Abstract
Asymmetric messenger RNA (mRNA) localization facilitates efficient translation in cells such as neurons and fibroblasts. However, the extent and importance of mRNA polarization in epithelial tissues are unclear. Here, we used single-molecule transcript imaging and subcellular transcriptomics to uncover global apical-basal intracellular polarization of mRNA in the mouse intestinal epithelium. The localization of mRNAs did not generally overlap protein localization. Instead, ribosomes were more abundant on the apical sides, and apical transcripts were consequently more efficiently translated. Refeeding of fasted mice elicited a basal-to-apical shift in polarization of mRNAs encoding ribosomal proteins, which was associated with a specific boost in their translation. This led to increased protein production, required for efficient nutrient absorption. These findings reveal a posttranscriptional regulatory mechanism involving dynamic polarization of mRNA and polarized translation.
Intracellular mRNA localization is an important determinant of various cellular functions (1–6). In mammals, localized translation of transcripts at the sites where their encoded proteins are needed is thought to confer cellular efficiency and a timely response (2, 7, 8). Epithelial tissues are inherently polarized with distinct functional specialization of basal and apical sides. The intestinal epithelium consists of a monolayer of enterocytes that absorb nutrients from the apical lumen and excrete them into the blood stream from the basal sides. Key transporters are specifically localized in these two compartments (9). Owing to the transient nature of nutrients in the gut, efficient and timely translation of such proteins might be important. The mRNA intracellular localization of a few enterocyte genes have been reported (10–12); however, we lack a global characterization of intracellular mRNA localization and its biological functions in epithelia.
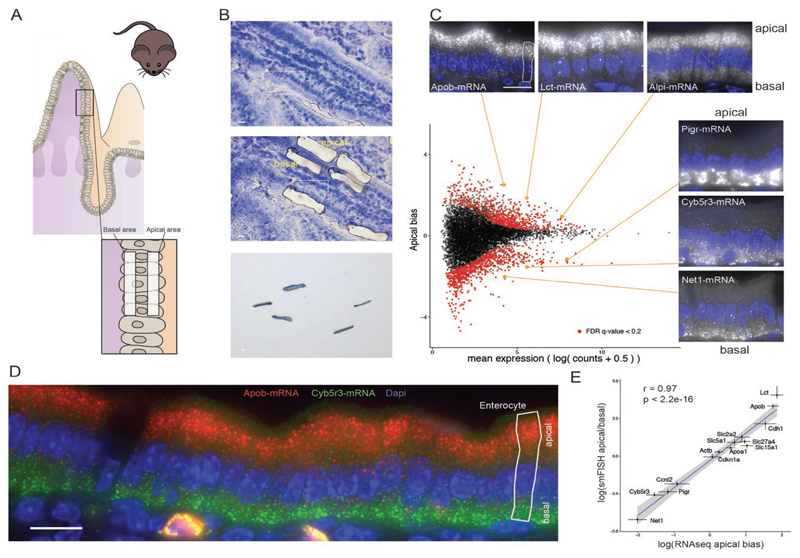
To obtain a transcriptome-wide view of mRNA localization in the intestinal epithelium, we isolated apical and basal subcellular fractions by laser capture microdissection (Fig. 1A) and performed RNA sequencing (RNA-seq) (13). Almost 30% of the most highly expressed 2000 transcripts, for which experimental noise levels were lower, were significantly polarized (Fig. 1B and table S1). To validate the sequencing results, we performed single-molecule fluorescence in situ hybridization (smFISH) (14) on intact intestinal tissues for 14 genes that span different polarization patterns (Fig. 1, B to D). We found an excellent correspondence between the apical bias of mRNA measured with smFISH and with the whole-transcriptome method (Spearman’s r = 0.97, P < 2.2 × 10–16, Fig. 1D and table S2).
Fig. 1. Global analysis of mRNA polarization in the intestinal epithelium.
(A) Laser capture microdissection of paired apical and basal fragments. (B) RNA-seq data of isolated subcellular areas. Insets show smFISH validation results of transcripts of interest. Four outlier data points are omitted from the plot. Of the transcripts, 645 of 9905 are significantly apical and 779 of 9905 are significantly basal. Dashed vertical line separates the 2000 most highly expressed transcripts, of which 392 genes are significantly apical and 194 are significantly basal. (C) smFISH staining of the apical Apob (red) and basal Cyb5r3 (green) mRNA. (D) Strong correlation of smFISH quantifications and RNAseq data for 14 representative genes (Spearman’s r = 0.97, P < 2.2 × 10–16); dots and error bars represent median and 95% confidence interval of smFISH and mean and SE of RNAseq. All scale bars are 10 μm.
