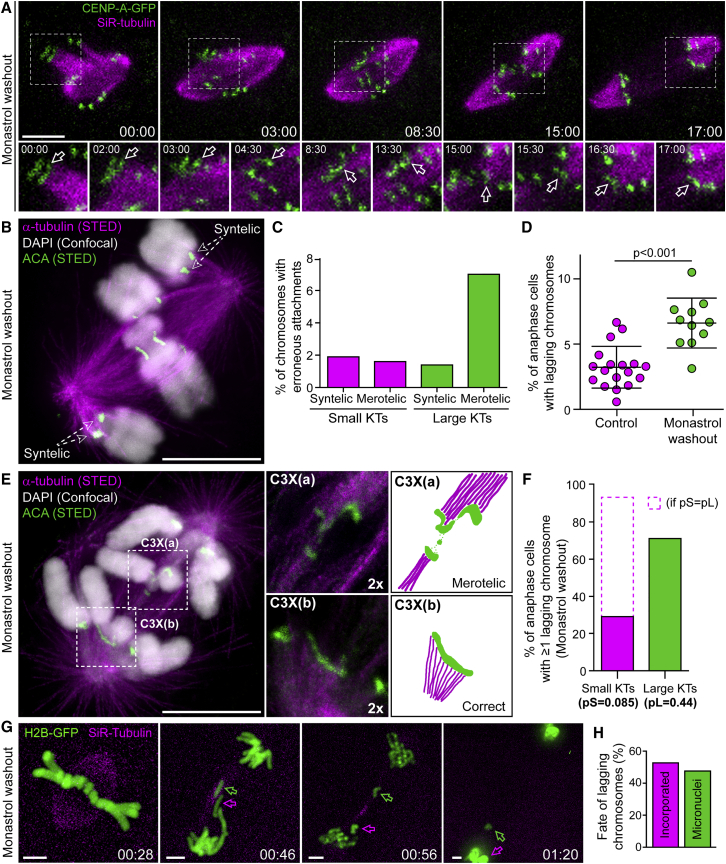
To directly investigate whether chromosomes with large kinetochores are more prone to establish erroneous attachments with spindle microtubules, we set up a monastrol treatment/washout assay in Indian muntjac fibroblasts (Figures 6A, 6B, S3, and S4; Video S4; see STAR Methods). Calculation of the fraction of each chromosome group (with small or large kinetochores) with merotelic (same kinetochore attached to microtubules from both poles) or syntelic (microtubules from the same pole attached to both sister kinetochores) attachments in fixed cells revealed a low frequency of syntelic attachments for chromosomes with either small or large kinetochores (1.9% versus 1.4%, respectively) (Figure 6C). However, the frequency of chromosomes with large kinetochores that established erroneous merotelic attachments was several-fold higher when compared with chromosomes with small kinetochores (7.0% versus 1.6%, respectively) (Figure 6C).
Figure 6.
Chromosomes with Larger Kinetochores Are More Prone to Establish Erroneous Merotelic Attachments that Result in Non-random Missegregation
(A) Error correction after monastrol washout in live Indian muntjac fibroblasts stably expressing CENP-A-GFP (green) and treated with 20 nM SiR-tubulin (magenta). Dashed boxes highlight a region with a chromosome with large kinetochores (arrows in lower panels that show 1.5× zoom images, plus additional time frames). Scale bar, 5 μm. Time, min:s.
(B) STED/confocal image of a prometaphase Indian muntjac fibroblast after monastrol washout showing syntelic attachments. Microtubules (α-tubulin, magenta), chromosomes (DAPI, white), and kinetochores (ACA, green) are indicated. Scale bar, 5 μm.
(C) Quantification of erroneous attachments on chromosomes with small or large kinetochores (KTs) (n = 207 cells, pool of three independent experiments).
(D) Frequency of anaphase cells with lagging chromosomes in controls and after monastrol washout (mean ± SD; each data point indicates an independent experiment; 2,099 control anaphase cells scored; 3,739 anaphase cells scored after monastrol washout; Mann-Whitney rank-sum test).
(E) STED/confocal image of an Indian muntjac fibroblast in anaphase after monastrol washout. Microtubules (α-tubulin, magenta), chromosomes (DAPI, white), and kinetochores (ACA, green) are indicated. Dashed boxes indicate a lagging chromatid (C3X(a)) containing a large kinetochore with merotelic attachments and the corresponding sister (C3X(b)). Scale bar, 5 μm. The images and graphical sketches on the right highlight the type of attachments in the two sisters (2× zoom).
(F) Frequency of anaphase cells with at least 1 lagging chromosome with small or large kinetochores after monastrol washout (n = 32 cells from nine independent experiments). Dashed bar represents theoretical values for the frequency of lagging chromosomes with small kinetochores, if the probability to lag was equal for chromosomes with large or small kinetochores.
(G) Live-cell imaging of an Indian muntjac fibroblast stably expressing H2B-GFP (green) and treated with 50 nM SiR-tubulin (magenta) illustrating missegregation of lagging chromosomes after monastrol washout. Scale bars, 5 μm. Time, hr:min. The green and magenta arrows indicate two lagging chromosomes that failed to integrate and reintegrated the main nucleus, respectively.
(H) Percentage of cells (from live-cell imaging) with lagging chromosomes incorporating or forming micronuclei after monastrol washout (n = 59 cells, pool of five independent experiments).
See also Figures S3–S5 and Videos S4, S5, and S6.
Because, when challenged, chromosomes with large kinetochores tended to establish merotelic attachments, we investigated whether they lagged more in anaphase. Immunofluorescence analysis in untreated Indian muntjac fibroblasts showed a low frequency (particularly at low passages) of spontaneously lagging chromosomes in anaphase (3.22% ± 1.60%, mean ± SD, different passages) (Figure 6D), suggesting that correction mechanisms are usually robust to prevent chromosome missegregation during normal mitosis. Nevertheless, despite the small number of anaphase cells where the exact number of chromosomes with small or large kinetochores could be unequivocally determined (n = 22 cells, pool of seven independent experiments) and the fact that 2/3 of all chromosomes in Indian muntjac have smaller kinetochores, we observed a higher probability (see STAR Methods) for chromosomes with large kinetochores (pL = 0.34) to lag in anaphase, when compared with chromosomes with small kinetochores (pS = 0.14).
To further evaluate the significance of the previous observations and increase our sample size, we promoted the formation of erroneous kinetochore-microtubule attachments by monastrol treatment/washout. As expected, this treatment doubled the frequency of anaphase cells with lagging chromosomes relative to unperturbed controls (6.62% ± 1.91%, mean ± SD) (Figure 6D). Stimulated emission depletion (STED) super-resolution microscopy indicated that the large kinetochores on anaphase lagging chromosomes were often found stretched and deformed due to the formation of merotelic attachments (Figure 6E). Most strikingly, we found that 73% of the anaphase cells after monastrol treatment and washout showed at least one lagging chromosome with a large kinetochore, whereas only 30% of the anaphase cells showed at least one lagging chromosome with small kinetochores (Figure 6F). This corresponds to a much higher probability of chromosomes with large kinetochores to lag in anaphase, when compared with chromosomes with small kinetochores (pL = 0.44; pS = 0.085) (see STAR Methods). In fact, if chromosomes with small or large kinetochores had equal probabilities to lag behind in anaphase, one would predict a much higher frequency of chromosomes with small kinetochores to lag in anaphase than the one observed experimentally (94% versus 30%, respectively; Figure 6F; see STAR Methods). Importantly, because chromosome 3+X in female Indian muntjac is smaller than chromosome 1 (which has a smaller kinetochore) but larger than chromosome 2 (also with a smaller kinetochore), these work as internal controls to exclude that the measured bias for chromosome 3+X to lag behind in Indian muntjac was related to chromosome size [36]. Finally, we tracked the fate of lagging chromosomes after monastrol washout in living fibroblasts and found that ∼50% resulted in the formation of micronuclei, a bona fide indicator of chromosome missegregation that has been implicated in chromosome rearrangements in human cancers [37] (Figures 6G and 6H; Video S5). We concluded that, despite robust error correction mechanisms during a normal mitosis, chromosomes with large kinetochores have a higher tendency to establish persistent merotelic attachments, resulting in a strong bias to lag behind in anaphase, potentially leading to missegregation.
