Figure 1.
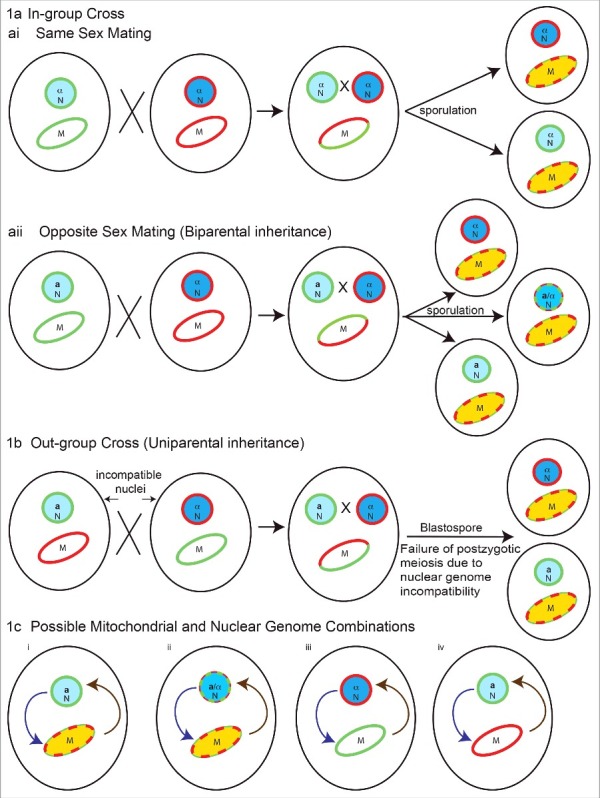
Possible nuclear and mitochondrial genetic genotypes in Cryptococcus species in progeny from crosses contributing to their virulence. 1a) In-group cross (within species), two possible scenarios: (ai) same sex mating (α-α), recombinant mitochondrial genomes, with isogenic nuclei, and (aii) opposite sex mating (a-α) with both recombinant mitochondrial and nuclear genomes generated in progeny. In these crosses mitochondrial inheritance is biparental, with contribution from single parent at a time. The exception is crosses between isogenic strains, where mitochondrial inheritance is uniparental. 1b) Outgroup cross (interspecies): the nuclear genome can be genetically incompatible leading to post-zygotic meiotic failure, but retaining the possibility of recombination amongst mitochondrial genomes. In such a scenario progeny will have recombinant mitochondria with nuclei isogenic to one of the parents. 1c) Possible mitochondrial and nuclear genome combinations: (i) recombinant mitochondrial genome in isogenic nuclear background, (ii) recombinant mitochondrial genome in a recombinant nuclear background, (iii) non-recombinant mitochondria from first parent with non-recombinant nuclei from another parent and (iv) vice versa of iii. Interorganelle (nuclear-mitochondrial) regulatory loops on each other are shown. Light blue circles with green outline and dark blue circles with red outline represents nuclei with different genetic background respectively. Blue circles with mixed dotted outline represents recombinant nuclei. The a and α represents mating types. Ellipse with green and red outline represents mitochondria with different genetic background respectively. Ellipse with red green dotted outline and yellow color represents recombinant mitochondria. Curved arrows in 1c represents interorganelle regulatory signaling loops for nucleus to mitochondria (purple) and mitochondria to nucleus (brown).