Given the functional polarization of enterocytes, we hypothesized that the observed global mRNA apical-basal polarization would match the location of the corresponding proteins. The most apically enriched gene sets included several nutrient transporter annotations, many of which encode apically localized proteins (fig. S1). Notably, however, the mRNAs that encode basolaterally localized proteins were strongly biased in localization toward the apical side, opposite to where their encoded proteins reside (Fig. 2, A and B, and fig. S2A). Examples of basolateral proteins with discordantly polarized apical mRNA included E-cadherin (Cdh1 gene, Fig. 2B) and integrins (fig. S2A). Thus, the global mRNA polarization in the intestinal epithelium does not generally correlate with the localization of the encoded proteins.
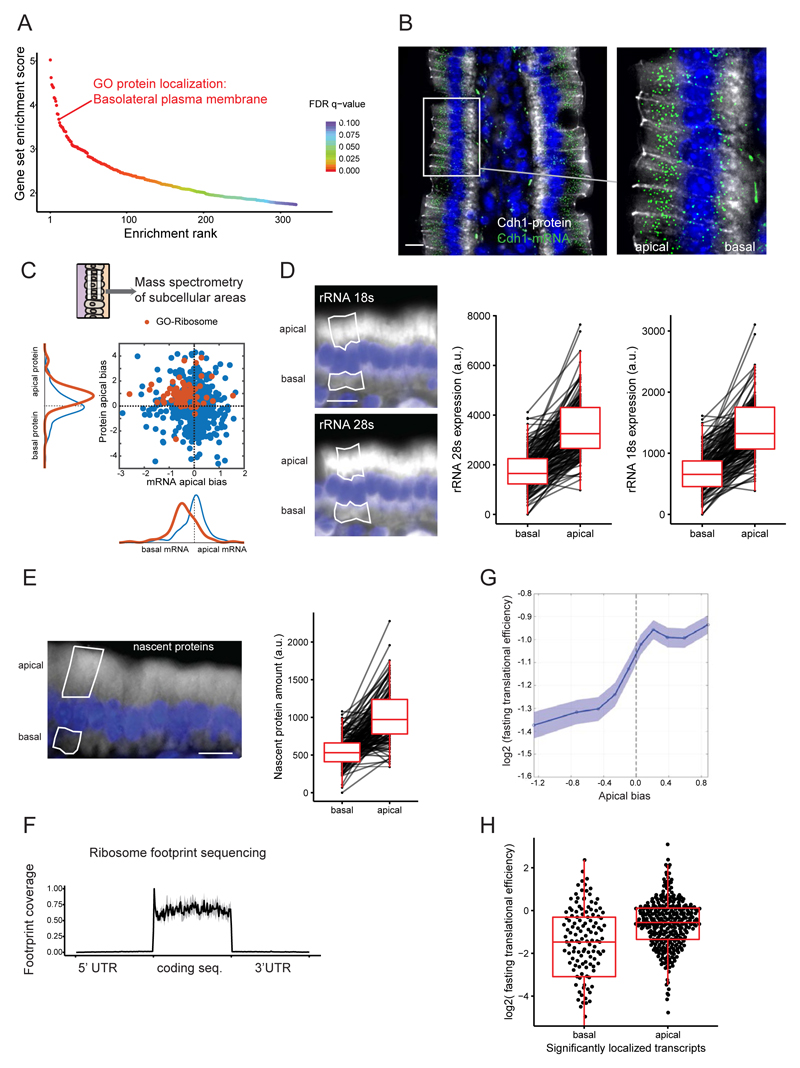
Fig. 2. Translational machinery is asymmetrically distributed.