Preventing Error Correction Also Generates a Missegregation Bias toward Chromosomes with Large Kinetochores
To test whether chromosomes with large kinetochores also missegregate at a higher frequency when error correction is prevented, we inhibited SAC activity with the Mps1 inhibitor Mps1-IN-1 [38]. Similar to its depletion by RNAi (Figures 3A and 3C), Mps1 inhibition with 20 μM Mps1-IN-1 forced cells to prematurely enter anaphase, resulting in a marked increase of cells with lagging chromosomes (10.3%, scored from fixed material) (Figures S5A–S5C; Video S6). Interestingly, we found that after Mps1 inhibition, 50% of the anaphase cells showed at least one lagging chromosome with a large kinetochore, whereas 56% of the anaphase cells showed at least one lagging chromosome with small kinetochores (Figure S5D). This corresponded to a higher probability of chromosomes with large kinetochores to lag in anaphase, when compared with chromosomes with small kinetochores (pL = 0.31 versus pS = 0.21; see STAR Methods). In other words, if chromosomes with small or large kinetochores had equal probabilities of lagging behind in anaphase after Mps1 inhibition, one would predict a frequency of 81% of anaphase cells with at least one lagging chromosome with small kinetochores (Figure S5D; see STAR Methods). Thus, preventing error correction also generates a missegregation bias toward chromosomes with large kinetochores.
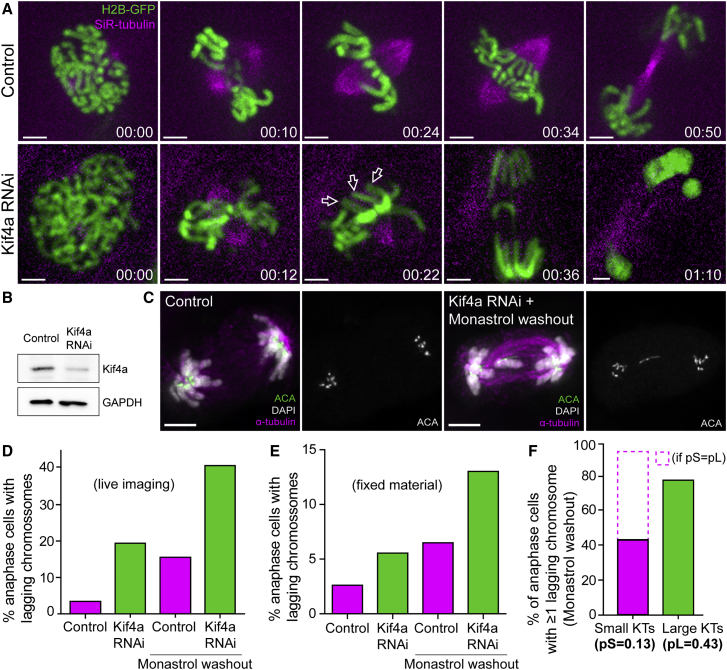
Polar Ejection Forces on Chromosome Arms Ensure Mitotic Fidelity but Are Not Implicated in the Observed Missegregation Bias for Chromosomes with Large Kinetochores
Previous reports in C. elegans have shown that loss of polar ejection forces after depletion of the kinesin-4 KLP-19 caused missegregation of holocentric chromosomes [39]. To test whether polar ejection forces acting on the long chromosome arms of Indian muntjac account for the observed missegregation bias, we have investigated chromosome segregation fidelity in fixed and living cells after RNAi against the chromokinesin Kif4a/kinesin-4 in Indian muntjac fibroblasts (Figures 7A–7C; Video S7). We found that experimental attenuation of Kif4a led to a striking increase in the frequency of lagging chromosomes in anaphase (Figures 7D and 7E; Video S7), consistent with a role of polar ejection forces in the modulation of kinetochore-microtubule attachments and chromosome segregation fidelity [39, 40, 41]. Importantly, chromosomes with large kinetochores still showed a strong bias to lag in anaphase after Kif4a RNAi (pL = 0.43 versus pS = 0.13) (Figures 7C and 7F). Conversely, if chromosomes with small or large kinetochores had equal probabilities to lag behind in anaphase after Kif4a RNAi, one would predict a frequency of 96% of anaphase cells with at least one lagging chromosome with small kinetochores, and not the experimentally observed value of 44% (Figure 7F; see STAR Methods). These results exclude the role of arm-associated forces in the observed missegregation bias and support that kinetochore size is the critical variable.
Figure 7.
Polar Ejection Forces on Chromosome Arms Ensure Mitotic Fidelity but Are Not Implicated in the Observed Missegregation Bias for Chromosomes with Large Kinetochores
(A) Live-cell imaging of Indian muntjac fibroblasts stably expressing H2B-GFP to visualize the chromosomes (green) in control (top) and Kif4a RNAi (bottom) cells treated with 50 nM SiR-tubulin to label spindle microtubules (magenta). Scale bar, 5 μm. Time, hr:min. White arrows point to the chromosome arms facing the spindle poles.
(B) Western blot to monitor Kif4a levels after RNAi. GAPDH was used as loading control.
(C) Chromosome missegregation after Kif4a RNAi (fixed cells). Kinetochores (anti-ACA), α-tubulin, and DNA (DAPI) are indicated. Scale bars, 5 μm.
(D and E) Comparison of the frequency of anaphase cells with lagging chromosomes in live (D) and fixed (E) material after Kif4a depletion and/or monastrol washout.
(F) Frequency of anaphase cells with at least 1 lagging chromosome with small or large kinetochores after monastrol washout in Kif4a-depleted fibroblasts. Dashed bar represents theoretical values for frequencies of lagging chromosomes with small kinetochores if the probability to lag was equal for chromosomes with small or large kinetochores.
See also Video S7.
Discussion
Here we show how kinetochore size impacts chromosome congression and bi-orientation, error formation and correction, as well as chromosome segregation fidelity during mitosis. Accordingly, we found that chromosomes with large kinetochores bi-orient and congress more efficiently and depend less on the kinetochore motor CENP-E. Because chromosomes with large kinetochores have an increased surface (and possibly a more favorable shape) for potential interaction with microtubules, these results help to explain why certain species with holocentric chromosomes, such as C. elegans, can complete congression in the absence of a CENP-E ortholog [5]. It is noteworthy that this does not seem to be a peculiarity of C. elegans, because 9/14 unrelated species with holocentric chromosomes and sequenced genomes also lack a bona fide CENP-E ortholog (D.D. and H.M., unpublished data).
Importantly, having a large kinetochore surface that facilitates chromosome bi-orientation comes with a price: chromosomes with large kinetochores have a much higher tendency to establish erroneous merotelic attachments and missegregate during anaphase. This implies that chromosomes that use the CENP-E pathway for congression are less prone to missegregate, offering a plausible explanation for why the CENP-E pathway emerged during evolution. On the other hand, species with holocentric chromosomes would be expected to be highly prone to chromosome missegregation, but in C. elegans only ∼1% of wild-type anaphases show lagging chromosomes [42]. In agreement, electron microscopy reconstructions of the C. elegans spindle failed to reveal merotelic attachments, against what would have been intuitively predicted for a species with the highest possible kinetochore/chromosome ratio [43, 44]. Thus, chromosome segregation fidelity might be ensured by a species-specific optimal kinetochore size.
Because chromosomes with large kinetochores also establish more errors, this would work as a negative selective pressure to maintain chromosomes with large kinetochores during evolution, suggesting that the errors resulting from incorrect merotelic attachments are unlikely to be propagated. Indeed, error correction mechanisms during normal mitosis appear to be very robust, in agreement with our findings of low missegregation rates in unperturbed cells. Moreover, based on direct live-cell imaging, we found that, even when cells were challenged, lagging chromosomes in Indian muntjac fibroblasts were able to re-integrate the main nuclei in ∼50% of cases. This most likely results from error correction mechanisms that are in place during anaphase and involve mechanical forces that stretch and deform merotelic-attached kinetochores, as shown in other systems [45, 46]. It is noteworthy that any potential loss of a single chromosome in Indian muntjac females would represent the loss of 1/3 of the haploid genome, which would seriously compromise cell viability. In agreement, previous work reported that chromosome missegregation and aneuploidy in Indian muntjac primary fibroblasts were essentially limited to the smallest Y2 chromosome in males [47].
Although kinetochore dimensions vary at least 2-fold among human chromosomes [12, 13, 14, 15, 16, 17, 20, 21, 22], a legitimate question is whether kinetochore size differences have any functional implications for chromosome segregation in humans. The length of α-satellite DNA arrays on human centromeres varies more than 25-fold, ranging from 200 kb in the Y chromosome to >5 Mb in chromosome 18 [48], and this has been proposed to contribute to CENP-A incorporation, at least in some chromosomes [21]. In agreement, the Y chromosome, which carries very little genetic information, was shown to recruit significantly less CENP-A compared with any other chromosome [20, 21, 22] and to missegregate at elevated frequencies in human cells [23]. Moreover, the loss of the Y chromosome is the most common somatic alteration in men and is associated with shorter survival and higher risk of cancer [49]. Thus, in addition to the low genetic pressure to keep the Y chromosome in men, its smaller kinetochore might compromise the establishment of competent microtubule attachments and contribute to the high missegregation rate. At the other extreme, CENP-A domain expansion and overexpression have been linked with chromosome missegregation and genomic instability in human cancer cell models [21, 50, 51]. Our finding that all centromere and kinetochore functional layers and respective microtubule binding capacity scale with centromere size suggests that any alterations at the foundations of kinetochore assembly will translate into architectural changes with functional implications for chromosome segregation. Because the level of CENP-A incorporation into human kinetochores also correlates with chromosome size [20], one prediction from our studies that has been recently validated is that larger human chromosomes missegregate at higher frequencies [52].
The microtubule binding capacity of human kinetochores in metaphase (excluding the Y chromosome) has been reported to range between 12 and 24 microtubules in one study [53] and 13 and 22 microtubules in another study [54]. This has been interpreted as though all kinetochores on human chromosomes bind, on average, to 17 microtubules. However, this 2-fold variability might instead reflect the structural variability in kinetochore size among different human chromosomes [12, 13, 14, 15, 16, 17, 20, 22]. More recently, adaptive changes in kinetochore architecture as cells progress into metaphase were also proposed to play a critical role in chromosome orientation and error prevention during spindle assembly in human cells [19]. A remarkable human condition in which constitutive differences in kinetochore size might bias chromosome missegregation is the occurrence of dicentric chromosomes that remain active during mitosis. As in the Indian muntjac, these chromosomes have a “compound” centromere and kinetochore and were shown to have a much higher tendency to lag behind in anaphase when compared with their normal counterparts [55]. Taken together, the findings reported here about the role of kinetochore size in non-random chromosome (mis)segregation have broad implications for our current understanding of chromosome segregation in metazoans and highlight the importance of adaptive changes in kinetochore size for mitotic fidelity in humans.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human anti - centromere (CREST) | Fitzgerald | Cat#90C-CS1058 |
| Mouse anti - α-tubulin (clone B512) | Sigma-Aldrich | Cat#T5168 |
| Mouse anti - CENP-A | [22] | Lars E.T. Jansen |
| Rabbit anti - Hec1 | Abcam | Cat#ab3613 |
| Mouse anti - cMad2 | Santa Cruz | Cat#sc-65492 |
| Rabbit anti - Mad2: | Bethyl Laboratories | Cat#A300-301A |
| Rabbit anti - pAuroraB, (pT232) | Rockland | Cat#600-401-677 |
| Rabbit anti – AuroraA | Novus Biologicals | Cat#NB100-267 |
| Mouse anti – GAPDH | Proteintech | Cat#60004-1-Ig |
| Rabbit anti - phospho-histone H3 (S10) | Cell Signaling | Cat#3377 |
| Rabbit anti – survivin | Novus Biologicals | Cat#NB500-201 |
| Rabbit anti - pKNL1 (clone 58A) | [31] | Iain Cheeseman |
| Sheep anti - CENP-E | [56] | William C. Earnshaw |
| Rabbit anti - kif4a | Thermo Fisher Scientific | Cat#pa5-30492 |
| Abberior donkey anti-human IgG STAR 580 | Abberior Instruments | Cat#D-08-2015Hp |
| Abberior goat anti- mouse IgG STAR 635p | Abberior Instruments | Cat#S-11-2015Hp |
| SiR-Tubulin | Spirochrome | Cat#CY-SC002 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Monastrol | Tocris bioscience | Cat#1305 |
| ZM447439 (Aurora B inhibitor) | Selleckchem.com | Cat#S1103 |
| MNL8054 (Aurora A inhibitor) | Selleck | Cat#869363-13-3 |
| AZ 3146 (Mps1 inhibitor) | Tocris bioscience | Cat#3994 |
| GSK923295 (CENP-E inhibitor) | Selleckchem.com | Cat#S7090 |
| BSA 98% | Sigma-Aldrich | Cat#A3294-50G |
| Experimental Models: Cell Lines | ||
| Immortalized female Indian muntjac fibroblasts | [29] | Jerry W Shay |
| Indian muntjac- H2B-GFP (female) | In this paper | N/A |
| Indian muntjac- CENP-A-GFP (female) | In this paper | N/A |
| Oligonucleotides | ||
| 5′-GCAGACATTGAGAGAATAA-3′ (Ndc80) | Sigma-Aldrich | N/A |
| 5′-GCAGAATGGTTATACAAGT-3′ (Mad2) | Sigma-Aldrich | N/A |
| 5′-GCTGCTGTTGCTGATGCTT-3′ (CLASP1) | Sigma-Aldrich | N/A |
| 5′-GCGTCTCCACGTTTAAGAA-3′ (Survivin) | Sigma-Aldrich | N/A |
| 5′-GAACCGTCAGCAAGACAA-3′ (Kif4a) | Sigma-Aldrich | N/A |
| 5′-GCCAGGGACCTCATTTCAA-3′ (Aurora A) | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| H2B-GFP | Addgene plasmid # 11680 | Geoff Wahl lab |
| pSV-IRESneo3-CENP-A-EGFP | [57] | Patrick Meraldi |
| Software and Algorithms | ||
| Fiji/ImageJ | ImageJ | N/A |
| CellProfiler | [58] | CellProfiler (http://cellprofiler.org) |
| CellProfilerAnalyzer | [58] | CellProfiler (http://cellprofiler.org) |
| MatLab8.1 | The MathWorks | https://www.mathworks.com/products/matlab.html |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Helder Maiato (maiato@i3s.up.pt).