(A) Ranking of gene sets that are significantly enriched on the apical cell sides. (B) Costaining of the discordantly localized basolateral protein E-cadherin, encoded by Cdh1 (gray staining) and its apically localized mRNA (green dots). (C) Mass spectrometry results of microdissected areas demonstrate apical enrichment of ribosomal proteins and a lack of positive correlation (Spearman’s r = –0.12, P = 8 × 10–4). Blue, all genes; red, genes with GO-Ribosome annotation. (D) Representative smFISH staining and quantification of intracellular distribution of rRNA 18S and 28S (n = 142 single cells, P < 2.2 × 10–16). (E) Representative staining and quantification of nascent proteins (n = 143 single cells, P < 2.2 × 10–16). (F) Normalized coverage plot of ribosome footprint sequencing data (n = 3 mice, gray area denotes SD). (G) TE increases with the apical bias of genes in fasting mouse samples. The y axis shows the mean TE over a sliding window of 1000 genes consecutively shifted from the most basal gene to the most apical gene, with a 500-gene overlap. The x axis is the mean apical bias of genes within each window. Patches are standard errors of the mean. (H) Translational efficiency of the significantly localized transcripts (false discovery rate adjusted P value <0.1, n apical = 346, n basal = 141, P = 8.4 × 10–10; 20 outlier data points are omitted from the plot). Data include only genes for which we obtained TE values and that are of epithelial origin (methods). All scale bars are 10 μm.
To obtain a broader view of the relative localization of mRNAs and their encoded proteins, we performed mass spectrometry of laser capture microdissected apical and basal subcellular fractions (Fig. 2C and table S3). The apical bias of proteins and of mRNA was weakly anticorrelated (Spearman’s r = –0.12, P = 8 × 10–4). This data set revealed additional examples of discordant localizations of proteins and their encoding mRNA, such as the basal protein sodium-potassium ATPase (adenosine triphosphatase), encoded by the highly apical mRNA Atp1b1 (fig. S2A). Notably, the mass spectrometry data revealed a significantly higher abundance of ribosomal proteins at the apical side (Fig. 2C and table S4) and mitochondrial proteins at the basal sides (fig. S3 and table S4). The mRNAs that encode ribosomal proteins were basally biased, again constituting a discordant set of genes, with opposite localizations of mRNA and their encoded proteins (Fig. 2C). Using smFISH, we identified a highly significant apical enrichment of ribosomal RNA 18S and 28S (Fig. 2D and fig. S2B, both twofold, P < 2.2 × 10–16), thus validating the apically polarized localization of the translational machinery. To assess whether the increased concentration of ribosomes on the apical sides yields higher translation at this subcellular compartment, we treated mice with O-propargyl-puromycin (OPP), a reporter that is efficiently incorporated into nascent peptides after a pulse-chase intraperitoneal injection (15) (fig. S4). Nascent peptides were significantly more abundant on the apical sides of the intestinal enterocytes (Fig. 2E, 1.82-fold higher concentration at the apical side, P < 2.2 × 10–16). Thus, translation in the intestinal enterocytes is also strongly polarized, with a twofold higher apical concentration of the translational machinery.
We next asked whether apical-basal mRNA polarization could regulate translation efficiency (TE) in this tissue. We performed ribosome profiling and RNA-seq of intestinal isolates from fasting mice and computed the TE of intestinal transcripts (Fig. 2F, fig. S5, and tables S5 and S6). Genes with significantly apical mRNA localization had almost twofold higher TE compared to genes with significantly basal mRNA (Fig. 2, G and H, P = 8.36 × 10–10). In several nonpolarized mouse primary cell data sets, the apical and basal groups were either indistinguishable or displayed much weaker apical enrichment of TE (fig. S6A). Thus, mRNA apico-basal polarization modulates TE in the intestinal epithelium.