Experimental Model and Subject Details
Cell culture
In all experiments we used hTERT-immortalized female Indian muntjac (IM) fibroblasts (kind gift from Jerry W Shay, University of Texas Southwestern Medical Center, Dallas, Texas) [29]. IM fibroblasts were grown in Minimum Essential Media (MEM) (GIBCO, Life Technologies), supplemented with 20% FBS (GIBCO, Life Technologies), 2mM L- Glutamine (Invitrogen) at 37°C in humidified conditions with 5% CO2. To collect IM fibroblasts we used Trypsin (GIBCO, Life Technologies). As all fibroblasts in general, hTERT-immortalized IM fibroblasts tended to become polyploid at higher passages. For this reason our analyses were focused only on diploid cells (2n = 6), which was controlled by counting the exact number of chromosomes or kinetochores, except for the determination of the absolute frequency of lagging chromosomes in anaphase.
Method Details
Cell transfection and transduction
IM fibroblasts were transfected either with human H2B-GFP (from Geoff Wahl lab, Addgene plasmid #11680) or pSV-IRESneo3-CENP-A-EGFP (kind gift from Patrick Meraldi, University of Geneva) [57] plasmids using Lipofectamine 2000 (Invitrogen) to generate stable cell lines. For this purpose, at day 1 cells were seeded in 6-well plates at 60%–70% confluence in MEM containing 20% FBS. The day after, cells were washed 3x with PBS and incubated with Optimem medium (GIBCO, Life Technologies) containing Lipofectamine 2000 (Invitrogen) and the respective DNA for 4 hr. Optimem with DNA and Lipofectamine 2000 were previously mixed and incubated for 20 min before adding to the cells. After 4 hr Optimem medium was exchanged to MEM supplemented with 20% FBS and transfected cells were selected with G418 (Merck Millipore) after 48 hr. For centriole labeling, cells were transfected with centrin1-GFP in LentiLox 3.7 [59]. Lentivirus were added to the standard culture media with 1:100 Polybrene (Sigma) for ∼12 hr. Stable lines with uniform level of expression and sufficient fluorescence intensity were selected by microscopy screening of individual clones generated by limited-dilutions.
Identification of Indian muntjac sequences
The protein sequences of human Ndc80, Mad2, Mps1, CLASP1, Survivin and Kif4a were obtained from NCBI and used as query for tblastn (version 2.2.29 [60]) using the IM genome scaffold sequences and predicted coding sequences as targets (H.A.L. and D.M.L., unpublished data). Sequence alignments with at least 80% identity, highest coverage of human genes, and with matching scaffold intervals from both tblastn runs were used to identify IM orthologs of gene human sequences.
Design of siRNAs for RNA interference (RNAi)
The design of the siRNA sequences was performed using the application BLOCK-ITTM RNAi Designer (ThermoFisher Scientific). We provided the nucleotide sequence of the genes of interest, selected an ideal CG percentage between 35%–55% and the recommended default motif pattern for the RNAi design. From the 10 designs generated, we selected the one with higher probability of knockdown.
RNAi experiments
In day 0, IM fibroblasts were cultured at 60%–70% confluence in 6-well plate/35 mm dishes. In day 1, the medium was changed to MEM supplemented with 5% FBS. Simultaneously, 5 μL of Lipofectamine RNAi Max (Invitrogen) and 50 nM of the respective siRNAs. The following target sequences were used: 5′-GCAGACATTGAGAGAATAA-3′ (Ndc80), 5′-GCAGAATGGTTATACAAGT-3′ (Mad2), 5′-CCAAGCAGTCACCACCAAT-3′ (Mps1), 5′-GCTGCTGTTGCTGATGCTT-3′ (CLASP1), 5′-GCGTCTCCACGTTTAAGAA-3′ (Survivin), 5′-GAACCGTCAGCAAGACAA-3′ (Kif4a) (Sigma-Aldrich). All siRNAs were diluted in 500 μL of Optimem and added to the cells. Mock transfection was used as control. The cells were analyzed 24 hr, 48 hr or 72 hr after depletion. Depletion efficiency was monitored by western blotting and phenotypic analysis.
Imunofluorescence
IM fibroblasts were seeded on poly-L-lysine-coated coverslips 2 days before the experiment. After fixation with ice-cold methanol (Invitrogen) for 4 min at −20°C or 4% paraformaldehyde (Electron Microscopy Sciences) for 10 min at room temperature (RT), cells were washed with PBS-0.05%Tween 20 (Sigma-Aldrich) or cytoskeleton buffer pH 6.1 (274 mM NaCl, 10 mM KCl, 2.2 mM Na2HPO4, 0.8 mM KH2PO4, 4 mM EGTA, 4 mM MgCl2, 10 mM Pipes, 10 mM Glucose). Extraction after paraformaldehyde fixation was performed using PBS-0.1%Triton (Sigma-Aldrich). Cells were incubated with primary antibodies in blocking solution (10% FBS diluted in PBS-0.05%Tween 20 (Sigma-Aldrich) or in cytoskeleton buffer pH 6.1) for 1 hr. The following primary antibodies were used: human anti-centromere antibodies (ACA) (1:2000; Fitzgerald), mouse anti-CENP-A (1:200, kind gift from Lars Jansen, Instituto Gulbenkian de Ciencia, Lisbon, Portugal) [22], mouse anti-α-tubulin (1:2000; B-512 clone, Sigma-Aldrich), rabbit anti-pH3(S10) (1:800, Santa Cruz Biotechnology), rabbit anti-pKNL1 58A (1:1000; kind gift from Iain Cheeseman, Whitehead Institute, MIT, Cambridge, MA, USA) [31], sheep anti-CENP-E (1: 1000, kind gift from William C. Earnshaw, Wellcome Trust Centre for Cell Biology, The University of Edinburgh, UK) [56], mouse anti-c-Mad2 (1:500; Santa Cruz), rabbit anti-pAuroraB (pT232) (1:1000; Rockland), mouse anti-Hec1/Ndc80 (9G3) (1:500, Abcam). Subsequently, cells were washed 3x with PBS-0.05% Tween or cytoskeleton buffer and incubated 45 min with the corresponding secondary antibodies Alexa 488, 568 and 647 (Invitrogen) or Abberior STAR 580 and Abberior STAR 635p (Abberior Instruments) for STED microscopy. For STED microscopy, both primary and secondary antibodies were used at 1:100 concentrations. After adding 1 μg/mL 4’6’-Diamidino-2-phenylindole (DAPI) (Sigma Aldrich) for 5 min, coverslips were washed in PBS and sealed on glass slides mounted with 20 mM Tris pH8, 0.5 N-propyl gallate, 90% glycerol.