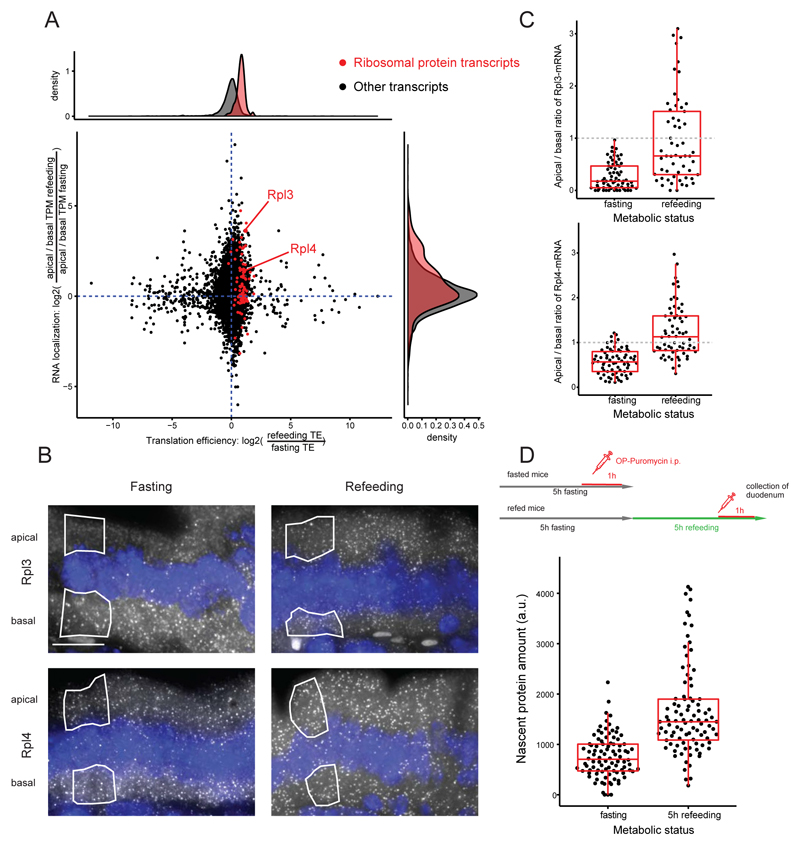
The intestinal lumen is a highly dynamic microenvironment that exhibits temporal oscillation in nutrient availability (16). Dynamic translocation of mRNA between the basal and apical sides could potentially modulate translational efficiency. To explore such dynamic responses, we performed our transcriptome-wide mRNA polarization measurements and ribosome profiling on duodenum tissues of fasting and refed mice (Fig. 3A). The translational machinery remained apical after refeeding, as evident by the apical polarization of ribosomal RNA (rRNA) (fig. S6B), and the TE of genes was higher the more apically polarized were their transcripts (fig. S6C). mRNAs that encode ribosomal proteins (RNA-RPs) shifted from the basal localization in fasted intestines to a more apical localization after 2 hours of refeeding (Fig. 3A, 1.36-fold for RNA-RPs versus 1.08-fold for other transcripts, P = 0.0001). We also observed a concomitant increase in TE for these transcripts upon shifting their localization to the apical side—the side in which the translation machinery is more abundant (Fig. 3A and fig. S7, 1.87-fold, P < 2 × 10–16). We validated these localization changes for Rpl3 and Rpl4 mRNA by smFISH (Fig. 3, B and C, Rpl3 P = 1.8 × 10–8, Rpl4 P = 2.9 × 10–13). The cellular mRNA expression levels of these genes were not increased (fig. S8, A to C). After 4 hours of refeeding, increased production of ribosomal components was associated with significantly higher protein synthesis compared to the fasting state (Fig. 3D, 2.05-fold, P < 2.2 × 10–16). TE of brush border proteins (17) also showed a slight, yet significant increase upon refeeding (1.1-fold, P = 0.02, fig. S8D). Thus, dynamic translocation of mRNAs encoding ribosomal proteins to the more translationally active apical side is associated with a specific translation of ribosomal components, facilitating a burst of protein production to meet the absorption demands upon refeeding.
Fig. 3. Dynamic shifts of localized transcripts are associated with differential translational efficiency.
(A) Scatterplot of TE and mRNA localization changes when comparing three fasting and three refed mice (n = 6282 transcripts). The x axis is log2 of the ratios between TEs in refed and in fasting states; the y axis is log2 of the ratios of apical biases between refed and fasting states, where apical bias for each condition is the ratio of apical and basal TPM (methods). Upon refeeding, mRNAs encoding ribosomal proteins (red dots) become more apically polarized and are translated more efficiently. (B) smFISH images of Rpl3 and Rpl4 mRNA across metabolic states. (C) Quantification of Rpl3 and Rpl4 smFISH analyses across metabolic states (n = 70 single cells, Rpl3 P = 1.8 × 10–8, Rpl4 P = 2.9 × 10–13). (D) Comparison of nascent protein content at fasting and 5-hour refeeding time point (n = 188 single cells, P < 2.2 × 10–16). All scale bars are 10 μm.