Chromosome spreads
IM fibroblasts were incubated with 3.3 μM of nocodazole (Sigma-Aldrich) for 6-7 h, then trypsinized and centrifuged for 5 min at 1200 rpm. The pellet was resuspended in 500 μL of the supernatant, and a hypotonic solution (medium:water 1:1 and 3.3 μM nocodazole) was added drop by drop until the final volume of 5 mL. The mixture was incubated at 37°C for 20 min. After centrifugation the supernatant was discarded and the cells were fixed with Carnoy solution (methanol (AppliChem Panreac): acetic acid (Millipore Corporation) - 3:1) overnight at −20°C. The following day, the Carnoy fixation was repeated and cells were subsequently spread drop-by-drop onto a glass slide. DNA was counterstained with 1 μg/mL DAPI (Sigma-Aldrich) for 10 min and the preparations were mounted on 20 mM Tris pH8, 0.5 N-propyl gallate, 90% glycerol. For chromosome spreads with antibody staining IM fibroblasts were incubated with 3.3 μM nocodazole for 6-7 h, then trypsinized and centrifuged for 5 min at 1200 rpm. The pellet was resuspended in 500 μL of the hypotonic solution containing sodium citrate (Sigma-Aldrich) and 10% BSA (Sigma-Aldrich) and incubated for 30 min at 37°C. Cells were then placed on glass slides using a cytospin 4 centrifuge (Thermo Scientific). Glass slides containing chromosome spreads were fixed with 4% paraformaldehyde and immunofluorescence was performed as indicated.
Measurement of intra-kinetochore distances
Selected centromeres with co-planar kinetochores (intensity peaks separated by no more than 1 plane in series recorded at 200-nm Z-steps) were line-scanned. Two Gaussian peaks were detected in the scans via a MATLAB function developed by Dr. O’Haver (https://www.mathworks.com/matlabcentral/fileexchange/23611-peakfit–command-line-peak-fitting-function). Delta values were calculated by subtracting the distance between the centers of CENP-A peaks from the distance between the centers of Ndc80/Hec1 peaks and dividing the result by 2. This approach automatically compensates potential chromatic aberrations [32]. CENP-A was visualized with 3-19 mouse monoclonal antibody (Abcam) and Ndc80/Hec1 with the 9G3 monoclonal antibody (Abcam), both at 1:200 dilution. Although both 3–19 (CENP-A) and 9G3 (Ndc80/Hec1) antibodies were mouse monoclonal, they have different isotypes. 3–19 was followed by a γ1b-specific secondary antibody conjugated to Alexa Fluor 488, and 9G3 was followed by a γ2a-specific secondary antibody conjugated to Alexa Fluor 594, both at 1:100 dilution (Life Technologies).
Error correction/formation assays
To promote error formation, IM fibroblasts were seeded on poly-L-lysine-coated (Sigma-Aldrich) coverslips 2 days before the experiment. Cells were incubated for 12 hr with 100 μM monastrol (Tocris bioscience) and subsequently washed out into MEM medium or MEM containing 1 μM Aurora B inhibitor (Selleckchem) and 20 μM MG-132 (Calbiochem) for the next 50 min before fixation, based on previous reports [61]. To prevent error correction, cells were incubated with 20 μM Mps1-IN-1 [38] (kind gift from N. Gray, Dana-Farber Cancer Institute, Boston, MA, USA) for 15 min, prior to fixation. This concentration was previously assessed for the formation of lagging chromosomes in IM fibroblasts by live cell imaging. When indicated, cells were fixed with ice-cold methanol (Invitrogen) for 4 min at −20°C or 4% paraformaldehyde (Electron Microscopy Sciences) for immunofluorescence analysis.
CENP-E inhibitor titration
IM fibroblast stably expressing H2B-GFP were seeded in 96-well plates (10.000 cells per well) two days before imaging and kept in culture medium at 37°C in humidified conditions with 5% CO2. On the day of imaging, CENP-E inhibitor, GSK923295 (35) (Selleckchem) was added in triplicates at the following concentrations: 5 nM, 10 nM, 20 nM, 40 nM, 80 nM, 160 nM and 320 nM. Control wells were treated with DMSO only. Live cell imaging was performed using In Cell Analyzer 2000 (GE Healthcare Life Sciences) 1 hr after addition of the inhibitor. Images were analyzed with CellProfiler 2.2.0. and CellProfiler Analyst [58]. In accordance with the results obtained from the CENP-E inhibitor titration, IM fibroblasts were treated with 20 nM GSK923295 1 hr before fixation or live-cell imaging.
Live-cell imaging
IM fibroblasts stably expressing human CENP-A-GFP or H2B-GFP were plated on fibronectin (Sigma-Aldrich) coated 35 mm glass-bottom dishes (14 mm, No 1.5, MatTek Corporation) 2 days before imaging. Before live-cell imaging, cells were cultured in Leibovitz 's-L15 medium (GIBCO, Life Technologies). For tubulin staining, we used 20-50 nM SiR-tubulin cell-permeable dye [34] (Spirochrome) and incubated cells for 6-12 hr. Live-cell imaging was performed on a temperature-controlled Nikon TE2000 microscope equipped at the camera port with a modified Yokogawa CSU-X1 spinning-disc head (Solamere Technology), an FW-1000 filter-wheel (ASI) and an iXon+ DU-897 EM-CCD (Andor). The excitation optics are composed of two sapphire lasers at 488 nm and 647 nm (Coherent), which are shuttered by an acousto-optic tunable filter (Gooche&Housego, model R64040-150) and injected into the Yokogawa head via a polarization-maintaining single-mode optical fiber (OZ optics). Sample position is controlled by a motorized SCAN-IM stage (Marzhauser) and a 541.ZSL piezo (Physik Instrumente). The objective was an oil-immersion 60x 1.4 NA Plan-Apo DIC CFI (Nikon, VC series), yielding an overall (including the pinhole-imaging lens) 190 nm/pixel sampling. A 1.5x tube lens (optivar) was also used (126 nm/pixel sampling). Eleven 1 μm separated z stacks were acquired every 2 min while recording IM fibroblasts stably expressing H2B-GFP. For 4D kinetochore tracking we used IM fibroblasts stably expressing CENP-A-GFP, recorded at 30 s or 60 s interval and 0.75 μm separated z stack. The system was controlled by NIS-Elements via a DAC board (National Instruments, PCI-6733).
STED super-resolution microscopy
For STED imaging we used a pulsed gated-STED microscope (Abberior Instruments) with excitation wavelengths at 561 nm and 640 nm doughnut-depleted with a single laser at 775 nm. All acquisitions were performed using a 1.4 NA oil-immersion and a pixel size set to 35 nm.
Serial section electron microscopy
IM fibroblasts were fixed with 2.5% glutaraldehyde (Sigma) in PBS, pH7.4 for 30 min, rinsed with PBS (3 × 5 min), and post-fixed with 2% OsO4 in dH2O for 60 min at 4°C. The coverslips were then rinsed in dH2O, treated with 0.25% tannic acid for 20 min, and stained with 2% uranyl acetate for 60 min. Dehydration was achieved by a series of ethanol solutions (30-50-70-80%–96%, 10 min in each solution) followed by acetone (10 min). After dehydration, cells were embedded in Epon 812 and cured for 48 hr at 60°C. Serial 70-nm thin sections were cut with a diamond knife (Diatome) on a Leica Ultracut UCT ultramicrotome and stained with lead citrate. Images were obtained on a JEOL 1400 microscope operated at 80 kV using a side-mounted 4.0 Megapixel XR401 sCMOS AMT camera (Advanced Microscopy Techniques Corp). Full series of images recorded at 12K magnification were used to reconstruct the volume of the cell and match orientation and superimpose this volume on the corresponding LM dataset. Higher-magnification images (30-40K) were then collected for individual kinetochores. These high-magnification images were subsequently used to trace microtubules end-on attached to the kinetochores. 3-D volumes occupied by the kinetochores and adjacent chromatin were visualized as isosurface models in Amira 5.3.3 (Visage Imaging).
Western Blotting
IM fibroblasts were collected after trypsinization and centrifuged at 1200 rpm for 5 min. The pellet was resuspended in PBS and centrifuged, the cells were resuspended in 30-50 μL of Lysis Buffer (NP-40: 20 nM HEPES/KOH pH 7.9; 1 mM EDTA pH 8; 1 mM EGTA; 150 nM NaCl; 0.5% NP40; 10% glycerol, 1:50 protease inhibitor; 1:100 Phenylmethylsulfonyl fluoride). The samples were flash frozen in liquid nitrogen and kept on ice for 30 min. After centrifugation at 14000crpm for 15 min at 4°C, protein concentration was determined by Bradford protein assay (Bio-Rad). Protein lysates were run on 7.5/10/15% SDS-PAGE (25-40 μg/lane) and transferred to a nitrocellulose Hybond-C membrane using an iBlot Gel Transfer Device (Thermo Scientific). Membranes were blocked in PBS 0.05% Tween with 5% milk and the primary antibodies were incubated overnight at 4c°C at the following dilutions: anti-Hec1/Ndc80 mouse anti-Hec1/Ndc80 (9G3); anti-Mad2 (rabbit, 1:500, Bethyl Laboratories); anti-CLASP1 (rat, 1:100 [62],), anti-Survivin (rabbit, 1:1000, Novus Biologicals), anti-Aurora A (rabbit, 1:1000, Novus Biologicals); anti-Kif4a (rabbit, 1:1000, Thermo Fisher Scientific); anti-GAPDH (mouse, 1:15000, Proteintech). After successive washes, the membrane was incubated with the secondary antibodies for 1 hr at RT (α-mouse-HRF; α-rabbit-HRF; α-sheep-HRP 1:5000). Detection was performed with Clarity Western ECL Substrate (Bio-Rad). Quantification of blots was performed with a Bio-Rad ChemiDoc XRS system using the IMAGELAB software and immunosignals were normalized to GAPDH expression.
Quantification and Statistical Analysis
Fixed image analysis and acquisition
Image acquisition (0.22 μm thick z stacks) was performed on a Zeiss AxioObserver Z1 wide-field microscope equipped with a plan-apochromatic (1.46 NA 60x) DIC objective and a cooled CCD (Hamamatsu Orca R2). Autoquant X (Media Cybernetics) was used for blind deconvolution. All images show maximum intensity projections. For classification of kinetochore-microtubule attachments, microtubules were traced through z stacks and the position of their ends determined relative to the kinetochore signal. In the case of merotelic attachments, kinetochore deformation and/or orientation were also used as secondary criteria. Protein levels (CENP-A, Ndc80/Hec1, CENP-E, Mad2, pKNL1 and pAuroraB) on chromosome spreads were analyzed using ROI manager in Fiji (ImageJ). For quantification of kinetochore protein levels in all chromosomes, fluorescence intensity for each protein was background subtracted and normalized for the levels obtained for chromosome X+3 in the same cell. Adobe Photoshop CS4 and Adobe Illustrator CS5 (Adobe Systems) were used for histogram adjustments and panel assembly for publication.
Frequency analysis and joint probability tables
Custom-made scripts were developed in MATLAB 8.1 (The MathWorks) to perform the frequency analysis for the number of chromosomes with small and large kinetochores staying at the pole upon CENP-E inhibition. Joint probability tables were calculated for six independent sets of mitotic cells, each with at least 100 cells. The tables were used to calculate the marginal and the conditional probabilities of the number of chromosomes with small/large kinetochores found at the pole. The random variables considered for the joint probability table were ‘S’, for the number of chromosomes with small kinetochores that stay at the pole, and ‘L’, for the number of chromosomes with large kinetochores that stay at the pole. Binomial distributions were fitted to both random variables, using information about the number of independent components (2 for large kinetochores, and 4 for small kinetochores) and the respective experimental values. All data are represented as the mean ± SD. Additional custom-made MATLAB scripts were developed to perform the frequency analysis on the number of lagging chromosomes during anaphase, according to the kinetochore size. Following a similar methodology as above, joint probability tables were calculated and used to obtain the marginal and the conditional probabilities of the number of lagging chromosomes with small and with large kinetochores. Experimental data was used to parameterize the associated probability distributions (binomial). The distributions were used to calculate the probability of having lagging chromosomes of a certain type, given that there was at least a lagging chromosome, under two different conditions: a) imposing equal values for the individual lagging probability, independently of the kinetochore size; and b) estimating the individual lagging probability for each chromosome type, constrained to the mean values of lagging chromosomes (of each type) observed experimentally. For control cells where the total number of chromosomes was not always 6 (4 chromosomes with small kinetochore and 2 with large kinetochore) the frequency analysis for the lagging chromosomes was not performed using joint probability tables. Given that a varying number of total chromosomes imposes important constraints to this approach, the frequency analysis was performed instead in terms of calculation of mean values for the fraction of chromosomes of each type that become lagging. This way, for each experiment, the total number of chromosomes with small and large kinetochores was accounted for to calculate a descriptive measurement, which is independent of the number of chromosomes.
Kinetochore tracking
Live-cell imaging of IM fibroblasts stably expressing CENP-A-GFP was performed as indicated, every 60 s, and analyzed after CENP-E inhibition using TrackMate Tool in Fiji (ImageJ). Initial kinetochore and pole positions at nuclear envelope breakdown were manually tracked in four dimensions (x,y,z,t) using Manual Tracking Tool. Further analyses and plotting were performed using MATLAB to assess the initial position of the chromosomes with large kinetochores relative to the spindle and spindle poles/equator. Data from different cells was pooled together by applying geometric affine transformations (without shear) to generate overlap for the poles location. Initial positions of the chromosomes with large kinetochores were plotted on a standardized geometrical representation of the mitotic spindle ellipsoid.
Statistical analysis
Statistical analysis was performed using SigmaStat 3.5 software. All data represent the mean ± SD. Statistical significance of differences between the population distributions was determined by Student’s t test. For data that did not follow a normal distribution, statistical analysis was performed using a Mann-Whitney Rank Sum test.
Acknowledgments
We would like to thank Sarah McClelland for discussions and for communicating results prior to publication. We also thank Geert Kops for sharing unpublished data on species with holocentric chromosomes, António Pereira for assistance with STED microscopy, and CID lab members for feedback on the manuscript and throughout this project. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement no. 681443) and FLAD Life Science 2020 (261/2014 to H.M.). A.K. was supported by NIH grant GM059363 and acknowledges the use of the Wadsworth Center’s Electron Microscopy Core Facility.