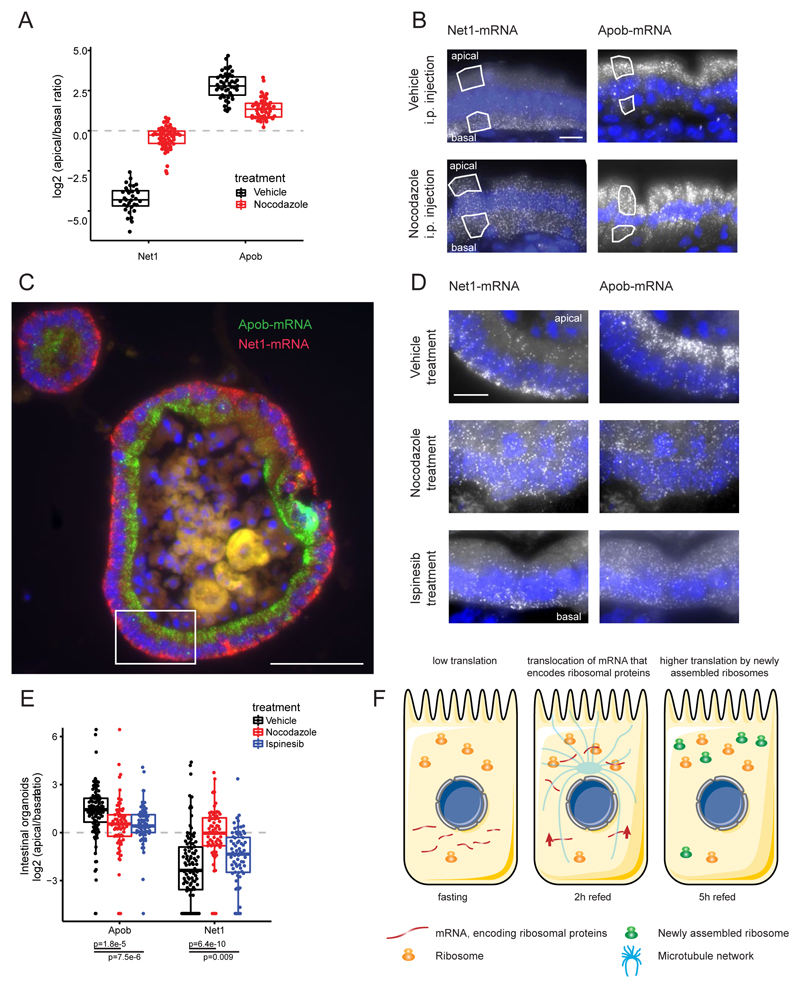
What facilitates this broad mRNA polarization? Active transport by motor proteins along the cytoskeleton, molecular anchors that bind and keep transcripts localized, or spatially varying RNA degradation rates within the cell have all been implicated in determining localization (1, 18). Additionally, our finding that the ribosomes are apically polarized in enterocytes could indicate that preferential retention of highly translated mRNA could lead to their apical polarization (1, 18). To investigate whether microtubules mediate the observed mRNA localization patterns, we depolymerized the microtubule network by injecting mice intraperitoneally with nocodazole (Fig. 4, A and B). The basally polarized Net1, Cyb5r3, Rpl3, and Rpl4 transcripts lost polarization, and the apically polarized Apob and Cdh17 transcripts were significantly less apical (Fig. 4, A and B, and fig. S9). Although ribosomal RNA remained significantly apically polarized in nocodazole-treated mice, the extent of rRNA polarization was smaller compared to that in vehicle-treated controls (fig. S10). To assess whether the difference in polarization of apical mRNA upon nocodazole treatment could stem from coordinated movement of ribosomes and their retained mRNA to the basal side, we measured the combined polarization of both rRNA and of four apical genes with high TE in nocodazole-treated mice and in controls, and stratified the results according to the single-cell rRNA polarization. Microtubule disruption decreased the apical polarization of these genes even when controlling for rRNA polarization changes (figs. S10 and S11A). Thus, the microtubule network mediates the asymmetric localization of these and potentially other transcripts in the intestinal epithelium, whereas additional anchoring mechanisms seem to render the apical localization of ribosomes less sensitive to microtubule disruption. Selective inhibition of kinesin 5 in intestinal organoids (19, 20) using ispinesib (21) decreased the polarization of Apob and Net1 (Fig. 4, C to E) but not of Cdh17 and Pigr (fig. S9, E and F).
Fig. 4. Intestinal mRNA localization is mediated by the microtubule network.