Author Contributions
D.D. established the Indian muntjac system. D.D. and A.C.A. performed or participated in all the experiments and analyzed the data, except the electron microscopy and Delta measurements. F.R. and A.K. performed the Delta measurements and electron microscopy. P.A. constructed the joint probability tables and performed frequency analysis and 4D tracking of kinetochores. H.M. conceived and supervised the project, designed the experiments, and analyzed the data. J.D., H.A.L., and D.M.L. provided Indian muntjac genome sequence information and identified conserved mitotic proteins in this system. D.D. and H.M. wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: April 26, 2018
Footnotes
Supplemental Information includes five figures and seven videos and can be found with this article online at https://doi.org/10.1016/j.cub.2018.03.023.
Supplemental Information
Live-cell imaging of hTERT-immortalized Indian muntjac fibroblasts stably expressing H2B-GFP to visualize the chromosomes (green) and treated with 50 nM SiR-tubulin to label spindle microtubules (magenta). Representative examples of control, Ndc80, Mad2, Mps1, Clasp1, and Survivin RNAi are shown. Time, hr:min.
Live-cell imaging of control (left) or GSK923295-treated (CENP-E inhibitor, 20 nM, 3 cells from the right) hTERT-immortalized Indian muntjac (IM) fibroblasts stably expressing CENP-A-GFP to visualize the kinetochores (green) and treated with 20 nM SiR-tubulin to label spindle microtubules (magenta). Time, hr:min.
Note that most kinetochore pairs (all green dots) occupy a position at the spindle periphery, but their distribution relative to the pole/equator (red dot) is random. One exceptional kinetochore pair that fell inside the spindle region is also indicated (black dot).
Monastrol treatment and washout in a live hTERT-immortalized Indian muntjac fibroblast stably expressing CENP-A-GFP to visualize the kinetochores (green) and treated with 20 nM SiR-tubulin to label spindle microtubules (magenta). Note the difference between chromosomes with a small or large kinetochore pair. One chromosome with a large kinetochore pair can be seen to change conformation and lag behind during anaphase for a short time, but eventually resolves and segregates to the correct daughter. Time, min:s.
Indian muntjac female fibroblast stably expressing H2B-GFP (green) treated with 20 nM SiR-tubulin (magenta) after monastrol washout. Time, min:s.
Control and Mps1-inhibited (20 μM Mps1-IN-1) Indian muntjac fibroblasts stably expressing H2B-GFP (green). Time, hr:min.
Control and Kif4a-RNAi-depleted Indian muntjac fibroblasts stably expressing H2B-GFP (green) with 50 nM SiR-tubulin (magenta). Time, hr:min.
References
- 1.Rutledge S.D., Cimini D. Consequences of aneuploidy in sickness and in health. Curr. Opin. Cell Biol. 2016;40:41–46. doi: 10.1016/j.ceb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Maiato H., Gomes A.M., Sousa F., Barisic M. Mechanisms of chromosome congression during mitosis. Biology (Basel) 2017;6:13. doi: 10.3390/biology6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barisic M., Aguiar P., Geley S., Maiato H. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 2014;16:1249–1256. doi: 10.1038/ncb3060. [DOI] [PubMed] [Google Scholar]
- 5.Maddox P.S., Oegema K., Desai A., Cheeseman I.M. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 6.McEwen B.F., Ding Y., Heagle A.B. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Res. 1998;6:123–132. doi: 10.1023/a:1009239013215. [DOI] [PubMed] [Google Scholar]
- 7.Maiato H., Hergert P.J., Moutinho-Pereira S., Dong Y., Vandenbeldt K.J., Rieder C.L., McEwen B.F. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 2006;115:469–480. doi: 10.1007/s00412-006-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comings D.E., Okada T.A. Fine structure of kinetochore in Indian muntjac. Exp. Cell Res. 1971;67:97–110. doi: 10.1016/0014-4827(71)90625-2. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro S.A., Gatlin J.C., Dong Y., Joglekar A., Cameron L., Hudson D.F., Farr C.J., McEwen B.F., Salmon E.D., Earnshaw W.C., Vagnarelli P. Condensin regulates the stiffness of vertebrate centromeres. Mol. Biol. Cell. 2009;20:2371–2380. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann P., Pavlíková Z., Koblížková A., Fuková I., Jedličková V., Novák P., Macas J. Centromeres off the hook: massive changes in centromere size and structure following duplication of CenH3 gene in Fabeae species. Mol. Biol. Evol. 2015;32:1862–1879. doi: 10.1093/molbev/msv070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malheiros N., de Castro D., Camara A. Cromosomas sem centrómero localizado. O caso da Luzula purpurea Link. Agron. Lusit. 1947;9:51–74. [Google Scholar]
- 12.Peretti D., Maraschio P., Lambiase S., Lo Curto F., Zuffardi O. Indirect immunofluorescence of inactive centromeres as indicator of centromeric function. Hum. Genet. 1986;73:12–16. doi: 10.1007/BF00292655. [DOI] [PubMed] [Google Scholar]
- 13.Cherry L.M., Johnston D.A. Size variation in kinetochores of human chromosomes. Hum. Genet. 1987;75:155–158. doi: 10.1007/BF00591078. [DOI] [PubMed] [Google Scholar]
- 14.Cherry L.M., Faulkner A.J., Grossberg L.A., Balczon R. Kinetochore size variation in mammalian chromosomes: an image analysis study with evolutionary implications. J. Cell Sci. 1989;92:281–289. doi: 10.1242/jcs.92.2.281. [DOI] [PubMed] [Google Scholar]
- 15.Sánchez L., Martínez P., Goyanes V. Analysis of centromere size in human chromosomes 1, 9, 15, and 16 by electron microscopy. Genome. 1991;34:710–713. doi: 10.1139/g91-109. [DOI] [PubMed] [Google Scholar]
- 16.Tomkiel J., Cooke C.A., Saitoh H., Bernat R.L., Earnshaw W.C. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nixon F.M., Honnor T.R., Clarke N.I., Starling G.P., Beckett A.J., Johansen A.M., Brettschneider J.A., Prior I.A., Royle S.J. Microtubule organization within mitotic spindles revealed by serial block face scanning electron microscopy and image analysis. J. Cell Sci. 2017;130:1845–1855. doi: 10.1242/jcs.203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., Salmon E.D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magidson V., Paul R., Yang N., Ault J.G., O’Connell C.B., Tikhonenko I., McEwen B.F., Mogilner A., Khodjakov A. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 2015;17:1134–1144. doi: 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irvine D.V., Amor D.J., Perry J., Sirvent N., Pedeutour F., Choo K.H., Saffery R. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 2004;12:805–815. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan L.L., Boivin C.D., Mravinac B., Song I.Y., Sullivan B.A. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19:457–470. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., Jansen L.E. The quantitative architecture of centromeric chromatin. eLife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fachinetti D., Han J.S., McMahon M.A., Ly P., Abdullah A., Wong A.J., Cleveland D.W. DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wynne D.J., Funabiki H. Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J. Cell Biol. 2015;210:899–916. doi: 10.1083/jcb.201506020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiato H., Fairley E.A., Rieder C.L., Swedlow J.R., Sunkel C.E., Earnshaw W.C. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- 26.Wurster D.H., Benirschke K. Indian muntjac, Muntiacus muntjak: a deer with a low diploid chromosome number. Science. 1970;168:1364–1366. doi: 10.1126/science.168.3937.1364. [DOI] [PubMed] [Google Scholar]