(A) Nocodazole strongly perturbs the polarization of Net1 and Apob (Net1 n = 94 cells, P = 1.1 × 10–15, Apob n = 96 cells, P = 2.9 × 10–12). (B) Representative smFISH images of strongly polar Net1 and Apob transcripts in vehicle- or nocodazole-injected animals. Scale bar, 10 μm. (C) Intracellular mRNA localization of Net1 and Apob in intestinal organoids phenocopies in vivo observations. Highlighted inset is shown in higher magnification in top row of (D). Scale bar, 50 μm. (D) smFISH images of Net1 and Apob in nocodazole-, ispinesib-, or vehicle-treated organoids. Scale bar, 10 μm. (E) Single-cell quantification of the nocodazole and ispinesib effects on transcript localization in smFISH images of intestinal organoids (vehicle n = 101, ispinesib n = 68, nocodazole n = 77 cells, costaining of Apob and Net1). (F) Transcripts that encode ribosomal proteins are stored in the less translationally active basal side of the intestinal epithelium in fasting mice. Refeeding induces a translocation of these transcripts into the more translationally active apical cell side. This translocation is associated with a concomitant increase in their translational efficiency. The increased ribosomal biogenesis is reflected in an increased total protein synthesis.
Given the transient nature of inputs in the gut, consistently high translation rates could be energetically inefficient (22). When nutrients arrive, however, there is a short temporal window in which they must be efficiently absorbed (9). Nutrient availability was shown to stimulate rapid ribosome biogenesis in other contexts (23, 24) via activation of translation initiation factors, rRNA transcription, and translation of RNAs that encode the translational machinery (24). Here, we found a similar burst of protein translation upon refeeding, associated with a rapid translocation of ribosomal-protein encoding transcripts, the nature of which is yet to be determined, into the translationally active apical side (Fig. 4F). The burst of ribosomal protein translation upon refeeding resembles a “bang-bang” control strategy, in which resources are first invested in making the ribosomal “machines,” to facilitate a rapid increase in total protein output (25, 26). Our approach, combining laser-capture microdissection and whole-transcriptome sequencing with smFISH, can be readily applied to characterize mRNA polarization in other tissues and organisms. Future studies will determine if mRNA mislocalizations are causatively involved in pathophysiology.
Supplementary Material
Acknowledgments
We thank K. Bahar Halpern, B. Toth, and S. Ben-Moshe for valuable comments on the manuscript. We thank the L Lokey animal facility (Weizmann Institute of Science, Rehovot, Israel), the Crown Institute for Genomics (Weizmann Institute of Science, Rehovot, Israel), and the Smoler Protein Research Center (Technion, Haifa, Israel) for help with experimental procedures. A.E.M. is supported by the Swiss National Science Foundation (grant 158999) and the European Molecular Biology Organization (EMBO) Long-Term Fellowship program (ALTF 306-2016). N.S.-G. research is funded by the European Research Council starting grant (StG-2014-638142). N.S.-G. is incumbent of the Skirball Career Development Chair in New Scientists. S.I. is supported by the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics, The Leir Charitable Foundations, Richard Jakubskind Laboratory of Systems Biology, Cymerman-Jakubskind Prize, The Lord Sieff of Brimpton Memorial Fund, the I-CORE program of the Planning and Budgeting Committee and the Israel Science Foundation (grants 1902/12 and 1796/12), the Israel Science Foundation grant no. 1486/16, the EMBO Young Investigator Program, and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)–ERC grant agreement no. 335122. S.I. is the incumbent of the Philip Harris and Gerald Ronson Career Development Chair. All data supporting the findings of this study and all analysis codes are available within the article and its supplementary materials or from the corresponding author upon request. The generated sequencing data have been deposited in the GenBank Gene Expression Omnibus database(www.ncbi.nlm.nih.gov/geo/) under accession code GSE95416.
References
- 1.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: Visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lécuyer E, et al. Global analysis of mrna localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife. 2015;4:e05003. doi: 10.7554/eLife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besse F, Ephrussi A. Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr Opin Cell Biol. 2008;20:144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frayn KN. Metabolic Regulation: A Human Perspective. ed. 3. Wiley-Blackwell; Chichester, UK: 2010. [Google Scholar]
- 10.Rings EHHM, Büller HA, de Boer PAJ, Grand RJ, Montgomery RK, Lamers WH, Charles R, Moorman AFM. Messenger RNA sorting in enterocytes. Co- localization with encoded proteins. FEBS Lett. 1992;300:183–187. doi: 10.1016/0014-5793(92)80192-J. [DOI] [PubMed] [Google Scholar]
- 11.Barth JA, Li W, Krasinski SD, Montgomery RK, Verhave M, Grand RJ. Asymmetrical localization of mRNAs in enterocytes of human jejunum. J Histochem Cytochem. 1998;46:335–343. doi: 10.1177/002215549804600307. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Krasinski SD, Verhave M, Montgomery RK, Grand RJ. Three distinct messenger RNA distribution patterns in human jejunal enterocytes. Gastroenterology. 1998;115:86–92. doi: 10.1016/S0016-5085(98)70368-3. [DOI] [PubMed] [Google Scholar]
- 13.Nichterwitz S, Chen G, Aguila Benitez J, Yilmaz M, Storvall H, Cao M, Sandberg R, Deng Q, Hedlund E. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun. 2016;7:12139. doi: 10.1038/ncomms12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, Tuganbaev T, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510.e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 17.McConnell RE, Benesh AE, Mao S, Tabb DL, Tyska MJ. Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol. 2011;300:G914–G926. doi: 10.1152/ajpgi.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pratt CA, Mowry KL. Taking a cellular road-trip: mRNA transport and anchoring. Curr Opin Cell Biol. 2013;25:99–106. doi: 10.1016/j.ceb.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt- villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 20.Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 21.Lad L, Luo L, Carson JD, Wood KW, Hartman JJ, Copeland RA, Sakowicz R. Mechanism of inhibition of human KSP by ispinesib. Biochemistry. 2008;47:3576–3585. doi: 10.1021/bi702061g. [DOI] [PubMed] [Google Scholar]
- 22.Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 23.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itzkovitz S, Blat IC, Jacks T, Clevers H, van Oudenaarden A. Optimality in the development of intestinal crypts. Cell. 2012;148:608–619. doi: 10.1016/j.cell.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madar D, Dekel E, Bren A, Zimmer A, Porat Z, Alon U. Promoter activity dynamics in the lag phase of Escherichia coli. BMC Syst Biol. 2013;7:136. doi: 10.1186/1752-0509-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, van Oudenaarden A. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2011;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyubimova A, Itzkovitz S, Junker JP, Fan ZP, Wu X, van Oudenaarden A. Single-molecule mRNA detection and counting in mammalian tissue. Nat Protoc. 2013;8:1743–1758. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
- 29.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 2013;8:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 31.Stern-Ginossar N, Weisburd B, Michalski A, Le VTK, Hein MY, Huang S-X, Ma M, Shen B, Qian S-B, Hengel H, Mann M, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome- protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 35.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA- seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 36.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, McManus J, Kingsford C. Isoform-level ribosome occupancy estimation guided by transcript abundance with Ribomap. Bioinformatics. 2016;32:1880–1882. doi: 10.1093/bioinformatics/btw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moor AE, Anderle P, Cantù C, Rodriguez P, Wiedemann N, Baruthio F, Deka J, André S, Valenta T, Moor MB, Győrffy B, et al. BCL9/9L-β-catenin signaling is associated with poor outcome in colorectal cancer. EBioMedicine. 2015;2:1932–1943. doi: 10.1016/j.ebiom.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: Identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169–e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network- based method for gene-set enrichment visualization and interpretation. PLOS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fatehullah A, Appleton PL, Näthke IS. Cell and tissue polarity in the intestinal tract during tumourigenesis: Cells still know the right way up, but tissue organization is lost. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130014. doi: 10.1098/rstb.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato T, Mushiake S, Kato Y, Sato K, Sato M, Takeda N, Ozono K, Miki K, Kubo Y, Tsuji A, Harada R, et al. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 2007;448:366–369. doi: 10.1038/nature05929. [DOI] [PubMed] [Google Scholar]
- 47.Singh B, Coffey RJ. Trafficking of epidermal growth factor receptor ligands in polarized epithelial cells. Annu Rev Physiol. 2014;76:275–300. doi: 10.1146/annurev-physiol-021113-170406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Röder PV, Geillinger KE, Zietek TS, Thorens B, Koepsell H, Daniel H. The role of SGLT1 and GLUT2 in intestinal glucose transport and sensing. PLOS ONE. 2014;9:e89977. doi: 10.1371/journal.pone.0089977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M, Praetorius J, Zheng W, Xiao F, Riederer B, Singh AK, Stieger N, Wang J, Shull GE, Aalkjaer C, Seidler U. The electroneutral Na+:HCO3– cotransporter NBCn1 is a major pHi regulator in murine duodenum. J Physiol. 2012;590:3317–3333. doi: 10.1113/jphysiol.2011.226506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.