- 27.Chi J.X., Huang L., Nie W., Wang J., Su B., Yang F. Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac chromosomes by BAC mapping. Chromosoma. 2005;114:167–172. doi: 10.1007/s00412-005-0004-x. [DOI] [PubMed] [Google Scholar]
- 28.Brinkley B.R., Valdivia M.M., Tousson A., Brenner S.L. Compound kinetochores of the Indian muntjac. Evolution by linear fusion of unit kinetochores. Chromosoma. 1984;91:1–11. doi: 10.1007/BF00286479. [DOI] [PubMed] [Google Scholar]
- 29.Zou Y., Yi X., Wright W.E., Shay J.W. Human telomerase can immortalize Indian muntjac cells. Exp. Cell Res. 2002;281:63–76. doi: 10.1006/excr.2002.5645. [DOI] [PubMed] [Google Scholar]
- 30.Yasui Y., Urano T., Kawajiri A., Nagata K., Tatsuka M., Saya H., Furukawa K., Takahashi T., Izawa I., Inagaki M. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 2004;279:12997–13003. doi: 10.1074/jbc.M311128200. [DOI] [PubMed] [Google Scholar]
- 31.Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zinkowski R.P., Meyne J., Brinkley B.R. The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukinavičius G., Reymond L., D’Este E., Masharina A., Göttfert F., Ta H., Güther A., Fournier M., Rizzo S., Waldmann H. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods. 2014;11:731–733. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 35.Wood K.W., Lad L., Luo L., Qian X., Knight S.D., Nevins N., Brejc K., Sutton D., Gilmartin A.G., Chua P.R. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl. Acad. Sci. USA. 2010;107:5839–5844. doi: 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spence J.M., Mills W., Mann K., Huxley C., Farr C.J. Increased missegregation and chromosome loss with decreasing chromosome size in vertebrate cells. Chromosoma. 2006;115:60–74. doi: 10.1007/s00412-005-0032-6. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwiatkowski N., Jelluma N., Filippakopoulos P., Soundararajan M., Manak M.S., Kwon M., Choi H.G., Sim T., Deveraux Q.L., Rottmann S. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat. Chem. Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powers J., Rose D.J., Saunders A., Dunkelbarger S., Strome S., Saxton W.M. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J. Cell Biol. 2004;166:991–1001. doi: 10.1083/jcb.200403036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cane S., Ye A.A., Luks-Morgan S.J., Maresca T.J. Elevated polar ejection forces stabilize kinetochore-microtubule attachments. J. Cell Biol. 2013;200:203–218. doi: 10.1083/jcb.201211119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 2012;198:847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stear J.H., Roth M.B. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 2002;16:1498–1508. doi: 10.1101/gad.989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redemann S., Baumgart J., Lindow N., Shelley M., Nazockdast E., Kratz A., Prohaska S., Brugués J., Fürthauer S., Müller-Reichert T. C. elegans chromosomes connect to centrosomes by anchoring into the spindle network. Nat. Commun. 2017;8:15288. doi: 10.1038/ncomms15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Toole E.T., McDonald K.L., Mäntler J., McIntosh J.R., Hyman A.A., Müller-Reichert T. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 2003;163:451–456. doi: 10.1083/jcb.200304035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cimini D., Moree B., Canman J.C., Salmon E.D. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 46.Cimini D., Cameron L.A., Salmon E.D. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 47.Vig B.K., Henderson A. Aneuploidy in male Indian muntjac cells is limited to the Y2 chromosome. Mutagenesis. 1998;13:33–37. doi: 10.1093/mutage/13.1.33. [DOI] [PubMed] [Google Scholar]
- 48.Rudd M.K., Willard H.F. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Forsberg L.A., Rasi C., Malmqvist N., Davies H., Pasupulati S., Pakalapati G., Sandgren J., Diaz de Ståhl T., Zaghlool A., Giedraitis V. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 51.Shrestha R.L., Ahn G.S., Staples M.I., Sathyan K.M., Karpova T.S., Foltz D.R., Basrai M.A. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget. 2017;8:46781–46800. doi: 10.18632/oncotarget.18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worrall J.T., Tamura N., Shaikh N., Mazzagatti A., van Lingen T., Bakker B., Spierings D.C.J., Vladimirou E., Foijer F., McClelland S.E. Non-random mis-segregation of human chromosomes. bioRxiv. 2018 doi: 10.1016/j.celrep.2018.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wendell K.L., Wilson L., Jordan M.A. Mitotic block in HeLa cells by vinblastine: ultrastructural changes in kinetochore-microtubule attachment and in centrosomes. J. Cell Sci. 1993;104:261–274. doi: 10.1242/jcs.104.2.261. [DOI] [PubMed] [Google Scholar]
- 54.McEwen B.F., Chan G.K., Zubrowski B., Savoian M.S., Sauer M.T., Yen T.J. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan B.A., Willard H.F. Stable dicentric X chromosomes with two functional centromeres. Nat. Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- 56.Maffini S., Maia A.R., Manning A.L., Maliga Z., Pereira A.L., Junqueira M., Shevchenko A., Hyman A., Yates J.R., III, Galjart N. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amaro A.C., Samora C.P., Holtackers R., Wang E., Kingston I.J., Alonso M., Lampson M., McAinsh A.D., Meraldi P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magidson V., He J., Ault J.G., O’Connell C.B., Yang N., Tikhonenko I., McEwen B.F., Sui H., Khodjakov A. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in Taxol-treated cells. J. Cell Biol. 2016;212:307–319. doi: 10.1083/jcb.201412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgulis A., Coulouris G., Raytselis Y., Madden T.L., Agarwala R., Schäffer A.A. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 62.Pereira A.L., Pereira A.J., Maia A.R., Drabek K., Sayas C.L., Hergert P.J., Lince-Faria M., Matos I., Duque C., Stepanova T. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live-cell imaging of hTERT-immortalized Indian muntjac fibroblasts stably expressing H2B-GFP to visualize the chromosomes (green) and treated with 50 nM SiR-tubulin to label spindle microtubules (magenta). Representative examples of control, Ndc80, Mad2, Mps1, Clasp1, and Survivin RNAi are shown. Time, hr:min.
Live-cell imaging of control (left) or GSK923295-treated (CENP-E inhibitor, 20 nM, 3 cells from the right) hTERT-immortalized Indian muntjac (IM) fibroblasts stably expressing CENP-A-GFP to visualize the kinetochores (green) and treated with 20 nM SiR-tubulin to label spindle microtubules (magenta). Time, hr:min.
Note that most kinetochore pairs (all green dots) occupy a position at the spindle periphery, but their distribution relative to the pole/equator (red dot) is random. One exceptional kinetochore pair that fell inside the spindle region is also indicated (black dot).
Monastrol treatment and washout in a live hTERT-immortalized Indian muntjac fibroblast stably expressing CENP-A-GFP to visualize the kinetochores (green) and treated with 20 nM SiR-tubulin to label spindle microtubules (magenta). Note the difference between chromosomes with a small or large kinetochore pair. One chromosome with a large kinetochore pair can be seen to change conformation and lag behind during anaphase for a short time, but eventually resolves and segregates to the correct daughter. Time, min:s.
Indian muntjac female fibroblast stably expressing H2B-GFP (green) treated with 20 nM SiR-tubulin (magenta) after monastrol washout. Time, min:s.
Control and Mps1-inhibited (20 μM Mps1-IN-1) Indian muntjac fibroblasts stably expressing H2B-GFP (green). Time, hr:min.
Control and Kif4a-RNAi-depleted Indian muntjac fibroblasts stably expressing H2B-GFP (green) with 50 nM SiR-tubulin (magenta). Time, hr:min.